Open Access Article
Open Access ArticleAdipogenesis or osteogenesis: destiny decision made by mechanical properties of biomaterials
Ting Su,
Mimi Xu,
Feng Lu* and
Qiang Chang *
*
Department of Plastic and Cosmetic Surgery, Nanfang Hospital, Southern Medical University, 510515, China. E-mail: doctorchq@hotmail.com
First published on 30th August 2022
Abstract
Regenerative medicine affords an effective approach for restoring defect-associated diseases, and biomaterials play a pivotal role as cell niches to support the cell behavior and decide the destiny of cell differentiation. Except for chemical inducers, mechanical properties such as stiffness, pore size and topography of biomaterials play a crucial role in the regulation of cell behaviors and functions. Stiffness may determine the adipogenesis or osteogenesis of mesenchymal stem cells (MSCs) via the translocation of yes-associated protein (YAP) and the transcriptional coactivator with a PDZ-binding motif (TAZ). External forces transmit through cytoskeleton reorientation to assist nuclear deformation and molecule transport, meanwhile, signal pathways including the Hippo, FAK/RhoA/ROCK, and Wnt/β-catenin have been evidenced to participate in the mechanotransduction. Different pore sizes not only tailor the scaffold stiffness but also conform to the requirements of cell migration and vessels in-growth. Topography guides cell geometry along with mobility and determines the cell fate ascribed to micro/nano-scale contact. Herein, we highlight the recent progress in exploring the regulation mechanism by the physical properties of biomaterials, which might lead to more innovative regenerative strategies for adipose or bone tissue repair.
1 Introduction
Regeneration has aroused human enthusiasm for much of recorded history and has become one of the most important projects in life science. The isolation techniques of stem cells from human tissues and the discovery of various differentiation inducers place artificial tissue and organs within the realm of possibility. The development and advancement of multifarious biomimetic materials bridge the gap between stem cells and fully functional tissue yet a single mesh of cells on 2D plates is too simple to be considered motile tissue. At present, deciphering the mechanism of stem cell proliferation and differentiation may replenish our arms arsenal against tissue defect-associated diseases, such as traumatic disfigurement, tumor removal, osteoporosis, arthritis, diabetes ulcers, and neurodegenerative diseases, etc.1MSCs are fibroblast-like or spindle-shaped pluripotent stem cells derived from bone marrow, adipose tissue, brain, synovium, and umbilical cord blood, ranking among the dominating seed cells in tissue engineering.2 Though MSCs with different origins demonstrate slightly various characteristics, over 95% of the cells must express CD105, CD73, and CD90, and with differentiation potential towards at least three different lineages such as adipocytes, osteoblasts, and chondrocytes.3 Osteogenesis or adipogenesis is always the bifurcated yet the fascinating fate determination of MSCs. The differentiation of stem cells is a highly synergistic procedure including two steps, namely lineage commitment and maturation. Multitudes of studies have revealed that critical signaling pathways, such as transforming growth factor-beta (TGFβ)/bone morphogenic protein (BMP) signaling, wingless-type MMTV integration site (Wnt)/β-catenin signaling, Notch, and Hedgehogs signaling, have been involved in the determination of MSCs differentiation.4
BMPs, are members of the transforming growth factor-β (TGF-β) superfamily. However, the concentration and type of BMPs display diverse effects on differentiation lineages in MSCs.5 MSCs are committed to preadipocytes when induced by bone morphogenic protein-4 (BMP-4). Then, the preadipocytes activate peroxisome proliferator-activated receptor gamma (PPARγ), CCAAT/enhancer-binding proteins (C/EBPs) and downstream early-adipogenesis biomarkers such as fatty acid-binding protein-2 (AP2), and glucose transporter-4 (GLUT4). With differentiation continuing, adiponectin, leptin, adipose triacylglyceride lipase (ATGL), lipoprotein lipase (LPL), and high level of perilipin-1 are expressed, known as indicators of maturation.6 The activation of Hedgehog signaling inhibits the expression of adipogenic differentiation markers by reducing the expression of C/EBPα.7
A high concentration of BMP2 has been proved osteoinductive in the C3H10T1/2 mesenchymal cell line.8 Activated BMP2 also interacts with another classical osteogenic Wnt/β-catenin signaling.9 Wnt/β-catenin signaling is essential in cell fate determination and proliferation and starts osteogenesis by stimulating the expression of transcription factor runt-related transcription factor 2 (RUNX2).10 RUNX2 is the master factor that regulates multiple steps throughout the osteoblast commitment. The level of RUNX2 peaks early in the process, inducing robust proliferation and expression of collagen, fibronectin, and transforming growth factor-β (TGF-β) receptor 1. In the further maturation, decreased RUNX2 activates downstream osterix and the expression of collagen type 1 alpha 1 chain (COL1A1) as a component of ECM and alkaline phosphatase (ALP) to mature the ECM. The process of matrix mineralization occurs with the expression of osteocalcin (OCN), osteopontin (OPN), osteonectin (ONN), and bone sialoprotein (BSP).11
Conventional tools for manipulating stem cell behaviors mainly rely on the special media or particular chemical inducers to arouse the pivotal signaling pathways branch in natural embryonic differentiation. Insulin and glucocorticoid are dominant ingredients in adipogenic culture medium to induce stable lipogenesis from preadipocytes.12 Fibroblast growth factors (FGF), hepatocyte growth factor (HGF), and VEGF are commonly used in adipose tissue engineering,13–15 while parathyroid hormone, estrogen, TGF-β, and osteogenic growth peptides are utilized to induce bone regeneration.16,17 However, investigation on physical cues, especially biomechanics, lends insights into the intricate signal transduction mechanisms between cells and the extracellular environment. All cells can actively sense and respond to external forces through a process called mechanical transduction, and exterior mechanical cues are then translated to interior biochemical signals within the cell to manipulate cell behaviors.18 The mechanical properties diversify the microenvironment via physical agents such as stiffness, elasticity, porosity, and topology, which influence the cellular contractility cell, adherent morphology, and final differentiation.19 For example, the wise choice for the scaffold stiffness did more with less in the lineage commitment of mesenchymal stem cells (MSCs).20 Stiff substrates encourage cell movement, while soft hydrogels attenuate the synthesis of cytoskeletal protein and lead to the spherical shape of stem cells;21 larger pore sizes are conducive to vascularization,22 and prefabricated grating patterns help with cell alignment.23
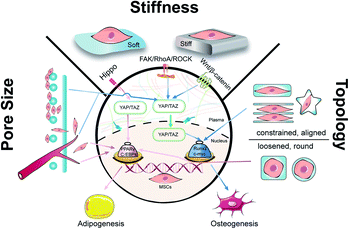
Biomaterials are of profound importance in instructing stem cell behaviors and functions by satisfying supportive structure, interactive microenvironment, and inductive bio-cues. It is well known that under physiological conditions, stem cells can release cellular chemicals and exchange substances in and out of cells to remodel an external environment in favor of different lineages.24 In order to harness the precise outcomes of cell behaviors in tissue engineering, intense efforts have been made to regulate the mechanical properties of biomaterials that include but are not limited to stiffness, pore size, and topography. By reviewing the recent progress of mechanical cues in manipulating the differentiation of MSCs, we would like to expatiate the underlying mechanisms and shed light on the significance of mechanical cues in regeneration medicine (Fig. 1).
2 Stiffness
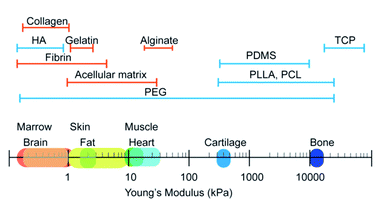
Stiffness, also called rigidity, is an extensive and elementary property of the solid object, representing the resistance ability of materials towards deformation under external force.18 In material mechanics, although many derived terms have been used under certain circumstances like elastic modulus/Young's modulus, compress modulus, and shear modulus, in essence, stiffness obeys the classical equation which is measured as load divided by deformation. When speaking to the biological context, the surrounding matrix exerts mechanical forces on cells and generates deformation, which is attributed to the stiffness of the extracellular matrix (ECM). The ECM is a collection of intricately interlocking collagens, fibrillar collagens and glycoproteins.25 The deposition, remodeling, and crosslinking of ECM composition determine the characteristic biomechanics of different cells. Hence, the stiffness of biogenetic tissue, from the softest blood to the most solid bone, can vary from 0.1 kPa to 2 GPa (Fig. 2).26 For better simulation the intrinsic tissue microenvironment, natural proteins or synthetic polymers are modulated by the subtle deployment of components, concentrations, and fabrication methods. Hydrogels like collagen and hyaluronic acid are usually soft materials with stiffness under 800 Pa, whilst the addition of fibrin stiffens the hydrogels to 5 kPa. For the commonest polyethylene glycol (PEG) gels, the stiffness can be varied from 200 Pa to 2 GPa by tuning the crosslinking density. The aforementioned materials are frequently applied in soft adipose tissue regeneration. On the contrary, hard materials (over 10 kPa) like alginate, polycaprolactone (PCL), polylactic acid and polydimethylsiloxane (PDMS) are more suitable in cartilage/bone tissue engineering.27,28 The appropriate adjustment of scaffold stiffness makes a great difference in leading stem cells to scheduled differentiation. Young et al. applied decellularized lipoaspirate with 0.03% polyacrylamide to fabricate gels (with Young's modulus of 2 kPa, close to 3–4 kPa of human adipose tissue), where adipose-derived stem cells (ADSCs) preferred lipid accumulation.29 Soft substrates also help to maintain the pluripotency of MSCs. MSCs cultured on soft substrates (1.5 kPa versus 15 kPa PDMS) presented decreased focal adhesion maturation, stress fiber content and nuclear stretching but enhanced the expression of pluripotency-related genes.30 Thriving studies have attempted to illuminate the involved mechanism about how the stiffness plays such a critical role in manipulating stem cell behaviors.2.1 The YAP/TAZ-related signaling pathways
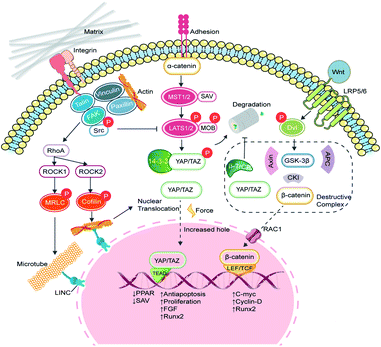
Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ) are two homologous transcriptional co-activator proteins that shuttle between the cytoplasm (inactive state) and nuclei (active state) to regulate target gene expression.31 The YAP/TAZ-relative signaling pathways are broadly divided into two categories, namely the Hippo-dependent and the Hippo-independent. YAP and TAZ are paralog proteins but YAP contains a PDZ-binding motif (PDZ-BM) in its C-terminus, and a proline-rich region in its N-terminus, which are absent in TAZ. Moreover, YAP protein contains a Tea Domain Transcription Factor (TEAD) binding domain, which facilitates the promoter binding.32 The Hippo pathway is considered an important inhibited signal in regulating cell proliferation, apoptosis, and stem cell self-renewal.33 In a general explanation, the upstream adhesion junction senses the inhibition signals, transduces to α-catenin and phosphorylates STE20 family protein kinases MST1/2 and the following large tumor suppressor proteins (Lats1/2). The activated Lats1/2 phosphorylates the serine residues in YAP/TAZ, leaving them to interact with cytoskeleton protein 14-3-3 and degradation.34 When the Hippo signaling is switched to ‘OFF’, YAP/TAZ enters the nucleus, and forms the functional transcriptional complexes with various transcriptional factors such as the TEAD family, Runx2, PPARγ, Smads, and p73 to regulate proliferation, migration, differentiation, antiapoptosis, and morphology (Fig. 3).35 In vertebrates, the upstream information includes apical–basal polarity, planar cell polarity (PCP), mechanical stress and G-protein-coupled receptor (GPCR) signaling, and actin cytoskeleton.36 With recent progress, YAP/TAZ are recognized as core mechanical force transducer factors of cytoskeletal organization, which contribute to the intricate regulation of cell reaction to the physiochemical cues from the extracellular matrix.37 Scaffolds with different elasticity moduli input different mechanical stresses, yielding either positive or negative signals. Dupont et al. built culture conditions with stiffness varying from 0.7 to 40 kPa by fibronectin-coated acrylamide hydrogels and found that YAP/TAZ were predominantly cytoplasmic on soft substrates and became a nuclear subset on harder substrates. Furthermore, by the knockdown of YAP/TAZ, adipogenesis of MSCs was achieved on stiff substrates.38 Fibronectin is a critically important ECM protein and was used to provide sequences for binding with cells via surface markers like integrins and heparin.39 Instead, overexpressed intranuclear TAZ could bind to RUNX2 and execute robust osteogenic procedures in adipose-derived MSCs.40 However, once the Hippo pathway is turned ‘ON’, MST1/2 stimulates the generation of PPARγ and salvador homolog 1 (SAV1) compound, which helps to phosphorylate TAZ and impairs the nuclear translocation of YAP/TAZ. Since TAZ will silence PPARγ, the cytoplasmic location of YAP/TAZ enhances the adipogenic process.41In addition to the Hippo pathway, Wnt/β-catenin signaling also participates in YAP/TAZ regulation. In cytoplasm, β-catenin and YAP/TAZ are bound with a destructive complex and ready for degradation. When membrane receptors Frizzled/LRP 5/6 recognize the secretion of Wnt glycoprotein, disheveled (Dvl) is activated to dissolve the destructive complex and release β-catenin and YAP/TAZ. The phosphorylated β-catenin bridges YAP/TAZ to its ubiquitin ligase β-TrCP, while free β-catenin impairs the degradation of YAP/TAZ.42,43 The remaining β-catenin can interact with E-cadherin and consists of adhesive bands. Otherwise, β-catenin is transported into the nucleus by RAC1, where it binds to the T-cell factor/lymphocyte enhancer factor (LEF/TCF) and initiates the transcription of c-myc, cyclin-D and Runx2.44
2.2 The integrins/FAK/RhoA/ROCK signaling
Integrins are a family of transmembrane heterodimers of non-covalently associated α and β subunits.45 It has been reported that 24 different integrins have been identified in vertebrates, comprised of 18 α and 8 β subunits.46 Each integrin demonstrates a similar shape that resembles a large “head” on two “legs”, with the head including the sites for ligand binding and subunit association.47 Intracellular proteins, such as talin and kindlin, bind to the cytoplasmic face of integrins to trigger a transformation to the activated state, which recruits the assembly of dynamic integrin adhesion complexes, namely focal adhesions (FAs).48 FAs mediate the connection between outside signaling and the cytoskeleton, with the assistance of proteins like vinculin, paxillin, focal adhesion kinase (FAK), and integrin-linked kinase (ILK).49 ILK acts as an adaptor protein at focal adhesion and was found to be a phosphorylase of GSK-3β and Akt.50,51 Mice lacking functional ILK exhibited reduced trabecular bone mass, together with impaired F-actin organization and downregulation of BMP/Smad and Wnt/β-catenin signalling pathways.52 FAK deficiency in mice also led to the downregulation of Wnt/β-catenin signaling and decreased osteoblast number.53 It is obvious that FAs play an important role in mediating cell morphology, migration and differentiation.Substrate stiffness can directly activate integrin β1 and FAK, which accelerates FAs maturation.54 Via FAs, extracellular forces can be transmitted to the cytoskeleton and relative signaling pathways. FAK is elevated with increasing stiffness and transduces external forces to the myosin cytoskeleton by inducing the downstream GTP-bound small GTPases (RhoA).55 For one thing, RhoA continues to activate mDia to enhance F-actin polymerization. For another, RhoA phosphorylates rho-associated protein kinase (ROCK). Two ROCK isoforms (ROCK1 and ROCK2) play distinctive roles in cell motility and mechanotransduction. ROCK1 phosphorylates the myosin regulatory light chain (MRLC) and generated traction force, while ROCK2 phosphorylates cofilin and stimulates the formation of actin bundles to regulate the cytoskeletal remodeling.56 On a soft PDMS matrix, the expression of RhoA and ROCK1/2 decreases in chondrocytes,57 while the activation of cytoskeleton-related adhesion proteins and cascades of paxillin, FAK, PKC, and RhoA were detected in osteoclasts on the stiffer PDMS substrates.58 RhoA/ROCK can not only respond to matrix stiffness but also act as intermediary molecules to mediate the cellular localization of YAP/TAZ. MSCs cultured in big fibronectin pattern have been found to entail the activation of the RhoA/ROCK signaling pathway and stimulate the formation of actin bundles, stress fibres and tensile actomyosin structures, which act as signals of the nuclear translocation of YAP/TAZ.38
2.3 Nuclear deformation
The cytoskeleton is composed of filamentous actin, intermediate filaments and microtubules and acts as a mechanical linker of plasma membranes, organelle membranes and nucleus membranes.59 During the force transduction, talin, the linker of the nucleoskeleton and cytoskeleton (LINC) complex exerts an indispensable effect in coupling ECM, focal adhesions, cytoskeleton, and nucleoskeleton. Integrins bind ligands on the extracellular side, whereas talin couples integrin receptors to the actin cytoskeleton.60 Talin unfolds and leads to focal adhesion and stress fiber formation.61 The LINC complex, as the name indicates, mechanically couples the nucleus to cytoplasmic stress fibers.62 Elosegui-Artola et al. also confirmed the importance of talin unfolding and LINC complex in delivering forces to the nucleus and driving YAP translocation.63 How did forces influence nucleus and even gene expression? By comparing the size of nuclear pores, they discovered that nuclei on stiff substrates showed a flatter geometry with greater size of nuclear pores than on soft substrates, which increased nuclear pore permeability and facilitated YAP nuclear import (Fig. 4A and B). The cellular ability to sense the environment and transmit forces into the nucleus eventually affects the phenotypic endpoints of MSCs.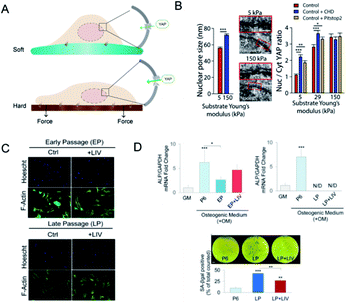 | ||
| Fig. 4 Nuclear deformation and YAP translocation. (A) The mechanism of how nucleus deformation influences the differentiation. In contrast to the soft matrix, more focal adhesions were formed on the hard matrix and transduced force to deform the nucleus and nucleopores, which may facilitate YAP transport. (B) SEM showed the increased nucleopore size when exposed to the stiff matrix. The ratio of nucleus/cytoplasm of YAP proteins was also significantly increased as compared to the soft matrix. (Reprinted from ref. 63 with permission from Elsevier, copyright 2017). (C) LIV increased the cell area of MSCs. (D) Osteogenic genes were promoted in the early passage but declined in the late passage of MSCs; via LIV treatment, SA-βgal positive cells were significantly lower when compared to LP. (Reprinted from ref. 68 with permission from Springer Nature, copyright 2020). | ||
On the other hand, nuclear deformation brings the alteration of chromosomal conformations, changes in genetic information, and even nuclear envelope rupture.64 The chromatin dynamics within the nucleus are also mediated by the cytoskeleton and nucleoskeleton.65 Nuclear levels of histone deacetylase 3 were higher when actomyosin contractility was inhibited.66 Besides, the disruption of the chromatin structure can decrease fibroblast mechanosensitivity and impair the nuclear anisotropy under extrinsic forces.67 Then, it is likely to utilize mechanical cues integrated with nuclear geometry and chromatin remodelling to manipulate gene expression and cell differentiation. It has been reported that regular low-intensity vibration (LIV) protects the early osteogenic and late adipogenic ability of aging MSCs and the recovery of oxidative reductase-related proteins, which indicated that mechanically induced genetic changes have been stored in the nuclei (Fig. 4C and D).68 The novel mechanism converts force into the importing of nuclear molecules and may influence the nucleocytoplasmic shuttling of any protein. However, whether the nuclear deformation induced by specific mechanic signals corresponds with particular molecules shuttling or interacts with mechanosensitive transcriptional regulators remains to be uncovered.
3 Pore size
Pores are the interconnected spaces of the bulk that are not occupied by solid materials but by gases, liquids, or even other microscopic particles. Pore size is the basic morphological property of porous materials, and the concomitant term porosity is defined as the percentage of void space in a solid.69 Pores come in a variety of sizes to address diverse applications (Fig. 5A). According to the IUPAC definition, pores with an internal width of less than 2 nm are referred to as micropores, those with a width between 2 to 50 nm as mesopores, and those larger than 50 nm are called macropores. If biomaterials constitute, metaphorically speaking, the beams of a house, then the crisscross pores provide room for cell motility and facilitate intercellular communication. For the cultivation of stem cells with different origins, choices of scaffold pore sizes were different. Based on polycaprolactone (PCL) scaffolds with various pore sizes, Han et al. found that chondrocytes are suitable for 100 and 200 μm, BMSCs for 200 μm, and tendon stem cells for 200 and 300 μm (Fig. 5C).70 Even for the same stem cell seeds, distinct pore sizes may imply different differential endpoints.71 Generally, compact and hard tissues like bone and skin require smaller pore sizes while the regeneration of cartilage and fat prefers scaffolds with larger pore sizes. When it comes to tissue engineering and regeneration, the optimal pore size should be consistent with the sizes of the initial cell morphing along with the eventual tissue. The minimal pore size is considered to be approximately 100 μm due to cell size, migration and transportation requirements.71 The activities of neurogliocytes and fibroblasts are ensured in such pores, yet 100–300 μm is recommended for osteoblasts and 200–400 μm for the growth of cartilage and vascularization.72 Adipose tissue engineering requires adequate pore size due to the fragility of adipocytes and the high demand for oxygen and blood supply. Adipogenic differentiation of MSCs seemed feasible in scaffolds within the pore size range of 200–600 μm, yet the largest pores proved to achieve the best adipogenesis.73 Beyond the primary concern of tissue volume, the relationship between pore size and other mechanical properties, and the effect of crosstalk along with synergistic effects on cell behaviors should also be taken into consideration.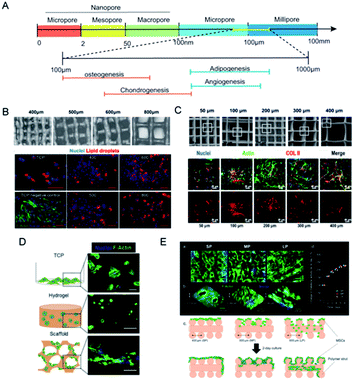 | ||
| Fig. 5 Pore sizes of scaffolds regulate cell movement and fluid penetration. (A) The classification of pore sizes and the optimal ranges for tissue regeneration. (B) Gelatin scaffold made with pore sizes from 400 μm to 800 μm, and better distribution and value of lipid staining were seen in the 800 μm group. (Reprinted from ref. 73 with permission from Wiley, copyright 2020). (C) PCL scaffolds with pore sizes ranging from 50 μm to 400 μm, and the 100 μm to 200 μm group exhibited the best osteogenic gene expression. (Reprinted from ref. 70 with permission from Frontiers, copyright 2021). (D) MSCs cultured in different matrixes made from alginates showed different morphologies. (Reprinted from ref. 82 with permission from Elsevier, copyright 2017). (E) Pore sizes of gelatin-based scaffolds determined whether MSCs would gather or expand. (Reprinted from ref. 84 with permission from American Chemical Society, copyright 2019). | ||
3.1 Pore size tailors stiffness
There comes a general agreement that bulk porosity is negatively correlated to stiffness. With particular raw materials, a larger pore size sacrifices the portion of solid materials, yields greater porosity, and results in weaker scaffold rigidity. As we elaborated before, scaffold stiffness renders significant physical cues to determine stem cell fate, which offers pore size access to indirectly regulate cellular functions through tailoring scaffold stiffness. Pore sizes vary considerably based on the method of fabrication. Slat leaching is used to generate scaffolds with chosen pore sizes ranging from 500 to 600 μm.74 Electrospun scaffolds had considerably smaller pore sizes (6–70 μm) and slightly lower porosity.75 With higher pressure pore-foaming agents, the average pore size of elastin hydrogels increased from 3.9 μm to 79.8 μm, yielding a compression modulus change from 18.8 to 8.6 kPa.76 When the printed Gel-MA scaffold achieved an increase in pore size from 400 to 800 μm, the compressive modulus significantly declined from 892 to 124 Pa.73 The introduction of engineered porosity allows the stiffness-to-weight ratio to optimize the desired mechanical properties. However, the present methods to fabricate porous hydrogels suffer from a deficiency of pore size and delicate structure. 3D print provides another strategy to precisely control the geometry and mechanical structure to create macroporous and multifunctional materials.77 For instance, PCL scaffolds were made with pore sizes of 215, 320 and 500 μm via fused deposition modelling. Among them, the 215 μm group exhibited the greatest tensile and compressive moduli, and finally improved the efficiency of MSCs at the aspect of proliferation and ECM deposition.69 Similarly, pore sizes larger than 200 μm were proved to support adipogenic differentiation from MSCs, better infiltration of cells and homogeneous distribution of lipid droplets were observed when the scaffold stiffness declined with the pore size being augmented (Fig. 5B).733.2 Pore size affects adhesion and penetration
Pore size that is approximate or slightly greater than cell diameter was proved to benefit initial cell attachment to a scaffold and lay a foundation for the subsequent cell migration and proliferation. Scaffolds with small pores (50 μm) provided more support and showed increased initial cell adhesion but restricted cell proliferation, while bigger pore sizes provided enough space for cell migration and cross-talks but led to weak mechanical strength (Fig. 5C).70 Osteoblasts in collagen–glycosaminoglycan scaffold with small pore size proliferated significantly on the first two days but the advantage was lost after 7 day culture,78 which was speculated that because of the morphogenesis of cells, a larger pore size allows space for cell attachment and proliferation in addition to the optimal diffusion of nutrients and vessels in-growth. MSCs cultured in big fibronectin islands showed cell spreading and stress fibre formation, which constitutes a mechanotransduction system of cell migration.38 Macroporous scaffolds were also accompanied by outstanding swelling properties,69 indicating feasible medium infiltration, substance exchange and signal transmission through the interconnected holes. Therefore, ASCs and the vascular endothelium prefer macroporous scaffolds with pore sizes of over 400 μm.79 On the contrary, in PCL layers with a pore size of 750 nm, ossification markers and microvessel markers were detected, and transcriptome sequencing revealed the activation of the HIF1α/FAK axis.22Apart from the multi-lineage differentiation function, MSCs are also acknowledged for remarkable paracrine action. A diversity of cytokines like IGF, FGF, PDGF, VEGF, ILs and MMPs are secreted to manipulate cell behaviors and influence tissue organization.80 The exosomes of MSCs have been proved to exert properties of neuroprotection, cardiac repair and anti-inflammation.81 An interconnected macroporous structure permits better interstitial fluid penetration and may render promoted paracrine action. Qazi et al. used alginates to fabricate nanoporous hydrogels (mean pore size: 5 nm) and macroporous scaffolds (mean pore size: 122 μm) sharing similar mean stiffness values of ∼20 kPa.82 MSCs were allowed to spread and contact on the scaffolds and exhibited the striking elevation of growth factor secretion (Fig. 5D). Intriguingly, they found that macrostructures promoted cell migration but hindered myotube formation, and in-depth investigation revealed the effect of N-cadherin that promotes cell–cell adhesion. For chondrocytes with relatively small size and stronger adhesion, smaller pore size seemed to promote cell movement and proliferation.83 In another study, Qazi and his colleagues printed collagen scaffolds with pore sizes distributed from about 200 μm to 382 μm. Smaller pores hampered cell infiltration, large ones caused cells to flow through the scaffolds and medium pores helped MSCs to aggregate and enhanced the angiogenic factors paracrine secretion (Fig. 5E).84 The optimal pore size is not “the bigger, the better” and should be wisely selected for different cells and target tissues.
4 Topology
Topology is the study of dots and lines by dissolving shapes and spaces. Topological materials preserve the shape under stretching or squeezing.26 Yet, in biomaterials engineering, topology is usually referred to as the macro-geometry and micro-surface features of the scaffolds.85 Efforts in designing and manufacturing scaffolds with topological structure serve to recapitulate the topographic heterogeneity of native tissues. For example, muscles, vessels, nerves, and tendons shared a homeomorphic microtubular structure. Hwangbo et al. designed a uniaxially aligned microtubular collagen scaffold, the so-called lotus-like structure, to induce aligned myoblasts distribution and significant myogenesis (Fig. 6A).23 Adipose tissue, in contrast, features loose connective tissue consisting of adipocytes filled with large globules of lipid and surrounded by fiber networks. Therefore, it is reasonable to construct a round, porous, and contractile environment for adipose tissue regeneration. By the use of micropatterned substrates, cell shape can be tightly controlled at the micro-and nanometer scale in vitro. Kilian et al., have already proved that MSCs planted on circular media represent the promoted expression of adipogenesis markers as compared to that on the holly leaf shape (Fig. 6B).86 With the assistance of soft lithography and photopolymerization, researchers can carve sophisticated structures like microgrooves, nanodots, ridges, etc. on scaffold surfaces.87 Meanwhile, a growing number of studies make the effort to probe the hidden mechanism behind topological regulation.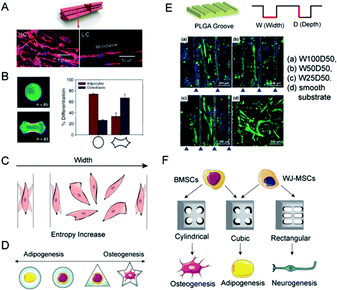 | ||
| Fig. 6 Topology determines the differential orientation of MSCs through micropatterns. (A) A lotus-like PVA/PCL scaffold induced the linear alignment of C2C12 cells. (Reprinted from ref. 23 with permission from American Chemical Society, copyright 2020). (B) MSCs planted on circular media showed the tendency for adipogenesis, while a holly leaf shape enhanced osteogenesis. (Reprinted from ref. 86 with permission from PNAS, copyright 2010). (C) The theory of entropy morphology increase for explaining the alignment of cells near the borders. (D) The osteogenic/adipogenic axis of a single MSC can be regulated by micropatterns. Circular and star shapes respectively supported the optimal adipogenic and osteogenic differentiations. (E) When cultured on micro-grooved PLGA with different w/d ratios, the alignment degrees of myotubes generated from C2C12 cells were different. The W100D50 group showed maximum alignment. (Reprinted from ref. 97 with permission from American Chemical Society, copyright 2021). (F) MSCs are inclined to express adipogenic genes while incubated in a restrained cubic scaffold, whereas the cylindrical and rectangular respectively facilitated the osteogenic and neurogenic differentiation. | ||
4.1 Contact guidance
Extracellular physical cues are continuously sensed and transmitted to the nuclei via mechanotransduction networks. This interaction between cells and the anisotropic environment fuels a cellular orientation response, viz. cellular organization. Cellular organization affects the microarchitecture of tissues and induces contact guidance, a phenomenon where cells are inclined to migrate along the direction of anisotropy (Fig. 6C).88 As a concrete manifestation, cells seeded on a substrate with a gradient of stiffness migrated from the soft towards the stiff side or aligned with the boundary to avoid the soft substrate.89 In addition, cells on stiffer substrates exhibited more elongated and spread geometry.90 Contact guidance is considered a symbol of recognition between cell morphology and matrix topographies. In the study of myogenic differentiation on patterned polystyrene, myoblasts showed a tendency toward alignment and elongation along the length of the grating patterns.91 According to the morphometric analysis of cells and the stress-fiber arrangements on fibronectin striped substrates, fibroblasts were aligned when stripe widths were smaller than cell sizes, whilst cell shapes fluctuated on larger stripe widths.88 It was proposed that cells tend to rotate and wander to achieve the maximization of morphological entropy. Nonetheless, the spatial constraint keeps the cells in line with the edges.The reconstruction of topological features generally occurs together with the introduction of polymeric coatings. Synthetic materials that have outstanding merits like manufacturability, easy modification, and tuneable physical properties, yet confront the problem of biocompatibility, are synthetic materials endowed with bioactive molecules, such as ligands for adhesion receptors, functional parts of natural growth factors, or synthetic regulators.92 Adhesive Arg-Gly-Asp (RGD) peptides are probably the most widely used and their effectiveness to promote cell adhesion has been verified on different substrates.93 Wong et al. found that high RGD tethered mobility delayed the early adhesion and spreading of MSCs, leading to compromised osteogenic differentiation, possibly due to additional time and effort for cells to develop a mechanical response, thereby delaying the maturation of FAs and activation of subsequent mechanotransduction signalling. In contrast, MSCs cultured on substrates with restricted RGD tethered mobility showed significantly better adhesion, spreading, and osteogenic differentiation.94
4.2 Contact transduction
Due to the micro- and nano-patterning techniques, elaborate engineered platforms are produced to confine cells to specific shapes and investigate cell behaviors at the individual scale. The initial physical contact between cells and scaffolds induces integrin assembly, and the follow-up focal adhesion formation transduces the biophysical cues into downstream cytoplasmic β-catenin accumulation and the resultant nuclear translocation.95 With shape constraint by micropatterns, contact brings cell deformation, and it seems that the aspect ratios of micropatterns play a key role in stem cell differentiation (Fig. 6D).96 Osteoblasts cultured in PLGA microgrooves with the highest width/depth ratio were discovered to exhibit robust myogenic differentiation and integrin-mediated FAK signaling activation (Fig. 6E).97 Similarly, scaffolds with cubic pores promoted the gene expression of adipogenesis and chondrogenesis, when MSCs in cylindrical pores expressed osteogenic markers (Fig. 6F).98 Muneekaew et al. applied this method to other differentiation and further corroborated its generality. They discovered that umbilical cord-derived MSCs cultured on rectangular collagen micropatterns showed neurogenic differentiation while the square ones induced adipogenesis (Fig. 6F).99 The mechanism of morphology-derived differentiation could be attributed to the interactions of F-actin, integrin, and extracellular environment, which transform physical cues into chemical cascades and thus changes in gene expression.5 Conclusions
Engineering stem cell culture systems offer an opportunity for the rapid progress of regenerative medicine. With an in-depth understanding of how the extracellular environment interacts with cell behaviors, scaffolds that capture and recapitulate in in vivo environments are a core factor for a satisfying outcome. In this article, we emphasize the outstanding effect of mechanical properties, mainly stiffness, pore size and topography, on stem cell mobility, functions and eventually the tissue differential endpoints. Stiffness plays a fundamental role in translating other mechanical cues and transducing mechanic force to nuclear, where the YAP/TAZ location, focal adhesion-related actin function, and nuclear deformation caused molecules to come into play. Pore sizes indirectly alter the mechanical strength of scaffolds and directly affect cell adhesion and material exchange. Topology is an emerging realm that has earned much expectation. The contact between cells and scaffolds transmits intriguing direction by confining cell/nucleus geometry and guiding adhesion and migration. All mechanical cues are united in the mechanotransduction system driven through information recognition by integrins, signaling cascades transmitted by molecules, gene expression launched by transcription factors, and deformation mediated by cytoskeleton arrangement.However, many puzzles remain to be unveiled. For example, whether any membrane protein has participated in recognizing mechanic cues, and if a direct correspondence exists between specific information input and gene expression output. Besides, other mechanical signals like viscoelasticity, fluid flow and tension are not expatiated here because these signals indirectly or synergistically function with the mentioned three and specific signaling pathways remain unclear. Sonam et al. have found that when fluid shear stress was applied, MSCs seeded on 1 μm wells underwent osteogenesis, whereas those on 2 μm gratings remained multipotent.100 The application of shear stress and contractile topography resulted in increased intracellular tension of MSCs and influenced the engagement of actin microfilaments and finally cell proliferation and differentiation.
Last but not least, not only stem cells can sense the mechanical signals of the environment, but also ECM can receive the intrinsic signals from the cell niche and extrinsic signals from other tissue. Scaffolds that can modulate their own properties to respond to environmental changes have great prospects after transplanting in living bodies. Understanding the interactions between environment mechanics and cell behaviors also provides novel insight for coping with progressive cancer, fibrosis and other diseases highly associated with ECM properties. Animal models have been extensively used in obesity research, yet dietary, genetic, and chemical modifications limit their applicability to human translation.101 The in-depth investigation of mechanical cues in controlling differentiation of pluripotent stem cells may pave the road to broader research in adipose tissue regeneration or obesity-related diseases.
Author contributions
Su Ting: conceptualization, writing original draft. Xu Mimi: data curation, revision. Lu Feng: supervision, validation. Chang Qiang: writing-review & editing, validation, supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the following foundation: National Natural Science Foundation of China (Grant number: 81772101, 82072196, 82002066). We also thank the support from the Science Fund for Distinguished Young Scholars of Southern Medical University (Grant number: 2020J009).Notes and references
- F. Berthiaume, T. J. Maguire and M. L. Yarmush, Annu. Rev. Chem. Biomol. Eng., 2011, 2, 403–430 CrossRef PubMed.
- Y. Han, X. Li, Y. Zhang, Y. Han, F. Chang and J. Ding, Cells, 2019, 8, 886 CrossRef CAS PubMed.
- C. Uder, S. Brückner, S. Winkler, H. M. Tautenhahn and B. Christ, Cytometry, Part A, 2018, 93, 32–49 CrossRef CAS PubMed.
- Q. Chen, P. Shou, C. Zheng, M. Jiang, G. Cao, Q. Yang, J. Cao, N. Xie, T. Velletri, X. Zhang, C. Xu, L. Zhang, H. Yang, J. Hou, Y. Wang and Y. Shi, Cell Death Differ., 2016, 23, 1128–1139 CrossRef CAS PubMed.
- D. Moseti, A. Regassa and W. K. Kim, Int. J. Mol. Sci., 2016, 17, 124 CrossRef PubMed.
- A. L. Ghaben and P. E. Scherer, Nat. Rev. Mol. Cell Biol., 2019, 20, 242–258 CrossRef CAS PubMed.
- J. M. Suh, X. Gao, J. McKay, R. McKay, Z. Salo and J. M. Graff, Cell Metab., 2006, 3, 25–34 CrossRef CAS PubMed.
- E. A. Wang, D. I. Israel, S. Kelly and D. P. Luxenberg, Growth Factors, 1993, 9, 57–71 CrossRef CAS PubMed.
- J. B. Lian, G. S. Stein, A. Javed, A. J. van Wijnen, J. L. Stein, M. Montecino, M. Q. Hassan, T. Gaur, C. J. Lengner and D. W. Young, Rev. Endocr. Metab. Disord., 2006, 7, 1–16 CrossRef CAS PubMed.
- L. Hu, C. Yin, F. Zhao, A. Ali, J. Ma and A. Qian, Int. J. Mol. Sci., 2018, 19, 10 Search PubMed.
- W. C. W. Chan, Z. Tan, M. K. T. To and D. Chan, Int. J. Mol. Sci., 2021, 22, 5445 CrossRef CAS PubMed.
- W. Jing, Y. Lin, L. Wu, X. Li, X. Nie, L. Liu, W. Tang, X. Zheng and W. Tian, Cell Tissue Res., 2007, 330, 567–572 CrossRef PubMed.
- Y. Kimura, W. Tsuji, H. Yamashiro, M. Toi, T. Inamoto and Y. Tabata, J. Tissue Eng. Regener. Med., 2010, 4, 55–61 CAS.
- L. S. Wang, H. Wang, Q. L. Zhang, Z. J. Yang, F. X. Kong and C. T. Wu, Hum. Gene Ther., 2018, 29, 413–423 CrossRef CAS PubMed.
- Q. Zhang, J. Hubenak, T. Iyyanki, E. Alred, K. C. Turza, G. Davis, E. I. Chang, C. D. Branch-Brooks, E. K. Beahm and C. E. Butler, Biomaterials, 2015, 73, 198–213 CrossRef CAS PubMed.
- Y. Qiao, X. Liu, X. Zhou, H. Zhang, W. Zhang, W. Xiao, G. Pan, W. Cui, H. A. Santos and Q. Shi, Adv. Healthcare Mater., 2020, 9, e1901239 CrossRef PubMed.
- D. S. Amarasekara, S. Kim and J. Rho, Int. J. Mol. Sci., 2021, 22, e8 Search PubMed.
- H. Lv, H. Wang, Z. Zhang, W. Yang, W. Liu, Y. Li and L. Li, Life Sci., 2017, 178, 42–48 CrossRef CAS PubMed.
- S. Wang, S. Hashemi, S. Stratton and T. L. Arinzeh, Adv. Healthcare Mater., 2021, 10, e2001244 CrossRef PubMed.
- Y. Liu, Z. Li, J. Li, S. Yang, Y. Zhang, B. Yao, W. Song, X. Fu and S. Huang, Burns & Trauma, 2020, 8, tkaa029 Search PubMed.
- L. R. Smith, S. Cho and D. E. Discher, Physiology, 2018, 33, 16–25 CrossRef CAS PubMed.
- Y. Sun, Q. Wu, Y. Zhang, K. Dai and Y. Wei, Nanomedicine, 2021, 37, 102426 CrossRef CAS PubMed.
- H. Hwangbo, W. Kim and G. H. Kim, ACS Appl. Mater. Interfaces, 2021, 13, 12656–12667 CrossRef CAS PubMed.
- W. L. Murphy, T. C. McDevitt and A. J. Engler, Nat. Mater., 2014, 13, 547–557 CrossRef CAS PubMed.
- A. D. Theocharis, D. Manou and N. K. Karamanos, FEBS J., 2019, 286, 2830–2869 CrossRef CAS PubMed.
- Y. Yang, K. Wang, X. Gu and K. W. Leong, Engineering, 2017, 3, 36–54 CrossRef CAS PubMed.
- C. D. Markert, X. Guo, A. Skardal, Z. Wang, S. Bharadwaj, Y. Zhang, K. Bonin and M. Guthold, J. Mech. Behav. Biomed. Mater., 2013, 27, 115–127 CrossRef CAS PubMed.
- S. Nemir and J. L. West, Ann. Biomed. Eng., 2010, 38, 2–20 CrossRef PubMed.
- D. A. Young, Y. S. Choi, A. J. Engler and K. L. Christman, Biomaterials, 2013, 34, 8581–8588 CrossRef CAS PubMed.
- H. Gerardo, A. Lima, J. Carvalho, J. R. D. Ramos, S. Couceiro, R. D. M. Travasso, R. Pires das Neves and M. Grãos, Sci. Rep., 2019, 9, 9086 CrossRef PubMed.
- A. Pocaterra, P. Romani and S. Dupont, J. Cell Sci., 2020, 133, jcs230425 CrossRef CAS PubMed.
- C. Webb, A. Upadhyay, F. Giuntini, I. Eggleston, M. Furutani-Seiki, R. Ishima and S. Bagby, Biochemistry, 2011, 50, 3300–3309 CrossRef CAS PubMed.
- S. A. Mosaddad, Y. Salari, S. Amookhteh, R. S. Soufdoost, A. Seifalian, S. Bonakdar, F. Safaeinejad, M. M. Moghaddam and H. Tebyanian, Cell. Physiol. Biochem., 2021, 55, 33–60 CrossRef CAS PubMed.
- J. Petzold and E. Gentleman, Front. Cell Dev. Biol., 2021, 9, 761871 CrossRef PubMed.
- B. C. Heng, X. Zhang, D. Aubel, Y. Bai, X. Li, Y. Wei, M. Fussenegger and X. Deng, Cell. Mol. Life Sci., 2021, 78, 497–512 CrossRef CAS PubMed.
- F. X. Yu and K. L. Guan, Genes Dev., 2013, 27, 355–371 CrossRef CAS PubMed.
- B. C. Heng, X. Zhang, D. Aubel, Y. Bai, X. Li, Y. Wei, M. Fussenegger and X. Deng, Front. Cell Dev. Biol., 2020, 8, 735 CrossRef PubMed.
- S. Dupont, L. Morsut, M. Aragona, E. Enzo, S. Giulitti, M. Cordenonsi, F. Zanconato, J. Le Digabel, M. Forcato, S. Bicciato, N. Elvassore and S. Piccolo, Nature, 2011, 474, 179–183 CrossRef CAS PubMed.
- J. Patten and K. Wang, Adv. Drug Delivery Rev., 2021, 170, 353–368 CrossRef CAS PubMed.
- Y. Zhu, Y. Wu, J. Cheng, Q. Wang, Z. Li, Y. Wang, D. Wang, H. Wang, W. Zhang, J. Ye, H. Jiang and L. Wang, Stem Cell Res. Ther., 2018, 9, 53 CrossRef CAS PubMed.
- B. H. Park, D. S. Kim, G. W. Won, H. J. Jeon, B. C. Oh, Y. Lee, E. G. Kim and Y. H. Lee, PLoS One, 2012, 7, e30983 CrossRef CAS PubMed.
- L. Azzolin, F. Zanconato, S. Bresolin, M. Forcato, G. Basso, S. Bicciato, M. Cordenonsi and S. Piccolo, Cell, 2012, 151, 1443–1456 CrossRef CAS PubMed.
- L. Azzolin, T. Panciera, S. Soligo, E. Enzo, S. Bicciato, S. Dupont, S. Bresolin, C. Frasson, G. Basso, V. Guzzardo, A. Fassina, M. Cordenonsi and S. Piccolo, Cell, 2014, 158, 157–170 CrossRef CAS PubMed.
- G. Hong, X. He, Y. Shen, X. Chen, F. Yang, P. Yang, F. Pang, X. Han, W. He and Q. Wei, Stem Cell Res. Ther., 2019, 10, 277 CrossRef PubMed.
- I. D. Campbell and M. J. Humphries, Cold Spring Harbor Perspect. Biol., 2011, 3, a004994 Search PubMed.
- M. Barczyk, S. Carracedo and D. Gullberg, Cell Tissue Res., 2010, 339, 269–280 CrossRef CAS PubMed.
- L. Wang, F. Zheng, R. Song, L. Zhuang, M. Yang, J. Suo and L. Li, Stem Cell Rev. Rep., 2022, 18, 126–141 CrossRef PubMed.
- L. Zhu, E. F. Plow and J. Qin, Protein Sci., 2021, 30, 531–542 CrossRef CAS PubMed.
- E. R. Horton, A. Byron, J. A. Askari, D. H. J. Ng, A. Millon-Frémillon, J. Robertson, E. J. Koper, N. R. Paul, S. Warwood, D. Knight, J. D. Humphries and M. J. Humphries, Nat. Cell Biol., 2015, 17, 1577–1587 CrossRef CAS PubMed.
- M. Maydan, P. C. McDonald, J. Sanghera, J. Yan, C. Rallis, S. Pinchin, G. E. Hannigan, L. J. Foster, D. Ish-Horowicz, M. P. Walsh and S. Dedhar, PLoS One, 2010, 5, e12356 CrossRef PubMed.
- A. A. Troussard, N. M. Mawji, C. Ong, A. Mui, R. St-Arnaud and S. Dedhar, J. Biol. Chem., 2003, 278, 22374–22378 CrossRef CAS PubMed.
- M. Dejaeger, A. M. Böhm, N. Dirckx, J. Devriese, E. Nefyodova, R. Cardoen, R. St-Arnaud, J. Tournoy, F. P. Luyten and C. Maes, J. Bone Miner. Res., 2017, 32, 2087–2102 CrossRef CAS PubMed.
- C. Sun, H. Yuan, L. Wang, X. Wei, L. Williams, P. H. Krebsbach, J. L. Guan and F. Liu, J. Bone Miner. Res., 2016, 31, 2227–2238 CrossRef CAS PubMed.
- G. Halder, S. Dupont and S. Piccolo, Nat. Rev. Mol. Cell Biol., 2012, 13, 591–600 CrossRef CAS PubMed.
- W. Zhang, S. Zhang, W. Zhang, Y. Yue, W. Qian and Z. Wang, Biochim. Biophys. Acta, Rev. Cancer, 2021, 1876, 188583 CrossRef CAS PubMed.
- Y. Peng, Z. Chen, Y. Chen, S. Li, Y. Jiang, H. Yang, C. Wu, F. You, C. Zheng, J. Zhu, Y. Tan, X. Qin and Y. Liu, Acta Biomater., 2019, 88, 86–101 CrossRef CAS PubMed.
- T. Zhang, T. Gong, J. Xie, S. Lin, Y. Liu, T. Zhou and Y. Lin, ACS Appl. Mater. Interfaces, 2016, 8, 22884–22891 CrossRef CAS PubMed.
- Q. Wang, J. Xie, C. Zhou and W. Lai, Cell Proliferation, 2022, 55, e13172 CAS.
- V. D. Tran and S. Kumar, Curr. Opin. Cell Biol., 2021, 68, 64–71 CrossRef CAS PubMed.
- S. Chakraborty, S. Banerjee, M. Raina and S. Haldar, Biochemistry, 2019, 58, 4677–4695 CrossRef CAS PubMed.
- A. Elosegui-Artola, R. Oria, Y. Chen, A. Kosmalska, C. Pérez-González, N. Castro, C. Zhu, X. Trepat and P. Roca-Cusachs, Nat. Cell Biol., 2016, 18, 540–548 CrossRef CAS PubMed.
- M. L. Lombardi, D. E. Jaalouk, C. M. Shanahan, B. Burke, K. J. Roux and J. Lammerding, J. Biol. Chem., 2011, 286, 26743–26753 CrossRef CAS PubMed.
- A. Elosegui-Artola, I. Andreu, A. E. M. Beedle, A. Lezamiz, M. Uroz, A. J. Kosmalska, R. Oria, J. Z. Kechagia, P. Rico-Lastres, A. L. Le Roux, C. M. Shanahan, X. Trepat, D. Navajas, S. Garcia-Manyes and P. Roca-Cusachs, Cell, 2017, 171, 1397–1410.e14 CrossRef CAS PubMed.
- Z. Jahed and M. R. Mofrad, Curr. Opin. Cell Biol., 2019, 58, 114–119 CrossRef CAS PubMed.
- M. T. Doolin, R. A. Moriarty and K. M. Stroka, Front. Physiol., 2020, 11, 365 CrossRef PubMed.
- M. Bao, J. Xie, A. Piruska and W. T. S. Huck, Nat. Commun., 2017, 8, 1962 CrossRef PubMed.
- K. Haase, J. K. Macadangdang, C. H. Edrington, C. M. Cuerrier, S. Hadjiantoniou, J. L. Harden, I. S. Skerjanc and A. E. Pelling, Sci. Rep., 2016, 6, 21300 CrossRef CAS PubMed.
- G. Bas, S. Loisate, S. F. Hudon, K. Woods, E. J. Hayden, X. Pu, R. Beard, J. T. Oxford and G. Uzer, Sci. Rep., 2020, 10, 9369 CrossRef CAS PubMed.
- Z. Z. Zhang, D. Jiang, J. X. Ding, S. J. Wang, L. Zhang, J. Y. Zhang, Y. S. Qi, X. S. Chen and J. K. Yu, Acta Biomater., 2016, 43, 314–326 CrossRef CAS PubMed.
- Y. Han, M. Lian, Q. Wu, Z. Qiao, B. Sun and K. Dai, Front. Bioeng. Biotechnol., 2021, 9, 629270 CrossRef PubMed.
- V. Karageorgiou and D. Kaplan, Biomaterials, 2005, 26, 5474–5491 CrossRef CAS PubMed.
- I. Bružauskaitė, D. Bironaitė, E. Bagdonas and E. Bernotienė, Cytotechnology, 2016, 68, 355–369 CrossRef PubMed.
- L. Tytgat, M. R. Kollert, L. Van Damme, H. Thienpont, H. Ottevaere, G. N. Duda, S. Geissler, P. Dubruel, S. Van Vlierberghe and T. H. Qazi, Macromol. Biosci., 2020, 20, e1900364 CrossRef PubMed.
- M. Flaibani and N. Elvassore, Mater. Sci. Eng., C, 2012, 32, 1632–1639 CrossRef CAS PubMed.
- J. Xue, D. Pisignano and Y. Xia, Adv. Sci., 2020, 7, 2000735 CrossRef CAS PubMed.
- N. Annabi, S. M. Mithieux, E. A. Boughton, A. J. Ruys, A. S. Weiss and F. Dehghani, Biomaterials, 2009, 30, 4550–4557 CrossRef CAS PubMed.
- K. J. De France, F. Xu and T. Hoare, Adv. Healthcare Mater., 2018, 7, 1700927 CrossRef PubMed.
- C. M. Murphy, M. G. Haugh and F. J. O'Brien, Biomaterials, 2010, 31, 461–466 CrossRef CAS PubMed.
- B. Feng, Z. Jinkang, W. Zhen, L. Jianxi, C. Jiang, L. Jian, M. Guolin and D. Xin, Biomed. Mater., 2011, 6, 015007 CrossRef PubMed.
- P. R. Baraniak and T. C. McDevitt, Regener. Med., 2010, 5, 121–143 CrossRef PubMed.
- C. Gwam, N. Mohammed and X. Ma, Ann. Transl. Med., 2021, 9, 70 CrossRef CAS PubMed.
- T. H. Qazi, D. J. Mooney, G. N. Duda and S. Geissler, Biomaterials, 2017, 140, 103–114 CrossRef CAS PubMed.
- Z. Tan, T. Liu, J. Zhong, Y. Yang and W. Tan, J. Biomed. Mater. Res., Part A, 2017, 105, 3281–3292 CrossRef CAS PubMed.
- T. H. Qazi, L. Tytgat, P. Dubruel, G. N. Duda, S. Van Vlierberghe and S. Geissler, ACS Biomater. Sci. Eng., 2019, 5, 5348–5358 CrossRef CAS PubMed.
- M. Bai, L. Cai, X. Li, L. Ye and J. Xie, J. Biomed. Mater. Res., Part B, 2020, 108, 2426–2440 CrossRef CAS PubMed.
- K. A. Kilian, B. Bugarija, B. T. Lahn and M. Mrksich, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4872–4877 CrossRef CAS PubMed.
- T. Gonzalez-Fernandez, A. J. Tenorio and J. K. Leach, 3D Print. Addit. Manuf., 2020, 7, 139–147 CrossRef PubMed.
- A. B. C. Buskermolen, H. Suresh, S. S. Shishvan, A. Vigliotti, A. DeSimone, N. A. Kurniawan, C. V. C. Bouten and V. S. Deshpande, Biophys. J., 2019, 116, 1994–2008 CrossRef CAS PubMed.
- E. A. Novikova, M. Raab, D. E. Discher and C. Storm, Phys. Rev. Lett., 2017, 118, 078103 CrossRef PubMed.
- J. Li, D. Han and Y. P. Zhao, Sci. Rep., 2014, 4, 3910 CrossRef PubMed.
- L. M. Murray, V. Nock, J. J. Evans and M. M. Alkaisi, J. Biomed. Mater. Res., Part A, 2016, 104, 1638–1645 CrossRef CAS PubMed.
- L. Yin, J. Cheng, T. J. Deming and M. J. Vicent, Adv. Drug Delivery Rev., 2021, 178, 113995 CrossRef CAS PubMed.
- S. Guo, X. Zhu and X. J. Loh, Mater. Sci. Eng., C, 2017, 70, 1163–1175 CrossRef CAS PubMed.
- D. S. Wong, J. Li, X. Yan, B. Wang, R. Li, L. Zhang and L. Bian, Nano Lett., 2017, 17, 1685–1695 CrossRef CAS PubMed.
- J. Xie, C. Zhou, D. Zhang, L. Cai, W. Du, X. Li and X. Zhou, ACS Appl. Bio Mater., 2018, 1, 792–801 CrossRef CAS PubMed.
- R. Peng, X. Yao and J. Ding, Biomaterials, 2011, 32, 8048–8057 CrossRef CAS PubMed.
- H. Gao, J. Xiao, Y. Wei, H. Wang, H. Wan and S. Liu, ACS Omega, 2021, 6, 20931–20940 CrossRef CAS PubMed.
- K. M. Ferlin, M. E. Prendergast, M. L. Miller, D. S. Kaplan and J. P. Fisher, Acta Biomater., 2016, 32, 161–169 CrossRef CAS PubMed.
- S. Muneekaew, M. J. Wang and S. Y. Chen, Sci. Rep., 2022, 12, 1812 CrossRef CAS PubMed.
- S. Sonam, S. R. Sathe, E. K. Yim, M. P. Sheetz and C. T. Lim, Sci. Rep., 2016, 6, 20415 CrossRef CAS PubMed.
- M. K. DeBari and R. D. Abbott, Int. J. Mol. Sci., 2020, 21, 6030 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |