Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Modulation of biliverdin dynamics and spectral properties by Sandercyanin†
Swagatha Ghosh‡
 *ab,
Sayan Mondal‡§
*ab,
Sayan Mondal‡§
 c,
Keerti Yadav
c,
Keerti Yadav ad,
Shantanu Aggarwalc,
Wayne F. Schaefer
ad,
Shantanu Aggarwalc,
Wayne F. Schaefer e,
Chandrabhas Narayana
e,
Chandrabhas Narayana c and
Ramaswamy Subramanian
c and
Ramaswamy Subramanian *af
*af
aInstitute for Stem Cell Science and Regenerative Medicine, Bangalore, 560065, India. E-mail: swagatha.ghosh@gu.se; subram68@purdue.edu
bDepartment of Chemistry and Molecular Biology, University of Gothenburg, Medicinaregatan 9C, 40530 Gothenburg, Sweden
cChemistry and Physics of Materials Unit, Jawaharlal Nehru Centre for Advanced Scientific Research, Bangalore 560064, Karnataka, India
dManipal Academy of Higher Education, Manipal University, Madhav Nagar, 576104, India
eDepartment of Biological Sciences, University of Wisconsin at Milwaukee, Washington County, West Bend, WI 53095, USA
fDepartment of Biological Sciences, Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN 47907, USA
First published on 13th July 2022
Abstract
Biliverdin IX-alpha (BV), a tetrapyrrole, is found ubiquitously in most living organisms. It functions as a metabolite, pigment, and signaling compound. While BV is known to bind to diverse protein families such as heme-metabolizing enzymes and phytochromes, not many BV-bound lipocalins (ubiquitous, small lipid-binding proteins) have been studied. The molecular basis of binding and conformational selectivity of BV in lipocalins remains unexplained. Sandercyanin (SFP)–BV complex is a blue lipocalin protein present in the mucus of the Canadian walleye (Stizostedion vitreum). In this study, we present the structures and binding modes of BV to SFP. Using a combination of designed site-directed mutations, X-ray crystallography, UV/VIS, and resonance Raman spectroscopy, we have identified multiple conformations of BV that are stabilized in the binding pocket of SFP. In complex with the protein, these conformers generate varied spectroscopic signatures both in their absorption and fluorescence spectra. We show that despite no covalent anchor, structural heterogeneity of the chromophore is primarily driven by the D-ring pyrrole of BV. Our work shows how conformational promiscuity of BV is correlated to the rearrangement of amino acids in the protein matrix leading to modulation of spectral properties.
Introduction
Biliverdin IX-alpha (BV) is a pigment responsible for characteristic blue-green coloration in many living organisms. Along with other tetrapyrroles, BV often acts as a natural chromophore of phytochromes (Phys) in plants, bacteria, fungi, and cyanobacteria. BV regulates various physiological responses, including chlorophyll synthesis, photoperiodic seed germination, flowering, and photo-taxis.1–4 BV is a metabolite of heme catabolism. Its presence is used to gain evolutionary and ecological advantages by marine fish,5 lizards,6,7 chlorotic tree-frogs,8 and insects.9,10Sandercyanin (SFP) is a BV-binding lipocalin protein found in the skin mucus of the Canadian walleye, Stizostedion vitreum (formerly Sander vitreus).11,12 We have previously reported the spectral properties and structures of tetrameric wild-type SFP–BV complex.11,13 BV-bound SFP shows an intense blue color and absorbances at 380 nm and 630 nm. BV associates non-covalently to each SFP monomer in a ZZZssa conformation and is stabilized by aromatic stacking, hydrophobic interactions, and water-mediated H-bonding inside the lipocalin-like beta-barrel fold of SFP. The binding of BV induces tetramerization of SFP via interaction between amino acid residues from adjacent monomeric subunits. The reported protein tetrameric structures suggest that the molecular properties are significantly modified if the protein is engineered into a monomeric form due to the loss of interactions between BV and the neighboring subunits.11,14 In other photoactive proteins that use BV as a chromophore (example, bacteriophytochromes (BPh)), the ethylidene side chain of the A-ring of BV is covalently linked to the protein chain via a thioether bond with a conserved cysteine residue. BV conformation remains unperturbed in the ground state on changing the native dimeric state of BPh to a monomeric chromophore binding domain.15,16 However, BPhs are well-studied systems for light-induced isomerization around the D-ring pyrrole of BV from a dark or red-absorbing state to a far-red absorbing state.17
In this report, we elucidate the influence of the oligomeric state of SFP on the conformational selection of BV. Based on the results from the monomeric variants of SFP, we created variants that altered the BV-binding pocket. These variants show interesting differences in their spectral properties compared to the wild-type protein. We use X-ray crystallography to determine structures of these variants and combine it with resonance Raman spectroscopy to provide a molecular explanation for the observed difference in spectral properties.
Materials and methods
Site-directed mutagenesis, protein expression, and purification of SFP variants
The gene for wild-type SFP cloned into pET21a(+) was used as a template for generating site-specific mutations. We performed a whole-vector polymerase chain reaction (PCR) of the template gene using high-fidelity Phusion DNA polymerase (New England Biolabs). The presence of the mutation was confirmed by sequencing the plasmid. Expression and purification of SFP variants were performed using methods published for the wild-type protein.11 All proteins were purified by size-exclusion chromatography (SEC) using a Superdex 200 10/300 GL column (Merck). BV-binding to SFP variants is confirmed by monitoring absorbance at 380 nm and 630 nm in the SEC profiles. For monomeric and tetrameric variants, blue-colored BV-bound fractions corresponding to the size of ∼18 kDa and ∼75 kDa, respectively, were identified by using a standard protein mixture (BioRAD gel filtration standard #1511901) used for calibration of the SEC column. Purified fractions were collected and used for spectroscopy and crystallographic studies. Apo-protein of each variant was purified using the same method except for the addition of BV.Absorbance and fluorescence spectroscopy
All experiments were performed with freshly purified protein samples (apo and BV-bound forms) at pH 7.5 and room temperature. UV-visible absorbance spectra of SFP variants were recorded on an Ultraspec 2100 pro spectrophotometer from Amersham Biosciences. Fluorescence and binding studies of SFP variants were monitored on Horiba Jobin Yvon Fluoromax-4 fluorimeter as described in the previous report.11 Briefly, the apoprotein of SFP was titrated with different concentrations of BV, and a change in fluorescence intensity was recorded on excitation at 380 nm and 630 nm. The emission values (at peak maxima) from both excitations were combined and analyzed by GraphPad Prism 9.4.0 to get a tentative estimate of the dissociation constant (Kd). OriginPro 8 software was used for plotting of binding curve. The equation for Kd calculation is the same as described in our previous paper.11Protein crystallization, data collection, and structural analysis
Purified SFP variants were mixed with excess BV before crystallization. Hanging drops were set up at 4 °C using a Mosquito high-throughput crystallization system from TTP life sciences, and crystals were grown by vapor diffusion. V71E protein bound to BV (11 mg mL−1) crystallized in 0.2 M calcium chloride dihydrate, 20% PEG 3350 at pH 5.1. The L135E protein bound to BV (26 mg mL−1) crystallized with 10% PEG 1000 and 10% PEG 8000 as precipitants. The Y142A protein bound to BV (9 mg mL−1) crystallized in 0.1 M Bis-Tris pH 5.5 and 3 M NaCl as the precipitant. The BV-bound protein crystals obtained in various conditions were flash-cooled in liquid nitrogen after soaking in 10% ethylene glycol as a cryo-protectant. Crystallography datasets of L135E, V71E, and Y142A SFP variants were collected at BM14, ID23-2, and ID30A-1 beamlines at ESRF (Grenoble, France). The images were indexed, integrated, and scaled using HKL2000 (ref. 18)/d*TREK19/MOSFLM.20 Structures were determined by molecular replacement method using Phaser21 and the wild-type SFP structure (PDB ID: 5EZ2) as starting model. All structures were refined with Refmac22 and PHENIX.23 Model building was carried out using the program Coot.24 All structural illustrations were generated using PyMOL.25 All parameters of data collection and refinement statistics are summarized in Table S1.† All the coordinates and structure factors have been deposited in the PDB.Resonance Raman spectroscopy
Resonance Raman (RR) spectra of BV-bound wild-type SFP and Y142A variants were recorded using LabRAM HR Evolution confocal Raman microscope (HORIBA Jobin Yvon S.A.S., France) equipped with a spectrograph of 800 mm focal length and a Peltier-cooled CCD to register the spectra. A holographic diffraction grating with 1800 gr per mm, and a 300 μm spectrograph entrance slit was used for high spectral resolution. All protein samples (250 μM concentration) were prepared in phosphate buffer at pH 7.4. Samples were loaded in EPR quality capillary made of Suprasil quartz of outer diameter of 3 mm (Wilmad-LabGlass, NJ, USA), and RR spectra were recorded in 180° backscattering geometry. Samples were resonantly excited with 405 nm or 532 nm solid-state laser operated in CW mode through a 50× objective with a numerical aperture (NA) of 0.5 (Model 1-U2N350, Olympus GmBh, Germany). The laser power on the sample was 4–5 mW for 405 nm and 8–9 mW for 532 nm wavelength. For the 405 nm excitation, each spectrum was recorded after 90 s exposure and accumulated for 270 s. In the case of 532 nm excitation, an exposure of 20 s and a total accumulation time of 100 s were used to acquire a spectrum. The final spectrum was an average of 3 to 5 spectra, each recorded from a fresh sample. Recorded RR spectra were analyzed using LabSpec 5 and Origin 7.Results
Engineering monomeric variants of SFP bound to BV
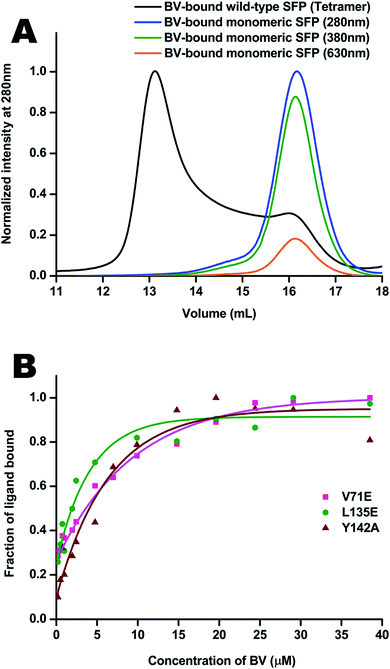
Molecular properties and BV-bound structures of wild-type SFP were reported by us earlier.11,13 In the wild-type protein, BV binding drives ligand-induced oligomerization from monomeric apoprotein to a BV-bound tetrameric form. Each monomer of the tetramer interacts with two other monomers. We picked residues in two interfaces to create site-directed mutations (Fig. 1). Both are buried hydrophobic residues in the interface of two subunits. We believed that mutating them into charged residues would disrupt the interface and create monomers. Two mutations – L135E (from interface 1) and V71E (from interface 2) stayed monomers on binding to BV (Fig. 2A) as identified by size-exclusion chromatography profiles. The overlap of the 280 nm chromatogram with the 380 nm and the 630 nm profiles confirms that BV is bound to the protein. The affinity of BV to the SFP variants are in similar order of magnitude to the wild-type protein (Fig. 2B). Similar binding affinities of monomer and wild-type protein suggest ligation of BV to SFP is independent of the oligomeric state of the protein. Mutating these hydrophobic residues into polar/charged residues disrupted the binding interface. The purified L135E and V71E variants of SFP (the BV-bound monomeric variants) showed a distinct blue-green coloration (ESI Fig. S1†) under physiological conditions, unlike wild-type protein that appears deep-blue in color.11 We characterized the two BV-bound monomeric variants by UV-VIS and fluorescence spectroscopy.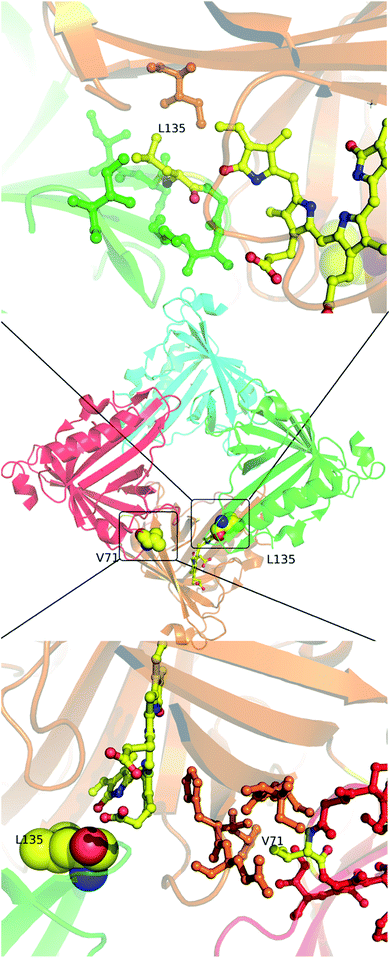 | ||
| Fig. 1 In the center is the tetramer of the published wild-type structure of the SFP–BV complex (PDB 5EZ2). The four monomers are shown in different colors. At the interface of the orange and green subunits (interface 1) is L135 – a zoomed-in view of the interface is shown above. L135 is from the green subunit. At the interface of the orange and the red subunits (interface 2) is V71 – a zoomed view of this interface is shown below. V71 is from the red subunit. The BV in both cases is from the orange subunit. Except for V71, L135, and BV, the residues are colored the same color as their subunits. | ||
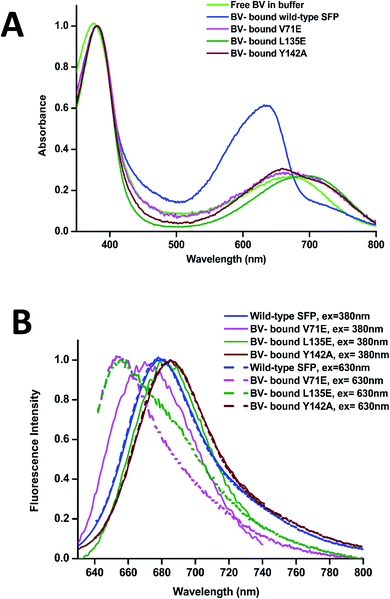
The absorbance spectra of BV-bound V71E and L135E show maxima at 380 nm and an attenuated red-shifted broad peak at the far-red wavelengths (∼680–700 nm) compared to the wild-type protein, where the red-absorbance is at 630 nm (Fig. 3A). The absorbance in the far-red wavelength resembles that of free BV, indicating that the loss of oligomerization enhances exchange between bound and free BV due to its non-covalent nature of interaction with SFP. In BV-binding BPhs, a red shift in absorbance has been associated with the flipping of the D-ring through a trans–cis isomerization of the double bond (C15![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C16) that connects ring C to ring D.26,27 This would suggest that, for BV-bound monomeric SFP variants, despite the conformational changes in the D-ring, the interactions of the BV to the protein are still preserved. We noticed that these variants show fluorescence on BV-binding. Excitation of these monomeric variants at 380 nm and 630 nm yields red fluorescence with maxima between 650 nm and 680 nm for different variants (Fig. 3B). A broad absorbance spectrum near the red wavelengths could indicate the presence of multiple conformers of BV inside these monomeric variants, which possibly contribute differently to the red emission on excitation at 380 nm and 630 nm. We also report a significantly diminished fluorescence quantum yield compared to the wild-type protein. The fluorescence quantum yield of the wild-type protein is 0.016. The V71E and L135E mutations showed an order of magnitude reduction, with fluorescence quantum yields of 0.003 and 0.0025, respectively (for excitation at 630 nm). The decrease in quantum yield was not surprising as we had predicted that BV would be more exposed to the aqueous environment in the monomers, providing paths for non-radiative loss of energy.11,28,29 Unlike wild-type SFP complex, which binds to a specific conformation of BV, decreased fluorescence quantum yield of monomeric complexes (measured at a specific wavelength) could be due to the presence of several conformers of BV showing variable fluorescence at different excitation wavelengths. The best method to elucidate the molecular basis of the observed spectral changes is by determining the structure of the V71E and L135E variants of the protein in complex with BV by X-ray crystallography.
C16) that connects ring C to ring D.26,27 This would suggest that, for BV-bound monomeric SFP variants, despite the conformational changes in the D-ring, the interactions of the BV to the protein are still preserved. We noticed that these variants show fluorescence on BV-binding. Excitation of these monomeric variants at 380 nm and 630 nm yields red fluorescence with maxima between 650 nm and 680 nm for different variants (Fig. 3B). A broad absorbance spectrum near the red wavelengths could indicate the presence of multiple conformers of BV inside these monomeric variants, which possibly contribute differently to the red emission on excitation at 380 nm and 630 nm. We also report a significantly diminished fluorescence quantum yield compared to the wild-type protein. The fluorescence quantum yield of the wild-type protein is 0.016. The V71E and L135E mutations showed an order of magnitude reduction, with fluorescence quantum yields of 0.003 and 0.0025, respectively (for excitation at 630 nm). The decrease in quantum yield was not surprising as we had predicted that BV would be more exposed to the aqueous environment in the monomers, providing paths for non-radiative loss of energy.11,28,29 Unlike wild-type SFP complex, which binds to a specific conformation of BV, decreased fluorescence quantum yield of monomeric complexes (measured at a specific wavelength) could be due to the presence of several conformers of BV showing variable fluorescence at different excitation wavelengths. The best method to elucidate the molecular basis of the observed spectral changes is by determining the structure of the V71E and L135E variants of the protein in complex with BV by X-ray crystallography.
Structures of BV-bound V71E and L135E variants
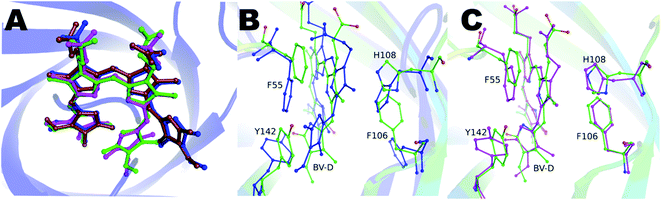
Blue-green crystals of the proteins were obtained after a few days at 4 °C (ESI Fig. S1†). The BV-bound structures of V71E and L135E were determined at 2.5 Å and 2.75 Å resolution, respectively, using wild-type protein structure (PDB ID: 5EZ2) as a model for molecular replacement. Data collection and refinement statistics of BV-bound structures of V71E and L135E are summarized in Table S1.† The crystal packing interactions of BV-bound monomeric variants do not show enough buried surface area to suggest oligomerization. The overall fold of the monomeric variant proteins is very similar to the wild-type protein, with BV bound at the center of the barrel. Loss of oligomerization created a vacant space in the monomer near the open/wider end of the barrel, exposing BV to an aqueous environment at the site of dimerization. N-terminal amino acids and protein backbone of loop residues (L50–E58, R80–I86, H108–V114, and I133–H139) show movement (ESI Fig. S2†). In all the refined structures, the electron density for the BV was well resolved, and the BV was modeled (ESI Fig. S3A and B†). Both structures show one pre-dominant conformation of BV, in spite of a broad absorbance spectrum at the red wavelengths. In the BV-binding pocket, both monomeric variants revealed a rotated D-ring pyrrole of BV around the C14–C15 single bond. Measured torsion angles were 122° and 125° (counter-clockwise) for V71E and L135E, respectively. The conformation of BV is ZZZsss in the variants compared to ZZZssa in wild-type SFP (Fig. 4A). The B and C rings show slight deviation from the native (ZZZssa) state, but the A-ring remains unperturbed even though there is no covalent linkage between the protein and BV like in the phytochromes.30 Relative B-factors of the different tetrapyrrole rings with respect to each other are a good indicator of the mobility of the different rings in the structure. The tetrapyrrole rings of the BV in the wild-type are all well-ordered and stabilized by interactions with the protein residues. However, the atoms of the D-ring of the L135E have larger B-factors suggesting that even in the crystal structure, this ring is more flexible. Light-induced isomerization of the C15![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C16 double bond of BV has been proposed in the Bphs.26 In the SFP–BV complex, the conformational changes in BV seem to be due to the rotation around the single bond C14–C15. This rotation suggests flexibility in the D-ring position in the ground state rather than a specific state induced by irradiation.
C16 double bond of BV has been proposed in the Bphs.26 In the SFP–BV complex, the conformational changes in BV seem to be due to the rotation around the single bond C14–C15. This rotation suggests flexibility in the D-ring position in the ground state rather than a specific state induced by irradiation.
Aromatic residues F55, F106, H108, and Y142, undergo concerted rearrangement of side chains to stabilize the interaction with BV in the protein (Fig. 4B and C). In the wild-type protein, the orientation of the D-ring is stabilized by interactions with residues in the neighboring subunit. However, in the monomeric forms, the orientation of the D-ring of BV is stabilized by interactions with the A-ring of BV (the D-ring moves closer to the A-ring of BV) and stacking interactions with the Y142 side chain (which rotates to stack with the D-ring). The changes are concerted on both sides of the BV, with F55 and Y142 on one side and F106 and H108 on the other. In the V71E structure, water molecules occupy the original space vacated by the reoriented side-chains of F55 and Y142. The lower resolution structure of L135E does not allow modeling of structured waters replacing the rotated residues; however, empty space possibly suggests randomly-oriented or the presence of low occupancy water molecules. In both structures, non-aromatic amino acids (such as D47 and Q56) near BV show subtle structural changes. The C-pyrrole of BV and its propionic chain get displaced with the overall chromophore moving slightly upwards from BV bound to the wild-type protein (Fig. 4A). Interestingly, with similar conformational freedom to the D-ring of BV, the non-covalently linked A-ring remains unperturbed and stabilized by surrounding amino acid residues: D47, A61, A63, Y65, N77, Y116, S144, and V146.
We infer from crystal structures of wild-type and monomeric variants that SFP can accommodate several ground state conformations of BV in the binding pocket. Further, we identified F55, F106, H108, and Y142 as crucial residues in stabilizing BV in ZZZsss conformation in the monomer mutants. A systematic mutation of these residues in the wild-type protein would possibly lead to increased conformational flexibility of BV in the binding pocket. We generated variants of all four amino acids, F55, F106, H108, and Y142, by mutating them into alanine to understand their role in the conformational flexibility of BV bound to SFP.
The tetrameric Tyr-142-Ala variant
We cloned, expressed, purified, and characterized the spectroscopic properties of the F55A, F106A, H108A, and Y142A variants in complex with BV. All variants are tetrameric on binding to BV (ESI Fig. 4A†). Recently published structure and properties of tetrameric F55A–BV complex revealed a rotated D-ring with a far-red absorbance and no observable fluorescence.14 However, F106A, H108A, and Y142A showed fluorescence spectra with slightly shifted peak maxima compared to the wild-type protein (Fig. 3B and ESI Fig. 4B†). Spectral properties of the Y142A–BV complex showed an absorbance maximum at 380 nm and a shift in absorption peak from 630 nm (in wild-type protein) to 660 nm, with a concomitant appearance of the 720 nm far-red band with similar intensity (Fig. 3A and ESI Fig. 4C†). On excitation with 380 nm and 630 nm, Y142A showed a red-shifted fluorescence with maxima at 685 nm and a fluorescence quantum yield (0.004) equivalent to the monomeric SFP variants (Fig. 3B).We hypothesized that the mutation of the large Tyr into Ala created enough space for the D-ring to rotate in the tetrameric protein giving rise to dual peaks in the red region of the absorbance spectrum. To test this, we determined the crystal structure of the Y142A–BV complex at 2.65 Å (ESI Table S1†). However, to our surprise, the BV adopted the ZZZssa conformation in the crystal structure (Fig. 4A), with a water molecule accommodated in the position of the mutated side chain of Y142 (similar to the V71E structure). The similarity of BV conformation to the wild-type protein could not explain the observed differences in the absorption spectra. To test if conformational heterogeneity did exist in the solution state producing the co-existing red (∼660 nm) and far-red (∼720 nm) absorbing states of Y142A, we decided to probe the wild-type and the Y142A variant using solution-state resonance Raman spectroscopy.
Conformational plasticity of BV in SFP studied by resonance Raman spectroscopy
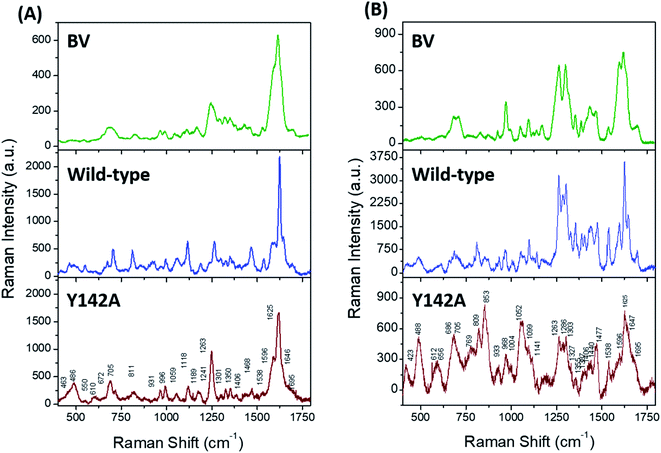
Solution state resonance Raman (RR) spectroscopy near the two absorbance states of the wild-type SFP–BV complex (Fig. 3A, blue) allowed us to probe conformations of BV in SFP.31,32 The RR spectra of free- and bound BV to wild-type protein at 405 nm (Fig. 5A) and 532 nm (Fig. 5B) reveal that binding to SFP leads to spectral narrowing of vibrational bands of BV in the high-frequency region. The high-frequency region primarily represents the resonance-enhanced in-plane vibrations coupled to D-ring (Table S2† for normal mode assignments of all bands).33 The loss of RR intensity in these bands implies a sudden decrease in polarizability of these modes – probably due to the disruption of the pi–electron conjugation, and suggests possible rotation of the D-ring of BV and stabilization of a specific BV conformation inside SFP.34 In contrast to the wild-type protein, BV bound to Y142A shows loss of fine structures of several RR bands in 532 nm excited spectra, specifically four bands at 1695, 1647, 1625, and 1596 cm−1 associated with D-ring in-plane stretching vibrations (Fig. 5). Significant broadening of these RR bands strongly suggests the occurrence of multiple BV conformers bound to Y142A in the solution state. Also, a significant reduction of RR intensity in the 1200–1750 cm−1 region indicates the loss of pi–electron conjugation between adjacent covalently linked rings and the disruption of non-covalent pi–pi interactions with aromatic residues stabilizing BV.Discussion
This paper combines spectroscopy (UV-VIS and RR) and X-ray crystallography to elucidate the influence of SFP on the dynamics of BV and its spectral properties. Our data on monomeric SFP and Y142A shows that the D-ring pyrrole of BV exhibits significant flexibility. Furthermore, monomeric SFP variants form a stable complex with BV but show different spectral properties compared to the wild-type protein. Interestingly, the presence of several ground state conformations suggests conformational promiscuity of BV bound to SFP.Dark (or resting state) ZZZssa conformation of BV of SFP is similar to BPhs that yields a red-absorption (called Pr in BPhs) state ∼630–645 nm.11,26,35 However, unlike BPhs, which undergo photo-isomerization to ZZEssa (or PFr) conformation, BV in SFP shows ground state conformers (ZZZsss) arising from single bond rotation around the C14–C15 single bond. This study refers to them as Sr (red) and SFr (far-red) photo-states.26,35–37 Light-induced photoconversion is not yet established in SFP. Nevertheless, both mechanisms (isomerization in Bphs and rotation in SFP) are driven primarily by the D-ring of the BV, despite the significant difference in the protein structures. Covalent bonding of A-ring pyrrole to a conserved cysteine residue in BPhs constraints the structural flexibility of the chromophore in the ground and illuminated state.38 In SFP, BV associates non-covalently, and the A-ring possesses similar conformational freedom as D-ring, yet it remains invariant in all the SFP variants (Fig. 4A). Comparison with other lipocalins bound to biliverdin isoforms in the PDB (Pieris brassicae (PDB: 1BBP) and Manduca sexta (PDB: 1Z24)) show many conserved residues interacting with the A-ring equivalent pyrrole that possibly stabilize the A-ring in SFP.
The RR studies of Y142A mutation reveal a flexible D-ring of BV in solution with two small shoulder peaks in the far-red absorbance, yet only one conformation is observed in the crystal structure. We have shown earlier that in SFP, absorbance peaks at 630 nm and 720 nm correspond to ZZZssa and ZZZsss forms of BV, respectively.11,14 The peak at 660 nm of Y142A could be an intermediate that consists of one or more conformers of BV overlapping together. Our earlier report showed that even the apo-protein (monomeric in solution) forms a tetramer at high concentration (PDB 5F1E).11 The increased stability provided to the ZZZssa conformation of BV by the residues of the neighboring subunit (Fig. 1) may be sufficient to shift the equilibrium towards this conformation in the high concentration of the protein in the crystals of Y142A. Hence, we conclude that both conformations of BV are possible in the Y142A variant in solution, even though only one is observed in the crystal structure.
There are only two well-characterized vertebrate fluorescent proteins (SFP and UnaG39). The reported monomeric forms of SFP are the smallest (∼18 kDa) far-red fluorescent proteins of vertebrate origin to date.11,40 Structural studies reveal several parameters responsible for reduced fluorescence quantum yield of monomeric SFP–BV complex. Firstly, exposure to the protic environment in the BV-binding pocket (due to increased water molecules) plays a significant role in quenching the fluorescence of chromophores.11,33,34,41,42 Crystal structures of BV complexes of V71E and Y142A reveal water molecules displacing side-changes of aromatic residues F55 and Y142 (absent in the Y142A variant), making it polar at the core of the binding pocket. We have earlier reported that increasing hydrophobicity around free BV and deuteration of wild-type SFP increases the fluorescence intensity, possibly due to reduced non-radiative excited-state proton transfer (ESPT) processes.11 Our recent discovery emphasizes that hydrophobicity and stacking of F55 with BV is a crucial factor for fluorescence of SFP–BV complex.14 Secondly, rotation of the D-ring of BV to adopt multiple ground-state conformations (ZZZssa and ZZZsss) could affect the quantum yield of the SFP–BV complex. Due to the higher flexibility of BV in monomeric protein (also evident by its broad absorbance spectra) and RR studies on Y142A variant with reduced quantum yield, only a fraction of BV conformers (mainly the ZZZssa isoform) may show fluorescence. This is further supported by studies on fluorescent proteins (FPs) developed from BV-binding BPhs that recognized conformational flexibility of BV as an important factor for reduced fluorescence quantum yield.28,29,43,44 Similar to BPhs, we could employ structure-based protein engineering techniques on monomeric SFP variants to generate bright monomeric far-red fluorescent proteins for in vivo applications. Our studies suggest that increasing the hydrophobicity of the binding site and attaining a sterically-locked ZZZssa conformer of BV would increase the fluorescence quantum yield of the SFP–BV complex.44,45 As SFP is a vertebrate protein, there are many opportunities for utilizing it for cellular applications, where BV is produced intrinsically as a metabolite.16,46,47
In conclusion, our work shows BV dynamics in the binding pocket of a lipocalin protein and postulates a dominant structural characteristic of the conserved D-ring pyrrole (despite the protein structure and nature of linkage present within) in driving chemistry of BV- or linear-tetrapyrroles binding proteins across diverse species.48,49
Data availability
The atomic coordinates and structure factors for BV-bound SFP variants have been deposited to the Protein Data Bank (http://www.pdb.org) with PDB IDs: 7O2Y (V71E), 7O3K (L135E), and 7YX1 (Y142A). All remaining data are contained within the article.Author contributions
SG and RS planned the research. SG designed and characterized the clones for detailed studies. SG and KY purified protein, performed UV-VIS spectroscopy, protein crystallization, and analyzed crystallography data. SM and SA carried out RR data collection; SM performed detailed data analysis and interpretation of RR data. RS, CN, WFS, and SG supervised the project. SG and SM wrote the manuscripts and received inputs from all authors.Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.Acknowledgements
The preparatory work for this project, spectroscopy, and crystallographic data analysis, was carried out at Institute for Stem Cell Science and Regenerative Medicine (inStem, Bangalore, India). We thank all members of our laboratory at inStem (India) for their support during this research. Further, we thank the beamline staff at ESRF for help in data collection. Support for this work was provided by the Department of Biotechnology India (Grant BT/PR5801/INF/22/156/2012) and WISYS Technology Foundation, Inc. (Madison, WI, USA). CN would like to acknowledge LSRET-JNC (BT/INF22/SP27679/2018) intramural funds for financial assistance.References
- K. A. Franklin and P. H. Quail, J. Exp. Bot., 2010, 11–24 CrossRef CAS PubMed.
- D. Duanmu, C. Bachy, S. Sudek, C. H. Wong, V. Jiménez, N. C. Rockwell, S. S. Martin, C. Y. Ngan, E. N. Reistetter, M. J. Van Baren, D. C. Price, C. L. Wei, A. Reyes-Prieto, J. C. Lagarias and A. Z. Worden, Marine algae and land plants share conserved phytochrome signaling systems, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 15827–15832, DOI:10.1073/pnas.1416751111.
- P. F. Devlin, Proc. Natl. Acad. Sci. U. S. A., 2016, 7301–7303 CrossRef CAS.
- K. A. Franklin and G. C. Whitelam, Ann. Bot., 2005, 169–175 CrossRef CAS PubMed.
- J. L. Bada and L.-S. Fang, The blue-green blood plasma of marine fish, Comp. Biochem. Physiol., Part B: Comp. Biochem., 1990, 97, 37–45 Search PubMed.
- K. W. Jessing and C. C. Austin, Green-blood pigmentation in lizards, Comp. Biochem. Physiol., Part A: Physiol., 1994, 109, 619–626 CrossRef.
- C. C. Austin, Z. B. Rodriguez and S. L. Perkins, Multiple origins of green blood in New Guinea lizards, Science, 2018, 4, 1–7 CrossRef.
- C. Taboada, A. E. Brunetti, M. L. Lyra, R. R. Fitak, A. F. Soverna, S. R. Ron, M. G. Lagorio, C. F. B. Haddad, N. P. Lopes, S. Johnsen, J. Faivovich, L. B. Chemes and S. E. Bari, Multiple origins of green coloration in frogs mediated by a novel biliverdin-binding serpin, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 18574–18581 CrossRef CAS PubMed.
- H. M. Holden, W. R. Rypniewski, J. H. Law and I. Rayment, The molecular structure of insecticyanin from the tobacco hornworm Manduca sexta L. at 2.6 Å resolution, EMBO J., 1987, 6, 1565–1570 CrossRef CAS PubMed.
- H. Kayser, R. Huber, M. Schneider, I. Mayr, R. Müller, R. Deutzmann, F. Suter, H. Zuber and H. Falk, Molecular structure of the bilin binding protein (BBP) from Pieris brassicae after refinement at 2.0 Å resolution, J. Mol. Biol., 1987, 198, 499–513 CrossRef.
- S. Ghosh, C.-L. Yu, D. J. Ferraro, S. Sudha, S. K. Pal, W. F. Schaefer, D. T. Gibson and S. Ramaswamy, Blue protein with red fluorescence, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 11513–11518 CrossRef CAS PubMed.
- W. F. Schaefer, M. H. Schmitz, V. S. Blazer, T. J. Ehlinger and J. A. Berges, Localization and seasonal variation of blue pigment (sandercyanin) in walleye (Sanders vitreus), Can. J. Fish. Aquat. Sci., 2014, 72, 1–9, DOI:10.1139/cjfas-2014-0139.
- C. L. Yu, D. Ferraro, S. Ramaswamy, M. H. Schmitz, W. F. Schaefer and D. T. Gibson, Purification and properties of Sandercyanin, a blue protein secreted in the mucus of blue forms of walleye, Sander vitreus, Environ. Biol. Fishes, 2008, 82, 51–58, DOI:10.1007/s10641-007-9252-3.
- S. Yadav, K. Ghosh, S. A. Barak, A. Schaefer and W. Ramaswamy, Phenylalanine stacking enhances the red fluorescence of biliverdin IXα on UV excitation in sandercyanin fluorescent protein, FEBS Lett., 2022, 596, 796–805, DOI:10.1002/1873-3468.14281.
- M. E. Auldridge, K. A. Satyshur, D. M. Anstrom and K. T. Forest, Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein, J. Biol. Chem., 2012, 287, 7000–7009, DOI:10.1074/jbc.M111.295121.
- X. Shu, A. Royant, M. Z. Lin, T. A. Aguilera, V. Lev-Ram, P. A. Steinbach and R. Y. Tsien, Mammalian Expression of Infrared Fluorescent Proteins Engineered from a Bacterial Phytochrome, Science, 2009, 324, 804–807 CrossRef PubMed.
- J. Abian, T. Lamparter, M. Carrascal, N. Michael, E. Martinez and G. Rottwinkel, The biliverdin chromophore binds covalently to a conserved cysteine residue in the N-terminus of Agrobacterium phytochrome Agp1, Biochemistry, 2004, 43, 3659–3669 CrossRef PubMed.
- M. Gilski, Acta Phys. Pol., A, 2012, 871–875 CrossRef CAS.
- J. W. Pflugrath, The finer things in X-ray diffraction data collection, Acta Crystallogr., Sect. D: Biol. Crystallogr., 1999, 55, 1718–1725, DOI:10.1107/S090744499900935X.
- T. G. G. Battye, L. Kontogiannis, O. Johnson, H. R. Powell and A. G. W. Leslie, iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 271–281, DOI:10.1107/S0907444910048675.
- A. J. McCoy, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2006, 32–41 Search PubMed.
- M. D. Winn, C. C. Ballard, K. D. Cowtan, E. J. Dodson, P. Emsley, P. R. Evans, R. M. Keegan, E. B. Krissinel, A. G. W. Leslie, A. McCoy, S. J. McNicholas, G. N. Murshudov, N. S. Pannu, E. A. Potterton, H. R. Powell, R. J. Read, A. Vagin and K. S. Wilson, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 235–242 CrossRef CAS PubMed.
- P. D. Adams, P. V. Afonine, G. Bunkóczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L. W. Hung, G. J. Kapral, R. W. Grosse-Kunstleve, A. J. McCoy, N. W. Moriarty, R. Oeffner, R. J. Read, D. C. Richardson, J. S. Richardson, T. C. Terwilliger and P. H. Zwart, PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 213–221, DOI:10.1107/S0907444909052925.
- P. Emsley and K. Cowtan, Coot: model-building tools for molecular graphics, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 2126–2132, DOI:10.1107/S0907444904019158.
- W. L. DeLano, The PyMOL Molecular Graphics System, Schrödinger LLC, 2002.
- X. Yang, J. Kuk and K. Moffat, Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 15639–15644, DOI:10.1073/pnas.0902178106.
- H. Takala, A. Björling, O. Berntsson, H. Lehtivuori, S. Niebling, M. Hoernke, I. Kosheleva, R. Henning, A. Menzel, J. A. Ihalainen and S. Westenhoff, Signal amplification and transduction in phytochrome photosensors, Nature, 2014, 509, 245–248, DOI:10.1038/nature13310.
- K. C. Toh, E. A. Stojković, I. H. M. Van Stokkum, K. Moffat and J. T. M. Kennis, Fluorescence quantum yield and photochemistry of bacteriophytochrome constructs, Phys. Chem. Chem. Phys., 2011, 13, 11985–11997, 10.1039/c1cp00050k.
- P. Hildebrandt, F. V. Escobar, T. Hildebrandt, T. Utesch, F. J. Schmitt, I. Seuffert, N. Michael, C. Schulz, M. A. Mroginski and T. Friedrich, Structural Parameters Controlling the Fluorescence Properties of Phytochromes, Biochemistry, 2014, 53, 20–29 CrossRef PubMed.
- X. Yang, E. A. Stojković, J. Kuk and K. Moffat, Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 12571–12576 CrossRef CAS PubMed.
- T. G. Spiro, Resonance Raman spectroscopy. New structure probe for biological chromophores, Acc. Chem. Res., 1974, 7, 339–344 CrossRef CAS.
- L. W. Chung, W. M. C. Sameera, R. Ramozzi, A. J. Page, M. Hatanaka, G. P. Petrova, T. V. Harris, X. Li, Z. Ke, F. Liu, H.-B. Li, L. Ding and K. Morokuma, The ONIOM Method and Its Applications, Chem. Rev., 2015, 115, 5678–5796 CrossRef CAS PubMed.
- F. Celis, M. M. Campos-Vallette, J. S. Gómez-Jeria, R. E. Clavijo, G. P. Jara and C. Garrido, Surface-enhanced Raman scattering and theoretical study of the bilichromes biliverdin and bilirubin, Spectrosc. Lett., 2016, 49, 336–342 CrossRef CAS.
- D. A. Long, The Raman Effect, John Wiley & Sons, Ltd, Chichester, UK, 2002 Search PubMed.
- K. C. Toh, E. A. Stojković, I. H. M. Van Stokkum, K. Moffat and J. T. M. Kennis, Proton-transfer and hydrogen-bond interactions determine fluorescence quantum yield and photochemical efficiency of bacteriophytochrome, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 9170–9175, DOI:10.1073/pnas.0911535107.
- K. C. Toh, E. A. Stojković, A. B. Rupenyan, I. H. M. Van Stokkum, M. Salumbides, M. L. Groot, K. Moffat and J. T. M. Kennis, Primary reactions of bacteriophytochrome observed with ultrafast mid-infrared spectroscopy, J. Phys. Chem. A, 2011, 115, 3778–3786, DOI:10.1021/jp106891x.
- J. Salewski, F. V. Escobar, S. Kaminski, D. Von Stetten, A. Keidel, Y. Rippers, N. Michael, P. Scheerer, P. Piwowarski, F. Bartl, N. Frankenberg-Dinkel, S. Ringsdorf, W. Gärtner, T. Lamparter, M. A. Mroginski and P. Hildebrandt, Structure of the biliverdin cofactor in the Pfr state of bathy and prototypical phytochromes, J. Biol. Chem., 2013, 288, 16800–16814, DOI:10.1074/jbc.M113.457531.
- H. Takala, H. K. Lehtivuori, O. Berntsson, A. Hughes, R. Nanekar, S. Niebling, M. Panman, L. Henry, A. Menzel, S. Westenhoff and J. A. Ihalainen, On the (un)coupling of the chromophore, tongue interactions, and overall conformation in a bacterial phytochrome, J. Biol. Chem., 2018, 293, 8161–8172, DOI:10.1074/jbc.RA118.001794.
- A. Kumagai, R. Ando, H. Miyatake, P. Greimel, T. Kobayashi, Y. Hirabayashi, T. Shimogori and A. Miyawaki, A bilirubin-inducible fluorescent protein from eel muscle, Cell, 2013, 153, 1602–1611, DOI:10.1016/j.cell.2013.05.038.
- O. S. Oliinyk, A. A. Shemetov, S. Pletnev, D. M. Shcherbakova and V. V. Verkhusha, Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing, Nat. Commun., 2019, 10, 279, DOI:10.1038/s41467-018-08050-8.
- S. E. Braslavsky, A. R. Holzwarth, H. Lehner and K. Schaffner, The fluorescence of biliverdin dimethyl ester, Helv. Chim. Acta, 1978, 67, 2219–2222, DOI:10.1002/hlca.19780610623.
- Y. Liu, Z. Chen, X. Wang, S. Cao, J. Xu, R. Jimenez and J. Chen, Ultrafast spectroscopy of biliverdin dimethyl ester in solution: pathways of excited-state depopulation, Phys. Chem. Chem. Phys., 2020, 22, 19903–19912, 10.1039/d0cp02971h.
- D. M. Shcherbakova, M. Baloban, S. Pletnev, V. N. Malashkevich, H. Xiao, Z. Dauter and V. V. Verkhusha, Molecular Basis of Spectral Diversity in Near-Infrared Phytochrome-Based Fluorescent Proteins, Chem. Biol., 2015, 22, 1540–1551, DOI:10.1016/j.chembiol.2015.10.007.
- A. A. Samma, C. K. Johnson, S. Song, S. Alvarez and M. Zimmer, On the origin of fluorescence in bacteriophytochrome infrared fluorescent proteins, J. Phys. Chem. B, 2010, 114, 15362–15369, DOI:10.1021/jp107119q.
- M. M. Sheehan, M. S. Magaraci, I. A. Kuznetsov, J. A. Mancini, G. Kodali, C. C. Moser, P. L. Dutton and B. Y. Chow, Rational Construction of Compact de Novo-Designed Biliverdin-Binding Proteins, Biochemistry, 2018, 57, 6752–6756, DOI:10.1021/acs.biochem.8b01076.
- A. A. Shemetov, O. S. Oliinyk and V. V. Verkhusha, How to Increase Brightness of Near-Infrared Fluorescent Proteins in Mammalian Cells, Cell Chem. Biol., 2017, 24, 758–766, DOI:10.1016/j.chembiol.2017.05.018.
- G. S. Filonov, K. D. Piatkevich, L. M. Ting, J. Zhang, K. Kim and V. V. Verkhusha, Bright and stable near-infrared fluorescent protein for in vivo imaging, Nat. Biotechnol., 2011, 29, 757–761, DOI:10.1038/nbt.1918.
- C. Song, G. Psakis, J. Kopycki, C. Lang, J. Matysik and J. Hughes, The D-ring, not the A-ring, rotates in Synechococcus OS-B′ phytochrome, J. Biol. Chem., 2014, 289, 2552–2562, DOI:10.1074/jbc.M113.520031.
- A. T. Ulijasz, G. Cornilescu, C. C. Cornilescu, J. Zhang, M. Rivera, J. L. Markley and R. D. Vierstra, Structural basis for the photoconversion of a phytochrome to the activated Pfr form, Nature, 2010, 463, 250–254, DOI:10.1038/nature08671.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra02880h |
| ‡ Authors contributed equally to this work. |
| § Current address: Department of Chemical Physics, School of Chemistry, Tel Aviv University, Tel Aviv 69978, Israel. |
| This journal is © The Royal Society of Chemistry 2022 |