Open Access Article
Open Access ArticleA label-free strategy for visual genotyping based on phosphate induced coloration reaction†
Jiaxing Zhang‡
 ab,
Hui Hui‡c,
Wei Xua,
Kai Hua
ab,
Hui Hui‡c,
Wei Xua,
Kai Hua a,
Yali Cui
a,
Yali Cui *a and
Xiaonan Liu
*a and
Xiaonan Liu *a
*a
aCollege of Life Sciences, Northwest University, Xi'an, 710069, China. E-mail: yalicui@nwu.edu.cn; xiaonanliuvip@163.com
bDepartment of Immunology, The Fourth Military Medical University, Xi'an, 710032, China
cShannxi Provincial Cancer Hospital, Xi'an, 710061, China
First published on 10th August 2022
Abstract
Genetic variation plays a crucial role in disease occurrence and development. However, current genotyping strategies not only require a long turnaround time for DNA purification, but also depend on sophisticated apparatus and complex data interpretation, which seriously limits their application in point of care diagnostic test scenarios. In this study, by integrating phosphate induced coloration reaction and loop-mediated isothermal amplification, a rapid and portable strategy for straightforward genotyping has been established to cater to the demand of precision medicine. By employing phosphate ions produced during the amplification as a signal generator, not only can the genotyping result be interpreted with only naked eye from a low-cost label-free strip, but also the amplification efficiency is increased to facilitate genotyping with a robust biological specimen ignoring DNA polymerase inhibitors. Moreover, the introduction of alkaline lysis for DNA release allows whole blood to be identified accurately avoiding DNA purification. As a proof of concept, the insertion/deletion polymorphisms of the angiotensin-converting enzyme, a crucial factor associated with cardiovascular and cerebrovascular diseases, has been selected as a model to evaluate the performance of this method. Accurate results can be obtained from as low as 1 ng genomic DNA within 30 min. For clinical specimen detection, a concordance rate up to 100% has been found compared with PCR-based electrophoresis. Thus, this novel strategy may serve as a promising tool for straightforward genotyping to provide timely diagnostic information, especially in resource-poor medical institutions.
1. Introduction
Rapid advances made in genetic variation research are providing us with a better understanding of genetic disease occurrence and development, which offers a new opportunity for personalized medicine. The insertion/deletion polymorphisms (indels) of the disease-associated gene have served as a significant biomarker for the diagnosis, prevention and treatment of genetic disease.1,2 A rapid and portable genotyping method with practicability and feasibility is the key step for implementing genetic variation-based personalized medicine in clinical routine, especially in point of care diagnostic test (POCT) scenarios. Although a wide variety of methods have been established for indels genotyping, such as real-time PCR, high-resolution melting and so on,3–7 the practical applications of these protocols have often been limited due to the labor-intensive process and sophisticated instrumentation, particularly in resource-limited medical institutions. In order to facilitate the popularization of precision medicine in clinical practice, a portable and rapid strategy for indels identification is highly desirable.As the key factor of the renin-angiotensin system, angiotensin-converting enzyme (ACE) plays a crucial role in regulating the body's blood pressure.8 The insertion/deletion of 287 bp Alu element located in the sixteenth intron of ACE significantly affects its expression.9,10 As the antihypertensive drug used routinely in clinical practice generally exists individual differences, the selection of appropriate therapeutic drugs based on clarifying ACE genotype of hypertensive patients is the fundamental guarantee of precision medication.
Target variant amplification is central to genotyping methods due to the sensitivity and specificity depend on the effective increase in the copy number for the target region.11 PCR is the most widely used method for target variant amplification, but it has some limitations for POCT applications, such as time consuming, thermal cycling as well as a specific device for strict temperature control.12 Thus, another simple and fast amplification method needs to be adopted to meet ever-growing clinical needs for precision medicine. Taking the advantage of high amplification efficiency under isothermal condition, loop-mediated isothermal amplification (LAMP) provides an alternative method for nucleotide amplification and several LAMP-based genotyping methods have been reported.13,14 However, the genotyping result interpretation always depends on exquisite instruments (such as fluorimeter) or time-consuming operations (such as electrophoresis). Hence, a straightforward and competent strategy for signal interpretation is highly demanded for the POCT scenarios.
Taking the advantage of straightforward, visual detection required only naked eye is preferred for signal judgment. Based on the combination of LAMP and lateral flow strip, some visual detection methods have been developed for pathogens determination.15–18 Although straightforward signal interpretation has been achieved, the label of antibody for functionalizing the lateral flow device highly increases the detection cost. Excitingly, phosphate induced coloration reaction provides a label-free strategy for LAMP amplicons visual interpretation and this strategy has been successfully applied to the identification of pathogens and transgenic crops.19,20 Based on phosphate induced coloration reaction, the label-free strip for visual detection not only facilitates rapid and portable genotyping, but also reduces the cost.
In the present study, a low-cost, rapid and portable strategy based on the combination of phosphate induced coloration reaction and LAMP has been established for straightforward genotyping to cater to the demand of precision medicine. Instead of amplicon, phosphate ion produced during the amplification has been employed as a signal generator, which not only allows the genotyping result to be interpreted with only naked eye from a low-cost label-free strip, but also increases the amplification efficiency to facilitate genotyping with robust biological specimen ignoring DNA polymerase inhibitors. By introducing alkaline lysis for DNA release, accurate results can be obtained from blood directly within 30 min avoiding DNA purification. Taking the indels of ACE as model, the excellent performance of this novel strategy has been verified with real clinical specimens, which indicates the great potential of this method for rapid and portable genotyping to facilitate clinical medication guidance.
2. Experimental
2.1. Oligonucleotides
As a demonstration, the indels of ACE was selected as a model for methodology establishment and LAMP primers were designed to distinguish the indels of ACE according to the principle described in the literature.13,21 For convenience, we denote insertion as “Ins” and deletion as “Del”. Two sets of primer for this method were designed using the Primer 5.0 software program (Primer-E Ltd., Plymouth, UK), including Ins primer set and Del primer set for recognizing eight distinct regions of target variant. Each primer set contains two out primers (F3 and B3), two loop primers (LF and LB) and two inner primers (FIP and BIP). Besides, another two primers were also designed for PCR-based electrophoresis as a reference to verify the result. The sequences of all primers were shown in Table S1,† and all primers were synthesized by TsingKe Biotech Ltd (Beijing, PRC).2.2. Clinical specimen and genomic DNA extraction
Fresh whole blood with normal and abnormal hematological indices (hyperbilirubinemia (>22.2 μM), hypercholesterolemia (>5.71 mM), hypertriglyceridemia (>1.7 mM), and hemolysis) was collected from 25 volunteers at the Shaanxi Provincial People's Hospital with informed consent. Five specimens of each blood type were taken for research. Genomic DNA from whole blood was extracted using a TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, PRC) according to the manufacturer's instructions. The concentration of genomic DNA was determined using NanoDrop 2000 UV-vis spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). This research was approved by the Ethics Committee of the College of Life Sciences, Northwest University (Xi'an, Shaanxi, PRC). All operations were carried out following the Helsinki declaration.2.3. Visual genotyping system design
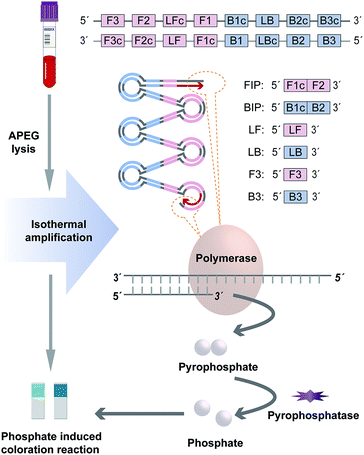
In order to simplify the procedure and break the dependence on sophisticated instruments, this proposed method employs two strategies to achieve rapid and portable genotyping to cater to the demand of POCT scenario, including genotype discrimination by LAMP based on target variant amplification and amplification signal interpretation based on phosphate induced coloration reaction.For target variant amplification and genotype discrimination, two individual amplification mixtures containing Ins primer set (Ins mixture for Ins variant amplification) and Del primer set (Del mixture for Del variant amplification) respectively were performed simultaneously to distinguish the indels of each specimen. Both of the reaction mixtures with a final volume at 20 μL containing 2 μL 10× buffer (New England Biolabs, Ipswich, MA, USA), 1.4 mM of each dNTPs, 8 mM MgSO4, 6.4 U Bst DNA Polymerase (New England Biolabs), 0.4 U pyrophosphatase (New England Biolabs), 1.2 μM FIP and BIP, 0.2 μM F3 and B3, 0.3 μM LF and LB and 1 μL genomic DNA were incubated at 65 °C via a heater for 25 min.
For amplification signal interpretation, the phosphate ion produced during amplification was employed as a signal generator for visual detection. After amplification, the whole reaction mixture (20 μL) of each tube was taken for the phosphate induced coloration reaction respectively, which was performed using Phosphate Test Strip (Lohand Biological, Hangzhou, PRC) according to the operation manual. As by-product, phosphate ion indicated the target variant amplification successfully and the existence of amplicons. Hence, the genotyping result could be interpreted with naked eye based on the chromatism of the strip. The reference DNA samples with the ACE genotypes of insertion homozygote (II), insertion-deletion heterozygote (ID), and deletion homozygote (DD) confirmed by sequencing were used to validate the method. Besides, we measured the chromatic difference (ΔE*) between the negative and positive results of the strategy through a CIELAB colorimeter (3nh, Shenzhen, China).
2.4. Direct genotyping from blood
In order to simplify the procedure and shorten the detection time further, direct genotyping to avoid DNA purification was achieved by introducing alkaline lysis for DNA release according to our previous report.22 Whole blood was mixed with alkaline polyethylene glycol in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 1
3, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 and 1
5 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 ratio respectively and the mixture was incubated at room temperature for 1 minute. Subsequently, 1 μL supernatant of the mixture was taken for amplification and the optimal ratio for DNA release was chosen according to the chromatism of the strip.
7 ratio respectively and the mixture was incubated at room temperature for 1 minute. Subsequently, 1 μL supernatant of the mixture was taken for amplification and the optimal ratio for DNA release was chosen according to the chromatism of the strip.
3. Results and discussion
3.1. Visual genotyping system establishment
In this approach, indels of ACE was discriminated by this novel visual genotyping system. In order to increase the sensitivity and specificity as high as possible, the primer sets for genotype distinguishing were tailor-made according to the target variant sequence. A thermodynamic analysis was used to select the candidate primers based on the parameter derived from the Primer 5.0 software program, which maximized the amplification for matched pairs (Ins primer set/Ins variant or Del primer set/Del variant) and hampered the coupling of unmatched pairs (Ins primer set/Del variant or Del primer set/Ins variant). The optimal primer set produced wide variation in standard free energies, expressing as the difference between the unmatched and matched pairs to allow the best discrimination of indels.For target variant amplification, two individual LAMP reactions containing Ins and Del primer sets were performed simultaneously to distinguish the indels of each specimen. During amplification, each phosphodiester bond formation between the DNA growing chain and complementary deoxyribonucleoside triphosphate would release a pyrophosphate group as by-products (Fig. 1). In the matched prime-variant reaction mixture, double stem-loop structure acted as the trigger for LAMP cycling was formed through strand displacement, which results in drastic DNA amplification and abundant pyrophosphate generation. Besides, pyrophosphatase was employed to hydrolyze pyrophosphate to phosphate for signal generation. While the unmatched prime-variant mixture failed to induce LAMP cycling and the trace of phosphate induced chromatism was negligible compared with the matched group. Hence, whether the amplicons produced can be directly judged through the phosphate induced coloration reaction, and the genotype could be determined accordingly.
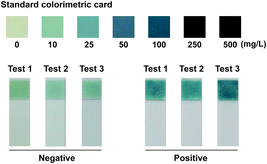
Phosphate ion could react with ammonium molybdate to form a blue molybdophosphate, which provided a colorimetric signal for visual detection. The indels of the ACE include insertion homozygote (II), insertion-deletion heterozygote (ID), and deletion homozygote (DD) respectively. The determination of genotype was based on the chromatism of both phosphate test strips from Ins and Del reaction mixtures. In addition, the standard colorimetric card from the commercial Phosphate Test Kid provides the signal intensities generated with different concentrations of phosphate ions, which was employed for positive and negative signal judgment (Fig. 2). ΔE* is employed to assess the difference between two colors, which is calculated as the Euclidean distance between two points in the CIELAB color space.23 The chromatic difference between the negative and positive results was measured through a CIELAB colorimeter with an average ΔE* value of 16.28 (Table S2†), which indicates the chromatic difference between the positive and negative results is significant for the human eye to distinguish.23,24
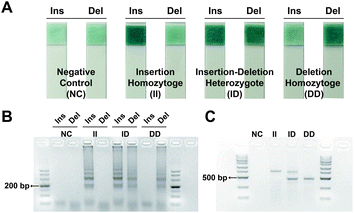
The II only generated positive signal on the strip from Ins reaction mixture. In contrast, the positive signal was only observed on the strip from Del reaction mixture for DD. While both strips generated positive signal indicating ID (Fig. 3A). In order to confirm the chromatism indicating amplification normally, the amplification kinetics curve was obtained from a real-time fluorometer (OptiGene Ltd., West Sussex, UK) and the amplicon of LAMP was analyzed via electrophoresis simultaneously. For matched sets of primer-variant, distinct amplification signal and typical ladderlike amplicon were observed while no amplification signal was detected in unmatched pairs (Fig. S1A† and 3B), which validated that the phosphate-based visual signal was not caused by spurious amplification. Furthermore, the PCR-based electrophoresis was employed as a reference for indels identification. The genotyping result was consistent between the visual genotyping system and PCR-based electrophoresis (Fig. 3C).
Genetic variation has been proved as a significant biomarker for disease diagnosis, prevention and treatment. In order to cater to the demand of precision medicine, the genotyping methods should be as straightforward as possible, as well as low-cost. Various strip-based molecular diagnostic methods have been established for visual detection. Lateral flow assay (LFA), for example, has been extensively applied in POCT, while the label of antibody for functionalizing the strip device highly increases the detection cost (Table 2).17,18,25–27 In the proposed research, by tracing phosphate ion produced during LAMP, a rapid and portable strategy for genotyping has been established. Compared with other visual-based molecular diagnostic methods, this strategy is free from specially designed labels, which enables cost-effective visual genotyping to facilitate clinical medication guidance.
3.2. Performance of visual genotyping system
Different combinations of primer set and variant were employed to evaluate the specificity of this method. For both primer sets, distinct positive signal was obtained with matched variants, while negative signal was judged with unmatched variants and deionized water as negative control (Fig. S2A†). Besides, a gradient dilution series of genomic DNA from 0 ng μL−1 to 50 ng μL−1 were prepared and 1 μL of each sample solution was taken to evaluate the sensitivity of this strategy. Positive signal could be still obtained by visual interpretation with as low as 1 ng genomic DNA (approximately 300 copies) (Fig. S2B†).Time consuming, thermal cycling as well as a specific device for strict temperature control make PCR unamiable for POCT scenarios. In this method, LAMP is integrated for target variant amplification, which results in high specificity and sensitivity. Moreover, enough phosphate ions can be produced for detection within 25 min just via a simple heater, which breaks the dependence on sophisticated instruments.
3.3. Direct genotyping avoiding DNA purification
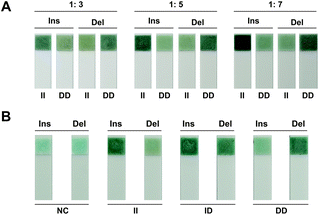
In order to genotyping from blood directly dispense with DNA extraction, alkaline lysis was introduced for DNA release. According to the chromatism from the strip, the best discrimination between positive and negative signals was observed with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 ratio for blood lysis (Fig. 4A). Hence, 1
5 ratio for blood lysis (Fig. 4A). Hence, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 was selected as the optimal ratio for direct genotyping based on the label-free strip. Accurate genotyping results were obtained from blood directly based on phosphate induced coloration reaction (Fig. 4B). The same result was also observed from amplification kinetics curve (Fig. S1B†), which demonstrated the excellent ability of this strategy for rapid and portable genotyping in clinical application.
5 was selected as the optimal ratio for direct genotyping based on the label-free strip. Accurate genotyping results were obtained from blood directly based on phosphate induced coloration reaction (Fig. 4B). The same result was also observed from amplification kinetics curve (Fig. S1B†), which demonstrated the excellent ability of this strategy for rapid and portable genotyping in clinical application.
As a typical and widely used biological specimen in clinical practice, whole blood was directly applied to genotyping with this method, which can provide timely diagnosis information and simplify the procedure dramatically. In addition, the amplification efficiency was increased owing to pyrophosphate serving as by-product was hydrolyzed by pyrophosphatase, which facilitates direct genotyping ignoring DNA polymerase inhibitors existed in blood. For visual interpretation, the color of blood could interfere with the colorimetric signal, which makes it equivocal for result judgment. In order to reduce the interference of blood on chromatism, the template for amplification should be as few as possible. Hence, a highly sensitive nucleic acid amplification technique is desired. By introducing LAMP into this method, not only trace of blood can result in drastic amplification for detection, but also the influence of blood is too weak to interfere with the colorimetric signal.
3.4. Clinical specimen application
For real specimen application, matched fresh whole blood and genomic DNA were used for ACE indels determination. The genotyping results derive from blood were consistent with those from genomic DNA as well as those obtained by PCR-based electrophoresis, which indicated that this genotyping system could accurately discriminate the ACE indels from both genomic DNA and whole blood (Tables 1 and S3†) Although whole blood with abnormal hematological indicators were used for genotyping, the accurate result could be still obtained ignoring pathological factors, which further proved the applicability of this method for varied specimens in clinical practice.| Genotyping with genomic DNA | Genotyping from whole blood directly | Total | Discrepant | Agreement | ||
|---|---|---|---|---|---|---|
| II | ID | DD | ||||
| II | 10 | 0 | 0 | 10 | 0 | 100% |
| ID | 0 | 10 | 0 | 10 | 0 | 100% |
| DD | 0 | 0 | 5 | 5 | 0 | 100% |
| Total | 10 | 10 | 5 | 25 | 0 | 100% |
| Conventional PCR3,4 | HRM5 | Real-time PCR6,7 | PCR-LFA26 | T-ARMS-PCR-LFA27 | LAMP phosphate induced coloration reaction | |
|---|---|---|---|---|---|---|
| a Note: T-ARMS-PCR: tetra-primer amplification-refractory-mutation-system PCR.b √ means DNA extraction is included, and × means DNA extraction is not included.c √ means the genotyping strategy is applicable for the POCT scenario, and × means the genotyping strategy is unamiable for POCT scenario and result detection requires interpretation by a professional. | ||||||
| Template | DNA | DNA | DNA | DNA/blood | DNA/blood | DNA/blood |
| Probes | None | None | None | None | None | None |
| Labels | None | None | None | Antibody/hapten | Antibody/hapten | None |
| DNA extraction | √ | √ | √ | √/× | √/× | √/× |
| Product detection | Agarose gel electrophoresis | Melting curve | Fluorescence | LFA | LFA | Phosphate induced coloration reaction |
| Total analysis time (min) | ∼160 | ∼145 | ∼130 | ∼135/∼80 | ∼135/∼80 | ∼85/∼30 |
| POCT | × | × | × | √ | √ | √ |
4. Conclusions
In this study, we developed an integrated strategy for miniaturizing simple process laboratory assays to shorten their complex steps and demonstrated that the entire contiguous sample-to-answer workflow could enable the genotyping within 30 min dispense with sophisticated instruments. We integrated two improved technologies into one system, a LAMP assay for target variant amplification and phosphate induced coloration reaction for result interpretation. These contributions address several bottlenecks of current methods while providing the advantages of simplicity, speediness and portability genotyping. Moreover, whole blood can be directly used for genotyping avoiding DNA purification. The excellent performance of this novel genotyping method allows the technique to be extrapolated to other genetic biomarkers. With further development for additional genetic biomarkers, this method could enable rapid clinical decision-making, improve management of disease prevention and facilitate personalized medicine, especially in resource-poor medical institutions.Author contributions
Jiaxing Zhang: conceptualization, methodology, investigation, data curation, writing-original draft. Hui Hui: methodology, investigation, data curation. Wei Xu: data curation. Kai Hua: investigation, funding acquisition. Yali Cui: resources, funding acquisition. Xiaonan Liu: conceptualization, methodology, writing-review and editing, supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81772289) and the Natural Science Basic Research Plan in Shaanxi Province of China (No. 2021JQ-443).References
- J. L. Weber, D. David, J. Heil, Y. Fan, C. Zhao and G. Marth, Am. J. Hum. Genet., 2002, 71, 854–862 CrossRef PubMed.
- R. E. Mills, C. T. Luttig, C. E. Larkins, A. Beauchamp, C. Tsui, W. S. Pittard and S. E. Devine, Genome Res., 2006, 16, 1182–1190 CrossRef CAS PubMed.
- B. Rigat, C. Hubert, P. Corvol and F. Soubrier, Nucleic Acids Res., 1992, 20, 1433 CrossRef CAS PubMed.
- V. Shanmugam, K. W. Sell and B. K. Saha, PCR Methods Appl., 1993, 3, 120–121 CrossRef CAS PubMed.
- P. H. Nissen, N. B. Campbell, C. S. Hojskov, A. Floe, H. J. Hoffmann, O. Hilberg, S. A. Ladefoged and H. J. Moller, Ann. Clin. Biochem., 2015, 52, 105–112 CrossRef CAS PubMed.
- M. H. Lin, C. H. Tseng, C. C. Tseng, C. H. Huang and C. P. Tseng, Clin. Biochem., 2001, 34, 661–666 CrossRef CAS PubMed.
- M. Hiratsuka, Y. Kishikawa, K. Narahara, T. Inoue, S. I. Hamdy, Y. Agatsuma, Y. Tomioka and M. Mizugaki, Anal. Biochem., 2001, 289, 300–303 CrossRef CAS PubMed.
- A. H. Chowdhury, M. M. Zaman, K. M. Haque, M. A. Rouf, A. T. Shah, T. Nakayama, T. Yokoyama, N. Yoshiike and H. Tanaka, Bangladesh Med. Res. Counc. Bull., 1998, 24, 55–59 CAS.
- B. Rigat, C. Hubert, F. Alhenc-Gelas, F. Cambien, P. Corvol and F. Soubrier, J. Clin. Invest., 1990, 86, 1343–1346 CrossRef CAS PubMed.
- H. E. Montgomery, R. Marshall, H. Hemingway, S. Myerson, P. Clarkson, C. Dollery, M. Hayward, D. E. Holliman, M. Jubb, M. World, E. L. Thomas, A. E. Brynes, N. Saeed, M. Barnard, J. D. Bell, K. Prasad, M. Rayson, P. J. Talmud and S. E. Humphries, Nature, 1998, 393, 221–222 CrossRef CAS PubMed.
- S. K. Vashist, P. B. Luppa, L. Y. Yeo, A. Ozcan and J. H. T. Luong, Trends Biotechnol., 2015, 33, 692–705 CrossRef CAS PubMed.
- H. Deng and Z. Gao, Anal. Chim. Acta, 2015, 853, 30–45 CrossRef CAS PubMed.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, e63 CrossRef CAS PubMed.
- S. Yongkiettrakul, J. Kampeera, W. Chareanchim, R. Rattanajak, W. Pornthanakasem, W. Kiatpathomchai and D. Kongkasuriyachai, Parasitol. Int., 2017, 66, 964–971 CrossRef CAS PubMed.
- A. B. Nurul Najian, E. A. R. Engku Nur Syafirah, N. Ismail, M. Mohamed and C. Y. Yean, Anal. Chim. Acta, 2016, 903, 142–148 CrossRef CAS PubMed.
- X. Liu, J. Zhang, Y. Cai, S. Zhang, K. Ma, K. Hua and Y. Cui, Sens. Actuators, B, 2021, 333, 129624 CrossRef CAS.
- R. Kumvongpin, P. Jearanaikoon, C. Wilailuckana, N. Sae-Ung, P. Prasongdee, S. Daduang, M. Wongsena, P. Boonsiri, W. Kiatpathomchai, S. S. Swangvaree, A. Sandee and J. Daduang, Mol. Med. Rep., 2017, 15, 3203–3209 CrossRef CAS PubMed.
- E. A. Phillips, T. J. Moehling, S. Bhadra, A. D. Ellington and J. C. Linnes, Anal. Chem., 2018, 90, 6580–6586 CrossRef CAS PubMed.
- F. Zhang, R. Wang, L. Wang, J. Wu and Y. Ying, Chem. Commun., 2014, 50, 14382–14385 RSC.
- F. Zhang, L. Wang, R. Wang, Y. Ying and J. Wu, Anal. Chem., 2015, 87, 1523–1526 CrossRef CAS PubMed.
- K. Nagamine, T. Hase and T. Notomi, Mol. Cell. Probes, 2002, 16, 223–229 CrossRef CAS PubMed.
- X. Liu, C. Zhang, K. Hua, J. Liang and Y. Cui, Anal. Biochem., 2019, 582, 113351 CrossRef CAS.
- A. Fullerton, T. Fischer, A. Lahti, K. P. Wilhelm, H. Takiwaki and J. Serup, Contact Dermatitis, 1996, 35, 1–10 CrossRef CAS PubMed.
- J. Y. Hardeberg, Acta Gr., 2003, 15, 89–104 Search PubMed.
- L. Ma, H. Wang, T. Zhang, Y. Xuan, C. Li, W. Chen, B. Liu and Y. Zhao, Sens. Actuators, B, 2019, 298, 126819 CrossRef CAS.
- X. Yang, J. Liang, X. Huang, X. Wang, Y. Sun, C. Dong, Y. Cui and W. Hui, Biol. Proced. Online, 2021, 23, 1–10 CrossRef.
- Y. Cai, S. Zhang, J. Zhang, X. Liu, K. Ma, W. Xu, X. Deng, J. Yang, T. Ma, C. Jiang, W. Hui and Y. Cui, Anal. Chem., 2022, 94, 4686–4694 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra03989c |
| ‡ Contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |