Open Access Article
Open Access ArticleCo-immobilized bienzyme of horseradish peroxidase and glucose oxidase on dopamine-modified cellulose–chitosan composite beads as a high-efficiency biocatalyst for degradation of acridine
Yaohua Gu†
 a,
Lin Yuan†a,
Mingming Li†b,
Xinyu Wanga,
Deyu Raoa,
Xiaoyan Baia,
Keren Shic,
Haiming Xu*a,
Shaozhang Hou*a and
Huiqin Yao
a,
Lin Yuan†a,
Mingming Li†b,
Xinyu Wanga,
Deyu Raoa,
Xiaoyan Baia,
Keren Shic,
Haiming Xu*a,
Shaozhang Hou*a and
Huiqin Yao *a
*a
aKey Laboratory of Environmental Factors and Chronic Disease Control, College of Public Health and Management, School of Basic Medicine, Ningxia Medical University, Yinchuan 750004, P. R. China. E-mail: huiqin_yao@163.com; Tel: +86-951-698-0689
bUrology Surgery, General Hospital of Ningxia Medical University, Yinchuan 750004, P. R. China
cState Key Laboratory of High-efficiency Coal Utilization and Green Chemical Engineering, Ningxia University, Yinchuan 750021, P. R. China
First published on 16th August 2022
Abstract
Co-immobilized bienzyme biocatalysts are attracting increasing interest in the field of wastewater treatment due to their widespread application. In this study, we successfully prepared a co-immobilized bienzyme biocatalyst by immobilizing horseradish peroxidase (HRP) and glucose oxidase (GOD) on dopamine (DA) modified cellulose (Ce)–chitosan (Cs) composite beads via covalent binding, designated as Ce–Cs@DA/HRP–GOD beads, and found that the bienzyme biocatalyst had a good ability to catalytically degrade acridine in wastewater. SEM, EPR, FTIR, and XRD were used to characterise the structure and properties of the Ce–Cs@DA/HRP–GOD beads. The co-immobilized bienzyme biocatalyst with a small amount of HRP exhibited better degradation efficiency for acridine (99.5%, 8 h) in simulated wastewater compared to the Ce–Cs@DA/HRP (93.8%, 8 h) and Ce–Cs@DA/GOD (15.8%, 8 h) beads alone. In addition, a reusability study showed that the co-immobilized bienzyme biocatalyst maintained a degradation rate of 61.2% after six cycles of acridine degradation. The good biodegradability and reusability of the biocatalyst might be due to the synergistic effect of bienzyme HRP–GOD, including the strong covalent bonding. Accordingly, the co-immobilized bienzyme biocatalyst based on the cascade reaction may pave the way for efficient and eco-friendly treatment of industrial wastewater.
1 Introduction
There has been an increase in applications of acridine, a typical nitrogenous heterocyclic compound, in different industries, such as dyes, pharmaceuticals, coking, and pesticides,1,2 due to its good biological activity and particular functional unit. Nowadays, more and more wastewater containing acridine has been discharged into the environment.3 Even at low concentrations below 10 mg L−1, it may gradually accumulate in organisms through bioenrichment, causing irreversible damage to cells and eventually leading to teratogenesis, mutagenesis and carcinogenesis among various life forms.4 In addition, it can cause intense irritation to the skin and mucous membranes.5 Acridine is difficult to degrade under natural conditions because of its relatively stable structure.6 Therefore, it is urgent to find an efficient, green and economical technique to degrade acridine in wastewater. In recent years, enzyme-catalysed degradation has attracted widespread attention thanks to its wide selectivity, high efficiency, and environmental friendliness.7 Among the various oxidoreductases utilised for wastewater degradation, horseradish peroxidase (HRP) has promising application prospects owing to its mild reaction conditions and good stability.8However, free HRP is easy to deactivate in the process of use, and difficult to reuse. Therefore, in practical application, HRP is generally used after immobilization.9,10 The immobilization of enzyme means that the enzyme is restricted or bound in a certain space with the carrier material, so that it retains the catalytic activity, improves the stability and obtains the reusability. Various techniques have been utilised to immobilize enzyme, such as encapsulation, cross-linking, adsorption, and covalent attachment. Currently, among the various immobilization methods, the most frequently used method is the covalent bonding of enzyme, due to advantages such as high strength and good stability under extreme conditions.11 For example, laccase was immobilized on polyamide 6/chitosan (Cs) nanofibers to degrade endocrine-disrupting chemicals,12 and the corresponding degradation efficiency of bisphenol A and 17α-ethinylestradiol by immobilized laccase was higher than that by the same amount of free laccase, that is, the immobilized laccase exhibited good mechanical stability and reusability. In addition, HRP was immobilized in chitosan–graphene oxide composite microspheres,13 which also exhibited superior stability and catalysis compared to that of its free counterpart. Sun14 et al. also investigated the degradation of malachite green by immobilized laccase in a hydrogel, and the degradation efficiency, activity, and stability of the immobilized enzyme improved significantly compared with free laccase.
In addition, studies have found that some redox mediators, including 1-hydroxybenzotriazole (HBT), 2,2-azino-bis-(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), and 2,2,6,6-tetramethylpiperidine-1-oxyl (TEMPO), were introduced into the enzyme-catalytic system to cooperate with enzymatic catalysis,8 which can expand the range of substrates, and promote the oxidoreductase catalytic performance.15,16 For example, Leng and co-workers17 investigated tetracycline transformation in wastewater using HRP and found that the tetracycline was hardly degraded without the mediator molecular ABTS, whereas the transformation rate increased dramatically to above 95.0% in the presence of ABTS. Chen and co-workers18 immobilized HRP in preformed multilayer polyelectrolyte microcapsules to degrade polycyclic aromatic hydrocarbons in water, and the degradation rate was 80.9% in the presence of HBT, compared to 48.7% in the absence of HBT. This suggests that redox mediators have a synergistic effect on the catalysis of HRP by significantly improving its catalytic performance. This application greatly improves the range of substrates catalyzed by HRP. Prior to this, HRP could only degrade compounds with low redox potential, such as phenolic and aniline compounds.19
The biggest problem of using HRP to degrade acridine in wastewater is that hydrogen donor—H2O2 must be added in the reaction process.20 H2O2 is a corrosive reagent that is difficult to handle and store. Excess H2O2 can lead to the partial inactivation of HRP, consequently resulting in a significantly reduced degradation rate during the reaction.21 In order to solve this problem, we conducted a study to reduce the harm of H2O2 by using a cascade reaction catalysed by bienzyme. Glucose oxidase (GOD) is an important enzyme that is widely used to produce H2O2 by oxidising β-D-glucose (Glu) in the presence of O2.22,23 Therefore, HRP and GOD were co-immobilized in this study, in the catalytic reaction, the H2O2 generated in situ by GOD-catalyzed oxidation of glucose in the presence of O2 that causes the further catalytic reduction by HRP, which not only weakens product inhibition in the first reaction (GOD + glucose + O2 → H2O2 + gluconolactone) but also weakens substrate inhibition in the second reaction (HRP + H2O2 + redox mediators → degradation product). In addition, balance is driven toward product formation because of the shift in the equilibrium of chemical reactions. This not only accelerates the catalytic rate of the bienzyme, improves their stability, but also reduces the use cost of HRP from their synergistic effects. Therefore, co-immobilized HRP and GOD on the same carrier appear to have promising applications. Many studies have reported the application of this system in biosensors in recent years.24 However, to the best of our knowledge, the utility of co-immobilized bienzyme biocatalyst for acridine high efficiency degradation in wastewater has not been reported.
The structure and properties of carrier materials are also crucial for the immobilized enzyme. Of the various carriers used for enzymatic immobilization, cellulose (Ce) and chitosan (Cs), which are abundant and renewable in nature, are generally used for this purpose.25,26 These materials possess similar structures and a mixture of them not only retains their respective properties, but also solves the problems of the poor mechanical strength and adsorption performance of chitosan and cellulose, respectively.27 Cellulose and chitosan can be dissolved and modified in an ionic liquid, which creates cost-effective conditions for the functional regeneration of the materials.28 Dopamine (DA) is an adhesive protein from mussels that contains catechol and amine functional groups.29 It can self-polymerize in alkaline environments to form polydopamine (PDA) films on various types of carriers, and then covalently bind biomacromolecules by aldehyde acid condensation or Schiff base reaction.30 In recent years, DA has been used as a modifier for the covalent binding of enzymes.
In the present work, HRP and GOD were co-immobilized on dopamine-modified cellulose–chitosan composite beads as a novel high-efficient biocatalyst for the degradation of acridine in wastewater. Compared to immobilized HRP or GOD alone, co-immobilized HRP–GOD exhibited a higher reaction efficiency, better stability, and lower reaction cost. Furthermore, the factors affecting the degradation rate of acridine, reusability, and storage stability of the biocatalyst were systematically evaluated.
2 Materials and methods
2.1 Chemicals and enzyme
Horseradish peroxidase (>200 U mg−1) from Wasabi was provided by TCI Development Co., Ltd. (Shanghai, China). Glucose oxidase (>100 U mg−1) from Aspergillus niger and acridine (purity 98.0%) were purchased from Sigma (St. Louis, MO, USA). β-D-Glucose was obtained from Sinopharm Chemical Reagent Co., Ltd. (Beijing, China). Dopamine hydrochloride and Tris were provided by Innochem Technology Co., Ltd. (Beijing, China). Microcrystalline cellulose (Ce), 1-ethyl-3-methylimidazolium acetate ([Emim][OAc]), HBT, ABTS, and chitosan (Cs) were all supplied by Hengyuanchuang Technology and Trade Co., Ltd. (Ningxia, China). HPLC-grade methanol was provided by Fisher Company (Morristown, NJ, USA). All the chemicals used in the experiments were of analytical pure grade and used without further purification, except H2O2, which needed to be diluted before use.2.2 Preparation of the Ce–Cs@DA beads
Cellulose (Ce, 0.3 g) and chitosan (Cs, 0.3 g) were completely dissolved in 10 mL of the ionic liquid ([Emim][OAc]) under mechanical stirring, and the reaction contents were maintained in an oil bath at 85 °C for 3 h. After the reaction ended, the contents were cooled to room temperature, and then added dropwise into ultrapure water via a peristaltic pump to obtain the Ce–Cs composite beads. The resulting product obtained by filtration was subsequently indurated for 2 h and washed several times with ultrapure water.Ce–Cs composite beads of 1.0 g (wet weight) and dopamine (DA) of 15 mg were dispersed in a 10 mL Tris–HCl buffer solution of 0.01 mol L−1 and pH 8.5.29 The conical flask was placed in a thermostatic oscillator for 3 h, 150 rpm at 30 °C, to form Ce–Cs@DA beads. Then, the beads obtained by filtration were washed three times with ultrapure water.
2.3 Immobilization of HRP and GOD on Ce–Cs@DA beads
The Ce–Cs@DA beads of 1.0 g (wet weight) and a certain amount of HRP and GOD were dissolved in a 10 mL PBS buffer solution of 0.1 mol L−1 and pH 7.0. Herein, the concentration of HRP–GOD was 1 mg mL−1, the ratios of HRP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GOD were 3
GOD were 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, and 1
2, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, respectively. The contents were placed in a thermostatic oscillator at 30 °C, 150 rpm for 24 h, to co-immobilize the bienzyme on the beads. Next, the beads obtained by filtration were washed 3 times with a PBS buffer solution of 0.1 mol L−1 and pH 7.0 and denoted as Ce–Cs@DA/HRP–GOD beads.
3, respectively. The contents were placed in a thermostatic oscillator at 30 °C, 150 rpm for 24 h, to co-immobilize the bienzyme on the beads. Next, the beads obtained by filtration were washed 3 times with a PBS buffer solution of 0.1 mol L−1 and pH 7.0 and denoted as Ce–Cs@DA/HRP–GOD beads.
In order to study the effect of different biocatalysts on acridine degradation efficiency, Ce–Cs@DA/HRP@GOD beads, Ce–Cs@DA/GOD@HRP beads, Ce–Cs@DA/HRP beads and Ce–Cs@DA/GOD beads were prepared, respectively. Their preparation process was similar to that of Ce–Cs@DA/HRP–GOD beads. The difference is that Ce–Cs@DA/HRP@GOD beads are prepared by first placing 1.0 g Ce–Cs@DA beads in PBS buffer solution (10 mL, 0.1 mol L−1, pH 7.0) containing HRP (6.7 mg) for 12 h and then placing them in PBS buffer solution containing GOD (3.3 mg) for 12 h after washing. Ce–Cs@DA/GOD@HRP beads are prepared by implanting 1.0 g Ce–Cs@DA beads in PBS buffer solution containing GOD (3.3 mg) for 12 h, then implanting them in PBS buffer solution containing HRP (6.7 mg) for 12 h after washing. Ce–Cs@DA/HRP beads are prepared by placing 1.0 g Ce–Cs@DA beads in PBS buffer solution containing HRP (10 mg) for 24 h. Ce–Cs@DA/GOD beads are prepared by implanting 1.0 g Ce–Cs@DA beads into PBS buffer solution containing GOD (10 mg) for 24 h. The enzyme loading of the Ce–Cs@DA/HRP–GOD beads was determined by the Bradford method.
2.4 Degradation of acridine by the biocatalyst
Degradation of acridine was performed at 30 °C in a PBS buffer solution (10 mL, pH 7.0) containing Ce–Cs@DA/HRP–GOD beads (wet weight, 1 g), HBT (20 mg), Glu (20 mg), and acridine (15 mg L−1). Ce–Cs@DA/HRP@GOD beads and Ce–Cs@DA/GOD@HRP beads degraded acridine under the same conditions, whereas the Ce–Cs@DA/HRP beads (wet weight, 1 g) degraded acridine in the presence of H2O2 (100 μL) and HBT (20 mg) at 30 °C, pH 7.0, and the Ce–Cs@DA/GOD beads (wet weight, 1 g) degraded acridine in the presence of Glu (20 mg) at 30 °C, pH 7.0.The residual concentration of acridine was determined by high-performance liquid chromatography equipped (HPLC) with a C18 column. Detection conditions: mobile phase: 80% methanol; detection wavelength: 250 nm; flow rate: 1 mL min−1; injection volume: 10 μL. Prior to injection into the column, the sample was filtered through a 0.45 μm syringe filter. The formula used to calculate the degradation efficiency of acridine was as follows:
| D (%) = [(C0 − Ct)/C0] ×100% |
In order to study the metabolic pathway of Ce–Cs@DA/HRP–GOD beads, the degradation products were identified by Liquid Chromatography-Mass Spectrometry (LC-MS). The acridine water samples degraded by Ce–Cs@DA/HRP–GOD beads are concentrated in rotary evaporation instrument, so that the final volume is about 2 mL, and then transferred to the sample bottle through 0.22 μm polyethersulfone filter membrane for determination. Operating conditions of LC-MS: the column: Agilent SBC-18 (4.6 × 150 mm, 5 μm); mobile phase: 20% methanol; flow rate: 1 mL min−1; injection volume: 30 μL; scanned speed: 50–500 m/z; drying temperature: 350 °C.
2.5 Determination of kinetic parameters
The Michaelis–Menten kinetic parameters, Km and Vmax, of the free HRP–GOD and Ce–Cs@DA/HRP–GOD beads were determined by measuring the HRP activity with ABTS as the substrate with an initial concentration in the range of 0.3–20.0 mmol L−1. The Km was calculated according to the Lineweaver–Burk double reciprocal model of the Michaelis–Menten equation as follows:| 1/V0 = Km/Vmax × 1/[S] + 1/Vmax |
2.6 Optimal temperature and pH of Ce–Cs@DA/HRP–GOD beads
In order to investigate the temperature effect on the degradation of acridine, the free HRP–GOD and Ce–Cs@DA/HRP–GOD beads were placed in acridine solution (pH 7.0) at different temperatures (10, 20, 30, 40, 50, 60 °C), and the reaction was carried out in the presence of HBT and Glu. To study the pH effect on the degradation of acridine, the free HRP–GOD and Ce–Cs@DA/HRP–GOD beads were placed in acridine solution at different pH (4.0, 5.0, 6.0, 7.0, 8.0, 9.0) at 30 °C, and then HBT and Glu were added for the reaction. The degradation rate of acridine was measured by HPLC, and the optimal temperature and pH value were obtained.2.7 Reusability and storage stability of Ce–Cs@DA/HRP–GOD beads
To investigate the reusability, the Ce–Cs@DA/HRP–GOD beads (wet weight, 1 g), HBT (20 mg), Glu (20 mg) and acridine solution (10 mL, 15 mg L−1) were added into a 25 mL conical flask, and then placing it in a thermostatic oscillator at 30 °C, 150 rpm for 12 h to complete the degradation. Next, the beads were washed with PBS buffer solution (0.1 mol L−1, pH 7.0), and reintroduced into a new acridine solution for another degradation. The operation was repeated six times. The concentration of acridine in the solution was detected by HPLC after each reaction.In order to explore the storage stability, the Ce–Cs@DA/HRP–GOD beads (wet weight) were incubated in PBS buffer solution (0.1 mol L−1, pH 7.0) and stored at 4 °C for 60 d. 1 g of beads were removed every 10 days to measure the degradation rate of acridine. Each reaction was carried out at 30 °C, pH 7.0. The concentration of acridine in the solution was detected by HPLC after each reaction.
2.8 Characterisation of the biocatalyst
The biocatalyst was characterised after freeze-drying. Scanning electron microscopy (SEM, JEOL JSM-6360LV) was used to observe the morphology of the beads. Fourier transform infrared (FTIR, Bruker, Karlsruhe, Germany) spectroscopy was performed to analyse the molecular structure and chemical composition of the biocatalyst. X-ray diffraction (XRD, K/Max-3c) was conducted to investigate the structure of the beads. Electron paramagnetic resonance (EPR, JES FA200, JEOL) was performed to monitor the electron transfer radical signals in the supernatants of the Ce–Cs@DA/HRP–GOD beads + HBT + Glu, Ce–Cs@DA/HRP–GOD beads + HBT, Ce–Cs@DA/HRP–GOD beads + Glu, free HRP + free GOD + HBT + Glu, raw Glu, and raw HBT. All experiments were performed at ambient temperature of 20 ± 2 °C.3 Results and discussion
3.1 Characterisation
The construction of Ce–Cs@DA/HRP–GOD beads was first confirmed by SEM (Fig. 1). The surface morphology of Ce–Cs composite beads, Ce–Cs@DA beads and Ce–Cs@DA/HRP–GOD beads were all regular spherical or elliptical shapes (Fig. 1a, d and g). There were many irregular gullies on the surface of the Ce–Cs composite beads (Fig. 1a), which could provide a large specific surface area for DA attachment and bienzyme co-immobilization. After the modification (Fig. 1d) and co-immobilization of the bienzyme (Fig. 1g), the gullies on the surface of the beads gradually became shallower or even flattened, which indicated that part of DA and bienzyme had successfully adhered to the beads. Fig. 1b, e and h shows the cross-sectional morphology of the beads. It was evident that the inner lamellar structure of the beads became significantly rough, indicating that some DA and bienzyme diffused through the water into the beads and adhered to the lamellar structure. In addition, the beads clearly possess abundant coral ultra-large pore structures with disordered channels that run through each other, which was beneficial for the co-immobilization of the bienzyme and could avoid space barriers caused by molecular accumulation. Fig. 1c, f and i shows the lamellar structure of the beads. The interior of the Ce–Cs composite beads exhibited a smooth lamellar structure. In contrast, the lamellar surfaces of the Ce–Cs@DA beads (Fig. 1f) and Ce–Cs@DA/HRP–GOD beads (Fig. 1i) were significantly rougher, suggesting that the DA attached successfully. Due to the small size of the enzyme molecule (35 Å for HRP, 60 Å for GOD),31 it was difficult to observe under a scanning electron microscope. Therefore, this characterization can only infer the successful immobilization of the bienzyme, but cannot provide definitive proof.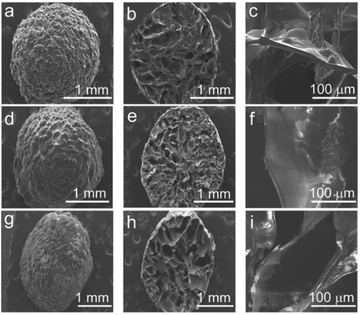 | ||
| Fig. 1 SEM images of different carriers: (a, b, c) the Ce–Cs composite beads; (d, e, f) Ce–Cs@DA beads; (g, h, i) Ce–Cs@DA/HRP–GOD beads. | ||
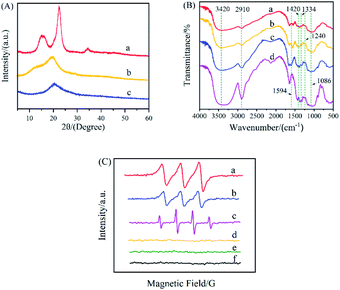
XRD spectra of raw Ce, raw Cs, and the Ce–Cs composite beads were shown in Fig. 2A. The XRD patterns of raw Ce (Fig. 2Aa) represented that there was a typical cellulose I diffraction angles, with a sharp peak at 2θ = 22.5°, a small peak at 2θ = 34.5°, and a broad peak between 2θ = 14.9° and 2θ = 16.2°, which was identical with the results reported.32 There was a strong diffraction peak at 2θ = 20.5° and a weak reflection peak at 2θ = 10.5° in the XRD patterns of raw Cs (Fig. 2Ab), as reported previously in the literature.33 After regenerated from ionic liquid ([Emim][OAc]), the XRD patterns of the Ce–Cs composite beads (Fig. 2Ac) exhibited a broad and overlapped peak at 2θ = 20.5°, which were in accordance with the characteristic peaks of cellulose II and chitosan. This conclusion was consistent with that previously reported. Zhang et al. studied the effect of the imidazolium structure on the membrane properties of chitosan, and the results inferred that the imidazolium ionic liquids decrease the crystallinity of chitosan membranes.34 Liu et al. used ionic liquids as a solvent for cellulose and chitosan dissolution and regeneration, and the XRD results showed that the diffraction angles of regenerated Ce and Cs was almost the same as that of the typical cellulose II structure.35 The XRD patterns indicated that there was no chemical change in Ce and Cs after the functional modification of the ionic liquid ([Emim] [OAc]).
The formation of Ce–Cs@DA/HRP–GOD beads were confirmed by FTIR spectroscopy in comparison with raw Ce, raw Cs, and Ce–Cs@DA beads (Fig. 2B). All characteristic FTIR peaks of raw Cs (Fig. 2Bc) and raw Ce (Fig. 2Bd) were observed at the same or similar positions for Ce–Cs@DA (Fig. 2Bb) and Ce–Cs@DA/HRP–GOD (Fig. 2Ba) samples at 1086 cm−1 (C–O bonds), 2910 cm−1 (C–H bonds), 3420 cm−1 (O–H and N–H bonds), 1420 cm−1 (C–H bonds), and 1594 cm−1 (N–H bonds), indicating that the dissolution of Ce and Cs by the ionic liquid was only physical.35 Furthermore, the characteristic spectrum observed at 1334 cm−1 (C–N bonds) and 1240 cm−1 (C–O bonds) were attributed to the typical absorption peaks of the DA structure, confirming that DA was successfully attached to the Ce–Cs@DA beads. From the spectrum of the Ce–Cs@DA/HRP–GOD beads (Fig. 2Ba), apart from the above-listed signals, the peaks also observed at 3420 cm−1 (N–H bonds) and 2910 cm−1 (C–H bonds), indicating that the bienzyme HRP–GOD was successfully co-immobilized on Ce–Cs@DA beads.4 Therefore, FTIR spectroscopy confirmed the successful preparation of the co-immobilized bienzyme biocatalyst.
The EPR spectra of Ce–Cs@DA/HRP–GOD beads + HBT + Glu, Ce–Cs@DA/HRP–GOD beads + HBT, Ce–Cs@DA/HRP–GOD beads + Glu, free HRP-GOD + HBT + Glu, raw HBT, and raw Glu were obtained to monitor the electron transfer radical signals during the reaction (Fig. 2C). There was no free radical signal in the Ce–Cs@DA/HRP–GOD beads + HBT system (Fig. 2Cd), raw HBT (Fig. 2Ce), and raw Glu (Fig. 2Cf), which suggested that there was no electron transfer in the reaction system. In contrast, a significant signal attributed to the OH˙ radical was observed for the Ce–Cs@DA/HRP–GOD beads + Glu system (Fig. 2Cc), which was similar to the previously reported OH˙ free radical peak pattern.36,37 This indicated that GOD was successfully immobilized on the beads because Glu could only be oxidised by GOD immobilized in the bienzyme system to produce H2O2, which was further transformed to OH˙. A clear free radical signal of HBT˙ was observed in the Ce–Cs@DA/HRP–GOD beads + HBT + Glu system (Fig. 2Ca), which proved that bienzyme was successfully immobilized on the beads, because HBT˙ was produced only when HRP and GOD were present together. The reaction process was that GOD oxidized Glu to produce H2O2, and HRP oxidized HBT to produce HBT˙ in the presence of H2O2.38 Furthermore, a similar free radical signal was also observed in the free HRP–GOD + HBT + Glu system (Fig. 2Cb), which further demonstrated that the HRP–GOD bienzyme was successfully co-immobilized on the beads and could react normally. The EPR spectra confirmed that the successful preparation of co-immobilized bienzyme biocatalyst and the cascade reactions occurring within the biocatalyst.
3.2 Acridine degradation by different biocatalysts
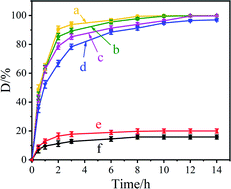
Fig. 3 shows the acridine degradation results by the Ce–Cs@DA/HRP–GOD beads, Ce–Cs@DA/HRP@GOD beads, Ce–Cs@DA/GOD@HRP beads, Ce–Cs@DA/HRP beads, and Ce–Cs@DA/GOD beads in wastewater. As illustrated in Fig. 3, in the presence of HBT, acridine could be almost completely degraded by the Ce–Cs@DA/HRP–GOD beads (Fig. 3a), Ce–Cs@DA/HRP@GOD beads (Fig. 3b), Ce–Cs@DA/GOD@HRP beads (Fig. 3d), and Ce–Cs@DA/HRP beads (Fig. 3c), although it was barely degraded by the Ce–Cs@DA/GOD beads (Fig. 3f). This indicated that acridine cannot be degraded by immobilized GOD alone. In addition, acridine was difficult to degrade by the Ce–Cs@DA/HRP–GOD beads in the absence of HBT (Fig. 3e), which indicated that HBT as a redox mediator enhanced the reaction process through the hydrogen transfer mechanism and promoted HRP degradation of acridine.It is worth noting that Ce–Cs@DA/HRP–GOD beads (Fig. 3a) show the best acridine degradation efficiency compared with the Ce–Cs@DA/HRP@GOD beads (Fig. 3b) and Ce–Cs@DA/GOD@HRP beads (Fig. 3d), which may be closely related to the spatial distribution of bienzyme of the three biocatalysts. The spatial distribution of HRP and GOD on Ce–Cs@DA/HRP–GOD beads may be more consistent with the sequence of cascade reactions in the catalytic process. Besides, compared with the Ce–Cs@DA/HRP beads (Fig. 3c), the Ce–Cs@DA/HRP–GOD beads (Fig. 3a) also exhibited a higher degradation rate, which may be due to the synergistic effect of HRP and GOD. Therefore, Ce–Cs@DA/HRP–GOD beads was selected as the subsequent research object.
In other words, the cascade reaction catalysed by HRP–GOD could avoid the accumulation of some harmful intermediates by in situ generation and consumption of the intermediate, and speeded up the reaction rate by improving the stability of the enzyme. This was similar to the results reported in previous literature. For example, Rocha-Martín39 et al. assembled three different bio-redox orthogonal cascades in vitro and found that the co-immobilized multi-enzyme system based on the cascades was more efficient compared to the two enzymes separately immobilized on two different carriers. Surprisingly, compared with the co-immobilized bienzyme (HRP–GOD) biocatalysts previously reported, our biocatalyst had a higher degradation rate. For instance, Razzaghi40 et al. investigated the decolorization properties of different dyes by co-immobilizing HRP and GOD on polyurethane. The results demonstrated that the degradation rates of azo dye, methylene blue and malachite green were 87.5%, 91.6% and 95.2%, respectively. Most importantly, the immobilized HRP–GOD system used the same amount of total enzyme as the immobilized HRP system, but the immobilized HRP–GOD system replaced part of HRP with GOD, and the price of GOD was much lower than that of HRP, which greatly reduced the experimental cost.
3.3 Kinetic parameters of Ce–Cs@DA/HRP–GOD beads and free HRP–GOD
Fig. 4A shows the initial reaction rate of increasing concentrations of ABTS for free HRP–GOD and Ce–Cs@DA/HRP–GOD beads and Table 1 shows the kinetic constants, maximum reaction rate (Vmax) and apparent Michaelis–Menten constant (Km), of free HRP–GOD and Ce–Cs@DA/HRP–GOD beads determined. The Km is a measure of the affinity that an enzyme has for a given substrate. The lower the value of Km, the higher is the affinity of the enzyme to the substrate. The lesser the value of Km, the higher is the affinity for the substrate. The Km value for the Ce–Cs@DA/HRP–GOD beads (4.46 mmol L−1) was found to be 3-fold more than that of the free HRP–GOD (1.36 mmol L−1) demonstrating a lower affinity for the substrate caused by diffusional limitations. However, the Vmax of the Ce–Cs@DA/HRP–GOD beads (0.48 mmol min L−1) was higher than that of free HRP–GOD (0.08 mmol min L−1), probably because of the diffusion limitation and steric hindrance in the Ce–Cs@DA/HRP–GOD beads.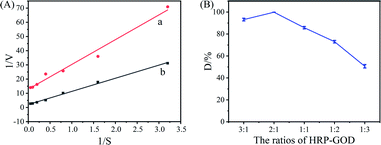 | ||
Fig. 4 (A) Michaelis–Menten parameters of (a) free HRP–GOD and (b) Ce–Cs@DA/HRP–GOD beads. (B) Effect of the concentration ratios of HRP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GOD in solution on the degradation efficiency of acridine. GOD in solution on the degradation efficiency of acridine. | ||
| Michaelis–Menten parameters | ||
|---|---|---|
| Km/(mmol L−1) | Vmax/(mmol min L−1) | |
| Free HRP–GOD | 1.36 | 0.07 |
| Ce–Cs@DA/HRP–GOD beads | 4.46 | 0.43 |
3.4 Effect of HRP–GOD ratio on degradation of acridine by Ce–Cs@DA/HRP–GOD beads
In Fig. 4B, we discuss the effect of the concentration ratios of HRP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GOD in solution on the degradation efficiency of acridine. The initial concentration of HRP–GOD was constant at 1 mg mL−1. The Ce–Cs@DA beads were added into PBS buffer solution containing different ratios of HRP
GOD in solution on the degradation efficiency of acridine. The initial concentration of HRP–GOD was constant at 1 mg mL−1. The Ce–Cs@DA beads were added into PBS buffer solution containing different ratios of HRP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GOD (3
GOD (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), and the contents were placed in a thermostatic oscillator at 30 °C, 150 rpm for 24 h, to form Ce–Cs@DA/HRP–GOD beads. It can be seen from the Fig. 4B that as the concentration ratios of HRP
3), and the contents were placed in a thermostatic oscillator at 30 °C, 150 rpm for 24 h, to form Ce–Cs@DA/HRP–GOD beads. It can be seen from the Fig. 4B that as the concentration ratios of HRP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GOD in the solution changed from 3
GOD in the solution changed from 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, the biocatalyst they formed had a first increase and then a decrease in the degradation efficiency of acridine. Clearly, Ce–Cs@DA/HRP–GOD beads had the highest degradation rate of acridine when the concentration ratios of HRP
3, the biocatalyst they formed had a first increase and then a decrease in the degradation efficiency of acridine. Clearly, Ce–Cs@DA/HRP–GOD beads had the highest degradation rate of acridine when the concentration ratios of HRP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GOD was 2
GOD was 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. At this point, the bienzyme amount loaded on beads was determined to be 7.52 mg (HRP–GOD) per g (beads) by Bradford method, and the loading yield of bienzyme reached 75.2%. When the concentration ratios of HRP
1. At this point, the bienzyme amount loaded on beads was determined to be 7.52 mg (HRP–GOD) per g (beads) by Bradford method, and the loading yield of bienzyme reached 75.2%. When the concentration ratios of HRP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GOD changed from 2
GOD changed from 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 3
1 to 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, degradation rate of acridine decreased, which might be due to less GOD immobilized on the beads, so that the generation of H2O2 by GOD cannot meet the needs of the enzymatic reaction of HRP. When the concentration ratios of HRP
1, degradation rate of acridine decreased, which might be due to less GOD immobilized on the beads, so that the generation of H2O2 by GOD cannot meet the needs of the enzymatic reaction of HRP. When the concentration ratios of HRP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GOD were further changed from 2
GOD were further changed from 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, the degradation rate of acridine still decreased, which might be due to the following two reasons. First, the immobilized HRP on the beads decreased, which reduced the HRP active centre and reduced the catalytic efficiency. Second, the beads were loaded with relatively more GOD, which produced too much H2O2, and the toxic effect of H2O2 would cause the deactivation of HRP. Therefore, the ratio of HRP
3, the degradation rate of acridine still decreased, which might be due to the following two reasons. First, the immobilized HRP on the beads decreased, which reduced the HRP active centre and reduced the catalytic efficiency. Second, the beads were loaded with relatively more GOD, which produced too much H2O2, and the toxic effect of H2O2 would cause the deactivation of HRP. Therefore, the ratio of HRP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GOD in the solution was optimized to be 2
GOD in the solution was optimized to be 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, indicating that the H2O2 generated by GOD could precisely meet the needs of HRP without causing the deactivation of HRP.41
1, indicating that the H2O2 generated by GOD could precisely meet the needs of HRP without causing the deactivation of HRP.41
3.5 Effect of temperature and pH on degradation of acridine by Ce–Cs@DA/HRP–GOD beads
In Fig. 5A, the effect of temperature on the degradation rate of free HRP–GOD and the Ce–Cs@DA/HRP–GOD beads in an acridine solution are investigated. Free HRP–GOD (1 mg mL−1, the concentration ratios of HRP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GOD was 2
GOD was 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and the Ce–Cs@DA/HRP–GOD beads (wet weight, 1 g) were reacted in acridine solution at pH 7.0 with a temperature range of 10–60 °C for 8 h. Obviously, both free HRP–GOD and the Ce–Cs@DA/HRP–GOD beads showed the highest degradation rate of acridine at 30 °C, and the heat resistance of the Ce–Cs@DA/HRP–GOD beads were higher than that of the free HRP–GOD. The degradation rate of acridine by the Ce–Cs@DA/HRP–GOD beads (Fig. 5Aa) reached 98.0% after 8 h at 20–30 °C, and remained above 88.5% even at a temperature range of 10–60 °C. However, the degradation rate of free HRP–GOD (Fig. 5Ab) to acridine was only 67.5% at extreme temperature conditions, with a degradation rate of 99.9% at 30 °C. This is attributed to the conformational change of immobilized bienzyme and the protective effect of the carrier. Therefore, the temperature sensitivity of immobilized bienzyme is obviously lower than that of free bienzyme.42
1) and the Ce–Cs@DA/HRP–GOD beads (wet weight, 1 g) were reacted in acridine solution at pH 7.0 with a temperature range of 10–60 °C for 8 h. Obviously, both free HRP–GOD and the Ce–Cs@DA/HRP–GOD beads showed the highest degradation rate of acridine at 30 °C, and the heat resistance of the Ce–Cs@DA/HRP–GOD beads were higher than that of the free HRP–GOD. The degradation rate of acridine by the Ce–Cs@DA/HRP–GOD beads (Fig. 5Aa) reached 98.0% after 8 h at 20–30 °C, and remained above 88.5% even at a temperature range of 10–60 °C. However, the degradation rate of free HRP–GOD (Fig. 5Ab) to acridine was only 67.5% at extreme temperature conditions, with a degradation rate of 99.9% at 30 °C. This is attributed to the conformational change of immobilized bienzyme and the protective effect of the carrier. Therefore, the temperature sensitivity of immobilized bienzyme is obviously lower than that of free bienzyme.42
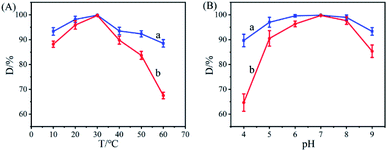 | ||
| Fig. 5 Effect of temperature (A), pH (B) on acridine degradation: (a) Ce–Cs@DA/HRP–GOD beads; (b) free HRP–GOD. | ||
Fig. 5B shows the effect of pH conditions on the degradation efficiency of free HRP–GOD and the Ce–Cs@DA/HRP–GOD beads in the acridine solution. Free HRP–GOD (1 mg mL−1, the concentration ratios of HRP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GOD was 2
GOD was 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and the Ce–Cs@DA/HRP–GOD beads (wet weight, 1 g) were reacted in acridine solution at 30 °C at a pH range of 4.0–9.0 for 8 h. The results revealed that, both free HRP–GOD and the Ce–Cs@DA/HRP–GOD beads showed the highest degradation rate of acridine at pH 7.0, and compared to free HRP–GOD, the Ce–Cs@DA/HRP–GOD beads exhibited a strong acid–base tolerance. The degradation rate of acridine by the Ce–Cs@DA/HRP–GOD beads (Fig. 5Ba) remained above 98.0% in a pH range of 6.0 to 8.0, and reached 88.0% in an acidic and basic solution of pH 4.0 and pH 9.0, respectively. Although the degradation rate of acridine by free HRP–GOD (Fig. 5Bb) decreased to 65.0% at extreme pH conditions, a degradation rate of 99.9% was achieved at pH 7.0. This is also due to the carrier protection and the altered structure of the protein of the immobilized bienzyme, which reduced the effect of pH factors.43
1) and the Ce–Cs@DA/HRP–GOD beads (wet weight, 1 g) were reacted in acridine solution at 30 °C at a pH range of 4.0–9.0 for 8 h. The results revealed that, both free HRP–GOD and the Ce–Cs@DA/HRP–GOD beads showed the highest degradation rate of acridine at pH 7.0, and compared to free HRP–GOD, the Ce–Cs@DA/HRP–GOD beads exhibited a strong acid–base tolerance. The degradation rate of acridine by the Ce–Cs@DA/HRP–GOD beads (Fig. 5Ba) remained above 98.0% in a pH range of 6.0 to 8.0, and reached 88.0% in an acidic and basic solution of pH 4.0 and pH 9.0, respectively. Although the degradation rate of acridine by free HRP–GOD (Fig. 5Bb) decreased to 65.0% at extreme pH conditions, a degradation rate of 99.9% was achieved at pH 7.0. This is also due to the carrier protection and the altered structure of the protein of the immobilized bienzyme, which reduced the effect of pH factors.43
3.6 Effect of HBT dosage on degradation of acridine by Ce–Cs@DA/HRP–GOD beads
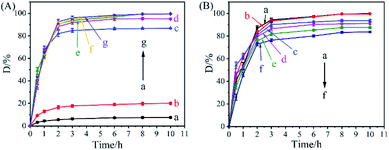
Fig. 6A shows the effect of the HBT dosage on the degradation rate of acridine by the co-immobilized biocatalyst Ce–Cs@DA/HRP–GOD beads. The results demonstrated that the degradation rate of acridine using free HRP–GOD (Fig. 6Aa) or the Ce–Cs@DA/HRP–GOD beads (Fig. 6Ab) without HBT was low, only 7.5% and 20.0% respectively, after treatment for 8 h. The degradation rate of acridine by the Ce–Cs@DA/HRP–GOD beads (Fig. 6Ab) increased slightly, possibly due to the physical adsorption of the beads to acridine. In the presence of HBT, the degradation rate gradually increased to 99.9% when the amount of HBT was increased from 0.5 to 2 mg mL−1 (Fig. 6Ac–Ae). However, there was no significant improvement in the degradation rate when HBT was increased to 3 mg mL−1 or more (Fig. 6Af and Ag), which ascribed to that 2 mg mL−1 HBT molecule could produce enough HBT˙ to degrade acridine. The experimental results demonstrated that the redox mediator HBT could remarkably promote the degradation efficiency of HRP on acridine. Nevertheless, an excess of HBT did not significantly contribute to the improvement of the degradation efficiency. Zeng44 et al. studied the degradation of the herbicide isoproturon by laccase–HBT systems, which showed that isoproturon was poorly degraded by laccase alone, whereas the laccase–HBT systems effectively improved the degradation rate of isoproturon by hydrogen transfer. These results were consistent with our experimental observations.3.7 Effect of concentration on acridine degradation by Ce–Cs@DA/HRP–GOD beads
The effect of different substrate concentrations (5, 15, 25, 35, 45, and 55 mg L−1) on the degradation rate of acridine by Ce–Cs@DA/HRP–GOD beads was investigated under the reaction conditions of 30 °C and pH 7.0 (Fig. 6B). Acridine was completely degraded with an increase in the acridine solution concentration from 5 to 15 mg L−1 (Fig. 6Ba and Bb). The degradation rates decreased significantly after a further increase in the acridine concentration. When the acridine concentration reached 35 mg L−1 (Fig. 6Bd), 91.1% of acridine was degraded. The degradation rate gradually decreased to 83.6% at a higher acridine concentration of 55 mg L−1 (Fig. 6Bf). These results suggest that a high concentration of acridine may have adverse effects on the enzymatic activity.8 However, as far as we know, the maximum allowable concentration of acridine in water was 0.5 mg L−1, even if the pollution was serious, it was not more than 10 mg L−1. Therefore, in practical application, the biocatalyst could keep its enzymatic activity not destroyed.3.8 Reusability and storage stability of Ce–Cs@DA/HRP–GOD beads
Reusability and storage stability are important factors to consider for industrial applications. To investigate the reusability, the Ce–Cs@DA/HRP–GOD beads were washed with PBS buffer solution (pH 7.0) after one catalysis run and reintroduced into a new acridine solution for another degradation at 30 °C. As shown in Fig. 7A, for the first three uses, the degradation rate of acridine was maintained at 99.0% due to HRP and GOD co-immobilized on the Ce–Cs@DA beads by covalent binding, which ensured a firm attachment of the enzyme. After six cycles, the removal efficiency reduced from 99.0% to 61.2%. This decrease might be because of the HBT mediator attacking the active amino acid groups on the surface of the bienzyme leading to the activity loss, and thus a certain number of enzymatic molecules would break off the beads during application and washing. Additionally, the harmful inhibition of acridine may also lead to reduced enzymatic activity.45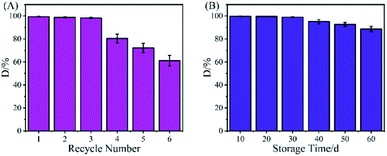 | ||
| Fig. 7 (A) Reusability and (B) storage stability of the Ce–Cs@DA/HRP–GOD beads for acridine degradation. | ||
The storage stability of the Ce–Cs@DA/HRP–GOD beads was examined by storing them at 4 °C for 60 d and then removing them every 10 d to determine the degradation rate of acridine. The results of this study are shown in Fig. 7B. The degradation rate of acridine by the Ce–Cs@DA/HRP–GOD beads was 88.8% after storing for 60 d, which indicated that the Ce–Cs@DA/HRP–GOD beads have good storage stability and excellent application prospects in wastewater treatment.
3.9 Acridine degradation mechanism using Ce–Cs@DA/HRP–GOD beads
A schematic diagram of HRP–GOD co-immobilized on the Ce–Cs@DA beads is shown in Fig. 8A. First, cellulose and chitosan were dissolved and modified by ionic liquid to form Ce–Cs composite beads. Then, DA adhered closely to the surface of Ce–Cs composite beads through self-polymerization to form polydopamine films, which were rich in dopamine quinone groups and catechol structures. Finally, the amino-functional groups of the HRP–GOD covalently binded to the dopamine quinone groups and catechol of polydopamine, and the bienzyme was successfully immobilized on the beads.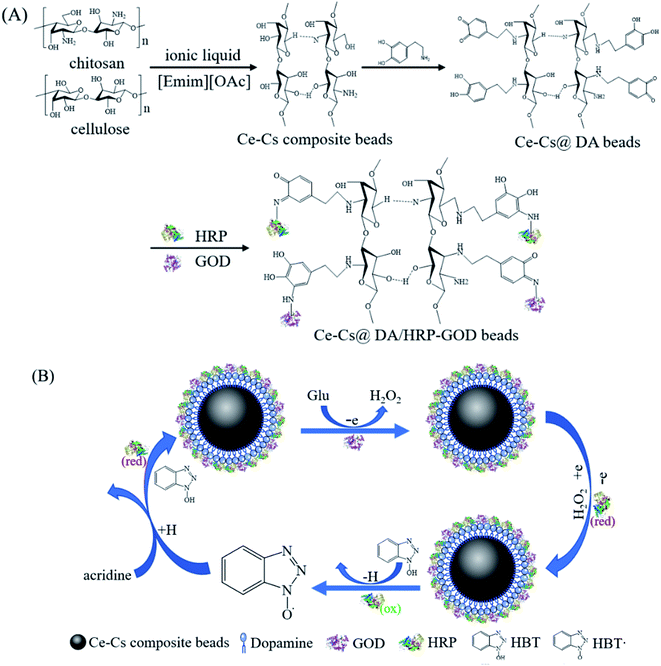 | ||
| Fig. 8 (A) A schematic diagram of HRP–GOD co-immobilized on the Ce–Cs@DA beads. (B) Schematic diagram of the degradation process of acridine catalysed by the Ce–Cs@DA/HRP–GOD beads. | ||
A schematic diagram of the degradation process of acridine catalysed by the Ce–Cs@DA/HRP–GOD beads is shown in Fig. 8B. Firstly, the immobilized GOD oxidised Glu in the presence of O2 to produce H2O2, which is required for the HRP reaction. Secondly, the immobilized reduced state HRP (red) was converted to the oxidised state HRP (ox) by reducing H2O2. Thirdly, the HBT (N–OH) was oxidised by HRP (ox) to form the free radical HBT˙ (N–O˙) with a higher redox potential. Finally, the HBT˙ oxidised the acridine via a hydrogen atom transfer mechanism, and the HBT˙ was simultaneously reduced to its initial state, thereby completing the single redox cycle of the Ce–Cs@DA/HRP–GOD beads.
3.10 Degradation pathways of acridine
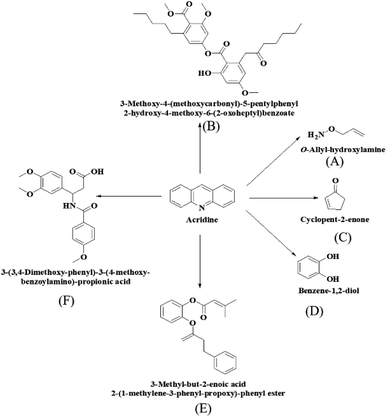
To investigate the biocatalytic degradation pathway of acridine by Ce–Cs@DA/HRP–GOD beads, the intermediates of acridine degradation were further identified by LC-MS analysis. Six possible intermediates were identified in Fig. 9, and their molecular structures were identified as compounds A (m/z = 74.06), B (m/z = 515.02), C (m/z = 83.06), D (m/z = 111.09), E (m/z = 325.11) and F (m/z = 360.15). Among them, compounds A, C and D belong to small molecular compounds, which may be formed by the gradual cleavage of benzene ring on acridine, while the molecular weight of B, E and F is larger, which may be caused by the polymerization of some small molecular products after the degradation of acridine. According to the results of intermediate analysis, the possible degradation pathway of acridine by Ce–Cs@DA/HRP–GOD beads was speculated as shown in Fig. 9.4 Conclusions
In this study, Ce–Cs@DA/HRP–GOD beads were prepared by the co-immobilisation bienzyme HRP–GOD on DA-modified cellulose–chitosan composite beads and used as a biocatalyst for the degradation of acridine. The degradation rate of the biocatalyst to acridine was 99.9% under the synergistic catalytic action of the mediator. Moreover, the degradation efficiency of the Ce–Cs@DA/HRP–GOD beads for acridine was higher than that of the single enzyme Ce–Cs@DA/HRP beads, which not only achieved the purpose of reducing the experimental cost by replacing part of the expensive HRP with cheap GOD, but also had better environmental protection effect for no additional H2O2 was needed. In addition, the Ce–Cs@DA/HRP–GOD beads demonstrated better acid–base stability, thermal stability, reusability, and storage stability than that observed for free HRP–GOD. Therefore, Ce–Cs@DA/HRP–GOD beads as a co-immobilized bienzyme biocatalyst demonstrated significant application potential for the degradation of acridine from industrial wastewater and were also beneficial for preventing the potential pollution of H2O2.Author contributions
Yaohua Gu: Conceptualization, Formal analysis, Investigation, Data curation, Writing – review & editing, Funding acquisition; Lin Yuan: Conceptualization, Formal analysis, Investigation, Data curation, Writing – original draft, Visualization; Mingming Li: Formal analysis, Investigation, Data curation, Writing – original draft, Visualization; Xinyu Wang: Data curation, Visualization; Deyu Rao: Data curation, Visualization; Xiaoyan Bai: Data curation, Visualization; Keren Shi: Characterization analysis; Haiming Xu: Data and image processing; Shaozhang Hou: Conceptualization, Formal analysis; Huiqin Yao: Conceptualization, Methodology, Writing – review & editing, Supervision, Funding acquisition. All the authors participated in the discussion, Yaohua Gu, Lin Yuan and Mingming Li wrote the paper with the inputs of all the co-authors.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was financially supported by the Scientific Research Project of the Ningxia Higher Education Department of China (NGY2020034), Key Research and Development Projects of Ningxia of China (2021BEB04055), School-level Special Talents Launching Project of Ningxia Medical University (XT2020017), Innovation and Entrepreneurship Training Program for College Students of Ningxia Medical University (S202110752004) and CAS “Light of West China Program” of China (XAB2020YW16).Notes and references
- R. M. Abdelhameed and H. E. Emam, Design of ZIF(Co & Zn)@wool composite for efficient removal of pharmaceutical intermediate from wastewater, J. Colloid. Interface Sci., 2019, 552, 494–505 CrossRef CAS PubMed.
- C. Bokun, Y. Siyu, W. Yangyang, S. Miyangzi and Q. Yu, Intensified phenols extraction and oil removal for industrial semicoking wastewater: a novel economic pretreatment process design, J. Clean. Prod., 2020, 242, 118453 CrossRef.
- P. Prasher and M. Sharma, Medicinal chemistry of acridine and its analogues, MedChemComm, 2018, 9, 1589–1618 RSC.
- P. Xue, X. Liu, Y. Gu, W. Zhang, L. Ma and R. Li, Laccase-mediator system assembling co-immobilized onto functionalized calcium alginate beads and its high-efficiency catalytic degradation for acridine, Colloids Surf., B, 2020, 196, 111348 CrossRef CAS PubMed.
- G. Gissel-nielsen and T. Nielsen, Phytotoxicity of acridine, an important representative of a group of tar and creosote contaminents, n-pac compounds, Polycycl. Aromat. Comp., 1996, 8, 243–249 CrossRef CAS.
- J. Shi, H. Han and C. Xu, Anovel enhanced anaerobic biodegradation method using biochar and Fe(OH)3@biochar for the removal of nitrogen heterocyclic compounds from coal gasification wastewater, Sci. Total Environ., 2019, 697, 134052 CrossRef CAS PubMed.
- H. Yi, M. Yan, D. Huang, G. Zeng, C. Lai, M. Li, X. Huo, L. Qin, S. Liu, X. Liu, B. Li, H. Wang, M. Shen, Y. Fu and X. Guo, Synergistic effect of artificial enzyme and 2D nano-structured Bi2WO6 for eco-friendly and efficient biomimetic photocatalysis, Appl. Catal. B Environ., 2019, 250, 52–62 CrossRef CAS.
- X. Liu, P. Xue, F. Jia, K. Shi, Y. Gu, L. Ma and R. Li, A novel approach to efficient degradation of indole using co-immobilized horseradish peroxidase-syringaldehyde as biocatalyst, Chemosphere, 2021, 262, 128411 CrossRef CAS PubMed.
- R. DiCosimo, J. McAuliffe, A. J. Poulose and G. Bohlmann, Industrial use of immobilized enzymes, J. Electroanal. Chem., 2013, 42, 6437–6474 CAS.
- R. A. Sheldon and P. C. Pereira, Biocatalysis engineering: the big picture, Chem. Soc. Rev., 2017, 46, 2678–2691 RSC.
- X. Qiu, S. Wang, S. Miao, H. Suo, H. Xu and Y. Hu, Co-immobilization of Laccase and ABTS onto amino-functionalized ionic liquid-modified magnetic chitosan nanoparticles for pollutants removal, J. Hazard. Mater., 2021, 401, 123353 CrossRef CAS PubMed.
- M. Maryskova, I. Ardao, C. A. Garcia-Gonzalez, L. Martinova, J. Rotkova and A. Sevcu, Polyamide 6/chitosan nanofibers as support for the immobilization of Trametes versicolor laccase for the elimination of endocrine disrupting chemicals, Enzyme Microb. Technol., 2016, 89, 31–38 CrossRef CAS PubMed.
- A. Suri, V. Khandegar and P. J. Kaur, Ofloxacin exclusion using novel HRP immobilized chitosan cross-link with graphene oxide nanocomposite, Groundw. Sustain. Dev., 2021, 12, 100515 CrossRef.
- H. Sun, W. Huang, H. Yang and S. Zhang, Co-immobilization of laccase and mediator through a self-initiated one-pot process for enhanced conversion of malachite green, J. Colloid Interface Sci., 2016, 471, 20–28 CrossRef CAS PubMed.
- Z. B. Guan, Q. Luo, H. R. Wang, Y. Chen and X. R. Liao, Bacterial laccases: promising biological green tools for industrial applications, Cell. Mol. Life Sci., 2018, 75, 3569–3592 CrossRef CAS PubMed.
- K. Sun, Q. Huang and S. Li, Transformation and toxicity evaluation of tetracycline in humic acid solution by laccase coupled with 1-hydroxybenzotriazole, J. Hazard. Mater., 2017, 331, 182–188 CrossRef CAS PubMed.
- Y. Leng, J. Bao, H. Xiao, D. Song, J. Du, S. Mohapatra, D. Werner and J. Wang, Transformation mechanisms of tetracycline by horseradish peroxidase with/without redox mediator ABTS for variable water chemistry, Chemosphere, 2020, 258, 127306 CrossRef CAS PubMed.
- Z. Chen, H. Li, A. Peng and Y. Gao, Oxidation of polycyclic aromatic hydrocarbons by horseradish peroxidase in water containing an organic cosolvent, Environ. Sci. Pollut. Res., 2014, 21, 10696–10705 CrossRef CAS PubMed.
- X. Ji, Z. Su, M. Xu, G. Ma and S. Zhang, TiO2−horseradish peroxidase hybrid catalyst based on hollow nanofibers for simultaneous photochemical−enzymatic degradation of 2,4-dichlorophenol, ACS. Sustain. Chem. Eng., 2016, 4, 3634–3640 CrossRef CAS.
- G. Bai, X. Xu, Q. Dai, Q. Zheng, Y. Yao, S. Liu and C. Yao, An electrochemical enzymatic nanoreactor based on dendritic mesoporous silica nanoparticles for living cell H2O2 detection, Analyst, 2019, 144, 481–487 RSC.
- A. N. Hiner, J. Hernandez-Ruiz, J. N. Rodriguez-Lopez, M. B. Arnao, R. Varon, F. Garcia-Canovas and M. Acosta, The inactivation of horseradish peroxidase isoenzyme A2 by hydrogen peroxide: an example of partial resistance due to the formation of a stable enzyme intermediate, J. Biol. Inorg Chem., 2001, 6, 504–516 CrossRef CAS PubMed.
- Q. H. Gibson, B. E. P. Swoboda and V. Massey, Kinetics and mechanism of action of glucose oxidase, J. Biol. Chem., 1964, 239, 3927–3934 CrossRef CAS PubMed.
- G. Fabregat, S. Lanzalaco, J. Aït Saïd, X. Muñoz-Pascual, J. Llorca and C. Alemán, Immobilization of glucose oxidase on plasma-treated polyethylene for non-invasive glucose detection, J. Electroanal. Chem., 2021, 895, 115509 CrossRef CAS.
- H. Yang, W. Wei and S. Liu, Monodispersed silica nanoparticles as carrier for co-immobilization of bi-enzyme and its application for glucose biosensing, Spectrochim. Acta, Part A, 2014, 125, 183–188 CrossRef CAS PubMed.
- G. Vitola, R. Mazzei and L. Giorno, Enzyme-loaded membrane reactor to degrade a pesticide in vegetative waters, J. Membr. Sci., 2021, 635, 119438 CrossRef CAS.
- M. Bilal, T. Rasheed, Y. Zhao and H. M. N. Iqbal, Agarose-chitosan hydrogel-immobilized horseradish peroxidase with sustainable bio-catalytic and dye degradation properties, Int. J. Biol. Macromol., 2019, 124, 742–749 CrossRef CAS PubMed.
- S. Peng, H. C. Meng, L. Zhou and J. Chang, Synthesis of novel magnetic cellulose-chitosan composite microspheres and their application in laccase immobilization, J. Nanosci. Nanotechnol., 2014, 14, 7010–7014 CrossRef CAS PubMed.
- S. Park, K. K. Oh and S. H. Lee, Biopolymer-based composite materials prepared using ionic liquids, Adv. Biochem. Eng. Biotechnol, 2019, 168, 133–176 CrossRef CAS PubMed.
- Y. Yang, P. Qi, Y. Ding, M. F. Maitz, Z. Yang, Q. Tu, K. Xiong, Y. Leng and N. Huang, A biocompatible and functional adhesive aminerich coating based on dopamine polymerization, J. Mater. Chem. B., 2015, 3, 72–81 RSC.
- K. Dumri and D. Hung Anh, Immobilization of lipase on silver nanoparticles via adhesive polydopamine for biodiesel production, Enzyme. Res, 2014, 2014, 389739 Search PubMed.
- F. E. Muller, Chemistry and biochemistry of flavoenzymes, CRC Press, Boca Raton, FL, 1991 Search PubMed.
- G. Yaohua, X. Ping, J. Feng and S. Keren, Recent developments of a co-immobilized laccase–mediator system: a review, J. Hazard. Mater., 2019, 365, 118–124 CrossRef CAS PubMed.
- M. Liu, Y. Zhou, Y. Zhang, C. Yu and S. Cao, Preparation and structural analysis of chitosan films with and without sorbitol, Food Hydrocoll., 2013, 33, 186–191 CrossRef CAS.
- H. Zhang, M. Kong, Q. Jiang, K. Hu, M. Ouyang, F. Zhong, M. Qin, L. Zhuang and G. Wang, Enzyme-inorganic hybrid nanoflowers: classification, synthesis, functionalization and potential applications, J. Mol. Liq., 2021, 340, 129075 Search PubMed.
- Z. Liu, H. Wang, B. Li, C. Liu, Y. Jiang, G. Yu and X. Mu, Biocompatible magnetic cellulose–chitosan hybrid gel microspheres reconstituted from ionic liquids for enzyme immobilization, J. Mater. Chem., 2012, 22, 15085 RSC.
- Y. Liu, Y. Feng, Y. Zhang, S. Mao, D. Wu and H. Chu, Highly efficient degradation of dimethyl phthalate from Cu(II) and dimethyl phthalate wastewater by EDTA enhanced ozonation: performance, intermediates and mechanism, J. Hazard. Mater., 2019, 366, 378–385 CrossRef CAS PubMed.
- Y. Yuan, C. Zhang, C. Zhao, B. Wang, X. Wang, B. Gao, S. Wang and J. Rinklebe, One-step preparation of a novel graphitic biochar/Cu0/Fe3O4 composite using CO2-ambiance pyrolysis to activate peroxydisulfate for dye degradation, J. Environ. Sci., 2023, 125, 26–36 CrossRef.
- L. Munk, M. L. Andersen and A. S. Meyer, Influence of mediators on laccase catalyzed radical formation in lignin, Enzyme Microb. Technol., 2018, 116, 48–56 CrossRef CAS PubMed.
- J. Rocha-Martín, B. d. l. Rivas, R. Muñoz, J. M. Guisán and F. López-Gallego, Rational co-immobilization of bi-enzyme cascades on porous supports and their applications in bio-redox reactions with in situ recycling of soluble cofactors, ChemCatChem, 2012, 4, 1279–1288 CrossRef.
- M. Razzaghi, A. Karimi, H. Aghdasinia and M.-T. Joghataei, Oxidase-Peroxidase sequential polymerization for removal of a dye from contaminated water by horseradish peroxidase (HRP)/glucose oxidase (GOx)/polyurethane hybrid catalyst, Korean J. Chem. Eng., 2017, 34, 2870–2878 CrossRef CAS.
- J. N. Vranish, M. G. Ancona, E. Oh, K. Susumu and I. L. Medintz, Enhancing coupled enzymatic activity by conjugating one enzyme to a nanoparticle, Nanoscale, 2017, 9, 5172–5187 RSC.
- H. Li, J. Hou, L. Duan, C. Ji, Y. Zhang and V. Chen, Graphene oxide-enzyme hybrid nanoflowers for efficient water soluble dye removal, J. Hazard. Mater., 2017, 338, 93–101 CrossRef CAS PubMed.
- M. Kiani, S. Mojtabavi, H. Jafari-Nodoushan, S. R. Tabib, N. Hassannejad and M. A. Faramarzi, Fast anisotropic growth of the biomineralized zinc phosphate nanocrystals for a facile and instant construction of laccase@Zn3(PO4)2 hybrid nanoflowers, Int. J. Biol. Macromol., 2022, 204, 520–531 CrossRef CAS PubMed.
- S. Zeng, X. Qin and L. Xia, Degradation of the herbicide isoproturon by laccase-mediator systems, Biochem. Eng. J., 2017, 119, 92–100 CrossRef CAS.
- J. Rong, T. Zhang, F. Qiu and Y. Zhu, Preparation of efficient, stable, and reusable laccase−Cu3(PO4)2 hybrid microspheres based on copper foil for decoloration of congored, ACS. Sustain. Chem. Eng., 2017, 5, 4468–4477 CrossRef CAS.
Footnote |
| † Contribute equally as the first author. |
| This journal is © The Royal Society of Chemistry 2022 |