Open Access Article
Open Access ArticleEnhanced antibacterial activity with increasing P doping ratio in CQDs †
Shuiqin Chai *ab,
Lijia Zhoub,
Yuting Chib,
Linshuo Chen
*ab,
Lijia Zhoub,
Yuting Chib,
Linshuo Chen b,
Shuchen Pei
b,
Shuchen Pei ab and
Bin Chen
ab and
Bin Chen *c
*c
aChongqing Key Laboratory of Industrial Fermentation Microorganism, Chongqing University of Science and Technology, Chongqing 401331, P. R. China. E-mail: chaisq0104@cqust.edu.cn
bCollege of Chemistry and Chemical Engineering, Chongqing University of Science and Technology, Chongqing 401331, P. R. China
cChongqing Key Laboratory of Non-linear Circuit and Intelligent Information Processing, College of Electronic and Information Engineering, Southwest University, Chongqing 400715, P. R. China. E-mail: chenbin121@swu.edu.cn
First published on 28th September 2022
Abstract
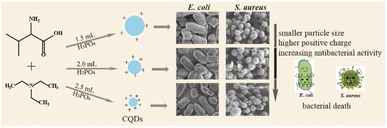
It is an urgent challenge to develop efficient antibacterial agents against resistant bacteria in the treatment of infectious diseases. Carbon quantum dots (CQDs) have attracted much attention owing to their good stability, low toxicity and excellent biocompatibility. In this work, CQDs doped with different contents of the element phosphorus (P) were prepared by a simple hydrothermal method using valine as a carbon source, triethylamine as a nitrogen source and different volumes of phosphoric acid as a phosphorus source. The average diameter and the surface charge could be regulated from 2.89 nm to 1.56 nm and +2.58 mV to +5.47 mV by increasing the content of the element P in these CQDs. Importantly, these CQDs showed effective bacterial inhibition against Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus). The minimal inhibitory concentration (MIC) decreased from 0.71, to 0.51 to 0.18 mg mL−1 on E. coli and S. aureus with the increase of P element content. Furthermore, the morphologies of E. coli cells and S. aureus were damaged and became irregular upon treatment with these CQDs. The results of singlet oxygen (1O2) detection demonstrated that intracellular 1O2 was generated during the antibacterial process. We speculated that bacterial inhibition induced by these CQDs was accompanied by disruption of permeability and structural integrity, owing to strong electrostatic interactions between negatively charged bacteria and positively charged CQDs and production of singlet oxygen of CQDs. Together, this study indicates that the CQDs can be a candidate to treat resistant bacterial infections and may improve the understanding of killing pathogens by antibacterial CQD drugs.
Introduction
Bacterial infections have been a growing problem worldwide and threaten human life and health.1 More concerning, super bacteria with superb resistance to antibiotics have risen dramatically due to the abuse of organic antibiotics, increasingly threatening our ability to treat common infections.2,3 The threat to health of multi-drug resistant bacteria has been extensively reported, suggesting that fatal infections from multi-drug resistant bacteria are predicted to exceed those from cancer by 2050.4 Thus, it is vital to create new and effective antibacterial agents against resistant bacteria. In the early stage of research, metal nanomaterials, such as gold nanoclusters,5–7 silver nanoparticles8–13 and Cu2O nanotubes,14 have generated a lot of attention due to their significant potential for treating illnesses caused by antibiotic-resistant bacteria. However, the poor environment and biocompatibility of metal nanomaterials has become an obstacle in the clinical application.As a novel type of carbon nanomaterials, carbon quantum dots (CQDs) with good stability, fluorescence, low toxicity and biocompatibility have been widely used in the fields of bioimaging, biosensing and drug delivery.15–24 Additionally, synthetic procedures of CQDs are fairly simple, and the raw materials are cost-efficient and widely available. Thus, CQDs as a new antibacterial agent for biomedical applications have been widely studied.25–28 Bing et al. prepared three kinds of carbon dots with different surface charge, which induced programmed bacteria death.29 Liu et al. used metronidazole to prepare carbon dots with selective antibacterial activity against obligate anaerobes.30 Furthermore, researchers have performed a series of experiments to explore the antibacterial mechanism of CQDs. For instance, Jian et al. proved the cell membranes of E. coli and S. aureus treated with CQDs for two hours were severely damaged and intact treated without CQDs by TEM and SEM.31 Lee et al. investigated the antibacterial mechanism of CQDs by measuring singlet oxygen (1O2, a type of ROS) generation using a singlet oxygen sensor green assay, which played an important role in the eradication of bacteria.32 The above studies indicate that CQDs destroy bacteria through different mechanisms.
In recent years, the element doping in CQDs has aroused widespread interests of scientists because the doping of elements could effectively adjust some of the available functional groups and chemical properties of prepared CQDs,33–36 transforming CQDs into functional nanomaterials. For instance, Zhao et al. prepared N, P co-doped yellow emitting carbon dots with strong inhibitory ability to bacteria.37 Travlou et al. obtained S- and N-containing CQDs with a capability against Gram-positive and Gram-negative bacterial strains by a simple hydrothermal approach and the N-CQDs exhibited stronger antibacterial ability involved electrostatic interactions.38
In this work, we synthesized a series of N, P doped-CQDs using valine as a carbon source, triethylamine as a nitrogen source and different volumes of H3PO4 as a phosphorus (P) source. The functional groups and chemical properties of CQDs were regulated by increasing the content of phosphorus. In addition, these CQDs showed different growth-inhibiting properties by rupturing the cell walls of E. coli and S. aureus, which might be related to strong electrostatic interactions between negatively charged bacteria and positively charged CQDs and production of 1O2 of CQDs, and the antibacterial activity was increased with the increase of P element in the CQDs (Scheme 1).
Experimental
Materials
Valine (99%), triethylamine (99%) and 1,3-diphenylisobenzofuran (DPBF) were purchased from Aladdin Reagent Co., Ltd. (Shanghai, China). Sodium chloride, yeast extract, agar powder, tryptone were purchased from Sinopharm Chemical Reagent Co., Ltd. All chemicals were used directly without further purification. All of solutions were prepared with deionized water from a Millipore water purification system (Milli-Q, Millipore, Billerica, USA).Apparatus
A RF-5301PC fluorescence spectrophotometer was used to collect all fluorescence spectra (Shimatzu Corp., Kyoto, Japan). Transmission electron microscopy (TEM) and hgh-resolution transmission electron microscopy (HRTEM) images were collected using a JEOL-2100F transmission electron microscope (Tokyo, Japan). The thickness of P-doped CQDs was carried out by a Dimension Icon Scan Asyst atomic force microscope (AFM, Bruker Co.). Structural analysis was characterized by D8 ADVANCE X-ray diffraction (XRD, Bruker Co.) using Cu Kα radiation (λ = 0.15406 nm). Elemental and functional group analyses were measured using an ESCALAB Xi+ X-ray photoelectron spectrometer (XPS, Thermo Fisher Scientific Inc.) and a Nicolet iS5 Fourier Transform Infrared spectrometer (FTIR, Thermo Fisher Scientific Inc.). Zeta potentials of P-doped CQDs were recorded using a Zetasizer Nano ZS90 System (Malvern, UK). The concentration of bacteria was determined by measuring the optical density at 600 nm (OD600) via UV-vis spectroscopy. The images of bacteria morphology were obtained under a S-3400N scanning electron microscope (SEM, Hitachi, Japan).Synthesis of CQDs
CQDs were prepared via a facile one-pot solvothermal method.39 Briefly, 0.5 g valine, 1 mL triethylamine and different volumes (1.5 mL, 2.0 mL, 2.5 mL) of H3PO4 were dispersed in different volumes (2.5 mL, 2.0 mL, 1.5 mL) of deionized water, respectively. 5.0 mL of the reaction mixture was transferred into a 25 mL poly(tetrafluoroethylene)-lined autoclave, and heated at 180 °C for 12 h, and subsequently cooled to ambient temperature. The solution was filtered using a 0.22 μm BIOSHARP membrane filter and dialyzed using a 500 Da dialysis membrane for 48 h. Finally, a series of CQDs powder was obtained by vacuum freeze-drying for 48 h and kept in cold storage at 4 °C. The three CQDs were named by CQDs-1, CQDs-2 and CQDs-3 according to volumes of H3PO4 from 1.5 to 2.5 mL, respectively.Cellular toxicity test
1 × 105 cells per mL of Human HCT-116 colon cancer cells were seeded on 96-well plates (100 μL per well) and grown in McCoy's F-12 cell medium with 10% fetal bovine serum (FBS) at 37 °C. After 24 h, the cells were treated with different doses of CQDs and were incubated with McCoy's F-12 medium with 2% FBS for an additional 24 h. The CQDs-free cells served as control groups. Following the removal of the medium, the cells were washed three times with PBS buffer before being cultured for an additional hour with a mixture of 10 μL MTT and 90 μL McCoy's F-12 medium. After discarding the MTT culture medium, 150 μL of DMSO was added and mixed for around 10 minutes. Finally, the optical density (OD) values of wells were measured at 538 nm. The cell viability was estimated with the eqn (1):| Cell viability = (ODtreated/ODcontrol) × 100% | (1) |
Photostability of CQDs
The pH stability was analyzed by mixing 30 μL of different pH of Britton–Robinson (BR) buffer with 20 μL of CQDs and 450 μL deionized water. For ionic strength and oxidation degree, 30 μL of various concentrations of NaCl (0–4.0 M) and H2O2 (0–1000 μM) solution were mixed with 20 μL of CQDs and 30 μL PBS buffer (pH 7.4), and then diluted to 500 μL with deionized water and mixed well. The fluorescence emission of the solutions was observed at a wavelength of 430 nm upon excitation of 320 nm.Antibacterial activity test
S. aureus and E. coli were cultured overnight in lysogeny broth (LB) medium at 37 °C in an orbital shaker (180 rpm). The medium contained 10.0 g of NaCl, 5.0 g of yeast extract powder and 10.0 g of tryptone per liter. Measurements of optical density at 595 nm (OD595) were used to estimate the bacterial concentration. The MIC of CQDs was determined by microdilution with a 96-well cell culture plate. Bacteria were adjusted to 1.5 × 107 CFU mL−1 with LB medium. First, 50 μL of bacterial cell suspension was added to each well. Subsequently, 50 μL of various concentrations of CQDs was dispersed in 96-well plate with a final volume of 100 μL suspension per well and mixed well. Bacteria suspension in an LB medium without CQDs was the control and only LB medium was the blank. The samples were incubated at 37 °C for 12 h with agitation at 180 rpm. After incubation for 12 h, the concentration of bacteria was measured at 595 nm. The experiment was repeated three times.Detection of singlet oxygen (1O2)
The ability of CQDs to generate 1O2 under visible light illumination was determined by measuring the absorbance of 1O2 quencher 1,3-diphenylisobenzofuran (DPBF) at 410 nm. In present work, an appropriate amount of DPBF and CQDs was added in methanol solution, illuminated upon white LEDs irradiation for 30 min, and the rate of DPBF oxidation was calculated by periodically recording UV-visible spectrum with the absorbance at 410 nm.Results and discussion
Characterizations of the P-doped CQDs
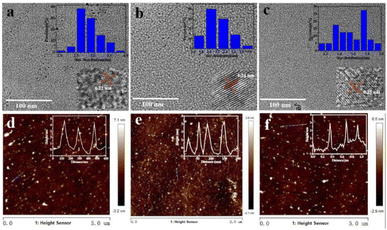
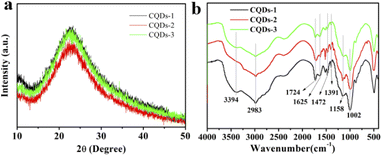
The morphology and structure of the three as-prepared CQDs, noted as CQDs-1, CQDs-2 and CQDs-3, were first characterized by TEM/HRTEM and AFM, respectively. As shown in Fig. 1, the average diameter of CQDs-1, CQDs-2 and CQDs-3 was 2.89 nm, 2.21 nm and 1.56 nm (Fig. 1a–c), respectively. And the average height of CQDs-1, CQDs-2 and CQDs-3 was 4.13 nm, 3.16 nm and 2.93 nm (Fig. 1d–f), respectively. The HRTEM images of these CQDs (inset in Fig. 1a–c) all displayed one in-plane lattice spacings of 0.23 nm which is attributed to the (100) facet of graphene. Furthermore, XRD pattern (Fig. 2a) exhibited a typical broad peak around 22.9° associated with the graphitic structure, further revealing that a typical graphite-like structure is formed during the hydrothermal treatment. The morphology results indicate a declining tendency in size distribution with the increase of the doping ratio of P element from CQDs-1 to CQDs-3.To investigate the elementary composition and the functional groups in these CQDs, FTIR spectra and XPS were performed. The functional groups on the surface of these CQDs doped with different ratio of P element were characterized by FTIR. As shown in Fig. 2b, the peaks at 3394 cm−1 and 2983 cm−1 were attributed to the stretching vibration of O–H/N–H and C–H,40,41 the peaks at 1724 cm−1, 1625 cm−1, 1472 cm−1 and 1391 cm−1, 1158 cm−1 were attributed to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C
O, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, C
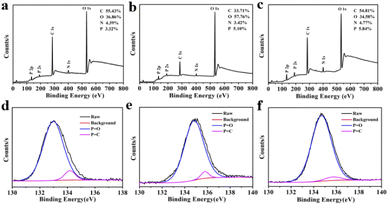
N, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–N and C–O stretching vibration,42,43 the peak at 1002 cm−1 was attributed to P–O stretching vibration.34,44 As shown in Fig. 3a–c, the full-scan XPS spectra of these CQDs confirmed that all of the CQDs contain four elements involving carbon, nitrogen, oxygen and phosphorus. The elements ratio of C, O, N, P in the three CQDs were 55.43%, 36.86%, 4.39%, 3.32% for CQDs-1 and 33.71%, 57.76%, 3.42%, 5.10% for CQDs-2 and 54.81%, 34.58%, 4.77%, 5.84% for CQDs-3, respectively. Obviously, the doping proportion of P element was increased with the increase of H3PO4 volumes from CQDs-1 to CQDs-3. As shown in Fig. 3d–f, the high-resolution P 2p spectra of the three CQDs showed two peaks around 134.7 eV and 135.6 eV, which belong to P
C, C–N and C–O stretching vibration,42,43 the peak at 1002 cm−1 was attributed to P–O stretching vibration.34,44 As shown in Fig. 3a–c, the full-scan XPS spectra of these CQDs confirmed that all of the CQDs contain four elements involving carbon, nitrogen, oxygen and phosphorus. The elements ratio of C, O, N, P in the three CQDs were 55.43%, 36.86%, 4.39%, 3.32% for CQDs-1 and 33.71%, 57.76%, 3.42%, 5.10% for CQDs-2 and 54.81%, 34.58%, 4.77%, 5.84% for CQDs-3, respectively. Obviously, the doping proportion of P element was increased with the increase of H3PO4 volumes from CQDs-1 to CQDs-3. As shown in Fig. 3d–f, the high-resolution P 2p spectra of the three CQDs showed two peaks around 134.7 eV and 135.6 eV, which belong to P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds and P
O bonds and P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds,45,46 respectively. As shown in Fig. S1a–c,† the high-resolution C 1s spectra of the three CQDs showed three peaks including 284.8 (C
C bonds,45,46 respectively. As shown in Fig. S1a–c,† the high-resolution C 1s spectra of the three CQDs showed three peaks including 284.8 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/C–C bonds), 285.8 eV (C–O/C–N/C–P bonds) and 288.3 eV (C
C/C–C bonds), 285.8 eV (C–O/C–N/C–P bonds) and 288.3 eV (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds).47 As shown in Fig. S1d–f,† the O 1s spectra of the three CQDs presented two peaks around 532.6 eV (C
O bonds).47 As shown in Fig. S1d–f,† the O 1s spectra of the three CQDs presented two peaks around 532.6 eV (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds) and 534.5 eV (C–O/P
O bonds) and 534.5 eV (C–O/P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds).40 As shown in Fig. S1g–i,† the high-resolution N 1s spectra of the three CQDs revealed the existence of C–N/N–N/P–N bonds (401.0 eV) and N–H bonds (402.8 eV).45 As demonstrated by the results of FTIR and XPS, the as-prepared three different ratio of P element doped CQDs have abundant functional groups, including –COOH, –OH, and a small number of N, P-containing groups.
O bonds).40 As shown in Fig. S1g–i,† the high-resolution N 1s spectra of the three CQDs revealed the existence of C–N/N–N/P–N bonds (401.0 eV) and N–H bonds (402.8 eV).45 As demonstrated by the results of FTIR and XPS, the as-prepared three different ratio of P element doped CQDs have abundant functional groups, including –COOH, –OH, and a small number of N, P-containing groups.
 | ||
| Fig. 3 The full-scan XPS spectra (a–c) the high-resolution P 2p XPS spectra (d–f) of CQDs-1 (a, d), CQDs-2 (b, e) and CQDs-3 (c, f). | ||
Optical properties of CQDs
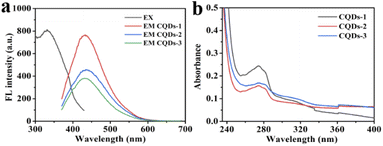
As shown in Fig. 4a, the maximum fluorescence (FL)emission was located at 430 nm when excited at 330 nm. While the FL intensity at 430 nm was decreased with the increase of P doping ratio from CQD-1 to CQDs-3. Besides, the maximum emission was independent of the excitation wavelength from 280 nm to 400 nm in these CQDs (Fig. S2†). Fig. 4b showed that all of these CQDs displayed a distinctive absorption peak at 275 nm owing to the π–π* transition of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C.48,49 In addition, the as-prepared CQDs showed excellent stability. The FL intensity slightly changed under continuous UV lamp excitation (330 nm) for 60 min, indicating these CQDs have excellent photostability (Fig. S3a†). Meanwhile, the FL intensity almost remained invariant even when the concentrations of NaCl and H2O2 was up to 4 M and 1000 μM, respectively, which indicating the stability of these CQDs in comparatively high ionic-strength and oxidation condition (Fig. S3b and c†). These CQDs also showed excellent stability in the pH range from 1.89 to 11.98 (Fig. S3d†).
C.48,49 In addition, the as-prepared CQDs showed excellent stability. The FL intensity slightly changed under continuous UV lamp excitation (330 nm) for 60 min, indicating these CQDs have excellent photostability (Fig. S3a†). Meanwhile, the FL intensity almost remained invariant even when the concentrations of NaCl and H2O2 was up to 4 M and 1000 μM, respectively, which indicating the stability of these CQDs in comparatively high ionic-strength and oxidation condition (Fig. S3b and c†). These CQDs also showed excellent stability in the pH range from 1.89 to 11.98 (Fig. S3d†).
Antibacterial activity of CQDs
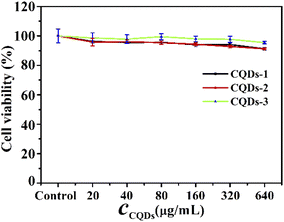
Before antibacterial activity experiments, we first evaluated the cytotoxicity of these CQDs by MTT method using human HCT-116 colon cancer cells. As depicted in Fig. 5, the cell viability remained greater than 90% after 24 hours of incubation even at concentrations as high as 640 μg mL−1, indicating that these CQDs have good biocompatibility. Gram-negative E. coli and Gram-positive S. aureus were used as the model pathogens to evaluate the antibacterial activity of these CQDs. Different concentrations of these CQDs were applied to treat equal amounts of bacterial cell suspension for 12 h. The bacterial vitality was evaluated by comparing OD595 values of the bacterial suspension. As shown in Fig. S4a and d,† the vitality of E. coli and S. aureus decreased with the increasing concentration of these CQD. The OD595 of E. coli and S. aureus were almost consistent with that of the blank group when the concentrations of CQDs-1, CQDs-2, CQDs-3 were respectively up to 0.71, 0.51, 0.18 mg mL−1, indicating that these CQDs have not selective inhibitory effect on Gram-negative and Gram-positive pathogens. Besides, to further confirm that these CQDs are not selective for bacteria, we measured the antibacterial activity on S. flexneri and P. aeruginosa bacteria. As shown in Fig. S4b, c† and Table 1, the MIC values were almost consistent with E. coli and S. aureus. However, the OD595 values of S. aureus were smaller than that of E. coli, S. flexneri and P. aeruginosa at the same concentration of CQDs, suggesting the inhibitory effect of Gram-positive is stronger than that of Gram-negative pathogens.| Bacterial type | MIC (mg mL−1) | ||
|---|---|---|---|
| CQDs-1 | CQDs-2 | CQDs-3 | |
| S. aureus | 0.71 | 0.51 | 0.18 |
| E. coli | 0.71 | 0.51 | 0.18 |
| S. flexneri | 0.71 | 0.51 | 0.18 |
| P. aeruginosa | 0.71 | 0.51 | 0.18 |
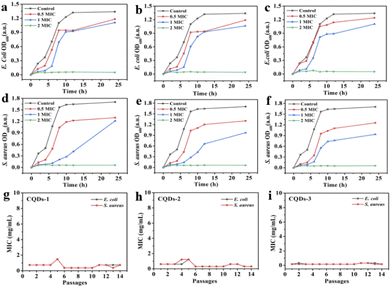
In addition, the diameter of the inhibition zone illustrates the bacterial viability of these CQDs. The better the antibacterial ability of CQDs, the larger the diameter of the zone of inhibition. As depicted in Fig. 6, the inhibition diameters of E. coli and S. aureus became clearly larger with the increase of the doping ratio of P element from CQDs-1 to CQDs-3 and even the inhibition diameter of S. aureus was larger than gentamicin (an aminoglycoside antibiotic). The plate colony formation assays of E. coli and S. aureus treated with these CQDs were shown in Fig. S5.† The results showed that CQDs-3 had the highest antibacterial activity against the tested bacteria, which is consistent with that of the zone of inhibition. These findings suggested that the particle size have a certain influence on the antibacterial activity against both E. coli and S. aureus. Next, the bacterial killing kinetics of these CQDs were studied. Fig. 7a–f showed these CQDs could rapidly eradicate bacteria and displayed a time-dependence at different doses.
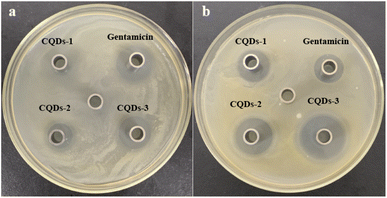 | ||
| Fig. 6 Photographs of inhibition zone containing E. coli (a) and S. aureus (b) treated with 0.71 mg mL−1 of CQDs-1, CQDs-2, CQDs-3 and gentamicin, respectively. | ||
To evaluate the potential of bacteria resistance to these CQDs after a continuous antibacterial treatment, bacteria were incubated with half inhibitory concentrations of these CQDs every day and the antibacterial activity was measured through 14 passages. Fig. 7g–i showed the MIC of these CQDs against both E. coli and S. aureus did not significantly change after 14 passages, indicating that these CQDs have no tendency to develop resistance to E. coli and S. aureus.
Antibacterial mechanism of CQDs
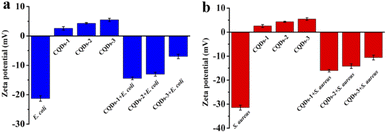
To further understand the antibacterial mechanism of these CQDs, the morphologies of bacteria, zeta potential and 1O2 production before/after CQDs treatment were studied. As shown in Fig. 8a–h, integrity of E. coli and S. aureus untreated with CQDs was kept well and smooth while the cell walls of E. coli were wrinkled and broken and some cytoplasmic components of S. aureus exudated from cells treated with CQDs, especially in the CQDs-3 groups. In addition, to further investigate the mechanism of antibacterial activity by these CQDs, we explored if 1O2 were generated. Fig. 9a–c showed the absorbance feature of DPBF at 410 nm was progressively decreased in the presence of these CQDs, suggesting intracellular singlet oxygen are generated during the antibacterial process. Furthermore, the absorbance of DPBF at 410 nm became lower at 30 min with increasing P element from CQDs-1 to CQDs-3. Meanwhile, the generation of 1O2 was further verified by measuring the electron paramagnetic resonance (EPR) of the samples. As shown in Fig. 9d, the signal intensity of 1O2 generation increased with the increase of P element. The results are consistent with that of the morphologies of bacteria. As shown in Fig. 10, zeta potential analysis showed that all of CQDs-1 (+2.58 mV), CQDs-2 (+4.33 mV) and CQDs-3 (+5.47 mV) were positively charged, and the positive charge was higher with the doping ratio of P element increased. We speculate that the surface charge of these CQDs is strongly related to antibacterial activity. Overall, these positively charged CQDs destroy the permeability and integrity of the bacterial plasma membrane, maybe owing to the production of reactive oxygen species. Importantly, the CQDs with higher P doping ratio in these carbon nanoparticles, which is smaller in sizes and have higher positive charge, have a significantly enhanced antibacterial activity compared to the others, which may be attributed to the difference in cellular uptake and distribution in the plasma membrane.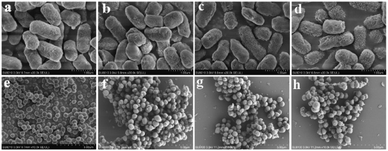 | ||
| Fig. 8 Bacterial morphology of E. coli and S. aureus without (a, e) or with CQDs-1 (b, f), CQDs-2 (c, g), CQDs-3 (d, h) treatment. | ||
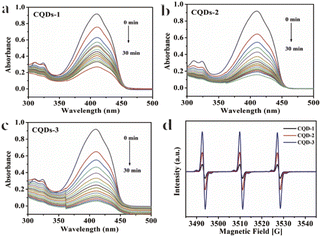 | ||
| Fig. 9 Absorbance spectra of DPBF obtained from different irradiation times in the presence of CQDs-1 (a), CQDs-2 (b), CQDs-3 (c). EPR spectra (d) of CQDs. | ||
Conclusions
In summary, a series of low toxic CQDs with different content of P element were prepared by a hydrothermal method. The average diameter and the surface charge of these CQDs could be regulated by increasing the content of P element. In addition, these positively charged CQDs exhibited effectively enhanced inhibition on Gram-negative and Gram-positive bacteria growth with increasing P doping ratio. The results of antibacterial mechanism demonstrated that bacterial death accompanied by disruption of the permeability and structural integrity of bacterial plasma membrane might be attributed to strong electrostatic interactions between negatively charged bacteria and positively charged CQDs and production of 1O2 of CQDs. This work provided a new insight into P-doped CQDs for the development of antibacterial nanomaterials as prospective alternative bacteria-resistant antibacterial agents.Author contributions
Data curation, writing-original draft, review, project administration, S. Q. Chai; validation, methodology, formal analysis, L. J. Zhou; formal analysis, investigation, funding acquisition, Y. T. Chi; software, funding acquisition, L. S. Chen; supervision, conceptualization, S. C. Pei; supervision, review and editing, project administration, B. Chen. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by Research Foundation of Chongqing University of Science and Technology, grant number ckrc2019031, and Open Fund of Chongqing Key Laboratory of Industrial Fermentation Microorganism (Chongqing University of Science and Technology), grant number GYFJWSW-07, Graduate Innovation Program Project of Chongqing University of Science and Technology, grant number YKJCX2120532, College Student Innovation Program of Chongqing University of Science and Technology, grant number s202211551015.References
- A. Ferreiro, N. Crook, A. J. Gasparrini and G. Dantas, Cell, 2018, 172, 1216–1227 CrossRef CAS PubMed.
- M. S. Morehead and C. Scarbrough, Prim. Care., 2018, 45, 467–484 CrossRef PubMed.
- S. B. Singh, K. Young and L. L. Silver, Biochem. Pharmacol., 2017, 133, 63–73 CrossRef CAS.
- G. F. Zha, H. D. Preetham, S. Rangappa, K. S. Sharath Kumar, Y. R. Girish, K. P. Rakesh, M. Ashrafizadeh, A. Zarrabi and K. S. Rangappa, Bioorg. Chem., 2021, 115, 105175 CrossRef CAS PubMed.
- Y. Z. Y. Xie, W. F. Zheng and X. Y. Jiang, ACS Appl. Mater. Interfaces, 2020, 12, 9041–9049 CrossRef CAS.
- J. C. Kuo, S. H. Tan, Y. C. Hsiao, C. Mutalik, H. M. Chen, S. Yougbaré and T. R. Kuo, ACS Sustainable Chem. Eng., 2022, 10, 464–471 CrossRef CAS.
- Y. K. Zheng, W. W. Liu, Y. Chen, C. M. Li, H. Jiang and X. M. Wang, J. Colloid Interface Sci., 2019, 546, 1–10 CrossRef CAS PubMed.
- S. Valsalam, P. Agastian, G. A. Esmail, A. K. M. Ghilan, N. A. Al-Dhabi and M. V. Arasu, J. Photochem. Photobiol., B, 2019, 201, 111670 CrossRef CAS PubMed.
- M. E. Khan, Nanoscale Adv., 2021, 3, 1887–1900 RSC.
- Z. M. Xiu, Q. B. Zhang, H. L. Puppala, V. L. Colvin and P. J. J. Alvarez, Nano Lett., 2012, 12, 4271–4275 CrossRef CAS PubMed.
- A. Ahmed Sharwani, K. Badri Narayanan, M. Ehtisham Khan and S. Soo Han, Arab. J. Chem., 2021, 14, 103456 CrossRef CAS.
- M. E. Khan, T. H. Han, M. M. Khan, M. R. Karim and M. H. Cho, ACS Appl. Nano Mater., 2018, 1, 2912–2922 CrossRef CAS.
- A. A. Sharwani, K. B. Narayanan, M. E. Khan and S. S. Han, Sci. Rep., 2022, 12, 10017 CrossRef CAS PubMed.
- H. Yang, H. Chen, L. Cao, H. Wang, W. F. Deng, Y. M. Tan and Q. J. Xie, Talanta, 2020, 212, 120797 CrossRef CAS PubMed.
- Y. Y. Cheng, Y. F. Xie, C. M. Li, Y. F. Li and C. Z. Huang, Anal. Chem., 2019, 91, 11023–11029 CrossRef CAS PubMed.
- Y. Y. Yao, G. Gedda, W. M. Girma, C. L. Yen, Y. C. Ling and J. Y. Chang, ACS Appl. Mater. Interfaces, 2017, 9, 13887–13899 CrossRef CAS.
- M. M. F. Baig and Y. C. Chen, J. Colloid Interface Sci., 2017, 501, 341–349 CrossRef CAS PubMed.
- J. J. Zhao, M. J. Huang, L. L. Zhang, M. B. Zou, D. X. Chen, Y. Huang and S. L. Zhao, Anal. Chem., 2017, 89, 8044–8049 CrossRef CAS.
- H. P. Liu, T. Ye and C. D. Mao, Angew. Chem., Int. Ed., 2007, 119, 6593–6595 CrossRef.
- J. Zhou, Y. Yang and C. Y. Zhang, Chem. Commun., 2013, 49, 8605–8607 RSC.
- X. Chu, P. Zhang, Y. Liu, B. Sun, X. Huang, N. Zhou, J. Shen and N. Meng, J. Mater. Chem. B., 2022, 10, 2865–2874 RSC.
- M. E. Khan, A. Mohammad, W. Ali, A. U. Khan, W. Hazmi, W. Zakri and T. Yoon, J. Cleaner Prod., 2022, 349, 131249 CrossRef CAS.
- M. E. Khan, A. Mohammad and T. Yoon, Chemosphere, 2022, 302, 134815 CrossRef CAS PubMed.
- M. Wang, A. Xia, S. Wu and J. Shen, Langmuir, 2021, 37, 7928–7935 CrossRef CAS PubMed.
- X. L. Dong, W. X. Liang, M. J. Meziani, Y. P. Sun and L. J. Yang, Theranostics, 2020, 10, 671–686 CrossRef CAS PubMed.
- F. Nichols, J. E. Lu, R. Mercado, M. D. Rojas-Andrade, S. Ning, Z. Azhar, J. Sandhu, R. Cazares, C. Saltikov and S. Chen, Langmuir, 2020, 36, 11629–11636 CrossRef CAS PubMed.
- F. C. Cui, Y. L. Ye, J. F. Ping and X. L. Sun, Biosens. Bioelectron., 2020, 156, 112085 CrossRef CAS PubMed.
- A. Anand, B. Unnikrishnan, S. C. Wei, C. P. Chou, L. Z. Zhang and C. C. Huang, Nanoscale Horiz., 2019, 4, 117–137 RSC.
- W. Bing, H. J. Sun, Z. Q. Yan, J. S. Ren and X. G. Qu, Small, 2016, 12, 4713–4718 CrossRef CAS PubMed.
- J. J. Liu, S. Y. Lu, Q. L. Tang, K. Zhang, W. X. Yu, H. C. Sun and B. Yang, Nanoscale, 2017, 9, 7135–7142 RSC.
- H. J. Jian, R. S. Wu, T. Y. Lin, Y. J. Li, H. J. Lin, S. G. Harroun, J. Y. Lai and C. C. Huang, ACS Nano, 2017, 11, 6703–6716 CrossRef CAS.
- C. H. Lee, S. Y. Song, Y. J. Chung, E. K. Choi, J. Jang, D. H. Lee, H. D. Kim, D.-U. Kim and C. B. Park, ACS Appl. Bio Mater., 2022, 5, 761–770 CrossRef CAS PubMed.
- S. Huang, E. L. Yang, Y. Liu, J. D. Yao, W. Su and Q. Xiao, Sens. Actuators, B, 2018, 265, 326–334 CrossRef CAS.
- B. F. Shi, Y. B. Su, L. L. Zhang, M. J. Huang, R. J. Liu and S. L. Zhao, ACS Appl. Mater. Interfaces, 2016, 8, 10717–10725 CrossRef CAS.
- Z. S. Sun, W. Y. Zhou, J. B. Luo, J. Q. Fan, Z. C. Wu, H. N. Zhu, J. Q. Huang and X. G. Zhang, J. Colloid Interface Sci., 2022, 607, 16–23 CrossRef CAS PubMed.
- M. Wang, Y. Su, Y. Liu, Y. Liang, S. Wu, N. Zhou and J. Shen, J. Colloid Interface Sci., 2022, 608, 973–983 CrossRef CAS.
- D. Zhao, Z. X. Zhang, X. M. Liu, R. Zhang and X. C. Xiao, Mater. Sci. Eng. C, 2021, 119, 111468 CrossRef CAS PubMed.
- N. A. Travlou, D. A. Giannakoudakis, M. Algarra, A. M. Labella, E. Rodríguez-Castellón and T. J. Bandosz, Carbon, 2018, 135, 104–111 CrossRef CAS.
- H. Wang, F. Lu, C. Ma, Y. Ma, M. Zhang, B. Wang, Y. Zhang, Y. Liu, H. Huang and Z. Kang, J. Mater. Chem. B., 2021, 9, 125–130 RSC.
- B. Kong, T. Yang, F. Cheng, Y. Qian, C. Li, L. Zhan, Y. Li, H. Zou and C. Huang, J. Colloid Interface Sci., 2022, 611, 545–553 CrossRef CAS.
- S. Q. Chai, J. H. He, L. Zhan, Y. F. Li, C. M. Li and C. Z. Huang, Talanta, 2019, 196, 100–108 CrossRef CAS.
- B. Sun, F. Wu, Q. Zhang, X. Chu, Z. Wang, X. Huang, J. Li, C. Yao, N. Zhou and J. Shen, J. Colloid Interface Sci., 2021, 584, 505–519 CrossRef CAS.
- Y. Q. Zhang, X. Y. Liu, Y. Fan, X. Y. Guo, L. Zhou, Y. Lv and J. Lin, Nanoscale, 2016, 8, 15281–15287 RSC.
- M. Algarra, D. Bartolić, K. Radotić, D. Mutavdžić, M. S. Pino-González, E. Rodríguez-Castellón, J. M. Lázaro-Martínez, J. J. Guerrero-González, J. C. G. Esteves da Silva and J. Jiménez-Jiménez, Talanta, 2019, 194, 150–157 CrossRef CAS PubMed.
- J. Zhao, F. T. Li, S. Zhang, Y. An and S. Q. Sun, New J. Chem., 2019, 43, 6332–6342 RSC.
- J. Zhou, X. Y. Shan, J. J. Ma, Y. M. Gu, Z. S. Qian, J. R. Chen and H. Feng, RSC Adv., 2014, 4, 5465–5468 RSC.
- H. Wang, F. Lu, C. Ma, Y. Ma, M. Zhang, B. Wang, Y. Zhang, Y. Liu, H. Huang and Z. Kang, J. Mater. Chem. B., 2020, 9, 125–130 RSC.
- J. Xu, Y. Zhou, G. Cheng, M. Dong, S. Liu and C. Huang, Luminescence, 2015, 30, 411–415 CrossRef CAS PubMed.
- B. C. M. Martindale, G. A. M. Hutton, C. A. Caputo, S. Prantl, R. Godin, J. R. Durrant and E. Reisner, Angew. Chem., Int. Ed., 2017, 56, 6459–6463 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: High-resolution XPS spectra of elements; FL emission spectra; stability of CQDs; the antibacterial ability of CQDs. See https://doi.org/10.1039/d2ra04809d |
| This journal is © The Royal Society of Chemistry 2022 |