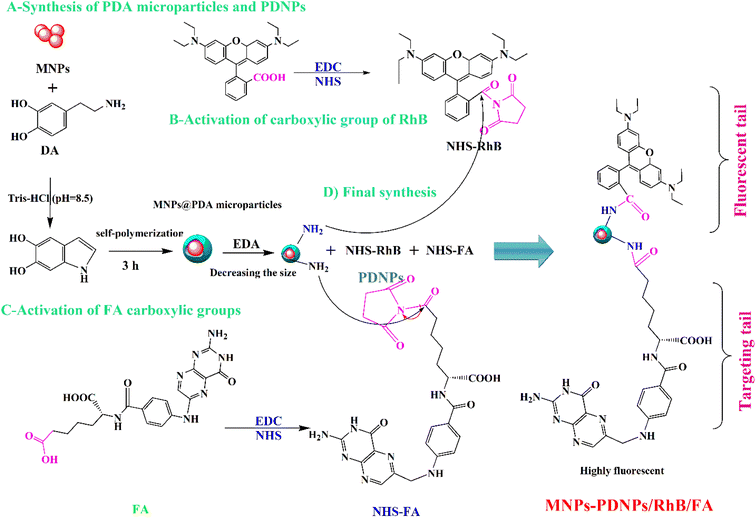
Open Access Article
Open Access ArticleSimple fluorescence chemosensor for the detection of calcium ions in water samples and its application in bio-imaging of cancer cells†
Maral Salek-Maghsoodia,
Zahra Golsanamlua,
Sanam Sadeghi-Mohammadib,
Masoud Gazizadehc,
Jafar Soleymani *a and
Reza Safaralizadehd
*a and
Reza Safaralizadehd
aPharmaceutical Analysis Research Center, Tabriz University of Medical Sciences, Tabriz, Iran. E-mail: jsoleymanii@gmail.com; soleymanij@tbzmed.ac.ir; Tel: +98 41 3337 5365
bTuberculosis and Lung Diseases Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
cLiver and Gastrointestinal Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
dDepartment of Biology, Faculty of Natural Sciences, Tabriz University, Tabriz, Iran
First published on 3rd November 2022
Abstract
This article describes the design, synthesis and characterization of a sensor suitable for practical measurement of ionized calcium in water samples and cancer cells. Calcium is an important ion in living organs and works as a messenger in several cellular functions. A lack of Ca ions interrupts the immune system and can lead to several diseases. A novel magnetic-polydopamine nanoparticle (PDNP)/rhodamine B (RhB)/folic acid (FA) nanoparticle was developed for the determination of calcium ions in MCF 7 cell lysates and water samples. Furthermore, the produced nanoparticle was employed for bioimaging of folate receptor (FR)-overexpressed cancer cells. This nanoprobe displayed a bright photoluminescence emission at 576 nm under an excitation wavelength of 420 nm. In the presence of calcium ions, the fluorescence emission of the MNPs-PDNPs/RhB/FA probe was proportionally decreased from 20 ng mL−1 to 100 ng mL−1 and 0.5 μg mL−1 to 20 μg mL−1 with a lower limit of quantification (LLOQ) of about 20 ng mL−1. The developed sensor showed a low-interference manner in the presence of possible coexistence interfering ions. In addition, this nanomaterial showed excellent biocompatibility with favorable differentiation ability to attach to the FR-positive cancer cells. The MNPs-PDNPs/RhB/FA nanoparticle has been utilized for bioimaging of the MCF 7 cell with favorable differentiation ability.
1. Introduction
Calcium ions (Ca2+) are the most ubiquitous signaling molecules in living organisms, regulating numerous biological functions. Calcium is a vital ion in living organs and elaborates as an extracellular messenger in several cellular functions. Lack of Ca2+ interrupts the immune system and leads to several diseases.1 The dysregulation of calcium concentration in cells is related to some neurodegenerative disorders of Alzheimer's disease (AD), Parkinson's disease (PD), and Huntington's disease (HD).2–4 Furthermore, some evidence proposes that an abnormal level of calcium is associated with the progress of cancer. Also, the concentration of Ca2+ in environmental sources must be controlled to regulate its concentrations in water and soil.5Recently, monitoring of intracellular concentrations of cationic and anionic ions has received great attention to be employed as an indicator for early-stage detection of the corresponded disorders.6,7 Hence, the design of a calcium sensing platform is extremely needed for its reliable detection in various biological and environmental samples. To date, there have been reported several analytical techniques for measuring and detection of Ca2+ including electrochemical method, NMR analysis, electrochemical, atomic spectroscopy, colorimetric (UV), and photoluminescence methods in diverse samples which offer specific, sensitive, and reliable detection.7–10 However, some disadvantages have been found for these techniques such as laborious detection approaches, high-cost instrumentation, and nonsufficient sensitivity and selectivity associated with atomic spectroscopy-based methods, fragile instrumentation and time consuming manipulations associated with electrochemical methods, the high detection limit with colorimetric techniques, and etc. These restrictions limit their uses for the fast detection of calcium ions. In recent years, fluorescence and electrochemical-based sensor systems for Ca2+ detection have attracted great attentions owing to their operational simplicity and real-time detection, etc., that have been widely utilized for the detection of various analytes due to their favorable analytical performance of high sensitivity, fast analysis time, and non-destructive detection approaches.8–15
Fluorescent materials not only showed potential fluorescent probes for the determination of ions in live cells and they also could be applied for the cancer cells bioimaging. There has been a growing request to use these materials in bioimaging applications alongside previously used techniques such as computed tomography (CT), single-photon emission computed tomography (SPECT), magnetic resonance imaging (MRI), etc. for in vivo bioimaging. The fluorescence-based bioimaging exhibited several beneficial features of high safety, simplicity, sensitivity, and biocompatibility nature toward living cells.16,17 The detection of biomolecular cellular functions using fluorescent materials is becoming much easier owing to their intensive development for biosensing and medical applications. Because, several human diseases, including various cardiomyopathies, neurodegenerative diseases, and cancer are known to be associated with altered Ca2+ concentration. There is a strong need to develop Ca2+ sensors capable of real-time quantitative Ca2+ concentration measurements in specific subcellular environments. Current estimates of Ca2+ have been derived using major types of Ca2+ indicators and sensors like (1) synthetic small molecule fluorescent indicators, (2) specifically modified derivatives of the chemiluminescent protein, (3) fluorescent indicators based on green fluorescence protein, and (4) some small molecule dyes accumulate in certain cellular compartments of cells.
Bioimaging of cancer cells is implemented by different approaches that the targeted imaging is the best one to differentiate cancer cells from normal cells. However, targeted bioimaging could be done by biological agents like antibody–antigen, aptamers, etc., or by chemical moieties like folic acid (FA), hyaluronic acid, etc. functionalized agents. Despite the high specificity of the biological agents for bioimaging, these substances are high-cost, low stable, and usually, severe conditions are needed to be applied for the storing of them. Employing simple techniques like fluorescence and electrochemical methods provide more versatile and facile approaches for bioimaging aims.18,19
FA molecules have been utilized as labeling molecules for the modification of various molecules and nanoparticles for different medical aims.20,21 FA-functionalized materials are very selective toward folate receptor (FR) of the surface membrane of some of the FRs-overexpressed cancer cells.22 To date, diverse new materials including metallic (gold, silver, etc.)-based, carbon-based, silica-based nanoparticles, etc. have been modified with FA molecules.23–26
Herein, the use of the magnetic-poly dopamine nanoparticles (PDNPs)/rhodamine B (RhB)/FA was presented for the facile determination of calcium ions in water samples, and cell lysates. Furthermore, the MNPs-PDNPs/RhB/FA nanoparticles were also employed for the bioimaging and measuring of intracellular calcium ions at trace levels. The MNPs-PDNPs/RhB/FA-based probe showed high selectivity towards calcium ions in the presence of metal ions. The developed fluorescent calcium sensor displayed good water dispersity and biocompatibility. Additionally, this novel nanoprobe has been utilized for the determination of calcium ions in MCF 7 cell lysates as well as the detection of calcium ions in water samples from different sources.
2. Experimental section
2.1. Materials
FA was obtained from Mehr Darou pharmaceutical company (Iran). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC, 98.0%), rhodamine B (RhB, 98.0%), and N-hydroxysuccinimide (NHS, 98.0%) were purchased from Sigma-Aldrich (USA). Dimethylsulfoxide (DMSO, 98.3%), ammonia solution (15%), sodium hydroxide (NaOH, 98.0%), and hydrochloric acid (HCl, 35.5%) were provided from Merck company (Germany). Also, sodium acetate, sodium dihydrogen phosphate, disodium hydrogen phosphate, acetic acid, and trisaminomethane hydrochloride (Tris-HCl) were purchased from Merck (Germany). Cell culture materials i.e. Roswell Park Memorial Institute (RPMI) 1640, trypsin–EDTA (25%), fetal bovine serum (FBS), and penicillin/streptomycin were obtained from Gibco Co. (UK). MCF 7 and HEK 293 cell lines were obtained from National Cell Bank of Iran (NCBI) (Iran).2.2. Apparatus
Fourier transform infrared (FTIR) spectra were recorded using Bruker model instrument (Billerica, Massachusetts, US). A JASCO FP 750 spectrofluorometer (Tokyo, Japan) was employed to record fluorescence emission spectra. The size and surface charge of the MNPs-PDNPs/RhB/FA nanoparticles were characterized by Malvern particle size analyzer (Malvern, UK). Field emission scanning electron microscopy (FE-SEM, FEG-SEM MIRA3 TESCAN, Brno, Czech Republic) was used to show the morphology and analyze the energy dispersive X-ray (EDX) spectrum of the MNPs-PDNPs/RhB/FA nanoparticles. Atomic force microscopy (AFM) was employed to study surface topography of the MNPs-PDNPs/RhB/FA through a Digital Instruments nanosurf mobile S (Grammetstrasse, Switzerland). Crystallographic structure of the MNPs-PDNPs/RhB/FA was verified using X-ray diffraction (XRD) technique using Siemens D 5000 X-ray diffractometer (Texas, USA) with a CuKα anode (λ = 1.54 Å, 40 kV, 30 mA). The magnetic property was determined by MDKB (Kashan, Iran). The cellular internalization of the MNPs-PDNPs/RhB/FA nanoparticles was recorded by fluorescent microscopy (Olympus BX64, Olympus, Japan) with U-MWU2 fluorescence filter (excitation filter BP 330e385, dichromatic mirror DM 400, emission filter LP 420). A microplate reader (Awareness Technology, Florida, USA) was utilized to measure absorbance of MTT assay.2.3. Synthesis
2.4. Biological section
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm). Then, the solution was treated with HCl (1 M). Finally, the supernatant was separated by a centrifuge and the amount of calcium was determined with the designed probe.
000 rpm). Then, the solution was treated with HCl (1 M). Finally, the supernatant was separated by a centrifuge and the amount of calcium was determined with the designed probe.2.5. Pretreatment of water samples
Tap water, commercial mineral, and Aras river water samples were collected from the local sources and then filtrated to delete possible particles. Then, suitable volumes were added to the probe i.e. the MNPs-PDNPs/RhB/FA nanoparticles at the optimized condition and fluorescence emissions were recorded by a fluorescence technique. The final concentrations of calcium ions were determined using a previously plotted calibration curve which was already provided in water media.3. Results and discussion
3.1. Characterization of MNPs-PDNPs/RhB/FA nanomaterials
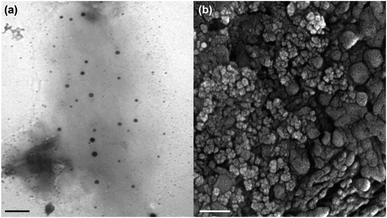
Fig. 1 displays the TEM and FE-SEM images of MNPs-PDNPs/RhB/FA nanoparticles. It is clear that the general shape of the MNPs-PDNPs/RhB/FA nanoparticles is spherical. The MNPs-PDNPs/RhB/FA nanoparticles size were computed from TEM and FE-SEM images using ImageJ 21.1 ± 6.3. Also, AFM analysis confirmed the topography of the nanoparticles as determined by FE-SEM and TEM techniques (Fig. S1†). The size of MNPs-PDNPs/RhB/FA nanoparticles is inconsistent with the hydrated diameter obtained by dynamic light scattering (DLS) techniques (Fig. S2a†). To observe the surface charge, the MNPs-PDNPs/RhB/FA zeta potential was determined as +17.9 mV (Fig. S2b†) which is largely affected by PDNPs with a zeta potential of +102.0 mV.29 Generally, amine groups turned to their cationic forms at low and neutral pHs. Also, the addition of FA molecules with negative zeta potential, influenced the final zeta potential to the negative values. The +17.9 mV zeta potential shows how stable during the colloidal phase. Stable colloidal dispersions are obtained at relatively high zeta. In other words, if the particles in any suspension possess large zeta potential values (negative or positive) then they tend to repulse each other with no desire to produce coagulation.The elemental composition analysis of the MNPs-PDNPs/RhB/FA nanoparticles was carried out using energy dispersive X-ray (EDX) analysis equipped with FE-SEM, and showed that the percentages of carbon, nitrogen, oxygen, and ferric/ferrous ions of 20.21%, 25.19%, 31.75%, and 22.85%, respectively (Fig. S3 and Table S1†). The obtained results confirm the presence of the elements which must be available on the final structure of the MNPs-PDNPs/RhB/FA nanoparticles.
The surface functional groups of the MNPs-PDNPs/RhB/FA nanoparticles and FA molecules were detected by FTIR (Fig. S4†). An absorption peak at around 550 cm−1 confirms the presence of the Fe–O bonds.30 The attachment of FA and RhB molecules to the surface of PDNPs could be observed between 1200–1800 cm−1 and 100–500 cm−1, respectively. The presence of amide groups is confirmed by the 3350 and 1651 cm−1 spectra which confirms the attachment of RhB and FA molecules to the amine-functionalized PDNPs.31,32 It is noteworthy that the formation of amide bonds is one of the main confirmations in the EDC/NHS coupling reactions.33
Magnetic property of the MNPs-PDNPs/RhB/FA was measured and shown in Fig. S5.† The saturation magnetization of the nanocomposite was found equal to 3.0 emu g−1, showing favorable magnetic property of the MNPs-PDNPs/RhB/FA. In Fig. S5,† it clear that by increasing the magnetic field −10 to +10 T, the magnetization enhanced severely with forming an S-like curve. Usually, materials with large magnetic properties showed higher magnetization value, however, the magnetic properties of the MNPs-PDNPs/RhB/FA is logically limited as compared with bare MNPs with magnetization value of around 39.5 emu g−1.34 The crystallographic structure of the MNPs-PDNPs/RhB/FA was also checked by the XRD technique at the 2θ range from 4–80° Fig. S6.† It is evidently observed that the MNPs-PDNPs/RhB/FA do not have sharp diffraction peaks which is a typical spectrum for amorphous or ultrafine crystalline materials. Hence, it is concluded that the iron oxide formed in the polymer-like structure which were the desired form of Fe3O4 particles.35
3.2. Study of spectrophotometric and fluorescence spectra of the MNPs-PDNPs/RhB/FA nanoparticles
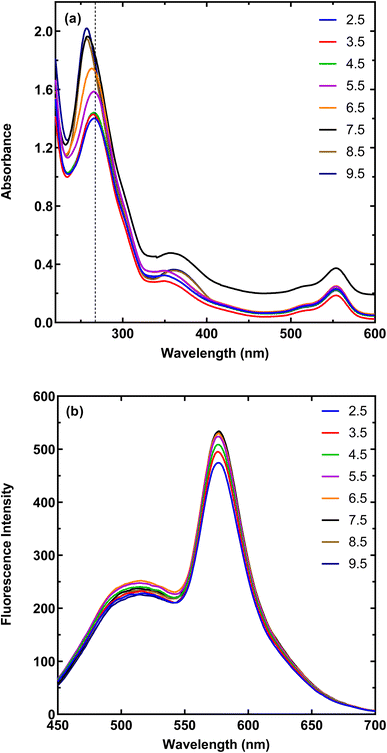
Spectrophotometric and fluorescence spectra of the MNPs-PDNPs/RhB and MNPs-PDNPs/RhB/FA nanoparticles are presented in Fig. 2. The MNPs-PDNPs/RhB have two shape absorption peaks at 259 nm and 555 nm and three broad peaks at 354 nm, 400 nm, and 513 nm. Peaks around 257 nm, 353 nm, and 553 nm are regarded as the characteristic absorption peaks of RhB molecules. However, the MNPs-PDNPs/RhB/FA nanoparticles showed absorption bands around 259 nm, 366 nm, 513 nm, and 555 nm. The absorbance band of 259 nm is arisen by the n–π* transition whereas the π–π* electron transition is produced a fluorescence peak at 553 nm (Fig. 2a). It seems that a sharp peak of RhB at the 259 nm overlaps the FA absorbance band and another technique (i.e. FTIR) is needed to confirm the attachment of FA to the MNPs-PDNPs/RhB (Fig. S4†).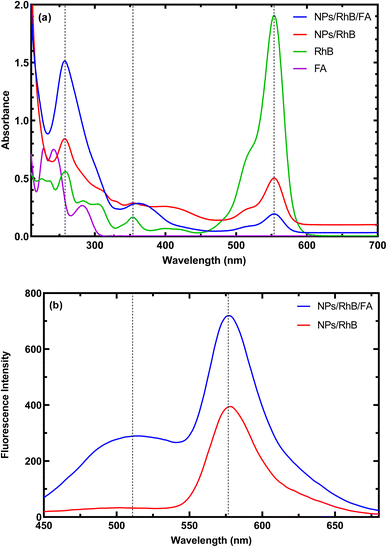 | ||
| Fig. 2 (a) Absorbance, and (b) fluorescence emission spectra of the MNPs-PDNPs/RhB and MNPs-PDNPs/RhB/FA nanoparticles. | ||
As above-mentioned, the π–π* electron transition with an absorbance wavelength of 555 nm is responsible for the photoluminescence of the MNPs-PDNPs/RhB/FA nanoparticles. The fluorescence spectra of MNPs-PDNPs/RhB/FA nanoparticles have two main peaks at 520 nm and 576 nm corresponding to the FA and RhB molecules, respectively. However, the RhB maximum peak was remained unchanged at both MNPs-PDNPs/RhB and MNPs-PDNPs/RhB/FA nanoparticles (Fig. 2b). The excitation and emission spectra of the MNPs-PDNPs/RhB/FA nanoparticles are presented in Fig. S7.† By altering the excitation wavelength in the range of 380 nm to 470 nm, the photoluminescence emission MNPs-PDNPs/RhB/FA nanoparticles were decreased and the maximum intensity was obtained at 420 nm. Furthermore, the emission peak of the MNPs-PDNPs/RhB/FA nanoparticles was 60 nm shifted to shorter wavelengths (Fig. S8†). Multiple excited states and the lower relaxation rates of the environments can result in the alteration of emission wavelength.36,37
3.3. Effect of pH value on the fluorescence and absorbance spectra of the MNPs-PDNPs/RhB/FA
The presence of different functional groups on the surface of MNPs-PDNPs/RhB/FA makes them be affected by the pH condition of the media. Therefore, both fluorescent and absorption spectra of MNPs-PDNPs/RhB/FA are changed by pH alerting (Fig. 3). The absorbances of MNPs-PDNPs/RhB/FA are enhanced upon increasing pHs up to 7.5 and then was constant up to about pH 9.5. However, a 10 nm blue shift was obtained at the maxima of 259 nm of the MNPs-PDNPs/RhB/FA, showing the participation of the functional groups in the formation of hydrogen bonds. In addition, fluorescence spectra of the MNPs-PDNPs/RhB/FA were influenced by pH. Fig. 3b and S9† show that the fluorescence emissions were enhanced up to pH 7.5, while the emission wavelength remained constant at all pHs. This trend is the same for both absorbance and fluorescence emission spectra. The π–π interactions are dominated at low pH values is a result of the amine groups protonation of the PDNPs. However, at the higher pHs, the π–π interactions are blocked at large scale by the free amine groups.383.4. Analytical performance of the established probe for calcium sensing
The type of buffer and its concertation has a massive effect on the amount of the quenched fluorescence intensity. Findings proposed that Tris buffer with a concentration of 25 mM is the best choice for the calcium ions and MNPs-PDNPs/RhB/FA interactions (Fig. S10b and c†). Fig. S10d† demonstrates the effects of the MNPs-PDNPs/RhB/FA concentrations on the measuring of Ca. The interactions between MNPs-PDNPs/RhB/FA and Ca ions may be decreased at the low concentrations of the nanoparticles.
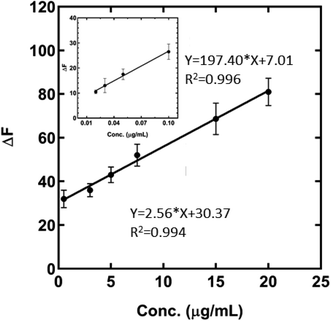
The calibration curve of the calcium determination is presented in Fig. 4 in which the fluorescence emission of the MNPs-PDNPs/RhB/FA has decreased upon the addition of calcium ions from 20 ng mL−1 to 100 ng mL−1 and 0.5 μg mL−1 to 20 μg mL−1. The calibration equations of the probe at the obtained two ranges are ΔF = 197.4 (±9.327) C (μg mL−1) + 7.007 (±0.5479) (p-value < 0.0022) and ΔF = 2.558 (±0.0969) C (μg mL−1) + 30.37 (±1.058) (p-value < 0.0001) with correlation coefficients (R2) of 0.9956 and 0.9943, respectively. The lower limit of detection of the probe is 20 ng mL−1 which covers the concentration of Ca in environmental samples. Also, the limit of detection (LOD) and limit of quantification (LOQ) of the probe were estimated to be 0.01 ng mL−1 and 0.033 ng mL−1, respectively, based on their classical definition as LOD = 3Sd/a and LOQ = 10Sd/a. Here, “Sd” and “a” are the slope of the calibration curve and standard deviation of the blank, respectively. The quenched fluorescence of the MNPs-PDNPs/RhB/FA probe is proportionally quenched with calcium concentrations. Table S2† lists the figures-of-merits of some of the spectroscopic probes for calcium measurement.40–46 As seen, the developed MNPs-PDNPs/RhB/FA-based probe can detect calcium ions at a wide dynamic range with acceptable sensitivity. However, the developed method is able to detect calcium ions in live-cell media.
Table 1 collects the repeatability accuracy and recovery finding of the developed probe for the determination of calcium ions. These tests were investigated at three concentration levels of 0.02, 5, and 15 μg mL−1. The repeatability of the probe is between 6.7–16.0% as the relative standard deviation (RSD%). However, the accuracy of the developed method ranged from 83.0 to 99.8%. In this work, all validations were done using the food and drug administration (FDA) guidelines.39
| Nominal concentration (μg mL−1) | Intraday precision (RSD%) | Interday precision (RSD%) | Intraday accuracy (RE%) | Interday accuracy (RE%) |
|---|---|---|---|---|
| 0.02 | 6.7 | 16.0 | 85.0 | 83.0 |
| 5.0 | 1.0 | 13.0 | 98.8 | 88.0 |
| 15.0 | 10.5 | 15.0 | 99.8 | 93.0 |
3.5. Specificity investigation
The specificity of the developed method was checked regarding FDA guidelines.39 Based on the FDA guidelines, the signal of the probe was recorded in the presence and absence of the con-existing agents and reported as RSD%. The effect of some agents of Al3+, Hg2+, Cd2+, Zn2+, Se4+, Cr2+, Ni2+, Fe3+, Cu2+, and Co2+ was investigated on the signal of the MNPs-PDNPs/RhB/FA probe to check the selectivity of the established method (Fig. S11†). The selectivity of the developed probe was measured by comparing the fluorescent intensity of the probe in the presence and absence of the interfering agents. Obtained results revealed that the fluorescence emission of the MNPs-PDNPs/RhB/FA probe was not significantly affected by the tested agents and all RSD% values are in the FDA-permitted range. The high specificity of the MNPs-PDNPs/RhB/FA probe toward calcium ions could be a consequence of interactions between the surface carboxylic acid and amine groups of MNPs-PDNPs/RhB/FA and vacant orbitals of calcium ions. Also, the size of ions and the type of vacant orbitals are other factors that affect the interaction between ions and nanoparticles.3.6. Application of the probe
| Type of water | Obtained concentration (μg mL−1) |
|---|---|
| Tap 1 | 22.92 (±1.8) |
| Tap 2 | 24.68 (±7.4) |
| Mineral | 24.68 (±1.3) |
| Aras river | 19.12 (±4.6) |
3.7. Bioimaging of cancer cells
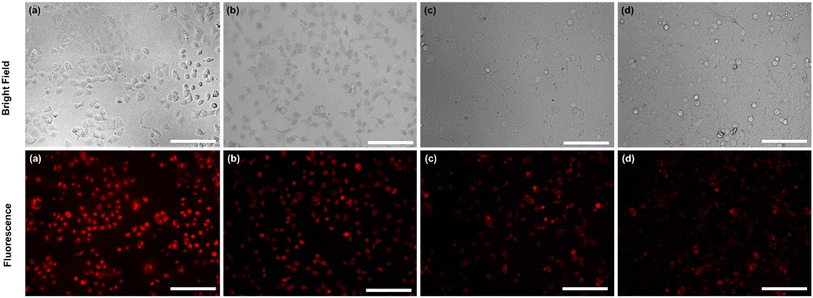
In addition, exclusively fluorescence bio-imaging of MCF 7 cells was carried out with NPs and obtained images are displayed in Fig. 5. The attachment of the FA molecules on the surface of MNPs-PDNPs/RhB makes them as a smart material that attaches to the FRs of the cancer cell membrane. This ability was evaluated by incubation of the cells at various concentrations and times. No fluorescence emission was observed for cells without treatment. Further, the addition of different concentrations of NPs with MCF 7 cells exhibits the difference in the fluorescence image (Fig. 5). Fig. 5 depicts the uptake of the MNPs-PDNPs/RhB/FA with concentrations of 25, 50, 100, 150, and 225 μg mL−1. From the results, it is clear that after incubation of the cells with 225 μg mL−1 of the MNPs-PDNPs/RhB/FA, the fluorescence intensity of the images reached its highest level. Fig. 6 shows the fluorescence bioimages of MCF 7 cells after contact with MNPs-PDNPs/RhB/FA nanoparticles in a time-dependent study. The 4 h of the incubation time shows bright fluorescence cell images when compared to 1 and 3 h of the incubation due to the high penetration NPs into the MCF 7 cells. The efficiency of MNPs-PDNPs/RhB/FA uptake to FR-positive cells was increased by the time of incubation. Bright red emission of MNPs-PDNPs/RhB/FA was mainly observed in the surface membrane of MCF 7 cells and cytoplasm, whole the nucleus was not significantly affected by the materials, demonstrating that the MNPs-PDNPs/RhB/FA is firstly attached to FRs and then gradually enter into the cells' cytoplasm of cells. These results confirmed that FRs are placed at the surface membrane of the FR-overexpressed cells. As seen, the cell morphology was not significantly affected by the MNPs-PDNPs/RhB/FA.
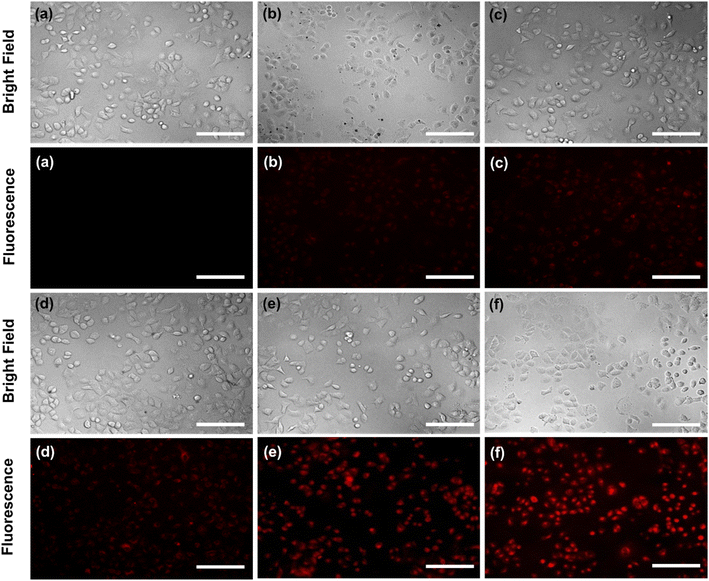 | ||
| Fig. 5 Effect of the concertation of the MNPs-PDNPs/RhB/FA ((a) control, (b) 25 μg mL−1, (c) 50 μg mL−1, (d) 100 μg mL−1, (e) 150 μg mL−1, and (f) 225 μg mL−1) (scale bar = 100 μm). | ||
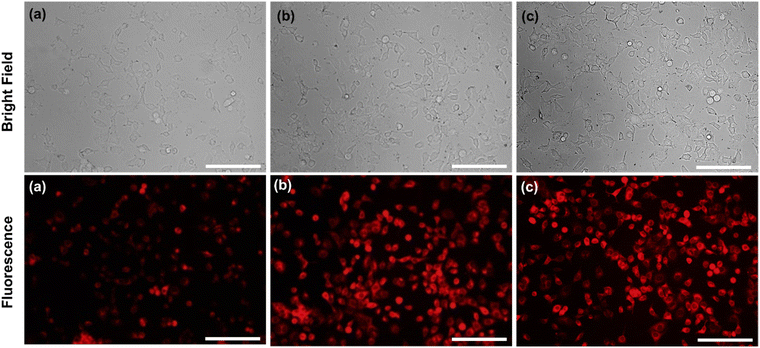 | ||
| Fig. 6 Effect of the time of incubation (a) 1 h, (b) 3 h, and (c) 4 h on the amount the cell uptake value (scale bar = 100 μm). | ||
The targeting specificity of the MNPs-PDNPs/RhB/FA was also studied by incubation of the HEK 293 cells with nanoparticles.47 Results proposed that the MNPs-PDNPs/RhB/FA failed to attach to the FR-negative cells due to the lower expression of FRs on the membrane of HEK 293 than MCF 7 cells (Fig. S14†). The extraordinary affinity of MNPs-PDNPs/RhB/FA to the MCF 7 cells is predominantly triggered by the high formation constant of FA/FR (10+8 to 10+9 M−1).48 The high formation constants enhance the targeted interactions and the specificity of the bioimaging of the cells.
4. Conclusion
A novel MNPs-PDNPs/RhB/FA nanocomposite was produced and used for the quantification of calcium ions in water samples and live cells. Also, it was employed for the targeting of FR-positive MCF 7 cancer cells. The cell viability of MNPs-PDNPs/RhB/FA nanoparticles was tested by the MTT assay with a biocompatible nature even at the 225 μg mL−1 concentration. Obtained results confirmed that the MNPs-PDNPs/RhB/FA nanoparticles can be uptake into the FR-positive cancer cells through FA/FR interactions with exceptional selectivity. In addition, the MNPs-PDNPs/RhB/FA was utilized for the detection of calcium ions in water samples and live cells with LLOQ of as low as around 20 ng mL−1. Bright and high fluorescence emission of the probe provides a more sensitive method for Ca detection. Furthermore, the emission wavelength is in the visible area, reducing the self-absorption of the fluorescence emission of the nanoparticles. Also, low-emission wavelengths reduce photobleaching and enhance the stability of the nanoparticles. In addition, high energy wavelengths may be harmful to the living cell. The applicability of this sensor was checked in the detection of calcium ions in the real samples which are collected from various sources and the detection of calcium ions in live cells. However, the application of this sensor may be limited in presence of some ions as studied in the main text.Conflicts of interest
There is no conflict of interest.Acknowledgements
The authors would like to acknowledge the financial supports from Tabriz University of Medical Sciences (Tabriz, Iran) (grant number: 67984).References
- A. T. Harootunian, J. P. Kao, S. Paranjape and R. Y. Tsien, Science, 1991, 251, 75–78 CrossRef CAS PubMed.
- R. Sangubotla and J. Kim, TrAC, Trends Anal. Chem., 2018, 105, 240–250 CrossRef CAS.
- E. Pchitskaya, E. Popugaeva and I. Bezprozvanny, Cell Calcium, 2018, 70, 87–94 CrossRef CAS PubMed.
- K. Reddy, C. L. Cusack, I. C. Nnah, K. Khayati, C. Saqcena, T. B. Huynh, S. A. Noggle, A. Ballabio and R. Dobrowolski, Cell Rep., 2016, 14, 2166–2179 CrossRef CAS PubMed.
- G. A. Weyhenmeyer, J. Hartmann, D. O. Hessen, J. Kopáček, J. Hejzlar, S. Jacquet, S. K. Hamilton, P. Verburg, T. H. Leach, M. Schmid, G. Flaim, T. Nõges, P. Nõges, V. C. Wentzky, M. Rogora, J. A. Rusak, S. Kosten, A. M. Paterson, K. Teubner, S. N. Higgins, G. Lawrence, K. Kangur, I. Kokorite, L. Cerasino, C. Funk, R. Harvey, F. Moatar, H. A. de Wit and T. Zechmeister, Sci. Rep., 2019, 9, 10450 CrossRef PubMed.
- Z. Golsanamlou, J. Soleymani, S. Abbaspour, M. Siahi-Shadbad, E. Rahimpour and A. Jouyban, Spectrochim. Acta, Part A, 2021, 256, 119747 CrossRef CAS PubMed.
- J. Soleymani, V. Shafiei-Irannejad, M. R. Hamblin, M. Hasanzadeh, M. H. Somi and A. Jouyban, Mater. Sci. Eng. C, 2021, 121, 111691 CrossRef CAS PubMed.
- M. Moirangthem, R. Arts, M. Merkx and A. P. H. J. Schenning, Adv. Funct. Mater., 2016, 26, 1154–1160 CrossRef CAS.
- A. Takahashi, P. Camacho, J. D. Lechleiter and B. Herman, Physiol. Rev., 1999, 79, 1089–1125 CrossRef CAS PubMed.
- J. Wu, L. Liu, T. Matsuda, Y. Zhao, A. Rebane, M. Drobizhev, Y.-F. Chang, S. Araki, Y. Arai, K. March, T. E. Hughes, K. Sagou, T. Miyata, T. Nagai, W. Li and R. E. Campbell, ACS Chem. Neurosci., 2013, 4, 963–972 CrossRef CAS PubMed.
- I. Isildak, F. Navaeipour, H. Afsharan, G. S. Kanberoglu, I. Agir, T. Ozer, N. Annabi, E. E. Totu and B. Khalilzadeh, Microchim. Acta, 2020, 187, 25 CrossRef CAS PubMed.
- B. Khalilzadeh, N. Shadjou, H. N. Charoudeh and M.-R. Rashidi, Microchim. Acta, 2017, 184, 3651–3662 CrossRef CAS.
- B. Khalilzadeh, M. Rashidi, A. Soleimanian, H. Tajalli, G. S. Kanberoglu, B. Baradaran and M.-R. Rashidi, Int. J. Biol. Macromol., 2019, 134, 695–703 CrossRef CAS PubMed.
- J. Soleymani, D. Perez-Guaita, M. Hasanzadeh, N. Shadjou and A. Jouyban, TrAC, Trends Anal. Chem., 2017, 86, 122–142 CrossRef CAS.
- J. L. Manzoori, M. Amjadi, J. Soleymani, E. Tamizi, A. Rezamand and A. Jouyban, Luminescence, 2012, 27, 268–273 CrossRef CAS PubMed.
- M. Hasanzadeh, M. Feyziazar, E. Solhi, A. Mokhtarzadeh, J. Soleymani, N. Shadjou, A. Jouyban and S. Mahboob, Microchem. J., 2019, 145, 778–783 CrossRef CAS.
- H. Yumak, B. Ozturk, J. Sheng and M. V Yigit, Langmuir, 2015, 31, 9943–9952 CrossRef PubMed.
- S. Jafarzadeh, N. Bargahi, H. B. Shamloo and J. Soleymani, RSC Adv., 2022, 12, 8492–8501 RSC.
- J. Soleymani, S. Azizi, S. Abbaspour-Ravasjani, M. Hasanzadeh, M. H. Somi and A. Jouyban, Microchem. J., 2021, 170, 106732 CrossRef CAS.
- X. Zhao, H. Li and R. J. Lee, Expert Opin. Drug Delivery, 2008, 5, 309–319 CrossRef CAS PubMed.
- X. Zhao, R. Shen, L. Bao, C. Wang and H. Yuan, Carbohydr. Polym., 2020, 245, 116509 CrossRef CAS PubMed.
- B. Farran, E. Pavitra, P. Kasa, S. Peela, G. S. Rama Raju and G. P. Nagaraju, Cytokine Growth Factor Rev., 2019, 45, 45–52 CrossRef CAS PubMed.
- J. Soleymani, M. Hasanzadeh, M. H. Somi and A. Jouyban, Mater. Sci. Eng. C, 2020, 107, 110320 CrossRef CAS PubMed.
- J. Soleymani, M. Hasanzadeh, N. Shadjou, M. H. Somi and A. Jouyban, J. Pharm. Biomed. Anal., 2020, 180, 113077 CrossRef CAS PubMed.
- M. Wang, Y. Liang, Z. Zhang, G. Ren, Y. Liu, S. Wu and J. Shen, Anal. Chim. Acta, 2019, 1086, 122–132 CrossRef CAS PubMed.
- S. Damiati, M. Peacock, R. Mhanna, S. Søpstad, U. B. Sleytr and B. Schuster, Sens. Actuators, B, 2018, 267, 224–230 CrossRef CAS.
- G. E. Gu, C. S. Park, H.-J. Cho, T. H. Ha, J. Bae, O. S. Kwon, J.-S. Lee and C.-S. Lee, Sci. Rep., 2018, 8, 4393 CrossRef PubMed.
- X. Liu, J. Cao, H. Li, J. Li, Q. Jin, K. Ren and J. Ji, ACS Nano, 2013, 7, 9384–9395 CrossRef CAS PubMed.
- Q. Lin, X. Huang, J. Tang, Y. Han and H. Chen, J. Nanopart. Res., 2013, 15, 2144 CrossRef.
- R. Nisticò, F. Cesano and F. Garello, Inorganics, 2020, 8, 6 CrossRef.
- W. Zhang, S. Wang, J. Ji, Y. Li, G. Zhang, F. Zhang and X. Fan, Nanoscale, 2013, 5, 6030–6033 RSC.
- Y. Fang, A. Zhou, W. Yang, T. Araya, Y. Huang, P. Zhao, D. Johnson, J. Wang and Z. J. Ren, Sci. Rep., 2018, 8, 229 CrossRef PubMed.
- M. J. E. Fischer, N. J. Mol and M. J. E. Fischer, Amine Coupling Through EDC/NHS: A Practical Approach BT - Surface Plasmon Resonance: Methods and Protocols, Humana Press, Totowa, NJ, 2010, pp. 55–73 Search PubMed.
- N. Mufti, D. R. P. Sari, A. F. Muyasaroh and A. Taufiq, Journal of Physics: Conference Series, IOP Publishing, 2020, vol. 1595, p. 12004 Search PubMed.
- Z. Ma, Y. Guan and H. Liu, J. Polym. Sci., Part A: Polym. Chem., 2005, 43, 3433–3439 CrossRef CAS.
- P. Suppan, Chemistry and Light, The Royal Society of Chemistry, London, UK, 1994 Search PubMed.
- M. Józefowicz and J. R. Heldt, J. Fluoresc., 2011, 21, 239–245 CrossRef PubMed.
- S. Bevers, T. P. O'De and L. W. McLaughlin, J. Am. Chem. Soc., 1998, 120, 11004–11005 CrossRef CAS.
- Food and Drug Administration (FDA), FDA guidelines, https://www.fda.gov/downloads/Drugs/Guidances/ucm070107.pdf, accessed 16 September 2021 Search PubMed.
- J. Yue, L. Li, L. Cao, M. Zan, D. Yang, Z. Wang, Z. Chang, Q. Mei, P. Miao and W.-F. Dong, ACS Appl. Mater. Interfaces, 2019, 11, 44566–44572 CrossRef CAS PubMed.
- E. Babaee, A. Barati, M. B. Gholivand, A. Arman, A. A. Taherpour, N. Zolfaghar and M. Shamsipur, J. Hazard. Mater., 2019, 367, 437–446 CrossRef CAS PubMed.
- S. R. Ankireddy and J. Kim, Highly Selective and Sensitive Detection of Calcium (II) Ions in Human Serum Using Novel Fluorescent Carbon Dots, Elsevier B.V., 2018, vol. 255 Search PubMed.
- L. Lu, X. An and W. Huang, Anal. Methods, 2017, 9, 23–27 RSC.
- H. Chen, J. Peng, L. Yu, H. Chen, M. Sun, Z. Sun, R. Ni, K. A. Alamry, H. M. Marwani and S. Wang, J. Fluoresc., 2020, 30, 463–470 CrossRef CAS PubMed.
- S. Ghosh, Y. Chen, A. George, M. Dutta and M. A. Stroscio, Front. Chem., 2020, 8, 1–9 CrossRef PubMed.
- Z. Liu, X. Jing, S. Zhang and Y. Tian, Anal. Chem., 2019, 91, 2488–2497 CrossRef CAS PubMed.
- K. Pal, A. Heinsch, A. Berkessel and A. L. Koner, Chem.–Eur. J., 2017, 23, 15008–15011 CrossRef CAS PubMed.
- M. D. Salazar and M. Ratnam, Cancer Metastasis Rev., 2007, 26, 141–152 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra04815a |
| This journal is © The Royal Society of Chemistry 2022 |