Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
An effective antimicrobial complex of nanoscale β-cyclodextrin and ciprofloxacin conjugated to a cell adhesive dipeptide†
Reza Taheri-Ledari a,
Farinaz Jalalia,
Leili Heidariab,
Fatemeh Ganjalia,
Fereshteh Rasouli Asla,
Simindokht Zarei-Shokata,
Mohadeseh Forouzandeh-Malatia,
Adibeh Mohammadia and
Ali Maleki
a,
Farinaz Jalalia,
Leili Heidariab,
Fatemeh Ganjalia,
Fereshteh Rasouli Asla,
Simindokht Zarei-Shokata,
Mohadeseh Forouzandeh-Malatia,
Adibeh Mohammadia and
Ali Maleki *a
*a
aCatalysts and Organic Synthesis Research Laboratory, Department of Chemistry, Iran University of Science and Technology, Tehran 16846-13114, Iran. E-mail: maleki@iust.ac.ir
bDepartment of Chemistry, Faculty of Basic Sciences, Ilam University, P. O. Box 69315-516, Ilam, Iran
First published on 15th December 2022
Abstract
Today, various drug delivery systems (DDS) are utilized to carry and deliver the desired drugs to the targeted action area to reduce potential side effects and negative interactions. Nanomaterials are an excellent candidate for the delivery of potent drugs, as they enhance pharmacokinetic and pharmacodynamic properties. Herein, we present a new ciprofloxacin (CPFX) delivery system based on a polymeric nanocarrier (β-cyclodextrin) conjugated to a cell-adhesive dipeptide structure. Cyclodextrin (CD) is an inexpensive, easily accessible, biodegradable, and biocompatible material. Also, the conjugation of cysteine–arginine (CR) dipeptide to the CPFX/β-CD particles is carried out to enhance cell adhesion growth. Through accurate analysis, the drug content and release for a final product have been estimated to be ca. 32%. Overall, the antimicrobial effects of CPFX were considerably raised through a low dose of CPFX. The growth zone inhibition of CPFX/β-CD–CR particles on the staphylococcus aureus and the Escherichia coli bacterial cells was 5.5 ± 0.2 cm and 3.5 ± 0.2 cm, respectively. Hence, this therapeutic nano bioconjugate is an excellent candidate to be applied in antimicrobial applications with the minimum incorporated CPFX.
1. Introduction
Antibiotics are regarded as the most significant developments in the modern era of medicine because of their tremendous effect on the reduction of mortality in humankind as well as economic harm. Nowadays, antimicrobial resistance (AMR) is the most critical concern in the fight against infectious diseases caused by drug abuse.1–3 Indeed, the mechanism of antibiotics action illustrates that antibiotics prevent the synthesis of the bacteria cell walls, disturb the cellular membrane, forbid the synthesis of nucleic acid,4 inhibit protein synthesis, block pathways and disrupt metabolism.5 Novel quinolones as antibiotics have been extended with enhanced effectiveness and spectrum over the years.6 CPFX is a second-generation fluoroquinolone used to treat various infections, including urinary infections, infections related to bone, cystic fibrosis, chronic otitis media, prostatitis, and so on.7,8 CPFX demonstrates more remarkable activity against Gram-negative bacteria (Enterobacteriaceae like Escherichia coli) than Gram-positive bacteria (Staphylococcus aureus).9 To clarify the antibacterial mechanism of the CPFX, it is crucial to know that CPFX is a DNA gyrase inhibitor. DNA gyrase is a tetramer consisting of two subunits GyrA and two GyrB subunits. DNA gyrase belongs to a group of enzymes called DNA topoisomerases, which is an essential part of the DNA replication system, and its function is to break the backbone of DNA. The GyrA subunit binds to DNA and hydrolyzes the backbone, while GyrB catalyzes the hydrolysis of ATP, which drives the supercoiling process. Topoisomerase IV is involved in resolving daughter chromosomes after DNA replication as well as the unscrewing of supercoils in the DNA. CPFX do not play a role in the breaking reaction of DNA strands catalyzed by these enzymes but inhibit the recombination step. CPFX are commonly bacteriostatic. However, high doses of CPFX can lead to double-stranded DNA breaks that are lethal to the affected cells.10In fact, one of the best ways to reduce interactions referred to drugs, diminish side effects, and increase the ability to kill bacteria is using appropriate DDS.11 Drug delivery means the delivery of pharmaceuticals to a target tissue to achieve safer and better therapeutic outcomes.12 DDS has some advantages and disadvantages, such as extending the duration of the action, reducing the frequency of dosage and ensuring the bioavailability of medicines, keeping the drug against degradation, minimize adverse drugs side effects, enhancement the compliance of patients and increase medication use through declining fluctuation plasma concentration. However, the disadvantages include the toxicity of the drug delivery device, the potential for hazardous degradation products, the need for surgery either on applying systems or removal, patients might be uncomfortable using the DDS device, and the cost and usage of DDS may be expensive.11
Nanotechnology affects everything from nanoscale gadgets to drug delivery frameworks.13–15 Drug delivery to the pain site can be active or passive via nanocarriers. In active targeting, peptides and antibodies are conjugated to the DDS via targeted tissue receptors, lipids, or antigens. The conduction of drugs via the self-assembled nanostructured and released drug encapsulation at the object is related to passive targeting.16–18 In the last few decades, the application of nanomaterials in a variety of scientific domains, including; catalysis,19–27 photocatalysis,28,29 gas adsorption,30 applications in environments,31 devices in photovoltaic and photoelectric,32,33 and drug delivery has grown. There are different types of nanocarriers like; polymeric nanoparticles (NPs), magnetic nanoparticles (MNPs), gold nanoparticles (AuNPs), and mesoporous silica nanoparticles (MSNPs).34 Preparing nanocarriers is crucial since many novel drugs have low solubility in water and low bioavailability.35 Drug delivery has had a lot of development over the last few decades. As proven, biodegradable and bioabsorbable polymers provide a magical option for many novel DDS. For instance, CD derivatives are one of the natural polymers for polymeric drug delivery carriers.36 CD-based nanocarriers can enhance the bioavailability, correct the metabolism of medication, toxicity decline, also boost the drug's half-life.37 β-CD has a versatile potential with its suitable geometry for forming inclusion complexes with innumerable drugs.38 Furthermore, β-CD is a good option for drug delivery because the size of the cavity (inner cavity diameter: 6.0–6.4 Å, outer diameter: 15.4 Å) is affordable, and the drug loading mechanism improves the bioavailability solubility and stability of cargo.39 However, to enhance the accomplishment of these DDSs, nanocarriers may be fitted with attached proteins or chains of synthetic peptides.38 Among various species of proteins, peptides do not damage the integrity of the cellular membrane.40 The attendance of the specific amino acids in the formation of peptides, such as arginine, is crucial because of its cationic guanidine groups, which are able to interact with anionic sulfate or phosphate groups over the cell membrane.41
The study represents an efficient DDS construct from β-CD nano carriers covered by 3-mercaptopropyl trimethoxy silane (MPTMS) cross-linker to deliver the antibiotic CPFX. Additionally, a new dipeptide structure, which has been synthesized in a solid phase, has been employed as an influential factor for penetrating the bacterial cells and better conjugating the structure. Notably, incorporating the peptides into the structure led to the CPFX dosage reduction, even in connection with Gram-negative bacteria. Briefly, the zone of inhibition (ZOI), minimum inhibitory concentration/minimum bactericidal concentration (MIC/MBC), and colony count experiments were accomplished to evaluate the in vitro cellular experiments of CPFX/β-CD–CR on Gram-negative (E. coli) and Gram-positive (S. aureus) bacteria. Nanocarrier integration with the drug content of 32% has enhanced the antibacterial activity of the prepared nanocarrier in comparison with the individual analogous dosage of CPFX. Notably, the derivatives of CD can use as anti-infective agents. Many CDs and their derivatives have low toxicity and resistance to enzymatic breakdown in biological fluids with GRAS (generally regarded as safe) status from the FDA. Moreover, we introduced the bacterial cellular uptake of CPFX by conjugating a dipeptide to CPFX/β-CD and its controlled release in different conditions.
2. Results and discussion
2.1. Preparation of CPFX/β-CD–CR nanocomplex
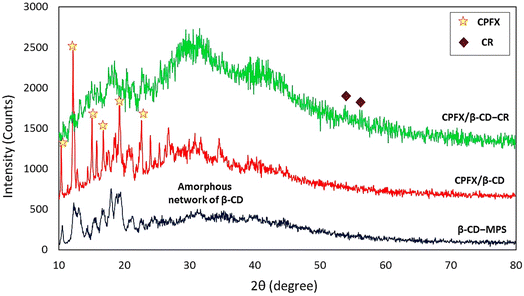
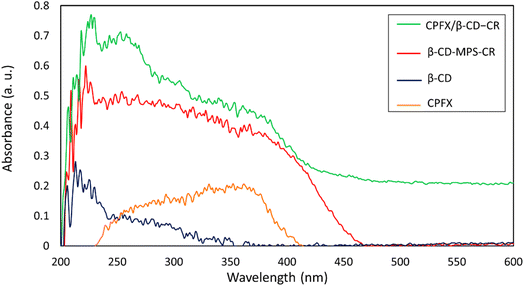
In the first stage, the surface of β-CD was modified with MPTMS to introduce –SH groups onto the surface. In the third stage, the synthesized CR dipeptide is connected to the surfaces via the foundation of a disulfide bond (Scheme 1). The dipeptide is created from 2-chlorotrityl chloride (CTC) resin through solid-phase synthesis.44 In this regard, the resin was washed with dimethyl formamide (DMF) and dichloromethane (DCM) to provide a pure and inflated CTC complex. Following this, cysteine amino acid and N,N-diisopropylethylamine (DIEA) were mixed in the DMF, and all contents were shaken at room temperature (25 °C). Then, the capping step was completed via methanol solution; subsequently, the protection group of Fmoc was eliminated with a piperidine solution in the DMF (25%). Then a blend of arginine amino acid, DIEA, and N,N,N′,N′-tetramethyl-O-(benzotriazole-1-yl)-uronium tetrafluoroborate (HBTU) has been appended to the system and shaken. Finally, the last deprotection was performed, and the CTC was separated via trifluoroacetic acid (TFA) (95%) from the complex. The structure of the synthesized CR dipeptide was confirmed by H-NMR spectroscopy (Fig. S1, in the ESI section†). To conjugate the CR dipeptide with the surface of the CPFX, a disulfide bond has been constructed between the mercaptopropyl silane (MPS) and the cysteine amino acid thiol end groups. In this step, β-CD–MPS particulates have been dispersed into distilled water through an ultrasonic ice bath. Next, dropwise ethanol was added to a mixture of CR dipeptide solution with partial quantities of hydrogen peroxide. The CPFX drug was loaded into the functionalized nanocarrier in the next stage. The inner cavity of β-CD is lipophilic, whiles the outer one is hydrophilic, which makes their complex formulation possible with lipophilic molecules. The β-CD can encapsulate molecules inside their cavity via non-covalent interactions such as electrostatic, van der Waals, hydrophobic, and hydrogen bonds to form an inclusion complex of host–guest type.42,43 Moreover, computational modeling has shown that van der Waals forces are the most important driving forces for the complexation and the moiety (piperazinyl) of the CPFX drug structure, which is included in the internal cavity of β-CD.45 Consequently, through centrifugation, the CPFX/β-CD–CR particulates were collected.46 In order to verify the prepared structures within the preparation route (presented in Scheme 1), H-NMR spectra of the neat β-CD, β-CD–MPS, and CPFX-included β-CD–MPS were prepared, as well. However, for evaluation of the CPFX inclusion (and also CR conjugation) into the structure, the H-NMR spectrum of CPFX/β-CD–CR was provided, but it was crowded and not quite interpretable. In fact, the signals in the same chemical shift areas extensively overlapped in the H-NMR spectrum. Therefore, the H-NMR spectrum of “CPFX/βCD–MPS” complex as a clue on the preparation of CPFX/βCD–CR complex was provided, as it is less crowded and the signals related to the included CPFX are quite interpretable. However, the H-NMR spectrum of CPFX/β-CD–CR has also confirmed the presence of CPFX into the structure, where the signals related to the aromatic protons of CPFX have appeared between 6.0 and 8.0 ppm. Also, the observed integrated amounts of the protons in the mentioned area can be another verification for the CPFX inclusion. As exhibited in Fig. S2–S8 (in the ESI section†), all of the shown structures in the scheme are corroborated by the NMR data.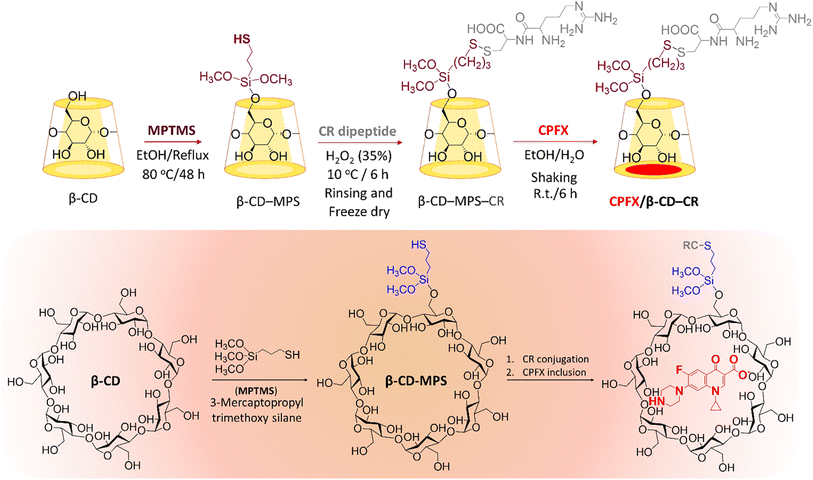 | ||
| Scheme 1 Schematic preparation of CPFX/β-CD–CR nanocomplex. Note: the “MPS” has been eliminated form final formulation (CPFX/β-CD–MPS–CR), as it does not play any significant role in the application. | ||
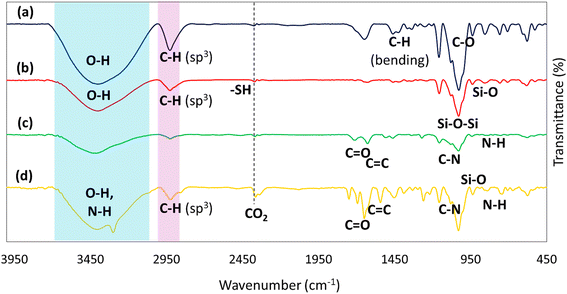
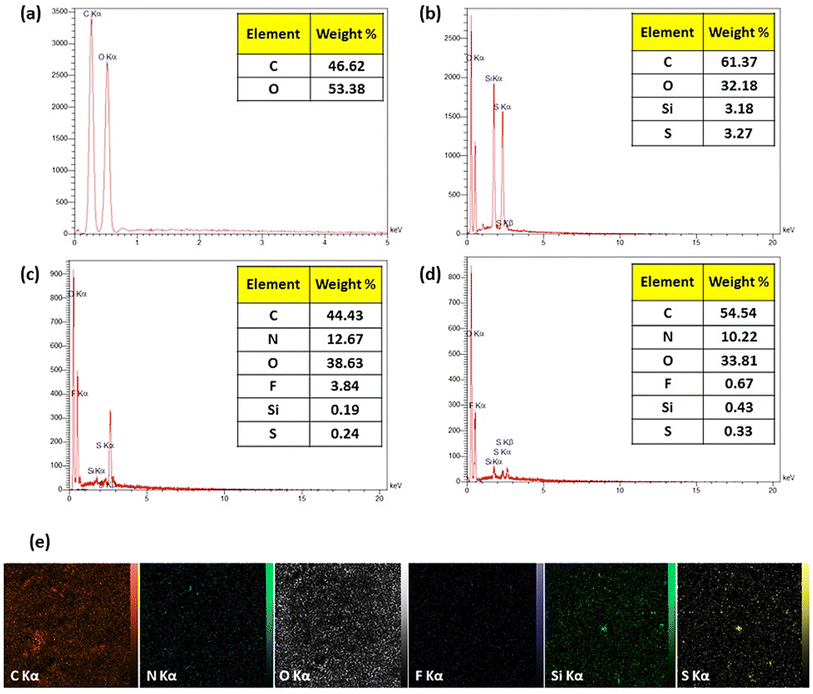
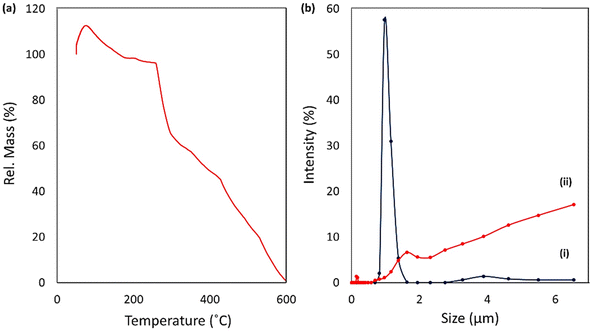
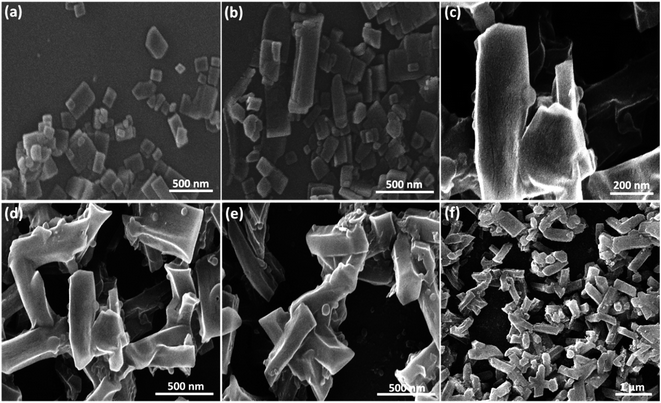
2.2. Characterization of CPFX/β-CD–CR nanocomplex
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (carbonyl) and C
O (carbonyl) and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching.49 Among the vibrations, those at 1100 cm−1 and 700 cm−1 are assigned to the vibration absorption of C–N stretching and secondary amine, respectively.50
N stretching.49 Among the vibrations, those at 1100 cm−1 and 700 cm−1 are assigned to the vibration absorption of C–N stretching and secondary amine, respectively.50
2.3. Antimicrobial properties of CPFX/β-CD–CR nanocomplex
The significant advantage of nanocarriers for targeted DDS is increasing the consistency of medicine and optimizing the half-life in blood serum.60 The carriers decline side effects disturbance and boost drug resistance against degradation. Moreover, nanocarriers exhibit more therapy advantages with low dosages of drugs, and low doses of the drug delivery to the target cell could be faster and more controlled.61 As mentioned in this literature, β-CD is ideal for drug delivery, and the drug loading mechanism improves cargo's bioavailability, solubility, and stability. Also, peptides are regarded as very effective and safe methods that do not damage the integrity of the cellular membrane. They can pass via different cell types of membranes. Some amino acids' attendance in forming the peptides, such as arginine, is crucial because it can interact with anionic sulfate or phosphate groups over the cell membrane. The MPTMS is the agent to create a disulfide bond between cysteine amino acids and can increase the antimicrobial effects. This study provided an efficient DDS construct from β-CD NPs covered by an MPTMS cross-linker to deliver the antibiotic CPFX. Furthermore, a new dipeptide chain, which has been synthesized in the solid phase, was utilized as a more efficient factor for entering the bacteria cells and conjugating them to the structure.| X = (A + 0.001)/(0.0845 × 0.05) | (1) |
| C = X × 1/1000 | (2) |
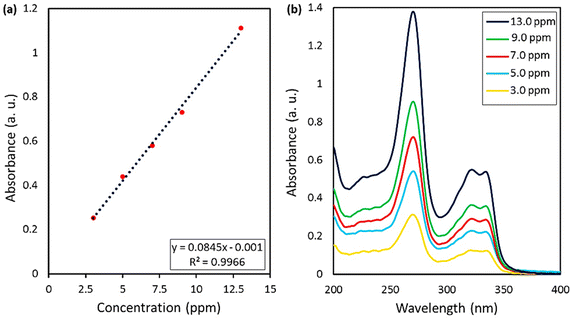 | ||
| Fig. 7 (a) Calibration curve of the standard CPFX solutions, obtained by UV-vis spectrophotometry, (b) UV-vis spectra of CPFX at various concentration values. | ||
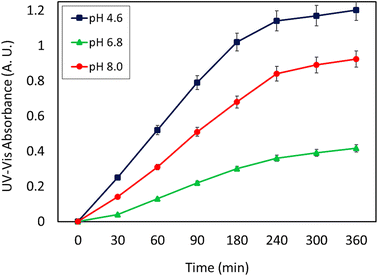
As can be seen, the maximum value of the released CPFX has been shown in Table 1, which was regarded as the approximation of the CPFX content (ca. 30 ± 0.2) wt% in the CPFX/β-CD–CR system. It can be deduced that inside the bacteria cell (following endocytosis), releasing the encapsulated CPFX has occurred in the acidic condition, which is a foundation of the entire released quantity (100%), and the remaining extents were compared to this measure. As presented in Fig. 8, the maximum drug release of CPFX from CPFX/β-CD–CR nano-cargo occurred when the particles were dispersed in an acidic medium.
| Entry | Conditiona | Absorbance (a.u.) | Concentration (mg mL−1) | CPFX release (%) | Rel. Eb (%) |
|---|---|---|---|---|---|
| a CPFX release was investigated at 37 °C and 4 h.b % Rel. E: relative error percentage for three identical samples.c Complete value of release (100%) was considered for most of the drugs released, and the remaining values were compared to the main value. | |||||
| 1 | ACB (0.1 M, pH = 4.6) | 1.203 | 0.301 | 100c | 3.5 |
| 2 | PBS (0.1 M, pH = 6.8) | 0.416 | 0.104 | 34.55 | 5.2 |
| 3 | PBS (0.1 M, pH = 8.0) | 0.924 | 0.231 | 76.74 | 4.6 |
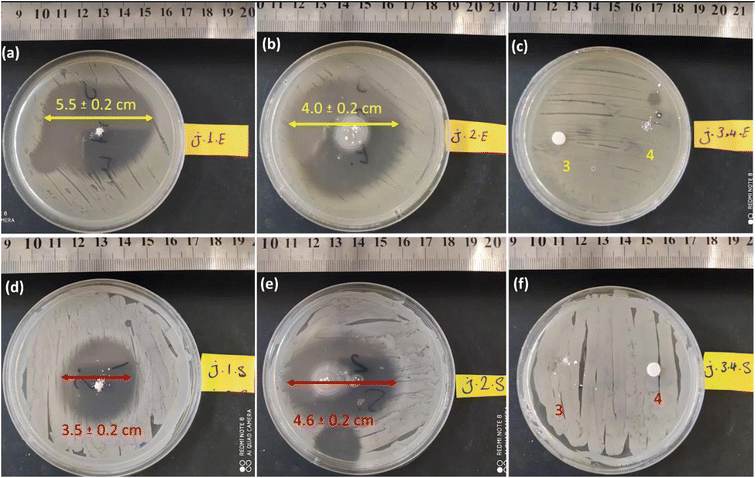
This assay tested similar dosages of CPFX/β-CD–CR, β-CD, CR, and CPFX. The powdery water-undissolved samples were situated on the agar plates containing bacterial cells. In this regard, 2.0 mg of powder samples were transformed onto agar gels containing S. aureus and E. coli. The disks were incubated at 37 °C, with a humidity of 95% for 24 h. After incubation, the inhibition zone diameter was measured. Fig. 9a displays that the ZOI of E. coli was larger than S. aureus, which is attributed to the CPFX/β-CD–CR therapeutic nano-bioconjugate. The growth zone inhibitory of CPFX/β-CD–CR particles on the E. coli and the S. aureus bacterial cells was 5.5 ± 0.2 cm and 3.5 ± 0.2 cm, respectively.
In contrast, as shown in Fig. 9b, the ZOI measurements for the individual CPFX on the E. coli were reduced to 4.0 ± 0.2 cm. Concludingly, the CPFX/β-CD–CR therapeutic nano-bioconjugate with about 30% drug content has higher antibacterial activity than the individual CPFX with the same dosage. Significantly, due to the thicker, strong walls of Gram-negative E. coli bacteria, CPFX hardly penetrates the membrane; hence, the ZOI of the E. coli samples is larger than in the case of S. aureus. In addition, the dipeptide sequence and polymer effects on the ZOI test have been evaluated. In this respect, the individual dipeptide and polymer did not demonstrate the ZOI onto S. aureus and E. coli cell lines, indicating no antibacterial activity of the dipeptide and polymer (Fig. 9c and f).
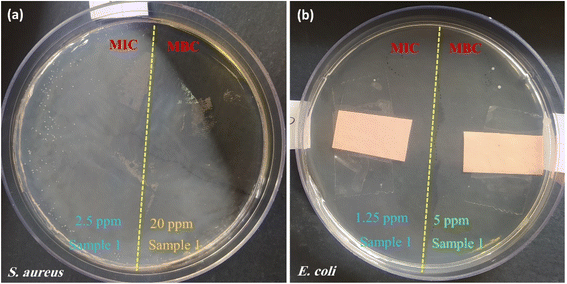
| Bacterial strain | Sample 1 (CPFX/β-CD–CR) | Sample 2 (ciprofloxacin) |
|---|---|---|
| S. aureus | MIC (μg mL−1): 2.5 | MIC (μg mL−1): 0.5 |
| MBC (μg mL−1): 20.0 | MBC (μg mL−1): 2.0 | |
| E. coli | MIC (μg mL−1): 1.25 | MIC (μg mL−1): 0.02 |
| MBC (μg mL−1): 5.0 | MBC (μg mL−1): 0.04 |
Further assessments on the antibacterial characteristics of the CPFX/β-CD–CR therapeutic nano-bioconjugates were accomplished through the colony count assay.66 In this account, the linear cell culture pattern was implemented on the agar gel. Then, the sample was spread throughout the disks and incubated at 37 °C, with 95% humidity for 48 h. The results revealed that the S. aureus population on the CPFX/β-CD–CR therapeutic nano-bioconjugates-containing disk after 30, 60 and 180 minutes was 250.0, 150.0 and 5.0 (×108 CFU mL−1), respectively, and E. coli bacteria population on the CPFX/β-CD–CR therapeutic nano-bioconjugates-containing disk during those times was 10.0, 5.0 and diminished to zero. Notably, the CPFX/β-CD–CR nano-cargo's cell-killing activity with about 30% drug content was analogous to the individual CPFX with a similar concentration. As expected, no bacterial population difference was seen between the control sample and the CR-containing disk.
3. Experimental section
3.1. Materials and instruments
All chemicals and devices utilized in this study are listed in Tables 3 and 4.| Materials | Brand & purity |
|---|---|
| Beta cyclodextrin | Sigma-Aldrich |
| (3-Mercaptopropyl)trimethoxy silane (MPTMS) | Sigma-Aldrich, 97.0% |
| Ciprofloxacin | Sigma-Aldrich |
| N,N,N′,N′-Tetramethyl-O-(benzotriazol-1-yl)uranium tetrafluoroborate (TBTU) | Sigma-Aldrich, ≥97.0% |
| Fmoc-Arg(Pbf)-OH | Sigma-Aldrich, ≥98.0% |
| Fmoc-Cys(Trt)-OH | Sigma-Aldrich, ≥95.0% |
| 2-Chlorotrityl chloride (CTC) resin | Sigma-Aldrich, 97.0% |
| N,N-Diisopropylethylamine (DIPEA) | Sigma-Aldrich, ≥99.0% |
| N,N-Dimethylformamide (DMF) | Sigma-Aldrich, 99.8% |
| Dichloromethane (DCM) | Sigma-Aldrich, 99.9% |
| Piperidine | Sigma-Aldrich, ≥99.0% |
| Trifluoroacetic acid (TFA) | Sigma-Aldrich, ≥99.0% |
| Synthetic ethanol | Merck, 99% |
| Synthetic methanol | Merck, 99% |
| Equipment | Brand |
|---|---|
| FT-IR spectroscopy | Shimadzu FT-IR-8400S |
| EDX spectroscopy | VEGA-TESCAN-XMU |
| TGA analysis | Bahr-STA 504 |
| DLS analysis | Horiba (SZ-100) |
| XRD | DRON-8 X-ray diffractometer |
| FESEM | Zeiss Sigma |
| Solid state UV-vis spectroscopy | Shimadzu-UV-2550/220v |
| UV-vis spectroscopy | Beckman DU640 |
| Ultrasonic cleaner bath | BANDELIN electronic GmbH & Co. KG |
| Centrifuge | Beckman Coulter GmbH |
| Oven | Genlab Ltd |
| Freeze drier | KASSEL Machinery (Zhejiang) Co., Ltd |
| Rotary evaporator | Heidolph Instruments GmbH & Co. KG |
| Incubator | Sh, Noor Sanat Ferdos |
| Shaker | VWR 5000 |
| Autoclave (for sterilization) | Reyhan Teb, 2KW-220v |
| Vacuum pump | Heidoph persia LQ1 |
3.2. Preparation methods
3.3. Cellular experiments
3.4. Sample preparation for characterization
For electron microscopy, the particles were dispersed in minimum amount of pure ethanol, and ultrasonicated with a cleaner bath (50 kHz, 100 W L−1) for 5 min, at an ambient temperature. Afterward, 50 μL of each sample was poured onto a glass laminate, dried in vacuum oven, and then sent to the SEM imaging laboratory. The same procedure was followed for the biological tests, with the difference that the environment in which the whole sample was dispersed in was DMEM instead of ethanol, and the temperature was maintained in a range of 0–5 degrees Celsius (ice bath). The samples were maintained on ice and subjected to living cells in an incubator. For the UV-vis studies, the nanoparticles were dispersed in buffered media, stirred, and finally centrifuged (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min) to collect the carrier coils. Then, they were filtered with a Whatman filter paper. Also, the blank sample was filtered with Whatman filter paper so as to eliminate the error related to the particulate matter of the paper filter. At last, the solution containing 5.0 mg of CPFX was diluted for a correct UV-vis spectroscopy study.
000 rpm, 10 min) to collect the carrier coils. Then, they were filtered with a Whatman filter paper. Also, the blank sample was filtered with Whatman filter paper so as to eliminate the error related to the particulate matter of the paper filter. At last, the solution containing 5.0 mg of CPFX was diluted for a correct UV-vis spectroscopy study.
4. Conclusions
We designed a novel DDS which is a complex of nanoscale β-CD with CPFX conjugated to a cell adhesive dipeptide (CR). The nanocomplex's biological activities versus Gram-positive (S. aureus) and Gram-negative (E. coli) bacteria were studied. The surface of β-CD was initially modified with MPTMS to introduce –SH groups onto the surface to provide this biocompatible DDS. In addition, the CPFX was loaded to the nanoscale β-CD, and finally, via the formation of a disulfide bond, the synthesized CR dipeptide is linked to the surfaces of CPFX/β-CD cargo. This dipeptide sequence was synthesized in the solid phase on CTC resin with high purity. The peptide structures are highly effective, safe techniques and can penetrate cells non-invasive. Also, the usage of this novel-designed strategy prepared a plethora of benefits compared to the usage of free CPFX for bacterial growth prohibition. The primary benefit was using lower doses of antibiotics for similar therapeutic effects. Herein, every essential characterization, including; FT-IR, EDX, TGA, zeta potential, UV-DRS, XRD, FESEM and UV-vis absorbance, was accomplished the received results confirmed with the designed structure.Furthermore, the drug content and release were investigated with precise analytical methods. The drug loading efficacy was 32%, and the main release was revealed in an acidic environment. Overall, according to the antimicrobial results, it can be deduced that the efficacy of the CPFX is remarkably gained via the complexation of nanoscale materials and engineered dipeptides. The growth zone inhibitory of CPFX/β-CD–CR particles on the E. coli and the S. aureus bacterial cells was 5.5 ± 0.2 cm and 3.5 ± 0.2 cm, respectively. Moreover, the half-life of the provided cargo related to the degradability of the used components in the system ought to be assessed. Following complete degradation, the magnitude of released components' toxicity may be another significant challenge.
Author contributions
Reza Taheri-Ledari: conceptualization, reviewing, and editing of the main draft, supervision and project administration, and software; Farinaz Jalali: performed all practical sections and also in preparation of the initial draft; Leili Heidari: performed all antimicrobial sections and interpreted the obtained results; Fatemeh Ganjali: wrote the initial draft and interpreted the obtained results; Fereshteh Rasouli Asl: participated in partial parts of the bench work and also in preparation of the initial draft; Simindokht Zarei-Shokat: participated in partial parts of the bench work; Mohadeseh Forouzandeh-Malati: graphics and software; Adibeh Mohammadi: participated in the revision stage; Ali Maleki: managed all sections of the work, supervision, project administration, and financial support.Funding
This work was partially supported by the Iran University of Science and Technology.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
The authors acknowledge the partial support from the Research Council of the Iran University of Science and Technology (IUST).References
- V. Soltaninejad, M. R. Ahghari, R. Taheri-Ledari and A. Maleki, Langmuir, 2021, 37, 4700–4713 CrossRef CAS PubMed.
- M. Tian, X. He, Y. Feng, W. Wang, H. Chen, M. Gong, D. Liu, J. L. Clarke and A. van Eerde, Antibiotics, 2021, 10, 539 CrossRef CAS PubMed.
- A. Maleki, R. Taheri-Ledari, R. Eivazzadeh-Keihan, M. de la Guardia and A. Mokhtarzadeh, Curr. Org. Synth., 2019, 16, 765–771 CrossRef CAS PubMed.
- K. Drlica and X. Zhao, Expert Rev. Anti-Infect. Ther., 2021, 19, 601–618 CrossRef CAS PubMed.
- W. Duan, H. Cui, X. Jia and X. Huang, Sci. Total Environ., 2022, 153178 CrossRef CAS PubMed.
- T. D. Pham, Z. M. Ziora and M. A. Blaskovich, Medchemcomm, 2019, 10, 1719–1739 RSC.
- M. Vidyavathi and G. Srividya, Int. J. Appl. Pharm., 2018, 10, 6 CrossRef CAS.
- A. K. Al-Buriahi, M. M. Al-shaibani, R. M. S. R. Mohamed, A. A. Al-Gheethi, A. Sharma and N. Ismail, Journal of Water Process Engineering, 2022, 47, 102725 CrossRef.
- G.-F. Zhang, X. Liu, S. Zhang, B. Pan and M.-L. Liu, Eur. J. Med. Chem., 2018, 146, 599–612 CrossRef CAS PubMed.
- A. Rehman, W. M. Patrick and I. L. Lamont, J. Med. Microbiol., 2019, 68, 1–10 CrossRef CAS PubMed.
- R. Taheri-Ledari, W. Zhang, M. Radmanesh, S. S. Mirmohammadi, A. Maleki, N. Cathcart and V. Kitaev, Small, 2020, 16, 2002733 CrossRef CAS.
- R. Taheri-Ledari, W. Zhang, M. Radmanesh, N. Cathcart, A. Maleki and V. Kitaev, J. Nanobiotechnol., 2021, 19, 239 CrossRef CAS PubMed.
- A. Patil, V. Mishra, S. Thakur, B. Riyaz, A. Kaur, R. Khursheed, K. Patil and B. Sathe, Curr. Nanosci., 2019, 15, 137–146 CrossRef CAS.
- Z. A. Bhutta, M. Shoaib, A. Kanwal, A. Ashar, A. Mahfooz, R. Ahmed, M. Ali, M. F.-e.-A. Kulyar and K. Li, in Advancements in Controlled Drug Delivery Systems, IGI Global, 2022, pp. 97–133 Search PubMed.
- R. Taheri-Ledari, E. Zolfaghari, S. Zarei-Shokat, A. Kashtiaray and A. Maleki, Commun. Biol., 2022, 5, 995 CrossRef CAS PubMed.
- H. Lu, J. Wang, T. Wang, J. Zhong, Y. Bao and H. Hao, J. Nanomater., 2016, 2016, 5762431 Search PubMed.
- W. Zhang, R. Taheri-Ledari and F. Ganjali, et al., Heliyon, 2022, 8, e09577 CrossRef CAS PubMed.
- R. Taheri-Ledari, in Heterogeneous Micro and Nanoscale Composites for the Catalysis of Organic Reactions, Classification of micro and nanoscale composites, Elsevier, 2022, pp. 1–21 Search PubMed.
- R. Taheri-Ledari, M. Saeidirad, F. S. Qazi, A. Fazeli, A. Maleki and A. E. Shalan, RSC Adv., 2021, 11, 25284–25295 RSC.
- R. Taheri-Ledari, J. Rahimi, A. Maleki and A. E. Shalan, New J. Chem., 2020, 44, 19827–19835 RSC.
- A. Maleki, R. Taheri-Ledari and M. Soroushnejad, ChemistrySelect, 2018, 3, 13057–13062 CrossRef CAS.
- R. Kore, A. D. Sawant and R. D. Rogers, ACS Sustainable Chem. Eng., 2021, 9, 8797–8802 CrossRef CAS.
- R. Taheri-Ledari, J. Rahimi and A. Maleki, Ultrason. Sonochem., 2019, 59, 104737 CrossRef PubMed.
- R. Taheri-Ledari, S. S. Mirmohammadi, K. Valadi, A. Maleki and A. E. Shalan, RSC Adv., 2020, 10, 43670–43681 RSC.
- R. Taheri-Ledari and A. Maleki, New J. Chem., 2021, 45, 4135–4146 RSC.
- W. Zhang, R. Taheri-Ledari, F. Ganjali, F. H. Afruzi, Z. Hajizadeh, M. Saeidirad, F. S. Qazi, A. Kashtiaray, S. S. Sehat and M. R. Hamblin, Heliyon, 2022, e09577 CrossRef CAS PubMed.
- F. Ganjali, A. Kashtiaray, S. Zarei-Shokat, R. Taheri-Ledari and A. Maleki, Nanoscale Adv., 2022, 4, 1263–1307 RSC.
- R. Taheri-Ledari, K. Valadi, S. Gharibi and A. Maleki, Mater. Res. Bull., 2020, 130, 110946 CrossRef CAS.
- R. Taheri-Ledari and A. Maleki, in Magnetic Nanoparticle-Based Hybrid Materials, Elsevier, 2021, pp. 619–636 Search PubMed.
- X. Zhang, Z. Chen, X. Liu, S. L. Hanna, X. Wang, R. Taheri-Ledari, A. Maleki, P. Li and O. K. Farha, Chem. Soc. Rev., 2020, 49, 7406–7427 RSC.
- Z. Hajizadeh, K. Valadi, R. Taheri-Ledari and A. Maleki, ChemistrySelect, 2020, 5, 2441–2448 CrossRef CAS.
- R. Taheri-Ledari, K. Valadi and A. Maleki, Prog. Photovoltaics, 2020, 28, 956–970 CAS.
- K. Valadi, S. Gharibi, R. Taheri-Ledari, S. Akin, A. Maleki and A. E. Shalan, Environ. Chem. Lett., 2021, 19, 2185–2207 CrossRef CAS.
- A. Banerjee, J. Qi, R. Gogoi, J. Wong and S. Mitragotri, J. Controlled Release, 2016, 238, 176–185 CrossRef CAS PubMed.
- K. E. Bremmell and C. A. Prestidge, Drug Dev. Ind. Pharm., 2019, 45, 349–358 CrossRef CAS.
- I.-H. Cho, D. H. Kim and S. Park, Biomater. Res., 2020, 24, 1–12 CrossRef PubMed.
- G. Crini, S. Fourmentin, É. Fenyvesi, G. Torri, M. Fourmentin and N. Morin-Crini, Environ. Chem. Lett., 2018, 16, 1361–1375 CrossRef CAS.
- V. J. Stella and R. A. Rajewski, Int. J. Pharm., 2020, 583, 119396 CrossRef CAS PubMed.
- T. Jia, S. Huang, C. Yang and M. Wang, Mol. Pharm., 2017, 14, 2529–2537 CrossRef CAS PubMed.
- W. Kauffman, S. Guha and W. C. Wimley, Nat. Commun., 2018, 9, 1–10 CrossRef CAS PubMed.
- C. Bechara and S. Sagan, FEBS Lett., 2013, 587, 1693–1702 CrossRef CAS PubMed.
- B. Khan, S. Kumar, N. Sanbhal, S. Abdullah, J. Hussain, N. Chowdhry, A. Q. Ansari and M. M. Surahio, Biomed. Mater. Devices, 2022, 1–12 Search PubMed.
- R. Jelić, M. Tomović, S. Stojanović, L. Joksović, I. Jakovljević and P. Djurdjević, Monatsh. Chem., 2015, 146, 1621–1630 CrossRef.
- W. Zhang, R. Taheri-Ledari, Z. Hajizadeh, E. Zolfaghari, M. R. Ahghari, A. Maleki, M. R. Hamblin and Y. Tian, Nanoscale, 2020, 12, 3855–3870 RSC.
- Z. Aytac, S. Ipek, I. Erol, E. Durgun and T. Uyar, Colloids Surf., B, 2019, 178, 129–136 CrossRef CAS PubMed.
- R. Taheri-Ledari, A. Fazeli, A. Kashtiaray, S. Salek Soltani, A. Maleki and W. Zhang, Langmuir, 2021, 38, 132–146 CrossRef.
- R. Xu, X. Lin, J. Xu and C. Lei, Ionics, 2020, 26, 3359–3365 CrossRef CAS.
- M. Hezarjaribi, G. Bakeri, M. Sillanpää, M. J. Chaichi, S. Akbari and A. Rahimpour, Environ. Sci. Pollut. Res., 2021, 28, 51808–51825 CrossRef CAS PubMed.
- R. Nithya and N. Meenakshi Sundaram, Int. J. Nanomed., 2015, 10, 119–127 CAS.
- B. A. Mohamed and P. Janaki, J. Appl. Nat. Sci., 2021, 13, 110–123 CrossRef CAS.
- H. Sadaquat and M. Akhtar, J. Inclusion Phenom. Macrocyclic Chem., 2020, 96, 333–351 CrossRef CAS.
- M. Sajid, M. Asif, N. Baig, M. Kabeer, I. Ihsanullah and A. W. Mohammad, Journal of Water Process Engineering, 2022, 47, 102815 CrossRef.
- H. Butt, M. U. Minhas, K. U. Khan, M. Sohail, I. Khalid, S. Rehmani and M. Suhail, Polym. Bull., 2022, 1–22 Search PubMed.
- M. Zarif, A. Afidah, J. Abdullah and A. Shariza, Biomed. Res., 2012, 23, 513–520 CAS.
- J. Souri, H. Almasi, H. Hamishehkar and S. Amjadi, Lwt, 2021, 151, 112174 CrossRef CAS.
- J. Chen, X. Qin, S. Zhong, S. Chen, W. Su and Y. Liu, Molecules, 2018, 23, 1179 CrossRef.
- D. Macocinschi, D. Filip, S. Vlad, C. G. Tuchilus, A. F. Cristian and M. Barboiu, J. Mater. Chem. B, 2014, 2, 681–690 RSC.
- Y. Yang, D. Nie, Y. Liu, M. Yu and Y. Gan, Drug Discovery Today, 2019, 24, 575–583 CrossRef CAS PubMed.
- T. Knoell, J. Safarik, T. Cormack, R. Riley, S. Lin and H. Ridgway, J. Membr. Sci., 1999, 157, 117–138 CrossRef CAS.
- J. Siepmann and F. Siepmann, Int. J. Pharm., 2008, 364, 328–343 CrossRef CAS PubMed.
- P. Misiak, K. H. Markiewicz, D. Szymczuk and A. Z. Wilczewska, Polymers, 2020, 12, 2620 CrossRef CAS PubMed.
- T. Kaewchomphunuch, T. Charoenpichitnunt, V. Thongbaiyai, N. Ngamwongsatit and K. Kaeoket, BMC Vet. Res., 2022, 18, 1–13 CrossRef.
- P. Singh, A. U. Mirza, A. H. Mondal, K. Mukhopadhyay and N. Nishat, J. Appl. Polym. Sci., 2022, 139, 51749 CrossRef CAS.
- R. Sonohara, N. Muramatsu, H. Ohshima and T. Kondo, Biophys. Chem., 1995, 55, 273–277 CrossRef CAS PubMed.
- P. Parvekar, J. Palaskar, S. Metgud, R. Maria and S. Dutta, Biomater. Invest. Dent., 2020, 7, 105–109 CAS.
- S. Bala Subramaniyan, S. Ramesh, S. Rajendran and A. Veerappan, Bioconjugate Chem., 2021, 32, 1823–1833 CrossRef CAS PubMed.
- R. Taheri-Ledari, M. R. Ahghari, F. Ansari, M. Forouzandeh-Malati, S. S. Mirmohammadi, S. Zarei-Shokat, S. Ramezanpour, W. Zhang, Y. Tian and A. Maleki, Nanoscale Adv., 2022, 4, 4418–4433 RSC.
- A. E. Mekky and S. M. Sanad, Bioorg. Chem., 2020, 102, 104094 CrossRef CAS PubMed.
- M. Shahbuddin, S. S. Mahamad and R. A. Raus, Biological and Natural Resources Engineering Journal, 2021, 5, 13–24 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: H-NMR spectra of the synthesized CR dipeptide structure and successively modified beta cyclodextrin. See DOI: https://doi.org/10.1039/d2ra05822g |
| This journal is © The Royal Society of Chemistry 2022 |