Open Access Article
Open Access ArticleFacile, rapid and efficient isolation of circulating tumor cells using aptamer-targeted magnetic nanoparticles integrated with a microfluidic device†
Abolghasem Abbasi Kajani‡
 a,
Laleh Rafiee‡b,
Mohamadmahdi Samandaribc,
Masoud Ayatollahi Mehrgardi
a,
Laleh Rafiee‡b,
Mohamadmahdi Samandaribc,
Masoud Ayatollahi Mehrgardi d,
Bahare Zarrinb and
Shaghayegh Haghjooy Javanmard
d,
Bahare Zarrinb and
Shaghayegh Haghjooy Javanmard *b
*b
aDepartment of Biotechnology, Faculty of Biological Science and Technology, University of Isfahan, Isfahan, 81746-73441, Iran
bApplied Physiology Research Center, Cardiovascular Research Institute, Isfahan University of Medical Sciences, Isfahan, 81746-73461, Iran. E-mail: sh_haghjoo@med.mui.ac.ir; Fax: +98-3136692836; Tel: +98-3137929128
cDepartment of Biomedical Engineering, University of Connecticut, Farmington, CT 06030, USA
dDepartment of Chemistry, University of Isfahan, Isfahan, 81746-73441, Iran
First published on 16th November 2022
Abstract
Facile and sensitive detection and isolation of circulating tumor cells (CTCs) was achieved using the aptamer-targeted magnetic nanoparticles (Apt-MNPs) in conjugation with a microfluidic device. Apt-MNPs were developed by the covalent attachment of anti-MUC1 aptamer to the silica-coated magnetic nanoparticles via the glutaraldehyde linkers. Apt-MNPs displayed high stability and functionality after 6 months of storage at 4 °C. The specific microfluidic device consisting of mixing, sorting and separation modules was fabricated through conventional photo- and soft-lithography by using polydimethylsiloxane. The capture efficiency of Apt-MNPs was first studied in vitro on MCF-7 and MDA-MB-231 cancer cell lines in the bulk and microfluidic platforms. The cell capture yields of more than 91% were obtained at the optimum condition after 60 minutes of exposure to 50 μg mL−1 Apt-MNPs with 10 to 106 cancer cells in different media. CTCs were also isolated efficiently from the blood samples of breast cancer patients and successfully propagated in vitro. The isolated CTCs were further characterized using immunofluorescence staining. The overall results indicated the high potential of the present method for the detection and capture of CTCs.
1. Introduction
Circulating tumor cells (CTCs), including viable or apoptotic cancer cells released from the primary and metastatic tumor sites into the peripheral blood or lymphatic system, play a critical role in cancer occurrence and metastasis.1,2 This type of cancer cells has been considered recently as an important indicator or biomarker for the early detection and prognosis of cancers.3,4 In this regard, the noninvasive isolation of CTCs, a kind of liquid biopsy, has received widespread attention for detection, characterization, and monitoring of cancers.5,6 However, the rapid and accurate isolation of CTCs is a serious challenge due to their extremely low abundance and heterogeneous nature. Therefore, tremendous efforts have been conducted toward the development of simple, convenient and efficient methods to solve these limitations.7To isolate CTCs from the other cells present in patient circulation, the difference between physical and biological properties of the tumor cells compared to the normal blood cells can be targeted. The physical properties which have been utilized for isolation of CTCs are density, size, and surface electrical properties.8 Generally, the density of CTCs are lower than red blood cells while it is close to mononuclear white blood cells' density. Additionally, most of the circulating tumor cells are larger than blood cells and have higher level of surface foldings, causing a differential surface ion accumulation compared to the blood cells, when they are located in an electric field.9 Although all of these physical properties have been extensively used for separation of CTCs from the patients' circulation,10 in many conditions, mononuclear white blood cells are also separated with CTCs due to their similar physical properties, which makes clinical decision challenging.2 On the other hand, specific biological properties of CTCs can be targeted using affinity-based approaches. In affinity-based methods, CTCs are captured by the surfaces or particles tagged with specificly designed biomolecules toward the specific surface markers of CTCs.11 Despite physical separation approaches, high specifity of biological methods made them more successful in clinical trials. As a result, the only FDA approved liquid biopsy platform by this time is CellSearch, which utilizes immunomagnetic isolation of CTCs.12
The immune-magnetic isolation of CTCs is one of the most attractive methods developed until now. The established method of magnetic separation has found widespread clinical applications due to its simplicity, low cost and minimal adverse effects on the captured cells.13,14 Owing to the use of specific and high-affinity interactions between the recognition agents and cancer cell receptors as well as the efficient magnetic fields, this method is appropriate for the facile, specific and precise detection of CTCs.15,16 Generally, antibodies with high affinity against specificly expressed receptors on the surface of CTCs are tagged into magnetic micro- or nano-particles and mixed with the body fluid to bind into the tumor cells.17–21 Subsequent application of a magnetic field can attract the CTC-attached magnetic particles and separate them from the normal cells. However, the clinical application of antibodies has been considerably limited due to their high cost and instability. On the other hand, the multiple batch processing of large volume blood samples with numerus normal blood cells and rare CTC may result in depletion of CTCs and reduced accuracy. Therefore, tremendous efforts have been conducted toward the development of alternative recognition agents and processing methods with comparable sensitivity and specificity.13
To address the challenge associated with application of antibodies, DNA aptamers have been considered as an attractive alternative for the reliable detection of various targets such as biomolecules, ions and whole cells.22–24 This newly emerged class of nucleic acid ligands, known as the antibody-like molecules, have found growing applications in diagnosis and therapy due to their advantages such as small size, facile synthesis and manipulation, high thermal and chemical stability, low immunogenicity, and high binding affinity and specificity.25 The increasing reports about the development and use of specific aptamers against different cancer cell receptors such as EGFR,26 EpCAM,27,28 PSMA,29 PTK7,30,31 sgc8c32,33 and sLex-AP,34 clearly indicate high potential of aptamers as the recognition agent.
Mucin 1 (MUC1) is a high molecular weight and glycosylated transmembrane protein belonging to the mucin family that is expressed in the majority of human epithelial cells with a dramatic overexpression in epithelial tumor cells, especially breast adenocarcinoma that has been attributed to their tumor aggressiveness.35–38 The circulating MUC1 fragments in the bloodstream of cancer patients are commonly used as a biomarker for determination of cancer stage and monitoring of therapy responses.39 Among 75 tumor-associated antigens studied by the National Cancer Institute (NCI), MUC1 has been reported as the second-best potential target for the development of vaccines against cancers.40,41 Furthermore, a short single-stranded DNA aptamer with high affinity for the MUC1 receptor has been reported previously.42 Due to these unique properties, MUC1 receptor was selected in this study as the target for detection and capture of CTCs based on this anti-MUC1 aptamer.
To harness the full potential of CTCs separation approaches with high accuracy, microfluidic devices have been implemented.10 The continuous flow of body fluids inside microchannels reduce the size scale of separation systems down to the dimentions comparable to the cell size, enhancing the possibility of the cells exposure to the separation field and consequently their isolation in designated microchannels.43,44 Furthermore, the continuous nature of assays performed using microfluidic tools provides the opportunity of their serial integration for multiple cell manipulation steps to increase the accuracy of CTCs isolation and detection without several batch processing steps.45 However, the serial integration usually comes with a cost of system complexity and higher chance of clogging due to the presence of multiple connections.
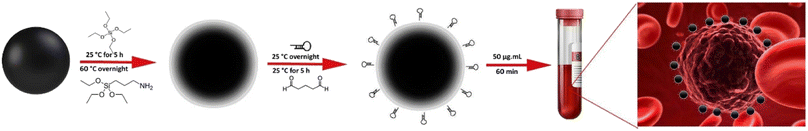
In the present study, a novel CTCs capture platform was developed based on CTCs targeting with the MUC1-aptamer conjugated MNPs within a microfluidic device. A new and simple approach including the covalent attachment of MUC1 aptamer into the surface of silica coated MNPs through glutaraldehyde linker was employed for development of the stable and functional nanoplatforms. An microfluidic network with integrated mixing, sorting and separation modules in a single microchip was also developed and used to investigate the efficiency of the integrated system for CTCs isolation. Considering the overexpression of MUC1 receptors on the surface of breast cancer cells, MCF-7 and MDA-MB-232 cell lines were used as the positive controls. Regarding the expression of MUC1 by the normal epithelial cells and fibroblast cells,46 the potential false detection of these cells should be considered. Therefore, the capture efficiency of normal human umbilical vein endothelial cells (HUVEC) and cancer-associated fibroblast (CAF) cells was also investigated, as the typical epithelial and fibroblast cells, respectively, with the critical role in the invasion and metastasis of cancer cells.47,48 In our opinion, this new platform has the potential to develop a simple, fast, precise and cost-effective method for CTCs isolation.
2. Experimental section
2.1. Materials
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 3-aminopropyltriethoxysilane (APTES), Hoechst 33258 solution, dimethyl sulfoxide (DMSO), red blood cell (RBC) lysis buffer, tetraethyl orthosilicate (TEOS) and Triton X-100 were obtained from Sigma (Germany). Absolute ethanol, ammonia solution (25%), ferric chloride hexahydrate (FeCl3·6H2O), ferrous chloride tetrahydrate (FeCl2·4H2O), and glutaraldehyde were purchased from Merck (Darmstadt, Germany). 0.25% (w/v) trypsin-0.03% (w/v) EDTA solution, antibiotic (penicillin-streptomycin) solution, fetal bovine serum (FBS), phosphate buffered saline (PBS) and RPMI 1640 medium were purchased from Gibco (Invitrogen, Grand Island, NY). FITC anti-human CD326 (EpCAM) antibody, and PE anti-CD45 antibody were purchased from BioLegend (San Diego, CA). The sequence of anti-MUC1 aptamer (5′ NH2C6-TTTGCAGTTGATCCTTTGGATACCCTGG 3′) was designed based on the previously report42 and synthesized by Bioneer (Daejeon, Republic of Korea). EasySep™ Direct Human CTC Enrichment Kit was purchased from STEMCELL Technologies Inc. (Vancouver, Canada). SU-8 negative photoresist was purchased from Microchem (Sunnyvale, CA, USA). Sylgard 184 was purchased from Dow Corning (Midland, Michigan, USA). All of the aqueous solutions were prepared with double-distilled water.2.2. Synthesis and surface engineering of MNPs
Magnetite nanoparticles were synthesized based on the chemical co-precipitation method according to the previous report.49 Briefly, FeCl3 and FeCl2 (molar ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were dissolved in 200 mL distilled water and degassed under nitrogen atmosphere at 60 °C for 20 min. After the addition of 6 mL ammonium hydroxide (25% w/w), the reaction was proceeded under sonication and stirring at 60 °C for 60 min. The resulted nanoparticles were separated magnetically and washed with deionized water to decrease the pH of the solution to 7. For silica coating, the resulted nanoparticles were added into 40 mL absolute ethanol and completely dispersed by sonication under the nitrogen atmosphere. Then, 6 mL deionized water, 3 mL ammonium solution (25% w/w), and 400 μL TEOS were sequentially added into the solution under vigorous stirring and the reaction continued for 5 h at room temperature. The silica-coated MNPs (SiMNPs) were separated, washed with deionized water and dried at 40 °C overnight before functionalization with amine groups. For this aim, 20 mg MNPs were first dispersed in 20 mL ethanol under sonication and 200 μL APTES was added into the solution and refluxed at 60 °C, overnight. The resulted amine-functionalized MNPs were separated, washed with deionized water and dispersed in 10 mL phosphate buffer (50 mM, pH 7.0) containing glutaraldehyde (2%, v/v) and incubated at 25 °C under stirring for 5 h. MNPs were then separated magnetically, washed with PBS and dispersed in 1 mL phosphate buffer (50 mM, pH 8.5). The NH2-terminated anti-MUC1 aptamers were subsequently added into the solution and incubated under gentle mixing at 25 °C overnight. The aptamer targeted MNPs (Apt-MNPs) were then separated magnetically, washed and dispersed in PBS (pH 7.4) and stored at 4 °C before use. The efficiency of the reaction was determined by measuring the concentration of aptamer in the solution before and after conjugation using an Epoch micro-volume spectrophotometer system (BioTek, USA).
1) were dissolved in 200 mL distilled water and degassed under nitrogen atmosphere at 60 °C for 20 min. After the addition of 6 mL ammonium hydroxide (25% w/w), the reaction was proceeded under sonication and stirring at 60 °C for 60 min. The resulted nanoparticles were separated magnetically and washed with deionized water to decrease the pH of the solution to 7. For silica coating, the resulted nanoparticles were added into 40 mL absolute ethanol and completely dispersed by sonication under the nitrogen atmosphere. Then, 6 mL deionized water, 3 mL ammonium solution (25% w/w), and 400 μL TEOS were sequentially added into the solution under vigorous stirring and the reaction continued for 5 h at room temperature. The silica-coated MNPs (SiMNPs) were separated, washed with deionized water and dried at 40 °C overnight before functionalization with amine groups. For this aim, 20 mg MNPs were first dispersed in 20 mL ethanol under sonication and 200 μL APTES was added into the solution and refluxed at 60 °C, overnight. The resulted amine-functionalized MNPs were separated, washed with deionized water and dispersed in 10 mL phosphate buffer (50 mM, pH 7.0) containing glutaraldehyde (2%, v/v) and incubated at 25 °C under stirring for 5 h. MNPs were then separated magnetically, washed with PBS and dispersed in 1 mL phosphate buffer (50 mM, pH 8.5). The NH2-terminated anti-MUC1 aptamers were subsequently added into the solution and incubated under gentle mixing at 25 °C overnight. The aptamer targeted MNPs (Apt-MNPs) were then separated magnetically, washed and dispersed in PBS (pH 7.4) and stored at 4 °C before use. The efficiency of the reaction was determined by measuring the concentration of aptamer in the solution before and after conjugation using an Epoch micro-volume spectrophotometer system (BioTek, USA).
2.3. Characterization of the nanoparticles
The size and shape of MNPs were studied using a field emission scanning electron microscope (FE-SEM, Zeiss Sigma VP-500) at an accelerating voltage of 15 kV. The elemental composition of MNPs was determined using X-ray energy dispersive spectroscopy (EDS, Oxford Instrument, UK). Fourier transform infrared spectroscopy (FTIR-6300 spectrometer, Jasco, Japan) was used in a wavelength range of 4000 cm−1 to 400 cm−1 to investigate the surface functional groups of MNPs. The magnetic activity of nanoparticles was studied using a vibration sample magnetometer (VSM; Lake Shore Model 7400, Japan) at 30 °C. The particle size distribution of the nanoparticles was determined using ZEN 3600 Zetasizer (Malvern, Worcestershire, UK).2.4. Design and fabrication of the microfluidic device
The microfluidic device was fabricated using the conventional approach of photo- and soft-lithography as described previously.50 Briefly, the master mold was fabricated by patterning SU-8 photoresist on a silicon wafer. Subsequently, the microchannels were formed by molding Sylgard 184 reagents, according to the manufacturer's instructions (Dow Corning, USA). A 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of base and curing agent was poured on the master, degassed and baked for 30 min at 80 °C on a hot plate (RH digital, IKA, Germany). The cured elastomer was then peeled, punched to open the desired inlets and outlets and cleaned with adhesive tapes. The microchannels were then sealed by bonding the polydimethylsiloxane (PDMS) substrate to a glass microscope slide using the plasma activation (BD-20AC, Electro-technic Products, USA). Finally, the bonded devices were baked on the hotplate for 60 min at 80 °C to strengthen the bonding and retrieve the hydrophobicity of PDMS surfaces. For CTC capture experiments, the microfluidic device was connected to the solution loaded syringes using Tygon® tubing (Cole-Parmer, USA) and metallic connectors, while the injection flow rates were controlled using the syringe pumps (AL-1000, World Precision Instruments, USA).
1 mixture of base and curing agent was poured on the master, degassed and baked for 30 min at 80 °C on a hot plate (RH digital, IKA, Germany). The cured elastomer was then peeled, punched to open the desired inlets and outlets and cleaned with adhesive tapes. The microchannels were then sealed by bonding the polydimethylsiloxane (PDMS) substrate to a glass microscope slide using the plasma activation (BD-20AC, Electro-technic Products, USA). Finally, the bonded devices were baked on the hotplate for 60 min at 80 °C to strengthen the bonding and retrieve the hydrophobicity of PDMS surfaces. For CTC capture experiments, the microfluidic device was connected to the solution loaded syringes using Tygon® tubing (Cole-Parmer, USA) and metallic connectors, while the injection flow rates were controlled using the syringe pumps (AL-1000, World Precision Instruments, USA).
2.5. In vitro studies
The biocompatibility of MNPs was first studied on the HUVEC and MCF-7 breast cancer cells by MTT assay, according to the previous report.51 Briefly, 104 cells were added into each well of a 96-well plate containing 200 μL RPMI 1640 medium supplemented with 10% FBS and 1% antibiotic solution and incubated in a humidified incubator at 37 °C under 5% CO2. After 24 h, different concentrations of MNPs (0, 5, 10, 20, 50, and 100 μg mL−1) were added into the individual wells and incubated again for 48 h in the same condition. The medium was then completely removed and 100 μL MTT solution (0.5 mg mL−1) was added into each well and incubated again for 4 h at 37 °C. Finally, the solution was completely removed and the precipitated formazan crystals were dissolved in 150 μL DMSO and the absorbance was measured at 570 nm. The viability was determined as the absorbance ratio of each treatment and control.The cell capture efficiency of Apt-MNPs was then evaluated in vitro in the bulk and microfluidic platforms. First, the capture efficiency of MCF-7 cells was studied using different incubation periods (10, 20, 30, 60, and 120 min), various concentrations of Apt-MNPs (10, 20, 50, 100 and 200 μg mL−1) and two media (PBS buffer or RPMI 1640 medium). The capture efficiency was then determined at the optimum condition using different numbers (10, 50, 102, 103, 104, 105, 106 cells in 1 mL PBS) of MCF-7 and MDA-MB-231 cancer cells. The number of captured and non-captured cells were counted using an optical microscope (Leica, UK). The selectivity of Apt-MNPs was also studied in vitro on HUVEC and CAF cells at the same condition. The cell capture efficiency of Apt-MNPs was also studied in conjugation with a microfluidic system by using 50 μg MNPs and 104 to 106 MCF-7 cancer cells in PBS buffer. The captured MCF-7 cells were then washed with PBS buffer and cultured in RPMI 1640 medium supplemented with 10% FBS and 1% antibiotic solution and incubated in a humidified incubator at 37 °C under 5% CO2 to evaluate their viability and proliferation potential.
2.6. CTCs detection and capture from the blood samples of patients
The capture efficiency and specificity of Apt-MNPs were studied using peripheral blood samples of breast cancer patients and healthy donors and compared with EasySep™ Direct Human CTC Enrichment Kit. After incubation of 5 mL blood samples with Apt-MNPs (50 μg mL−1) for 60 min, the labeled CTCs were magnetically isolated, washed with PBS and enumerated. The immunofluorescence (IF) staining was used for CTCs identification according to the previous report52 with some modifications. Briefly, the captured cells were first fixed and permeabilized via incubation with 2% paraformaldehyde and 0.1% Triton-X 100, respectively, for 10 min. The cells were then incubated with Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 258 solution, FITC-labeled anti-human CD326 (EpCAM) and PE-labeled anti-CD45 antibodies at 37 °C for 30 min. The stained cells were then washed with PBS and imaged by an inverted fluorescence microscope (Nikon, Eclipse Ti-U) to identify the captured CTCs. Hoechst+/EpCAM +/CD45− cells were considered as CTCs while Hoechst+/EpCAM−/CD45+ cells were considered as the leukocytes.
258 solution, FITC-labeled anti-human CD326 (EpCAM) and PE-labeled anti-CD45 antibodies at 37 °C for 30 min. The stained cells were then washed with PBS and imaged by an inverted fluorescence microscope (Nikon, Eclipse Ti-U) to identify the captured CTCs. Hoechst+/EpCAM +/CD45− cells were considered as CTCs while Hoechst+/EpCAM−/CD45+ cells were considered as the leukocytes.
2.7. Statistical analysis
The in vitro cell studies were carried out in triplicate and the results were presented as the mean value ± standard deviation (SD). The data analysis was performed by Student's t-test (Microsoft Excel, Microsoft Corporation, USA) and P values of less than 0.05 were considered as statistically significant.3. Results and discussion
3.1. Synthesis and characterization of MNPs
In the present study, MNPs were used for CTCs isolation due to their advantages including simple synthesis and functionalization, biocompatibility, superparamagnetic behavior, and also their simple use under the magnetic fields. These unique properties make MNPs an excellent candidate for various biomedical applications such as cell isolation and labeling, targeted drug delivery, imaging, diagnostics and therapeutics.53 CellSearch®, the only FDA approved kit for CTCs isolation, has also been developed based on the magnetic beads functionalized with a specific antibody against the epithelial cell-adhesion molecules (EpCAM). However, some related challenges with the CellSearch system, especially the limited sensitivity, time-consuming operation, and considerable cost, still need to be solved.54 Therefore, in the present study, we tried to use the simple and cost-effective methods with the minimum toxicity issues for development of a new approach of CTCs isolation.The successful application of MNPs mainly depends on the appropriate selection of their physicochemical properties such as size, shape, chemical composition, surface charge and chemistry.14 As illustrated in Scheme 1, the synthesized MNPs using the co-precipitation method were first coated with a silica layer to provide the appropriate surface chemistry for further conjugation to the amine-terminated aptamers. The surface chemistry is one of the most important criteria that significantly affect the biocompatibility, molecular interactions, further functionalization and efficiency of nanoparticles especially in biological fluids.14 Among various polymeric and inorganic materials that are commonly used for surface coating of MNPs, silica is a good candidate due to its biocompatibility, easy modification, less adverse effects on the magnetization and also low costs that make it suitable for commercial applications.55
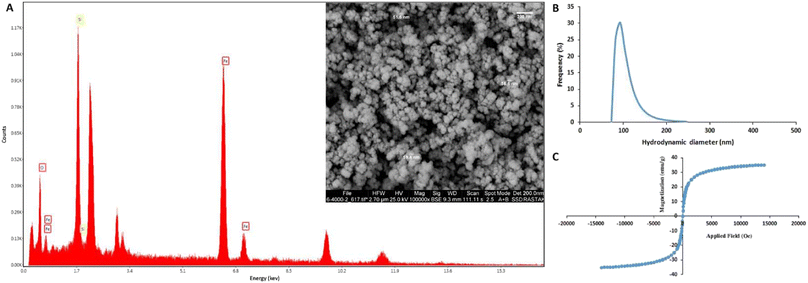
The results of the physicochemical characterization of SiMNPs are presented in Fig. 1. FE-SEM image (Fig. 1A) indicates the relatively monodisperse and semi-spherical shape of MNPs with sizes larger than 50 nm. The high surface area of nanoparticles could improve their interaction with the cell surfaces and leads to the efficient capture of cancer cells.56 The elemental analysis (Fig. 1B) revealed that the nanoparticles are composed of iron, silica and oxygen with the weight percentages of 57.31, 27.23, and 15.46, respectively. The VSM analysis (Fig. 1C) confirmed the high magnetic activity of MNPs, as the magnetic saturation value of 35.1 emu g−1 was obtained at 30 °C without any significant magnetic hysteresis following the application of an external magnetic field. These results clearly indicate the super-paramagnetic behavior of MNPs. DLS analysis revealed the average hydrodynamic size of 96.8 nm and polydispersity index (PDI) of 0.69 for the synthesized MNPs that is consistent with the results of FE-SEM.
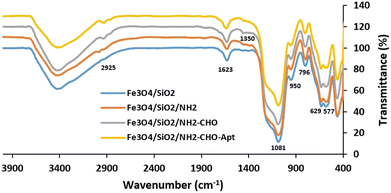
FT-IR spectra of MNPs after stepwise surface functionalization with TEOS (Fe3O4/SiO2), APTES (Fe3O4/SiO2/NH2), glutaraldehyde (Fe3O4/SiO2/NH2-CHO), and aptamer (Fe3O4/SiO2/NH2-CHO-Apt) have been shown in Fig. 2. The characteristic absorption bands of Fe–O are clearly seen at 577.5 cm−1 and 629.6 cm−1 while the absorption band at 1623.7 cm−1 could be ascribed to the bending vibration of water in Fe3O4 nanoparticles. The absorption bands at 796.4 and 950.7 cm−1 corresponding to the Si–O–H bending and Si–O stretching vibrations of silanol groups, respectively, as well as the significant peak located around 1081.8 cm−1 corresponding to the Si–O–Si asymmetric stretching vibration of siloxane, indicate the successful formation of silica layer.57 The absorption bands located at 1350.8 and 2925 cm−1 are attributed to the stretching vibrations of C–N and C–H bonds of APTES molecules, respectively.58
 | ||
| Fig. 2 FTIR spectra of MNPs after stepwise surface modification with TEOS (Fe3O4/SiO2), APTES (Fe3O4/SiO2/NH2), glutaraldehyde (Fe3O4/SiO2/NH2-CHO) and aptamer (Fe3O4/SiO2/NH2-CHO-Apt). | ||
In order to covalently bind the amine-functionalized aptamers to the surface amine groups of MNPs, a highly reactive dialdehyde reagent, glutaraldehyde, was used as the homobifunctional crosslinker. The new and weak band at 1685.4 cm−1 is attributed to the formation of an amide bond between the amine groups on the surface of nanoparticles and the aldehyde group of glutaraldehyde.59 The subsequent reaction between another terminal aldehyde group of glutaraldehyde and amine group of aptamers led to the covalent attachment of aptamers to the surface of MNPs. This facile and mild Schiff-base reaction using glutaraldehyde linker is commonly used in the biological assays especially for covalent conjugation of antibodies.60 The concentration of DNA aptamers in the solution before (61 ng μL−1) and after (14 ng μL−1) reaction indicated the participation of 77% of total aptamers in the reaction. The calculated efficiency of aptamer conjugation was 2.35 μg aptamer per mg of MNPs.
3.2. In vitro one-step capturing of cancer cells
The biocompatibility of MNPs was first studied in vitro on the HUVEC and MCF-7 cell lines. The survival rates of as much as 90.99 ± 1.16% and 91.38 ± 3.27% were obtained after 48 h exposure of HUVEC and MCF-7 cells, respectively, with 100 μg mL−1 MNPs (Fig. S1†) indicating the high biocompatibility of MNPs. Therefore, the nanoparticles are suitable for biological applications especially cell labeling and isolation.To achieve the best condition for capturing cancer cells, the concentration of Apt-MNPs and incubation time were optimized in the first step. To this aim, 106 MCF-7 cells were first incubated with different concentrations of Apt-MNPs (10 to 200 μg mL−1) and also different incubation times (10 to 120 min) in 1 mL of PBS buffer and then separated magnetically and enumerated under the optical microscope. Based on the results (Fig. S2A†), the highest capture efficiency (95.33 ± 2.52%) was obtained after 60 min incubation of MCF-7 cancer cells with Apt-MNPs and no significant increase in the capture efficiency was observed by further increases in the incubation times. The optimum concentration of Apt-MNPs was 50 μg mL−1, as the capture efficiency of 95 ± 2.65% was obtained after 60 min incubation of 106 MCF-7 cells with 50 μg mL−1 Apt-MNPs. Further increase of Apt-MNPs concentration had no significant effect on the cell capture efficiency (Fig. S2B†). In the next step, the effect of using PBS buffer and RPMI 1640 medium and also the number of MCF-7 cells was studied on the capture efficiency of Apt-MNPs. Based on the results (Fig. 3A), no significant difference was observed between PBS buffer and RPMI 1640 medium in terms of the cell capture efficiency. Moreover, Apt-MNPs were able to capture more than 90% of the cancer cells in aboundance as little as 10 cells. The efficient detection and capture of MDA-MB-231 cancer cells were also obtained by using Apt-MNPs, similar to the MCF-7 cells (Fig. 3B). The results were comparable with the previously reported efficiency of ∼90% after 2 h incubation with 100 μg anti-EpCAM antibody conjugated multifunctional magnetic upconversion nanoparticles.43
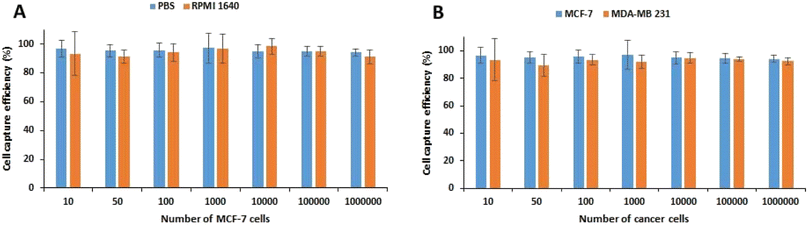 | ||
| Fig. 3 The capture efficiency of MCF-7 cells using APT-MNPs from PBS buffer and RPMI 1640 medium (A). The capture efficiency of MCF-7 and MDA-MB-231 cancer cells from PBS using APT-MNPs (B). | ||
The in vitro cell capture studies on HUVEC and CAF cells were also conducted in the optimum condition to evaluate the selectivity of Apt-MNPs. The capture yields of 53.7 ± 3.52% and 63.4 ± 4.04% were obtained after 60 min incubation of 50 μg mL−1 Apt-MNPs with 106 CAF and HUVEC cells, respectively, that were much lower than the capture yields of MCF-7 (95.33 ± 2.52%) and MDA-MB-231 (94.7 ± 3.52%) cancer cells, in the similar condition. Regarding the expression of MUC1 by the normal epithelial cells and also fibroblast cells,46 these results indicate the less capture efficiency of Apt-MNPs for CAF and HUVEC cells in comparison with MCF-7 and MDA-MB-231 cells, due to the lower expression level of MUC1 receptor. The results confirm the direct relationship between capture efficiency of cells and the expression level of receptor. MUC1 receptor is overexpressed on the surface of different cancer cells especially breast cancer cells.61,62 Anti MUC1 aptamer was selected and used in this study due to its high selectivity and affinity (Kd value of 0.135 nM) for the MUC1 receptor42 in comparison with the corresponding antibody (Kd values of 3–400 nM).63 As the positive selection of cancer cells is mainly dependent on the frequency of specific receptors on the cell surface, the capture of normal cells by Apt-MNPs is not expected due to the low expression of MUC1 receptors in these cells.
The microscopic observation of captured cells revealed the formation of a continuous and thick layer of MNPs around the cancer cells (Fig. S3†). This observation indicated the efficient capture of cancer cells with Apt-MNPs which may be due to the enhanced surface area of MNPs and the high affinity of MUC1 aptamers toward the corresponding receptor. The efficient CTCs capture by formation of a stable patch-like multivalent matrix nano-net via the complexes of antibody-targeted MNPs and graphene oxide sheets have also been reported by Lai et al.,64 recently. Since the conjugation method and the type of linker could significantly affect the structure and appropriate function of DNA aptamers, this result also confirmed that glutaraldehyde is a suitable linker for the aptamer conjugation to nanoparticles. Based on the results, Apt-MNPs could be an excellent alternative to the conventionally used immunomagnetic microbeads for the efficient capture of cancer cells.
The proliferation ability of magnetically separated MCF-7 cells was also investigated in vitro. Most of the captured cells were viable and successfully attached to the surface of culture plate (Fig. S4A†) and proliferated, however, it took several days for the cells to recover and begin to regrow. The proliferated cells displayed a normal morphology similar to the untreated cells (Fig. S4B†). These results also indicate the biocompatibility of MNPs in agreement with the results of MTT assay.
The high stability was another interesting feature of Apt-MNPs such that their capture efficiency was preserved even after more than 6 months' storage at 4 °C. The capture efficiency of 93.67 ± 2.52% was obtained for MCF-7 cells after 6 months storage of Apt-MNPs at 4 °C. Moreover, the long time storage didn't affect the biocompatibility of Apt-MNPs, as the cell viability of 90.7 ± 2.39% after exposure of 104 MCF-7 cells with 100 μg mL−1 Apt-MNPs for 48 h. Therefore, the simple and fast nature of glutaraldehyde-mediated conjugation along with the high stability of the resulted Apt-MNPs make the present method appropriate for widespread applications. Moreover, the glutaraldehyde-functionalized MNPs could be simply targeted by the aptamer immediately before use to achieve the maximum CTCs capture efficiency. It would be mentioned that the storage of Apt-MNPs at −20 °C led to the significant loss of their function and inhibition of cancer cell capture. The use of Apt-MNPs has also been reported recently by Bayramoglu et al.65 for the fast and sensitive detection of a pathogenic bacterium, Salmonella enterica, in the milk samples. A sensitive colorimetric method has also been reported recently by Shen et al.66 for the detection of circulating tumor-related materials by using aptamer-functionalized MNPs and alkaline phosphatase signal amplification.
3.3. Microfluidic assisted capture of cancer cells
The implementation of microfluidic platforms demonstrated promising improvements in accuracy of cell separation and sorting assays.67 Here, we developed a novel integrated microfluidic device for aptamer-based magnetic capture of CTCs. The integrated device can be used as a point of care system, without the requirement of previous magnetic labeling step. The platform of the microfluidic chip used in the present study is illustrated in Fig. 4. The microfluidic network consisted of three main modules including a tesla mixer, an inertial focusing channel, and the main magnetic separation section. Tesla mixer was selected as it works in a wide range of flow rates and is designed based on the previously optimized dimensions68 to maximize the mixing. Cells and Apt-MNPs were first introduced into the microfluidic chip and mixed homogeneously in the tesla mixer module. As a result, CTCs were specifically tagged with MNPs via the MUC1 aptamers. The inertial focusing module then sorts all the cells at the center of the microchannel, enabling separation of magnetically tagged CTCs from the sorted cell stream. An optimized asymmetric curving design69 based on the applied range of flow rates and average cell diameter was used for inertial focusing. Finaly,the magnetic separation module deviated the trajectory of the magnetically labeled cells and enabled their collection in the bottom outlet while the remained fraction of cells was discarded in the waste upper outlet (Fig. 4).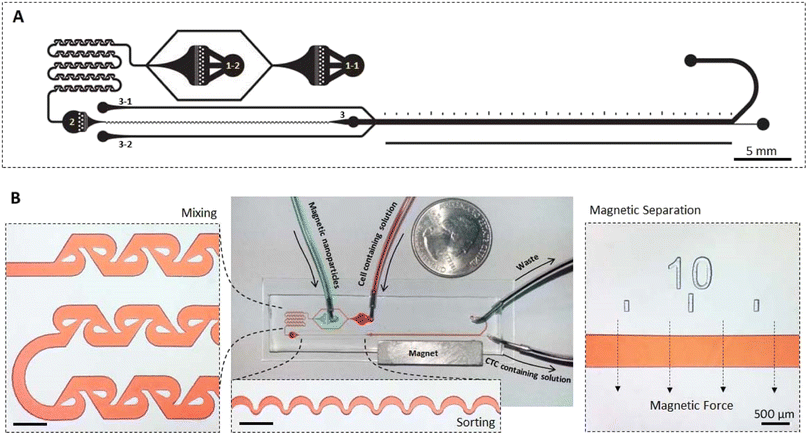 | ||
| Fig. 4 Microfluidic channels design. (A) The mask design (reversed) and (B) fabricated microfluidic device containing mixer module, sorting module and magnetic separation module. | ||
As one of the specific advantages of the developed microfluidic device, microchanel network was designed somehow that the device work either as an integrated system or with independent modules. To use all three modules, 1–1 and 1–2 inlets were used for the cell containing solution and MNPs solution, respectively, while other inlets were kept plug. For elimination of the mixer module, when MNPs and cells were previously mixed, only inlet 2 was used and other inlets were kept plug. For the application of only magnetic separation module, when lower flow rates are preferred and therefore inertial sorting module is not effective, inlet 3 was implemented. In this condition, sheath flow entering the microfluidic network through inlets 3–1 and 3–2 were applied to confine cells containing stream in the center of the magnetic separation microchannel.
The efficiency of the microfluidic chip to magnetically isolate the cancer cells was examined either by using the mixer module or without it (after initial exposure of MCF-7 cells and MNPs for 60 min). The results were almost similar as up to 90% capture yield was obtained when the cells were isolated directly using the microfluidic chip or after primary incubation with Apt-MNPs for 60 minutes. However, when the mixer module is used, a slow flow rate of less than 20 μL min−1 is necessary for the efficient capture of cancer cells as the higher flow rates lead to the accumulation of MNPs and cancer cells and subsequent blocking of microchannels. Therefore, the use of a mixer module is only suitable for the processing of low volume samples as the long process may result in the degeneration of samples. After subjecting to the magnetic field established by a permanent magnet, the cancer cells and MNPs were separated rapidly from the stream and moved toward the field (Movie S1 and S2†). This observation further confirmed the efficient and tight binding of Apt-MNPs to the cancer cells as well as the excellent magnetic activity of MNPs. Similar results were also obtained by Wu et al.70 regarding the integrated use of magnetic and size-selective separation of CTCs and also by Tang et al.71 with the integrated use of anti-EpCAM antibody functionalized MNPs and microfluidic device. A more than 90% capture efficiency of EpCAM positive tumor cells has also been reported recently via the integrated use of a microfluidic device with the aptamers functionalized magnetic nanoclusters.72
3.4. CTCs capture from the clinical samples of cancer patients
The efficiency and specificity of the present method for CTCs isolation from the peripheral blood samples of breast cancer patients were also investigated and compared with the EasySep™ Direct Human CTC Enrichment Kit. Unlike the negative selection methods that require multiple centrifugation steps, the present method is based on the positive selection and simply isolates CTCs magnetically in one-step without any centrifugation. Besides, based on the results, no preliminary steps of sample preparation such as the removal of red blood cells (RBC), are needed before CTCs capture with Apt-MNPs. The results (Table S1†) confirmed the comparable efficiency of present method with EasySep™ Kit. The microscopic observations showed the isolation of a few other blood cells along with CTCs (Fig. S5A†), indicating the sensitivity of the present method. These cells that resemble the size and morphology to the lymphocites, seems to be physically entangled between the MNPs during the process of magnetic separation. The density of Apt-MNPs on the surface of these small blood cells is much lower than CTCs. The isolation of mononuclear white blood cells along with CTCs is usually observed due to their similar physical properties.2 Regarding the direct isolation of CTCs from blood without any preliminary purification and also the frequency of white blood cells, this observation could be expected. This observation may be attributed to the overexpression of the MUC1 receptor on the surface of CTCs that is the basis of positive selection of cancer cells. A few similar blood cells were also isolated following the incubation of blood samples of healthy donors with Apt-MNPs but no CTCs were identified in these samples (Fig. S5B†). These results indicated the specific and efficient detection of MUC1 receptors on the surface of cancer cells via the MUC1 aptamers. With regard to the extremely low abundance of CTCs in the clinical samples, the high affinity and specificity of Apt-MNPs toward the target receptors on the surface of cancer cells are essential. Based on the results, direct exposure of Apt-MNPs with the blood samples for 60 min is appropriate for the sensitive and effective capture of CTCs. No need to pre-treat the blood samples and also use of the complex and expensive equipment are some of the advantages of the present method.For the identification of CTCs, the captured cells were fluorescently stained with Hoechst dye as well as PE-anti-CD45 and FITC-anti-EpCAM antibodies. While the anti-EpCAM antibody recognizes its specific receptors on the surface of cancer cells, the anti-CD45 antibody specifically detects the corresponding receptors on the white blood cells. Therefore, only cells that are detected and stained with the anti-EpCAM antibody are considered as CTCs. Hoechst was also used to identify and to stain the nuclear DNA of live cells.20 The fluorescence immunostaining confirmed that the captured cells are CTCs due to their green fluorescence signal originated from the attachment and staining with FITC-labeled EpCAM antibody (Fig. 5). The overall results indicated the high efficiency and specificity of Apt-MNPs for the rapid and direct isolation of CTCs from the blood.
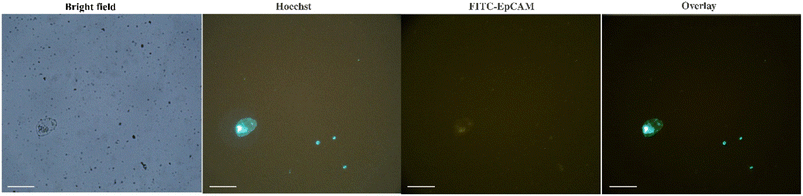 | ||
| Fig. 5 The captured CTC from the blood of breast cancer patients using Apt-MNPs after IF staining (scale bar 50 μm). | ||
The non-invasive and efficient detection and capture of CTCs with the minimum adverse effects on their viability and functions is the ultimate goal of current researches. This is essential for the subsequent molecular and cellular analysis of captured CTCs to profile the heterogeneous CTCs and also to understand the mechanism of metastasis and tumor evolution which finally leads to personalized cancer therapy. The promising results of the present study indicated the high potential of Apt-MNPs, to achieve this valuable goal.
4. Conclusions
Achievement of a simple and rapid method with the ability of efficient and sensitive detection and isolation of viable CTCs was the main focus of the present study. The integrated use of the microfluidic device and the biomimetic nanostructure of aptamer targeted MNPs was selected as the main idea to achieve this highly desirable goal. The facile and cost-effective synthesis, excellent magnetic activity, appropriate biocompatibility, colloidal stability and high surface to volume ratio of MNPs facilitate the simple and rapid isolation of viable CTCs. The microfluidic platform provides an appropriate condition for distinguishing the physical and biochemical properties of CTCs as well as the continuous and high throughput separation of cancer cells. The use of a high-affinity anti-MUC1 aptamer further improves the specificity and sensitivity of CTCs capture allowing take the advantages of DNA aptamers over the conventionally used antibodies. The simple use of glutaraldehyde linker leads to the stable conjugation of aptamers to MNPs with appropriate and prolonged functionality. Moreover, the use of Apt-MNPs makes possible the nondestructive isolation of viable CTCs for the subsequent in vitro culture and molecular analysis. The efficiency of the present method for CTCs capture from the clinical samples is comparable with the examined commercial kit. The overall results indicate the high potential of the present method for clinical applications. Based on the results, the integrated use of microfluidic chip with Apt-MNPs is useful to achieve a more automated and efficient method, especially for the analysis of lots of large volume samples. The simplicity and cost-effectiveness of manufacturing are other remarkable characteristics of the present device. In conclusion, the simple, rapid and sensitive nature of the present method makes it a suitable candidate for widespread use in clinical purposes.Ethical statement
The research related to human use has complied with all the relevant national regulations and institutional policies, follows the tenets of the Helsinki Declaration, and has been approved by the ethical committee of Isfahan University of Medical Sciences (IR.MUI.MED.REC.1395.2.233).Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The financial supports of Isfahan University of Medical Sciences are gratefully acknowledged.References
- M. Tang, C. Y. Wen, L. L. Wu, S. L. Hong, J. Hu, C. M. Xu, D. W. Pang and Z. L. Zhang, Lab Chip, 2016, 16, 1214–1223 RSC.
- J. M. Jackson, M. A. Witek, J. W. Kamande and S. A. Soper, Chem. Soc. Rev., 2017, 46, 4245–4280 RSC.
- J. Yang, X. Huang, C. Gan, R. Yuan and Y. Xiang, Biosens. Bioelectron., 2019, 143, 111604 CrossRef CAS PubMed.
- L. Wu, L. Zhu, M. Huang, J. Song, H. Zhang, Y. Song, W. Wang and C. Yang, TrAC, Trends Anal. Chem., 2019, 117, 69–77 CrossRef CAS.
- Y. Zhang, Y. Li and Z. Tan, Analyst, 2021, 146, 7048–7069 RSC.
- Y. Song, T. Tian, Y. Shi, W. Liu, Y. Zou, T. Khajvand, S. Wang, Z. Zhu and C. Yang, Chem. Sci., 2017, 8, 1736–1751 RSC.
- H. Safarpour, S. Dehghani, R. Nosrati, N. Zebardast, M. Alibolandi, A. Mokhtarzadeh and M. Ramezani, Biosens. Bioelectron., 2020, 148, 111833 CrossRef CAS PubMed.
- M. Samandari, M. G. Julia, A. Rice, A. Chronopoulos and A. E. del Rio Hernandez, Transl. Res., 2018, 201, 98–127 CrossRef PubMed.
- R. A. Harouaka, M. Nisic and S. Y. Zheng, J. Lab. Autom., 2013, 18, 455–468 CrossRef CAS PubMed.
- H. Pei, L. Li, Z. Han, Y. Wang and B. Tang, Lab Chip, 2020, 20, 3854–3875 RSC.
- Z. Lin, G. Luo, W. Du, T. Kong, C. Liu and Z. Liu, Small, 2019, 1903899, 1–21 Search PubMed.
- S. Riethdorf, H. Fritsche, V. Muller, T. Rau, C. Schindlbeck, B. Rack, W. Janni, C. Coith, K. Beck, F. Janicke, S. Jackson, T. Gornet, M. Cristofanilli and K. Pantel, Clin. Cancer Res., 2007, 13, 920–928 CrossRef CAS PubMed.
- N. Xia, D. Wu, H. Yu, W. Sun, X. Yi and L. Liu, Talanta, 2021, 221, 121640 CrossRef CAS PubMed.
- Y. Zhu, K. Kekalo, C. NDong, Y.-Y. Huang, F. Shubitidze, K. E. Griswold, I. Baker and J. X. J. Zhang, Adv. Funct. Mater., 2016, 26, 3953–3972 CrossRef CAS.
- P. Chen, Y. Y. Huang, K. Hoshino and X. Zhang, Lab Chip, 2014, 14, 446–458 RSC.
- X. Fan, C. Jia, J. Yang, G. Li, H. Mao, Q. Jin and J. Zhao, Biosens. Bioelectron., 2015, 71, 380–386 CrossRef CAS PubMed.
- N. Muhanna, A. Mepham, R. M. Mohamadi, H. Chan, T. Khan, M. Akens, J. D. Besant, J. Irish and S. O. Kelley, Nanomed.: Nanotechnol. Biol. Med., 2015, 11, 1613–1620 CrossRef CAS PubMed.
- L. Chen, L. L. Wu, Z. L. Zhang, J. Hu, M. Tang, C. B. Qi, N. Li and D. W. Pang, Biosens. Bioelectron., 2016, 85, 633–640 CrossRef CAS PubMed.
- B. Kwak, J. Lee, D. Lee, K. Lee, O. Kwon, S. Kang and Y. Kim, Biosens. Bioelectron., 2017, 88, 153–158 CrossRef CAS PubMed.
- L. L. Wu, C. Y. Wen, J. Hu, M. Tang, C. B. Qi, N. Li, C. Liu, L. Chen, D. W. Pang and Z. L. Zhang, Biosens. Bioelectron., 2017, 94, 219–226 CrossRef CAS PubMed.
- Y. Yu, Y. Yang, F. Wang, J. Ding, S. Meng, C. Li, D. Tang and X. Yin, Anal. Chim. Acta, 2018, 1044, 162–173 CrossRef CAS PubMed.
- H. K. Kordasht and M. Hasanzadeh, Talanta, 2020, 220, 121436 CrossRef CAS PubMed.
- H. Motaghi, S. Ziyaee, M. A. Mehrgardi, A. A. Kajani and A. K. Bordbar, Biosens. Bioelectron., 2018, 118, 217–223 CrossRef CAS PubMed.
- Y. Chen, W. Wang, D. Tyagi, A. J. Carrier, S. Cui, S. He and X. Zhang, Nanoscale, 2019, 11, 5879–5883 RSC.
- N. Kacherovsky, I. I. Cardle, E. L. Cheng, J. L. Yu, M. L. Baldwin, S. J. Salipante, M. C. Jensen and S. H. Pun, Nat. Biomed. Eng., 2019, 3, 783–795 CrossRef CAS PubMed.
- Y. Wan, Y. Liu, P. B. Allen, W. Asghar, M. A. I. Mahmood, J. Tan, H. Duhon, Y. T. Kim, A. D. Ellington and S. M. Iqbal, Lab Chip, 2012, 12, 4693–4701 RSC.
- Y. K. Jung, M. A. Woo, H. T. Soh and H. G. Park, Chem. Commun., 2014, 50, 12329–12332 RSC.
- F. Zhang, L. Wu, W. Nie, L. Huang, J. Zhang, F. Li and H. Y. Xie, Anal. Chem., 2019, 91, 15726–15731 CrossRef CAS PubMed.
- O. Boyacioglu, C. H. Stuart, K. George and W. H. Gmeiner, Mol. Ther.–Nucleic Acids, 2013, 2, e107 CrossRef PubMed.
- W. Sheng, T. Chen, W. Tan and Z. H. Fan, ACS Nano, 2013, 7, 7067–7076 CrossRef CAS PubMed.
- W. Sheng, T. Chen, R. Kamath, X. Xiong, W. Tan and Z. H. Fan, Anal. Chem., 2012, 84, 4199–4206 CrossRef CAS PubMed.
- Z. Li, G. Wang, Y. Shen, N. Guo and N. Ma, Adv. Funct. Mater., 2018, 28, 1–11 Search PubMed.
- Y. Lin, L. Jiang, Y. Huang, Y. Yang, Y. He, C. Lu and H. Yang, Chem. Commun., 2019, 55, 5387–5390 RSC.
- S. Wang, C. Zhang, G. Wang, B. Cheng, Y. Wang, F. Chen, Y. Chen, M. Feng and B. Xiong, Theranostics, 2016, 6, 1877–1886 CrossRef CAS PubMed.
- B. L. Santini, M. Zúñiga-Bustos, A. Vidal-Limon, J. B. Alderete, S. A. Águila and V. A. Jiménez, J. Chem. Inf. Model., 2020, 60, 786–793 CrossRef CAS PubMed.
- M. S. Nabavinia, A. Gholoobi, F. Charbgoo, M. Nabavinia, M. Ramezani and K. Abnous, Med. Res. Rev., 2017, 37, 1518–1539 CrossRef CAS PubMed.
- J. Z. Zaretsky, I. Barnea, Y. Aylon, M. Gorivodsky, D. H. Wreschner and I. Keydar, Mol. Cancer, 2006, 5, 1–14 CrossRef PubMed.
- D. W. Kufe, Oncogene, 2013, 32, 1073–1081 CrossRef CAS PubMed.
- S. Nath and P. Mukherjee, Trends Mol. Med., 2014, 20, 332–342 CrossRef CAS PubMed.
- M. A. Cheever, J. P. Allison, A. S. Ferris, O. J. Finn, B. M. Hastings, T. T. Hecht, I. Mellman, S. A. Prindiville, J. L. Viner, L. M. Weiner and L. M. Matrisian, Clin. Cancer Res., 2009, 15, 5323–5337 CrossRef PubMed.
- F. Charbgoo, M. Alibolandi, S. M. Taghdisi, K. Abnous, F. Soltani and M. Ramezani, Nanomed.: Nanotechnol. Biol. Med., 2018, 14, 685–697 CrossRef CAS PubMed.
- C. S. M. Ferreira, C. S. Matthews and S. Missailidis, Tumor Biol., 2006, 27, 289–301 CrossRef CAS PubMed.
- C. Wang, M. Ye, L. Cheng, R. Li, W. Zhu, Z. Shi, C. Fan, J. He, J. Liu and Z. Liu, Biomaterials, 2015, 54, 55–62 CrossRef CAS PubMed.
- C. L. Chang, W. Huang, S. I. Jalal, B. D. Chan, A. Mahmood, S. Shahda, B. H. O'Neil, D. E. Matei and C. A. Savran, Lab Chip, 2015, 15, 1677–1688 RSC.
- E. Ozkumur, A. M. Shah, J. C. Ciciliano, B. L. Emmink, D. T. Miyamoto, E. Brachtel, M. Yu, P. I. Chen, B. Morgan, J. Trautwein, A. Kimura, S. Sengupta, S. L. Stott, N. M. Karabacak, T. A. Barber, J. R. Walsh, K. Smith, P. S. Spuhler, J. P. Sullivan, R. J. Lee, D. T. Ting, X. Luo, A. T. Shaw, A. Bardia, L. V. Sequist, D. N. Louis, S. Maheswaran, R. Kapur, D. A. Haber and M. Toner, Sci. Transl. Med., 2013, 5, 1–20 Search PubMed.
- P. Kumar, J. Ji, T. L. Thirkill and G. C. Douglas, BioRes. Open Access, 2014, 3, 45–52 CrossRef CAS PubMed.
- Y. H. Wang, Y. Y. Dong, W. M. Wang, X. Y. Xie, Z. M. Wang, R. X. Chen, J. Chen, D. M. Gao, J. F. Cui and Z. G. Ren, J. Exp. Clin. Cancer Res., 2013, 32 Search PubMed.
- B. Aramini, V. Masciale, C. Arienti, M. Dominici, F. Stella, G. Martinelli and F. Fabbri, Cancers, 2022, 14, 1–20 Search PubMed.
- A. Abbasi Kajani, A. K. Bordbar, S. H. Zarkesh-Esfahani, A. Razmjou and J. Hou, J. Mol. Liq., 2017, 247, 238–245 CrossRef CAS.
- M. Samandari, F. Alipanah, S. Haghjooy Javanmard and A. Sanati-Nezhad, Sens. Actuators, B, 2019, 291, 418–425 CrossRef CAS.
- A. Abbasi Kajani and A. K. Bordbar, J. Hazard. Mater., 2019, 366, 268–274 CrossRef CAS PubMed.
- X. Y. Ma, L. L. Wu, L. Chen, Y. H. Qin, J. Hu, M. Tang, C.-M. Xu, C. Qi, Z. L. Zhang and D. W. Pang, ACS Appl. Nano Mater., 2018, 1, 4019–4027 CrossRef CAS.
- K. Hola, Z. Markova, G. Zoppellaro, J. Tucek and R. Zboril, Biotechnol. Adv., 2015, 33, 1162–1176 CrossRef CAS PubMed.
- K. C. Andree, G. van Dalum and L. W. M. M. Terstappen, Mol. Oncol., 2016, 10, 395–407 CrossRef CAS PubMed.
- W. P. Li, P. Y. Liao, C. H. Su and C. S. Yeh, J. Am. Chem. Soc., 2014, 136, 10062–10075 CrossRef CAS PubMed.
- T. Verano-Braga, R. Miethling-Graff, K. Wojdyla, A. Rogowska-Wrzesinska, J. R. Brewer, H. Erdmann and F. Kjeldsen, ACS Nano, 2014, 8, 2161–2175 CrossRef CAS PubMed.
- R. A. Bini, R. F. C. Marques, F. J. Santos, J. A. Chaker and M. Jafelicci, J. Magn. Magn. Mater., 2012, 324, 534–539 CrossRef CAS.
- Z. Chen, W. Xu, L. Jin, J. Zha, T. Tao, Y. Lin and Z. Wang, J. Mater. Chem. A, 2014, 2, 18339–18344 RSC.
- C. Miao, L. Yang, Z. Wang, W. Luo, H. Li, P. Lv and Z. Yuan, Fuel, 2018, 224, 774–782 CrossRef CAS.
- N. S. K. Gunda, M. Singh, L. Norman, K. Kaur and S. K. Mitra, Appl. Surf. Sci., 2014, 305, 522–530 CrossRef CAS.
- K. L. Rhinehardt, G. Srinivas and R. V. Mohan, J. Phys. Chem. B, 2015, 119, 6571–6583 CrossRef CAS PubMed.
- X. Han, Y. Jiang, S. Li, Y. Zhang, X. Ma, Z. Wu, Z. Wu and X. Qi, Nanoscale, 2019, 11, 339–347 RSC.
- H. Thie, L. Toleikis, J. Li, R. Von Wasielewski, G. Bastert, T. Schirrmann, I. T. Esteves, C. K. Behrens, B. Fournes, N. Fournier, C. de Romeuf, M. Hust and S. Dübel, PLoS One, 2011, 14, e15921 CrossRef PubMed.
- C. H. Lai, W. S. Tsai, M. H. Yang, T. Y. Chou and Y. C. Chang, Nanoscale, 2019, 11, 21119–21127 RSC.
- G. Bayramoglu, V. C. Ozalp, U. Dincbal and M. Y. Arica, ACS Biomater. Sci. Eng., 2018, 4, 1437–1444 CrossRef CAS PubMed.
- L. Shen, K. Jia, T. Bing, Z. Zhang, X. Zhen, X. Liu, N. Zhang and D. Shangguan, Anal. Chem., 2020, 92, 5370–5378 CrossRef CAS PubMed.
- F. S. Iliescu, D. P. Poenar, F. Yu, M. Ni, K. H. Chan, I. Cima, H. K. Taylor, I. Cima and C. Iliescu, Biomicrofluidics, 2019, 13, 041503 CrossRef PubMed.
- S. Hossain, M. A. Ansari, A. Husain and K. Y. Kim, Chem. Eng. J., 2010, 158, 305–314 CrossRef CAS.
- D. Di Carlo, D. Irimia, R. G. Tompkins and M. Toner, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 18892–18897 CrossRef CAS PubMed.
- L. L. Wu, M. Tang, Z. L. Zhang, C. B. Qi, J. Hu, X. Y. Ma and D. W. Pang, Anal. Chem., 2018, 90, 10518–10526 CrossRef CAS PubMed.
- M. Tang, C. Y. Wen, L. L. Wu, S. L. Hong, J. Hu, C. M. Xu, D. W. Pang and Z. L. Zhang, Lab Chip, 2016, 16, 1214–1223 RSC.
- F. Zhang, L. Wu, W. Nie, L. Huang, J. Zhang, F. Li and H. Y. Xie, Anal. Chem., 2019, 91, 15726–15731 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra05930d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |