Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A general strategy to the intracellular sensing of glycosidases using AIE-based glycoclusters†
Lei
Dong‡
ab,
Min-Yu
Zhang‡
a,
Hai-Hao
Han‡
a,
Yi
Zang
c,
Guo-Rong
Chen
a,
Jia
Li
*c,
Xiao-Peng
He
*a and
Sébastien
Vidal
 *bd
*bd
aKey Laboratory for Advanced Materials and Joint International Research Laboratory of Precision Chemistry and Molecular Engineering, Feringa Nobel Prize Scientist Joint Research Center, Frontiers Center for Materiobiology and Dynamic Chemistry, School of Chemistry and Molecular Engineering, East China University of Science and Technology, 130 Meilong Rd., Shanghai 200237, P. R. China. E-mail: xphe@ecust.edu.cn
bInstitut de Chimie et Biochimie Moléculaires et Supramoléculaires, Laboratoire de Chimie Organique 2-Glycochimie, UMR 5246, CNRS, Université Claude Bernard Lyon 1, Université de Lyon, 1 Rue Victor Grignard, F-69622 Villeurbanne, France. E-mail: sebastien.vidal@cnrs.fr
cNational Centre for Drug Screening, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 189 Guo Shoujing Rd., Shanghai 201203, P. R. China. E-mail: jli@simm.ac.cn
dUniversité Paris-Saclay, CNRS, Institut de Chimie des Substances Naturelles, UPR 2301, 91198, Gif-sur-Yvette, France
First published on 7th December 2021
Abstract
Glycosidases, which are the enzymes responsible for the removal of residual monosaccharides from glycoconjugates, are involved in many different biological and pathological events. The ability to detect sensitively the activity and spatiotemporal distribution of glycosidases in cells will provide useful tools for disease diagnosis. However, the currently developed fluorogenic probes for glycosidases are generally based on the glycosylation of the phenol group of a donor–acceptor type fluorogen. This molecular scaffold has potential drawbacks in terms of substrate scope, sensitivity because of aggregation-caused quenching (ACQ), and the inability for long-term cell tracking. Here, we developed glycoclusters characterized by aggregation-induced emission (AIE) properties as a general platform for the sensing of a variety of glycosidases. To overcome the low chemical reactivity associated with phenol glycosylation, here we developed an AIE-based scaffold, which is composed of tetraphenylethylene conjugated with dicyanomethylene-4H-pyran (TPE–DCM) with a red fluorescence emission. Subsequently, a pair of dendritic linkages was introduced to both sides of the fluorophore, to which six copies of monosaccharides (D-glucose, D-galactose or L-fucose) were introduced through azide–alkyne click chemistry. The resulting AIE-active glycoclusters were shown to be capable of (1) fluorogenic sensing of a diverse range of glycosidases including β-D-galactosidase, β-D-glucosidase and α-L-fucosidase through the AIE mechanism, (2) fluorescence imaging of the endogenous glycosidase activities in healthy and cancer cells, and during cell senescence, and (3) glycosidase-activated, long-term imaging of cells. The present study provides a general strategy to the functional, in situ imaging of glycosidase activities through the multivalent display of sugar epitopes of interest onto properly designed AIE-active fluorogens.
Introduction
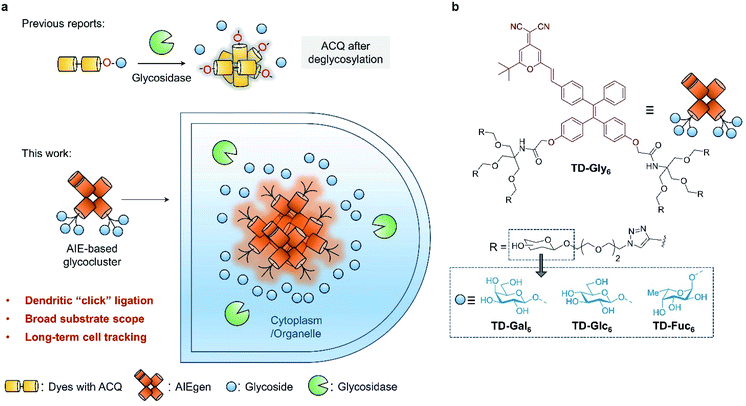
Glycosidases play critical roles in many biological processes through the hydrolysis of monosaccharides from polysaccharides and glycoconjugates (glycolipids or glycoproteins).1 Among them, β-D-galactosidase (β-Gal) is an essential enzyme involved in monitoring the efficiency of transcription and transfection of genes, and has also been demonstrated as a key biomarker for cell senescence, ovarian cancer and other pathological processes.2–4 α-Fucosidase (AFU) is commonly found in mammalian cells, which hydrolyzes an L-fucosyl residue from glycoconjugates under acidic pH (pH 4 to 6.5). AFU is involved in many biological processes such as inflammation, growth regulation, ligand–receptor recognition, antigenicity, and is also implicated in human diseases including fucosidosis5 and carcinoma.6–8 Therefore, the ability to accurately monitor the activity of endogenous glycosidases is of prime importance for the basic study of glycobiology as well as for disease diagnosis.Compared to traditional colorimetric and enzyme-linked immunosorbent assays, fluorescent probes have been viewed as a more privileged technique for the detection of enzymatic activities due to their high sensitivity, simplicity in design and preparation, and the ability for the in situ imaging of enzymatic activities in cells and in vivo.9–14 The rationale by which glycosidase probes are designed typically relied on the conjugation of one molecule of monosaccharide to the phenol group of a donor–acceptor (D–A) type fluorogen (Table S1†). This glycosylation reaction quenches the intrinsic fluorescence of the phenol dye through modulation of the intramolecular charge transfer (ICT) or photo-induced electronic transfer (PeT) process.15–18 Subsequently, hydrolysis of the glycosyl group from the dye by a selective glycosidase causes the fluorescence to recover (fluorescence off–on) (Fig. 1a). A number of fluorogenic glycosidase probes has been developed based on this strategy with good sensitivity and selectivity.18–21 To minimize background fluorescence and light scattering, the phenol glycosylation strategy has been extended to near-infrared (NIR) dyes for the monitoring of glycosidase activities in vivo.22–28 However, the previously used dyes for glycosidase detection generally suffer from aggregation-caused quenching (ACQ) in aqueous media,26,29 and are easily bound unselectively to biomacromolecules leading to possible false-positive signals.30–32
Here, we develop glycocluster-based probes characteristic of aggregation-induced emission (AIE) for the fluorogenic sensing and long-term cell imaging of a diverse range of glycosidases. We have previously developed a D–A-type AIE fluorogen (AIEgen) based on a tetraphenylethylene (TPE) core conjugated with a dicyano-methylene-4H-pyran (DCM) derivative (TPE–DCM or TD) with a reddish fluorescence emission (λemmax = 625 nm).33,34 TPE and DCM were conjugated to each other through an ethylene bridge to increase the delocalization of electrons in the system. Unlike previous reports that directly glycosylate phenol groups of conjugated dyes, we introduced a pair of dendritic arms on both phenol moieties of TPE (Fig. 1a). This enables the grafting of six copies of glycosyl epitopes pre-functionalized with an azido-triethyleneglycol to the fluorophore through azide–alkyne click chemistry (Fig. 1b). By comparison to a number of other synthetic glycoclusters bearing different linkages between the glycoside and the AIEgen, the dendritic strategy has proven to be the most effective yielding a sensitive fluorogenic response to glycosidases. The AIE-based glycoclusters were successfully used for (1) the fluorogenic detection of functionally diverse glycosidases including galactosidase, glucosidase and fucosidase, (2) the fluorescence imaging of the endogenous glycosidase activities in healthy and cancer cells, and during cell senescence, and (3) glycosidase-activated, long-term imaging of healthy and cancer cells.
Results and discussion
Synthesis and structure–activity relationship of TPE–DCM-based glycoclusters
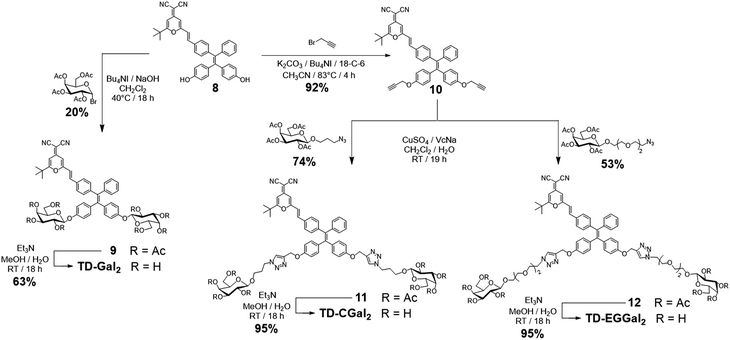
Four series of TPD–DCM-based glycoclusters with different linking patterns between TPE–DCM and monosaccharides were designed and synthesized; D-galactose was preliminarily used as a model for the study of structure–activity relationship.35–37 The galactosyl groups were introduced to the AIEgen by direct phenol glycosylation (linker free) (TD-Gal2) or azide–alkyne click chemistry conjugation through structurally different linker arms (short-arm, propyl: TD-CGal2; long-arm, triethyleneglycol: TD-EGGal2; dendritic-arm: TD-Gal6) (Schemes 1 and 2).The linker-free glycocluster TD-Gal2 was synthesized from intermediate 8. The known TPE-aldehyde 535 was coupled to DCM 638 through aldolization/crotonization. The resultant conjugate 7 with a red fluorescence emission showed a gradual fluorescence enhancement with increasing water fraction in THF, which is characteristic of AIE (Fig. S1†).39 Demethylation of 7 with BBr3 in CH2Cl2 afforded the bis-phenolic 8 (Scheme S1†). Subsequently, glycosylation under phase transfer conditions with bromo tetra-O-acetyl-D-galactose afforded the desired bis-glycosylated derivative 9. As expected, this bis-phenol glycosylation led to a poor yield of 20%; the stereoselectivity of the glycosylation was verified by analysis of the 1H NMR spectrum of the compound, suggesting a β-anomeric configuration (J1,2 = 8.2 Hz). Solvolysis of the acetyl protecting groups afforded the desired glycocluster TD-Gal2 (Scheme 1).
With TD-Gal2 in hand, we tested its fluorescence response to β-Gal (purified from E. coli)40 in a phosphate buffered saline (PBS) at pH 7.4 (Fig. S2a†). The fluorescence of the probe was observed to slightly decrease in the first 10 min, and then sharply increase from 10 to 40 min, in the presence of β-Gal. However, we determined that the fluorescence of TD-Gal2 in the absence of the enzyme also increased with time in PBS (pH 7.4). This suggests that (1) the initial fluorescence emission intensity of the probe in PBS was unstable probably due to a slow rate of molecular aggregation, and, thus exhibited a time-dependent AIE-based fluorescence enhancement, and (2) the two galactosyl residues modified on the phenol groups did not sufficiently quench the fluorescence of the AIEgen.
With the traditional phenol glycosylation strategy being unsatisfactory, we then sought to enhance the intrinsic AIE property of TPE–DCM through improving the water solubility of the glycoclusters. The better water solubility of the glycoclusters might lead to a lowered initial fluorescence emission of the system. When the glycosyl groups are removed from the dye, the decreased water solubility of TPE–DCM could lead to a drastically enhanced emission through AIE (Fig. 1a). To achieve this goal, additional linker arms (propyl, triethyleneglycol or dendritic) were introduced to TPE–DCM between the phenolic AIEgen and the galactosyl groups. The linker arms are retained on the dye scaffold even after hydrolysis of the glycosyl groups, thereby facilitating the formation of TPE–DCM-based amphiphilic micelles enhancing the AIE fluorescence.
The Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC) “click” chemistry was used for conjugation between the dye and monosaccharides.41–43 Starting from compound 8, propargylation was first attempted using potassium carbonate with propargyl bromide, which led to a poor chemical yield (<15%). Addition of Bu4NI and 18-crown-6 enhanced the reactivity of phenolates for a proper etherification in 92% yield to obtain the bis-propargyl compound 10 (Scheme 1). Galactosides with a 3-azidopropyl44 (short-arm) or azido-triethyleneglycol45 (long-arm) aglycon were then conjugated to compound 10 in the presence of CuSO4/sodium ascorbate (VcNa) as catalyst.33,46 Solvolysis of the ester protecting groups afforded the TD-CGal2 and TD-EGGal2 probes from compound 11 and 12, respectively (Scheme 1). The fluorescence of TD-CGal2 and TD-EGGal2 was then measured without β-Gal in PBS buffer at pH 7.4 (Fig. S2b and c†). Both glycoclusters showed a strong fluorescence emission, suggesting that the amphiphilic molecules are partially aggregated before reacting with the enzyme. Upon addition of the enzyme, the fluorescence of both probes decreased slightly. This might be the result of the formation of a poorly water-soluble AIE species after the enzymatic hydrolysis. These results suggest that the conjugation of only two glycosides onto the AIEgen is not sufficient to substantially enhance the water solubility of the dye.
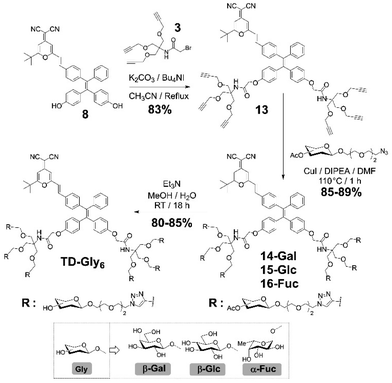
We then sought the possibility of introducing more than two glycosyl groups to the phenolic positions of the AIEgen in order to substantially enhance the water solubility, achieving an optimal “off–on” fluorescence response upon incubation with glycosidases. Consequently, a trivalent dendron was used to incorporate three glycosyl residues with each phenol group of TPE–DCM. Tris(hydroxymethyl)aminomethane (Tris) was converted into the tris-propargylated precursor 3 by a sequential synthetic route including Boc-protection, propargylation, removal of the Boc-carbamate and 2-bromo-acetamidation (Scheme 2). 2-Bromo-acetamide 3 was then conjugated to the TPE–DCM bis-phenol 8 to afford the hexakis-propargylated compound 13. CuAAC coupling of 13 with azido-triethyleneglycol galactoside was achieved under microwave activation to maximize the reactivity and completeness of the reaction.47,48 Removal of the protecting groups afforded the water-soluble hexavalent glycocluster TD-Gal6. The same synthetic strategy was also applied to the synthesis of the hexavalent glucosylated probe TD-Glc6 and fucosylated probe TD-Fuc6 (Scheme 2).
Subsequently, a comparative study of the AIE properties of TD-EGGal2, TD-Gal6, TD-Glc6 and TD-Fuc6 (10 μM) in a mixture solvent system of THF/PBS at different ratios (pH 7.4) was carried out (Fig. S3†). TD-EGGal2 displayed a typical fluorescence enhancement upon increase of PBS ratio in the solvent mixture (Fig. S3a†). This observation suggests that the presence of only two glycosides is not sufficient for TD-EGGal2 to remain well dispersible in water. In contrast, the fluorescence intensity of the hexakis-glycosylated probe TD-Gal6 in THF decreased with PBS (Fig. S3b†). The well quenched fluorescence of the probe in full PBS suggests that TD-Gal6 was hardly aggregated in aqueous solution. Other hexakis-glycosylated probes, TD-Glc6 (Fig. S3c†) and TD-Fuc6 (Fig. S3d†), showed a similar trend of fluorescence quenching with PBS ratio being increased. We noticed a slight fluorescence recovery of TD-Glc6 and TD-Fuc6 in 100% PBS, which we ascribe to a slightly different aggregation manner of the compounds in this fully aqueous environment. However, when the concentration of the probes was gradually decreased from 10 to 2.5 μM, the fluorescence enhancement became neglectable (Fig. S4†).
Glycosidase sensing with the AIE-based hexavalent glycoclusters
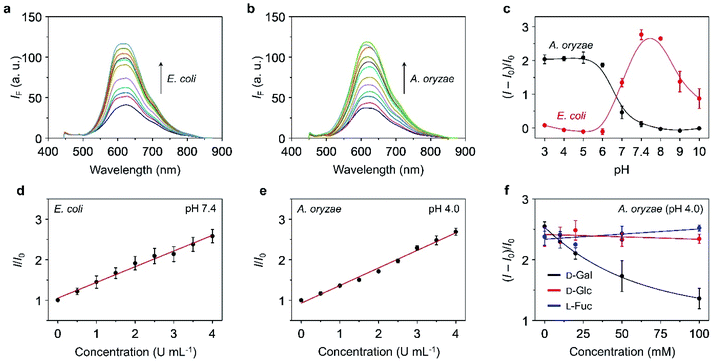
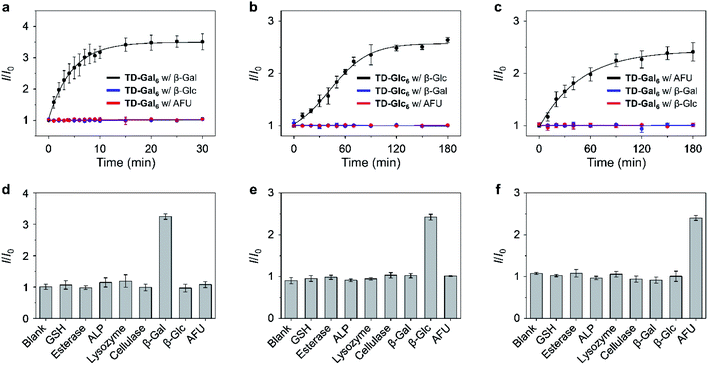
With the TPE–DCM-based hexavalent glycoclusters (TD-Gly6) in hand, we then tested their fluorescence responses to glycosidases in PBS. TD-Gal6 with six galactosyl epitopes was first used for the detection of β-Gal purified from bacteria. The β-Gal isolated from Escherichia coli (β-Gal/E. coli) has been extensively used for analysis of the sensing performances of fluorogenic β-Gal probes.28 However, the working pH of this enzyme is restrained to 6.5–7.5.40 Alternatively, Gao et al. reported the use of β-Gal purified from Aspergillus oryzae (β-Gal/A. oryzae) with a more acidic working pH range of 4.0–5.0, which is more similar to that of lysosomes.49,50 As a result, we attempted the detection of β-Gals isolated from different origins (bacterium or fungus) in order to examine the sensitivity of the AIE-based glycoclusters developed.TD-Gal6 was incubated separately with β-Gal/E. coli (pH 7.4) (Fig. 2a) and β-Gal/A. oryzae (pH 4.0) (Fig. 2b). We observed a concentration-dependent fluorescence enhancement of the probe for both galactosidases at their optimal pH. A good linearity was observed by plotting the fluorescence enhancement of TD-Gal6 as a function of the concentration (0–4.0 U mL−1) of β-Gal/E. coli (Fig. 2d) and β-Gal/A. oryzae (Fig. 2e) at pH 7.4 and 4.0, respectively. The limit of detection of the probe for β-Gal/E. coli and β-Gal/A. oryzae was determined to be 0.015 U mL−1 and 0.014 U mL−1, respectively. Comparing to previously reported fluorogenic probe, TD-Gal6 is among the most sensitive for β-Gal, and its unique structural feature of possessing multiple glycosyl epitopes endows the glycocluster with a better water solubility and potentially good biocompatibility for biological assays (Table S1†). In addition, we also determined that the AIE-based fluorescence enhancement is relevant to the specific working pH range of both enzymes (Fig. 2c). Within an acidic pH range (3–6), TD-Gal6 responded stably to β-Gal/A. oryzae, whereas the fluorescence of the probe only enhanced at a relatively neutral-basic pH (7–9) for β-Gal/E. coli. This suggests that the AIE-based glycocluster probe can differentiate the working pH of homogeneous glycosidases derived from different source (bacterium or fungus).
Next, the sensing properties of TD-Gal6 was tested in more detail. Owing to the sufficient water-solubility due to the presence of six galactosyl groups, the fluorescence of TD-Gal6 remained stable in PBS (pH 4.0 or 7.4) during 60 min of incubation (Fig. S5†). In the presence of β-Gal/E. coli, the fluorescence (ΦF = 0.005) of TD-Gal6 enhanced by ca. 5-fold (ΦF = 0.026) at pH 7.4 (Fig. S6†). Similarly, a drastic fluorescence enhancement of ca. 4.5-fold (ΦF from 0.006 to 0.027) was observed for the glycocluster with β-Gal/A. oryzae at pH 4.0 (Fig. S7†). This suggests that the cleavage of the glycosidic bonds in the probe by the enzyme reduced the hydrophilicity of the fluorescent dye, thereby leading to molecular aggregation. This action allowed for the detection of β-Gal through AIE effect in full PBS solution without the need for any organic solvent being added.
To better understand the sensing mechanism of TD-Gal6 for β-Gal, dynamic light scattering (DLS) and transmission-electron microscopy (TEM) were used to determine the hydrodynamic size and morphology of the molecular aggregates of TD-Gal6 in PBS solution before and after treatment with β-Gal, respectively (Fig. S8a and b†). The hydrodynamic diameter of the probe alone was determined to be around 10 nm in PBS buffer. In addition, the solution of TD-Gal6 displayed a negligible Tyndall effect, similar to that of control (distilled water) (Fig. S8c†). This observation agrees with the probe being well-dispersed in aqueous solution. After addition of β-Gal/E. coli at pH 7.4, the galactosyl groups on the probe were hydrolyzed to decrease the water-solubility of the dye, thereby producing more aggregated species with a particle size of 162 nm (Fig. S8b†). A bright light path was also observed, which we ascribed to the Tyndall effect, corroborating the aggregation with an enhanced fluorescence emission of the residues after enzyme hydrolysis (Fig. S8c†). In their representative TEM images, we also observed a morphological change of TD-Gal6 from amorphous to particle-like after being hydrolyzed by β-Gal (Fig. S8,† inset). The mass peak of the enzymatically hydrolyzed residue of TD-Gal6 with hexakis hydroxyl moieties was detected by mass spectrometry. Molecular ions of the residual products with one or two galactosides remaining on the dye scaffold could also be detected by mass spectrometry and are probably present in traces amounts (Fig. S9†). Moreover, a competition assay was also carried out. Pre-treatment of β-Gal/A. oryzae (pH 4.0) (Fig. 2f) and β-Gal/E. coli (pH 7.4) (Fig. S10a†) with free D-galactose (as β-Gal inhibitor) led to the concentration-dependent fluorescence suppression of TD-Gal6. In contrast, the pre-treatment of D-glucose and L-fucose did not cause the fluorescence of the probe to decrease in the presence of β-Gal/A. oryzae or β-Gal/E. coli. These data suggest that the enzymatic sensing was achieved by TD-Gal6 being selectively hydrolyzed by a galactosidase, leading to molecular aggregation with an enhanced fluorescence.
Next, we tested the fluorescence response of other hexavalent glycoclusters to their corresponding glycosidase. We observed a significant fluorescence enhancement of TD-Glc6 and TD-Fuc6 with a β-D-glucosidase (β-Glc, purified from almonds)51 and an α-L-fucosidase (AFU, purified from bovine kidney),12 respectively (Fig. S11†). By incubation of β-Glc (10 U mL−1) with TD-Glc6 for 2.5 h, a fluorescence enhancement of 3.7-fold (ΦF from 0.010 to 0.037) was achieved (Fig. S11a†). A good linearity was produced by plotting the fluorescence enhancement of TD-Glc6 as a function of increasing β-Glc from 0 to 2 U mL−1 (Fig. S12a†), and a LOD of 0.01 U mL−1 was determined (3δ/k, Fig. S12b†). This is lower than the previously reported glucosidase probes.51 Similarly, TD-Fuc6 displayed a fluorescence increase (1.6-fold, ΦF from 0.006 to 0.0096) after incubation with AFU (0.12 U mL−1) (Fig. S11b†). Meanwhile, the emission intensity of TD-Fuc6 enhanced gradually with an increase of AFU concentration from 0 to 0.12 U mL−1 (Fig. S13†).
A kinetic study showed that the fluorescence of the hexavalent AIE-based glycoclusters is enhanced in a time-dependent manner in the presence of the specific glycosidase, including TD-Gal6 with β-Gal (Fig. 3a/A. oryzae and Fig. S10b†/E. coli), TD-Glc6 with β-Glc (Fig. 3b) and TD-Fuc6 with AFU (Fig. 3c). In contrast, the long-time incubation of the glycoclusters with non-specific glycosidases TD-Gal6 with β-Glc/AFU (Fig. 3a and S10b†), TD-Glc6 with β-Gal/AFU (Fig. 3b) and TD-Fuc6 with β-Gal/β-Glc (Fig. 3c) did not cause the fluorescence of the probes to increase. Then, a broader range of relevant biomolecules including glutathione (GSH), esterase, alkaline phosphatase (ALP), lysozyme (Lyso) and cellulase (Cel) were used to assess the selectivity of the glycoclusters. To our delight, the glycoclusters showed a fluorescence enhancement only in the presence of their specific glycosidase (TD-Gal6 with β-Gal (Fig. 3d/A. oryzae and Fig. S10c†/E. coli), TD-Glc6 with β-Glc (Fig. 3e) and TD-Fuc6 with AFU (Fig. 3f)), corroborating the good enzyme specificity of the probes.
Fluorescence imaging of glycosidases in cells with the hexavalent glycoclusters
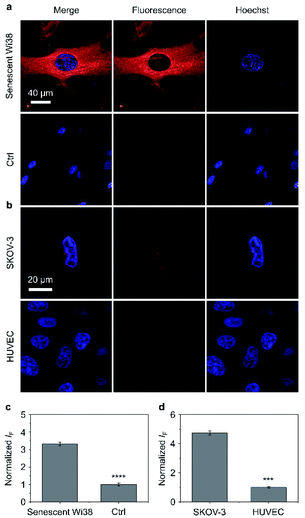
Having determined the outstanding sensitivity and selectivity of the AIE-based glycoclusters for glycosidases in solution, we turned our attention to the use of these probes for the fluorescence imaging of glycosidases activity in cells. β-Gal is a secretory protein that is over-expressed during cell senescence52 and in human ovarian cancer cells.53 The ability to accurately image β-Gal's activity at the cellular level can help advance glycobiology and offer useful tools for cancer diagnosis. Therefore, we examined the cell imaging performances of TD-Gal6 for endogenously produced β-Gal in Wi38 (fibroblasts derived from lung tissue) and SKOV-3 (human ovarian cancer) cells. We treated Wi38 cells with H2O2 leading to oxidative stress-induced cell senescence, and thus an overexpression of lysosomal β-Gal.3 For the cell imaging experiment with SKOV-3, HUVEC (human umbilical vein endothelial) cells that hardly express β-Gal were used as control (Fig. 4).54We determined that the fluorescence intensity of TD-Gal6 was negligible in untreated Wi38 cells (control) due to the absence of intracellular β-Gal. In contrast, in senescent Wi38 cells treated by H2O2, an intensive red fluorescence signal corresponding to the de-galactosylated AIE dye was observed (Fig. 4a). Similarly, TD-Gal6 produced a strong fluorescence in ovarian cancer cells (SKOV-3 cells, Fig. 4b), but not in HUVEC cells that lack the expression of β-Gal. A subsequent fluorescence quantification showed a 3.4-fold (Fig. 4c) and 4.7-fold (Fig. 4d) stronger fluorescence emission intensity of the glycocluster in senescent Wi38 and SKOV-3 cells than in untreated Wi38 and HUVEC cells, respectively. A cell viability assay showed that TD-Gal6 did not impact cell proliferation of SKOV-3 and Wi38 cells even at a concentration of 100 μM, which is 10-fold higher than that used for cell imaging (Fig. S14†). These data suggest the ability of the AIE-based glycocluster for the fluorescence-based imaging of intracellular glycosidase activity.
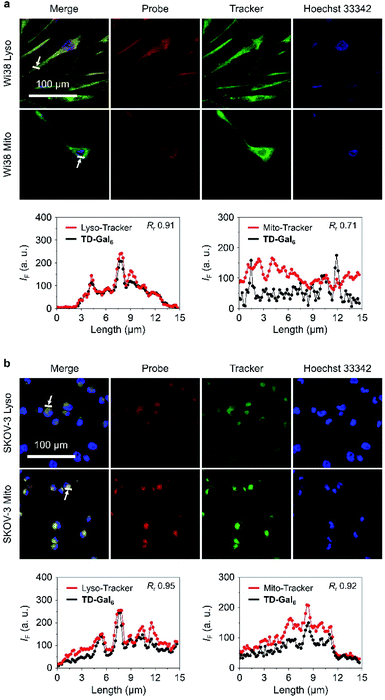
It is reported that the subcellular localization of endogenous β-Gal is different between senescent and ovarian cells.54 To test whether TD-Gal6 was able to differentiate the subcellular localization of β-Gal in different cell lines, a co-staining assay was carried out with commercial subcellular trackers including a Lyso-Tracker (for lysosomes), Mito-Tracker (for mitochondria) and Hoechst 33342 (for cell nucleus) (Fig. 5). We first observed that, the red fluorescence of the AIEgen produced by in situ β-Gal hydrolysis in senescent Wi38 cells overlapped well with the green fluorescence of Lyso-Tracker with a Pearson's coefficient of 0.91 (Fig. 5a). In contrast, a lower Pearson's coefficient of 0.71 was determined by overlapping the AIE fluorescence with that of the Mito-Tracker (Fig. 5a). By co-incubation of the subcellular trackers with TD-Gal6 in SKOV-3 cells, a good fluorescence overlap with both organelles was observed (Fig. 5b). A high Pearson's coefficient of 0.95 and 0.92 was determined for the probe with Lyso-Tracker and Mito-Tracker, respectively. These results prove the effectiveness of AIE-based glycoclusters to precisely localize β-Gal activity in cells with different pathological states.
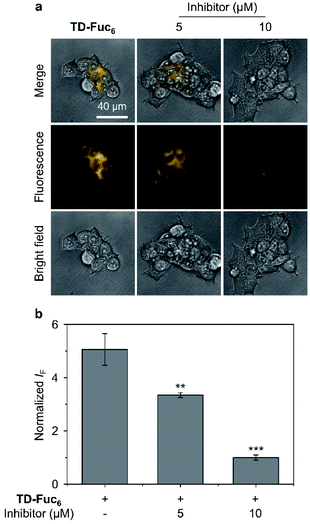
To demonstrate the scope of the AIE-based glycocluster for glycosidase sensing at the cellular level, we further used TD-Fuc6 for probing the endogenous AFU activity in 293T (human embryonic kidneys) cells.55 Cells pre-treated with a known AFU inhibitor (1-deoxyfuconojirimycin)56 were used as control. To our delight, an intensive fluorescence emission corresponding to the residual AIEgen of TD-Fuc6 after removal of the fucosyl epitopes was observed in the cytoplasm of 293T cells (Fig. 6a). However, the fluorescence intensity of the probe decreased gradually with increasing AFU inhibitor (5 μM and 10 μM) (Fig. 6). This suggests that the fluorescence production of TD-Fuc6 in 293T cells is dependent on the intracellular AFU activity. In addition, cell viability assay illustrated that TD-Fuc6 did not impact cell proliferation of 293T for 72 h incubation even at a concentration of 100 μM (Fig. S15†). A survey of previous literature reports demonstrates a scarcity in the achievement of fluorescence-based imaging of AFU activity in live cells. This is probably owing to the difficulty in the synthesis of fluorogenic α-L-fucosyl probes through direct phenol glycosylation. The successful development of TD-Fuc6 offers a powerful imaging tool for the in-depth study of AFU-relevant biological and pathological processes in different types of mammalian cells.
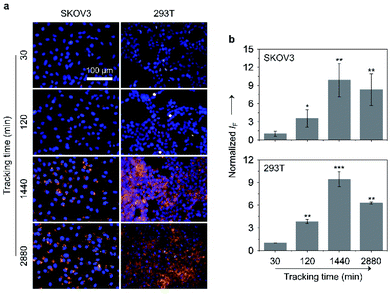
Large-sized aggregates and particles were shown to have a slower exocytotic rate than dispersed small molecules due to a prolonged intracellular retention thanks to the size effect.57–59 Given the AIE feature of the glycoclusters, we also tested their cell imaging capacity with time (Fig. 7). After treating SKOV-3 cells with TD-Gal6 for 30 min, a relatively weak fluorescence signal was observed probably due to the insufficient de-glycosylation of the glycoclusters intracellularly. This fluorescence was then enhanced when the incubation time was increased to 120 min, suggesting an increase in the proportion of de-glycosylated AIEgens. After a further prolonged incubation time of 1440 min (24 h), a ca. 9.9-fold increase in fluorescence intensity was observed with respect to the group with the incubation time of 30 min. Finally, the fluorescence was slightly reduced when the incubation time was increased to 2880 min (48 h). Likewise, a similar trend in fluorescence change was observed for TD-Fuc6 in 293T cells over time. The fluorescence of the probe maximized in 1440 min with a ca. 9.5-fold increase in fluorescence intensity with respect to the initial 30 min group, and then slightly decreased with a further prolonged incubation time of 2880 min. These data suggest that our glycosidase-responsive AIE-based glycoclusters are amenable to (1) monitoring the enzymatic activity of different glycosidases intracellularly, and (2) imaging of cells for more than 48 hours because of a slow exocytosis rate of the de-glycosylated AIEgen aggregates. The first point is particularly useful for the interrogation of the de-glycosylation process of a glycosidase of interest that is overly expressed during a given cellular process.
Conclusions
In summary, to overcome the synthetic limitations associated with previously reported fluorogenic glycosidase probes, we have developed AIE-based glycoclusters with excellent sensitivity and selectivity as a platform strategy for sensing the activity of diverse glycosidases in solution and in cells. A comparison of the sensing performances among four series of structurally different glycoclusters identified hexavalent TPE–DCM-based glycoclusters as the optimal structural motif for glycosidase sensing through AIE. The synthetic strategy avoids the direct glycosylation on phenol group of D–A-type fluorogens, leading to the acquisition of structurally more diverse glycosidase substrates based on the simple and efficient click coupling between an alkynylated TPE–DCM core and azido glycosides. Upon enzymatic hydrolysis of the glycosyl epitopes, the resultant amphiphilic AIEgens aggregated in water thereby producing a drastically enhanced, stable fluorescence signal.The AIE-based glycoclusters achieved (1) the fluorogenic detection of different glycosidases including galactosidase, glucosidase and fucosidase with excellent selectivity over a number of other enzymes that exist in the human body, (2) fluorescence-based imaging of the endogenous glycosidase activity in different cell types through intracellular de-glycosylation producing aggregated AIEgens, (3) differentiation of the subcellular localization of a galactosidase between cancer and senescent cells, and (4) study of the kinetics of the de-glycosylation action of different glycosidases, and the subsequent long-term fluorescent imaging of cells thanks to the slow exocytosis of AIEgen aggregates. Our research offers new insight into the development of AIE-based fluorogenic glycoclusters for glycosidase sensing, but addresses the scarcity in universal functional imaging tools for glycosidases, in particular fucosidases that have been identified to be useful cancer biomarkers.60
Data availability
Data for the synthesis and fluorescence properties of the designed molecules are available in the ESI.† Raw NMR data are available upon request.Author contributions
LG, MYZ, HHH synthesized the fluorescent probes and performed the fluorescence experiments in vitro and in cell assays, also described the experimental sections and drafted portions of the full text manuscript and ESI.† YZ, GRC, JL, XPH and SV directed the project, organized the data and discussion, drafted the text of the manuscript and ESI.†Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
S. V. thank the Université Claude Bernard Lyon 1 and the CNRS for financial support. The authors at ECUST are grateful to the financial support from the Natural Science Foundation of China (No. 21788102, 91853201 and 22107029), the Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX03) and the Fundamental Research Funds for the Central Universities (222201717003). L. D. thanks the China Scholarship Council for a PhD stipend (No. 201606740066). H.-H. H. would like to thank the Project funded by China Postdoctoral Science Foundation (No. 2020M681196). The authors are grateful to Fengming Liu, Yao Li, and Yang Yu of the Integrated Laser Microscopy System at the National Facility for Protein Science in Shanghai (NFPS), Zhangjiang Lab, SARI, China for their help in data collection.References
-
L. Weignerová, P. Bojarová and V. Křen, Glycosidases in synthesis, in Carbohydrate Chemistry, 2009, pp. 311–333 Search PubMed
.
- B. Y. Lee, J. A. Han, J. S. Im, A. Morrone, K. Johung, E. C. Goodwin, W. J. Kleijer, D. DiMaio and E. S. Hwang, Aging Cell, 2006, 5, 187–195 CrossRef CAS PubMed
.
- G. P. Dimri, X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj and O. Pereira-Smith, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 9363–9367 CrossRef CAS PubMed
.
- D. J. Spergel, U. Krüth, D. R. Shimshek, R. Sprengel and P. H. Seeburg, Prog. Neurobiol., 2001, 63, 673–686 CrossRef CAS PubMed
.
-
B. G. Winchester, Lysosomal metabolism of glycoconjugates, in Biology of the Lysosome, Springer, 1996, pp. 191–238 Search PubMed
.
- D. J. Becker and J. B. Lowe, Glycobiology, 2003, 13, 41R–53R CrossRef CAS PubMed
.
- H. Abdel-Aleem, A. Ahmed, A. Sabra, M. Zakhari, M. Soliman and H. Hamed, Int. J. Gynecol. Obstet., 1996, 55, 273–279 CrossRef CAS PubMed
.
- E. Waidely, A.-R. O. Al-Yuobi, A. Bashammakh, M. S. El-Shahawi and R. M. Leblanc, Analyst, 2016, 141, 36–44 RSC
.
- M. Safir Filho, P. Dao, M. Gesson, A. R. Martin and R. Benhida, Analyst, 2018, 143, 2680–2688 RSC
.
- E. Calatrava-Perez, S. A. Bright, S. Achermann, C. Moylan, M. O. Senge, E. B. Veale, D. C. Williams, T. Gunnlaugsson and E. M. Scanlan, Chem. Commun., 2016, 52, 13086–13089 RSC
.
- J. Y. Hyun, N. R. Kang and I. Shin, Org. Lett., 2018, 20, 1240–1243 CrossRef CAS PubMed
.
- X. Hou, J. Peng, F. Zeng, C. Yu and S. Wu, Mater. Chem. Front., 2017, 1, 660–667 RSC
.
- Y. L. Hsu, M. Nandakumar, H. Y. Lai, T. C. Chou, C. Y. Chu, C. H. Lin and L. C. Lo, J. Org. Chem., 2015, 80, 8458–8463 CrossRef CAS PubMed
.
-
(a) B. Thomas, K. C. Yan, X. L. Hu, M. Donnier-Marechal, G. R. Chen, X. P. He and S. Vidal, Chem. Soc. Rev., 2020, 49, 593–641 RSC
; (b) J. Zhang, X. Chai, X.-P. He, H.-J. Kim, J. Yoon and H. Tian, Chem. Soc. Rev., 2019, 48, 683–722 RSC
; (c) X. Chai, H.-H. Han, A. C. Sedgwick, N. Li, Y. Zang, T. D. James, J. Zhang, X.-L. Hu, Y. Yu, Y. Li, Y. Wang, J. Li, X.-P. He and H. Tian, J. Am. Chem. Soc., 2020, 142, 18005–18013 CrossRef CAS PubMed
; (d) W.-T. Dou, Z.-Y. Qin, J. Li, D.-M. Zhou and X.-P. He, Sci. Bull., 2019, 64, 1902–1909 CrossRef CAS
.
- X. Li, Y. Pan, H. Chen, Y. Duan, S. Zhou, W. Wu, S. Wang and B. Liu, Anal. Chem., 2020, 92, 5772–5779 CrossRef CAS PubMed
.
- Y. Zhang, C. Yan, C. Wang, Z. Guo, X. Liu and W. H. Zhu, Angew. Chem., Int. Ed., 2020, 59, 9059–9066 CrossRef CAS PubMed
.
- G. Jiang, G. Zeng, W. Zhu, Y. Li, X. Dong, G. Zhang, X. Fan, J. Wang, Y. Wu and B. Z. Tang, Chem. Commun., 2017, 53, 4505–4508 RSC
.
- X. X. Zhang, H. Wu, P. Li, Z. J. Qu, M. Q. Tan and K. L. Han, Chem. Commun., 2016, 52, 8283–8286 RSC
.
- H. M. Burke, T. Gunnlaugsson and E. M. Scanlan, Chem. Commun., 2015, 51, 10576–10588 RSC
.
- X. Cheng, Y. Huang, D. Li, C. Yuan, Z.-L. Li, L. Sun, H. Jiang and J. Ma, Sens. Actuators, B, 2019, 288, 38–43 CrossRef CAS
.
- M. Zhao, Z. Qian, M. Zhong, Z. Chen, H. Ao and H. Feng, ACS Appl. Mater. Interfaces, 2017, 9, 32887–32895 CrossRef CAS PubMed
.
- H. W. Liu, L. Chen, C. Xu, Z. Li, H. Zhang, X. B. Zhang and W. Tan, Chem. Soc. Rev., 2018, 47, 7140–7180 RSC
.
- H. W. Lee, C. H. Heo, D. Sen, H. O. Byun, I. H. Kwak, G. Yoon and H. M. Kim, Anal. Chem., 2014, 86, 10001–10005 CrossRef CAS PubMed
.
- J. Zhang, C. Li, C. Dutta, M. Fang, S. Zhang, A. Tiwari, T. Werner, F. T. Luo and H. Liu, Anal. Chim. Acta, 2017, 968, 97–104 CrossRef CAS PubMed
.
- E. J. Kim, R. Kumar, A. Sharma, B. Yoon, H. M. Kim, H. Lee, K. S. Hong and J. S. Kim, Biomaterials, 2017, 122, 83–90 CrossRef CAS PubMed
.
- K. Gu, W. Qiu, Z. Guo, C. Yan, S. Zhu, D. Yao, P. Shi, H. Tian and W. H. Zhu, Chem. Sci., 2019, 10, 398–405 RSC
.
- X. Li, W. Qiu, J. Li, X. Chen, Y. Hu, Y. Gao, D. Shi, X. Li, H. Lin, Z. Hu, G. Dong, C. Sheng, B. Jiang, C. Xia, C.-Y. Kim, Y. Guo and J. Li, Chem. Sci., 2020, 11, 7292–7301 RSC
.
- K. Gu, Y. Xu, H. Li, Z. Guo, S. Zhu, S. Zhu, P. Shi, T. D. James, H. Tian and W. H. Zhu, J. Am. Chem. Soc., 2016, 138, 5334–5340 CrossRef CAS PubMed
.
- X. Li, Y. Pan, H. Chen, Y. Duan, S. Zhou, W. Wu, S. Wang and B. Liu, Anal. Chem., 2020, 92, 5772–5779 CrossRef CAS PubMed
.
- F. Liu, J. Mu and B. Xing, Curr. Pharm. Des., 2015, 21, 1866–1888 CrossRef CAS PubMed
.
- Z. Liu and X. Chen, Chem. Soc. Rev., 2016, 45, 1432–1456 RSC
.
- J. Fan, W. Sun, Z. Wang, X. Peng, Y. Li and J. Cao, Chem. Commun., 2014, 50, 9573–9576 RSC
.
- L. Dong, G. R. Chen, X. P. He and S. Vidal, Org. Biomol. Chem., 2019, 17, 9251–9256 RSC
.
- L. Dong, M. Fu, L. Liu, H.-H. Han, Y. Zang, G.-R. Chen, J. Li, X.-P. He and S. Vidal, Chem.–Eur. J., 2020, 26, 14445–14452 CrossRef CAS PubMed
.
- X. Cai, C.-J. Zhang, F. Ting Wei Lim, S. J. Chan, A. Bandla, C. K. Chuan, F. Hu, S. Xu, N. V. Thakor, L.-D. Liao and B. Liu, Small, 2016, 12, 6576–6585 CrossRef CAS PubMed
.
- Z. Chen, W. Hu, M. Wang, L. Wang, G. Su and J. Wang, Carbohydr. Res., 2016, 429, 81–86 CrossRef CAS PubMed
.
- M. Rao, K. Kanagaraj, C. Fan, J. Ji, C. Xiao, X. Wei, W. Wu and C. Yang, Org. Lett., 2018, 20, 1680–1683 CrossRef CAS PubMed
.
- G. Zhao, Y. Zhu, S. Guang, F. Ke and H. Xu, New J. Chem., 2018, 42, 555–563 RSC
.
- L. Dong, H.-Q. Peng, L.-Y. Niu and Q.-Z. Yang, Top. Curr. Chem., 2021, 379 DOI:10.1007/s41061-021-00330-0
.
- G. R. Craven, E. Steers and C. B. Anfinsen, J. Biol. Chem., 1965, 240, 2468–2477 CrossRef CAS
.
- V. K. Tiwari, B. B. Mishra, K. B. Mishra, N. Mishra, A. S. Singh and X. Chen, Chem. Rev., 2016, 116, 3086–3240 CrossRef CAS PubMed
.
- X.-P. He, Y.-L. Zeng, Y. Zang, J. Li, R. A. Field and G.-R. Chen, Carbohydr. Res., 2016, 429, 1–22 CrossRef CAS PubMed
.
- M. Meldal and C. W. Tornøe, Chem. Rev., 2008, 108, 2952–3015 CrossRef CAS PubMed
.
- M. Rosselin, Z. S. Chinoy, L. M. Bravo-Anaya, S. Lecommandoux and E. Garanger, ACS Macro Lett., 2021, 10, 65–70 CrossRef CAS
.
-
S. Cecioni, M. Almant, J.-P. Praly and S. Vidal, Synthesis of Azido-Functionalized Carbohydrates for the Design of Glycoconjugates, in Carbohydrate Chemistry: Proven Synthetic Methods, ed. P. Kováč, CRC Press, Boca Raton, 2012, vol. 1, ch. 18, pp. 175–180 Search PubMed
.
- M. Donnier-Marechal, S. Abdullayev, M. Bauduin, Y. Pascal, M. Q. Fu, X. P. He, E. Gillon, A. Imberty, E. Kipnis, R. Dessein and S. Vidal, Org. Biomol. Chem., 2018, 16, 8804–8809 RSC
.
- C. Bouillon, A. Meyer, S. Vidal, A. Jochum, Y. Chevolot, J.-P. Cloarec, J.-P. Praly, J.-J. Vasseur and F. Morvan, J. Org. Chem., 2006, 71, 4700–4702 CrossRef CAS PubMed
.
- S. Cecioni, R. Lalor, B. Blanchard, J.-P. Praly, A. Imberty, S. E. Matthews and S. Vidal, Chem.–Eur. J., 2009, 15, 13232–13240 CrossRef CAS PubMed
.
- Y. Gao, Y. Hu, Q. Liu, X. Li, X. Li, C.-Y. Kim, T. D. James, J. Li, X. Chen and Y. Guo, Angew. Chem., Int. Ed., 2021, 60, 10756–10765 CrossRef CAS PubMed
.
- M. J. Bailey and M. Linko, J. Biotechnol., 1990, 16, 57–66 CrossRef CAS
.
- Y. Li, H. Wang, J. Li, J. Zheng, X. Xu and R. Yang, Anal. Chem., 2011, 83, 1268–1274 CrossRef CAS PubMed
.
- H. W. Lee, C. H. Heo, D. Sen, H.-O. Byun, I. H. Kwak, G. Yoon and H. M. Kim, Anal. Chem., 2014, 86, 10001–10005 CrossRef CAS PubMed
.
- D. Asanuma, M. Sakabe, M. Kamiya, K. Yamamoto, J. Hiratake, M. Ogawa, N. Kosaka, P. L. Choyke, T. Nagano, H. Kobayashi and Y. Urano, Nat. Commun., 2015, 6, 6463 CrossRef CAS PubMed
.
- X. Chai, H.-H. Han, A. C. Sedgwick, N. Li, Y. Zang, T. D. James, J. Zhang, X.-L. Hu, Y. Yu, Y. Li, Y. Wang, J. Li, X.-P. He and H. Tian, J. Am. Chem. Soc., 2020, 142, 18005–18013 CrossRef CAS PubMed
.
- K. Miura, T. Tsukagoshi, T. Hirano, T. Nishio and W. Hakamata, ACS Med. Chem. Lett., 2019, 10, 1309–1313 CrossRef CAS PubMed
.
- W. Haijun, S. Yin, Z. Lei and Y. Youfan, Curr. Med. Chem., 2021, 28, 628–643 CrossRef PubMed
.
- L. Zhou, F. Lv, L. Liu, G. Shen, X. Yan, G. C. Bazan and S. Wang, Adv. Mater., 2018, 30, 1704888 CrossRef PubMed
.
- J. Panyam and V. Labhasetwar, Pharm. Res., 2003, 20, 212–220 CrossRef CAS PubMed
.
- T. S. Wehrman, G. von Degenfeld, P. O. Krutzik, G. P. Nolan and H. M. Blau, Nat. Methods, 2006, 3, 295 CrossRef CAS PubMed
.
- E. Waidely, A.-R. O. Al-Yuobi, A. S. Bashammakh, M. S. El-Shahawi and R. M. Leblanc, Analyst, 2016, 141, 36–44 RSC
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc05057e |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2022 |