Open Access Article
Open Access ArticleGeneration of bright monomeric red fluorescent proteins via computational design of enhanced chromophore packing†
Sandrine
Legault
a,
Derek P.
Fraser-Halberg
 a,
Ralph L.
McAnelly
b,
Matthew G.
Eason
a,
Michael C.
Thompson
b and
Roberto A.
Chica
a,
Ralph L.
McAnelly
b,
Matthew G.
Eason
a,
Michael C.
Thompson
b and
Roberto A.
Chica
 *a
*a
aDepartment of Chemistry and Biomolecular Sciences, University of Ottawa, 10 Marie-Curie, Ottawa, Ontario K1N 6N5, Canada. E-mail: rchica@uottawa.ca
bDepartment of Chemistry and Biochemistry, University of California, Merced, Merced, California 95343, USA
First published on 11th January 2022
Abstract
Red fluorescent proteins (RFPs) have found widespread application in chemical and biological research due to their longer emission wavelengths. Here, we use computational protein design to increase the quantum yield and thereby brightness of a dim monomeric RFP (mRojoA, quantum yield = 0.02) by optimizing chromophore packing with aliphatic residues, which we hypothesized would reduce torsional motions causing non-radiative decay. Experimental characterization of the top 10 designed sequences yielded mSandy1 (λem = 609 nm, quantum yield = 0.26), a variant with equivalent brightness to mCherry, a widely used RFP. We next used directed evolution to further increase brightness, resulting in mSandy2 (λem = 606 nm, quantum yield = 0.35), the brightest Discosoma sp. derived monomeric RFP with an emission maximum above 600 nm reported to date. Crystallographic analysis of mSandy2 showed that the chromophore p-hydroxybenzylidene moiety is sandwiched between the side chains of Leu63 and Ile197, a structural motif that has not previously been observed in RFPs, and confirms that aliphatic packing leads to chromophore rigidification. Our results demonstrate that computational protein design can be used to generate bright monomeric RFPs, which can serve as templates for the evolution of novel far-red fluorescent proteins.
Introduction
Red fluorescent proteins (RFPs) are genetically-encoded fluorophores that have found widespread application in chemical and biological research due to their longer emission wavelengths that allow deeper tissue imaging1 and multicolor experiments.2,3 For most applications, bright monomeric RFPs are desirable. RFP brightness is typically improved by directed evolution involving multiple rounds of random mutagenesis followed by high-throughput screening of mutant libraries.4,5 Although directed evolution has successfully yielded many monomeric RFPs displaying enhanced brightness, it requires screening of large mutant libraries (≥103 sequences per round) due to its low-frequency and random sampling of sequence space, as well as its propensity to get trapped in local fitness optima.6,7 Thus, a computational methodology for increasing RFP brightness that can sample sequence space combinatorially could help to escape local optima and generate bright variants without the need for screening large mutant libraries, while yielding novel templates for directed evolution of brighter variants.Brightness of fluorescent proteins is defined as the product of their molar extinction coefficient and quantum yield, which are the ability of their chromophore to absorb light and its efficiency to convert the absorbed light into emitted light, respectively. Although increasing either of these two properties will proportionally increase brightness, it is not well understood how changes to the RFP structure can beneficially impact their extinction coefficient, complicating the prediction of beneficial mutations by rational design. On the other hand, it is known that the quantum yield of fluorophores is directly related to their conformational flexibility,8–10 as motions can dissipate the absorbed energy as heat instead of as photons. In the case of fluorescent proteins, it has been shown that twisting of the chromophore p-hydroxybenzylidene moiety via torsions in the methine bridge causes non-radiative decay.10,11 Therefore, it should be possible to enhance RFP brightness by designing mutations to restrict the conformational flexibility of the p-hydroxybenzylidene moiety, resulting in higher quantum yield.
Here, we used computational protein design with the Triad software12 to optimize packing of the chromophore pocket in the dim monomeric RFP mRojoA (quantum yield = 0.02), which we hypothesized would rigidify the chromophore and thereby enhance quantum yield. To do so, residues surrounding the p-hydroxybenzylidene moiety of the chromophore were mutated in silico to various combinations of aliphatic amino acids. The best mutant identified, mSandy1, contains six mutations that together cause an 11-fold enhancement to quantum yield relative to mRojoA, including the P63L mutation that has not previously been observed in natural RFPs or their engineered monomeric variants. We next used directed evolution to further increase brightness, which yielded mSandy2 (quantum yield = 0.35), the brightest Discosoma sp. derived monomeric RFP with an emission maximum greater than 600 nm reported to date. Crystallographic analysis confirmed that the chromophore p-hydroxybenzylidene moiety of mSandy2 was rigidified due to tight packing by aliphatic residues. Our results demonstrate the utility of computational protein design for increasing RFP quantum yield by enhancing chromophore packing, and generating novel templates for directed evolution of brighter variants. The approach developed here could be applied to other fluorescent proteins in order to increase their quantum yield and overall brightness.
Results
Computational design of enhanced chromophore packing
Algorithms for computational protein design (CPD) optimize amino-acid side-chain rotamers on a backbone template to stabilize a protein fold by improving non-bonding interactions such as van der Waals, hydrogen bonding, and electrostatic interactions,13,14 leading to increased atomic packing.15 These methods can therefore be used to predict amino-acid combinations to enhance packing of the chromophore p-hydroxybenzylidene moiety without destabilizing the protein fold, which we hypothesize will restrict its conformational flexibility and thereby increase quantum yield. CPD has successfully been used to design RFPs displaying red-shifted emission wavelengths,16 and create monomeric variants from oligomeric ones.17,18 CPD has also been used to repack the core of the blue fluorescent protein,19 yielding a brighter variant called Azurite.20 However, this required the high-throughput screening of a large computationally-designed combinatorial library comprising 3.3 × 105 sequences. Here, we build on this study and use CPD to repack the RFP chromophore pocket for enhanced quantum yield.As a starting template for design, we selected mRojoA, a variant of the widely-used monomeric RFP mCherry that we previously engineered for red-shifted fluorescence.16 mRojoA contains the V16T, R125H, Q163L, V195A, I197Y, and A217C mutations (numbering according to the mCherry structure, PDB ID: 2H5Q21) that together cause a 23 nm emission wavelength bathochromic shift that is accompanied by a 10-fold decrease in quantum yield (Table 1). We selected mRojoA as our design template because it is dim and its crystal structure is available (PDB ID: 3NEZ16). In mRojoA, the chromophore is sandwiched between the side chains of Tyr197, an aromatic residue that we introduced to red-shift the emission wavelength via pi-stacking interaction with the p-hydroxybenzylidene moiety, and Pro63, a residue that is conserved among all RFPs evolved from DsRed,22 the original RFP isolated from Discosoma sp.23 Presence of Pro63 results in suboptimal chromophore packing due to its conformationally-restricted cyclic side chain, which cannot adopt a rotamer able to form tight packing interactions against the planar p-hydroxybenzylidene moiety, resulting in a small cavity directly above the chromophore (Fig. 1a, ESI Fig. 1†). Therefore, it should be possible to enhance chromophore packing for efficient rigidification by mutating Pro63 and other residues surrounding the p-hydroxybenzylidene moiety to non-cyclic aliphatic amino acids (e.g., Val, Leu, Ile, and Met), whose bulky and/or branched aliphatic side chains could adopt rotameric configurations able to tightly pack the chromophore pocket.
| Proteina | λ ex (nm) | λ em (nm) | Quantum yield | Extinction coefficient (mM−1 cm−1) | Brightnessb (mM−1 cm−1) | Mutations relative to mRojoA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T16 | P63 | H125 | W143 | I161 | L163 | L165 | D174 | N194 | A195 | Y197 | L199 | C217 | ||||||
| a n ≥ 3 independent experiments for mRojoA, mRojoA-Y197I, mSandy1, mSandy2, and mCherry (mean ± s.d). n = 1 independent experiment for all other variants. Each independent experiment consists of triplicate measurements performed on an individual protein batch. b Brightness is the product of the quantum yield and extinction coefficient mean values. | ||||||||||||||||||
| mRojoA | 596 ± 2 | 632 ± 2 | 0.023 ± 0.001 | 68 ± 9 | 2 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| mRojoA-Y197I | 587 ± 1 | 618 ± 1 | 0.076 ± 0.003 | 75 ± 2 | 6 | — | — | — | — | — | — | — | — | — | — | I | — | — |
| mSandy0.1 | 585 | 608 | 0.13 | — | — | V | L | R | S | V | — | — | — | — | — | V | — | — |
| mSandy0.7 | 584 | 612 | 0.16 | 94 | 15 | V | L | R | S | — | — | — | — | — | — | V | — | — |
| mSandy0.9 | 585 | 608 | 0.13 | — | — | V | L | R | S | V | V | — | — | — | — | V | — | — |
| mSandy1 | 584 ± 1 | 609 ± 2 | 0.26 ± 0.02 | 81 ± 5 | 21 | V | L | R | S | V | — | — | — | — | — | I | — | — |
| mSandy2 | 581 ± 1 | 606 ± 1 | 0.35 ± 0.03 | 79 ± 9 | 28 | V | L | — | S | V | V | F | V | Y | — | I | M | — |
| mSandy1-L163V/L199M | 580 | 606 | 0.28 | — | — | V | L | R | S | V | V | — | — | — | — | I | M | — |
| mCherry | 586 ± 2 | 609 ± 1 | 0.23 ± 0.01 | 87 ± 3 | 20 | V | — | R | — | — | Q | — | — | — | V | I | — | A |
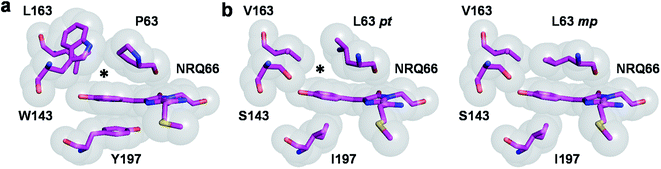 | ||
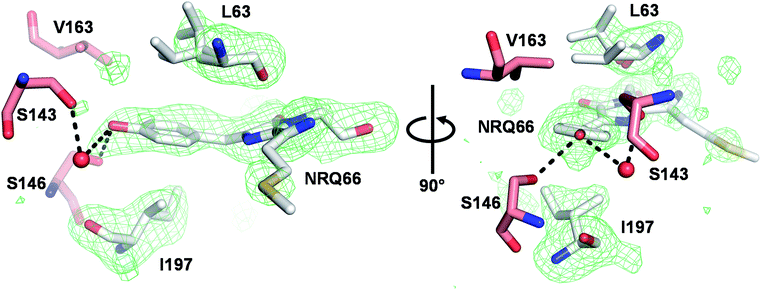
| Fig. 1 RFP chromophore pocket. (a) The chromophore pocket in mRojoA (chain A, PDB ID: 3NEZ) contains a small cavity above the chromophore p-hydroxybenzylidene moiety (indicated by an asterisk) due to the cyclic structure of P63. (b) This cavity is filled in mSandy2 (chain H, PDB ID: 7RY2) by aliphatic residue L63 when it adopts the mp but not pt rotamer. Occupancy of the mp rotamer varies between 34–53% in the 8 molecules of the crystal asymmetric unit. All images generated using PyMOL 2.5.0. | ||
Using multistate design with an ensemble of four mRojoA templates to approximate backbone flexibility (Methods), which has been shown to increase prediction accuracy compared to single-state design with a single fixed backbone template,24,25 we designed six first-shell residues surrounding the chromophore (ESI Fig. 2a†) and allowed only aliphatic amino acids at these positions with the exception of position 143, where we also allowed Ser because this amino acid has been shown to contribute to enhanced brightness in related RFPs.26 We also reverted the V16T mutation of mRojoA because polar residues at this position have been shown to hinder red chromophore maturation,27 and the R125H surface mutation to enhance solubility in the monomeric state.4 In contrast with the previous computational design of Azurite,20 which applied a library design algorithm to construct a large combinatorial library (>105 mutant sequences) based on optimal degenerate codons computed for each designed residue position, we directly analyzed the top-ranked individual sequences output by Triad. Our approach has the benefit of focusing exclusively on sequences predicted to be stable, in contrast with the Azurite library design approach where sequences were generated by combining design-specified degenerate codons, which can yield destabilized sequences if mutations are incompatible together. The top 10 designed sequences (ESI Fig. 2b†) contained between 4 and 7 mutations, including P63L directly on top of the chromophore, and W143S, which is likely required to accommodate the bulky Leu63 side chain since Ser is the smallest amino acid that we allowed at this position during design. At the other four design positions, various combinations of aliphatic residues were observed in the top 10 ranked sequences, which were selected for experimental characterization.
Spectral characterization of designs
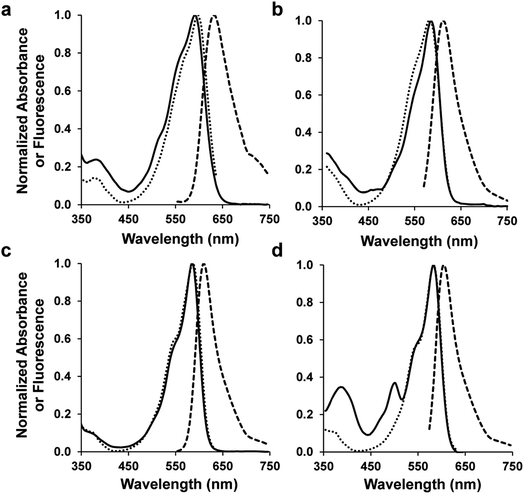
Four of the 10 designed sequences showed strong fluorescence with emission maxima around 610 nm (ESI Fig. 3†), which represent hypsochromic shifts of approximately 20 nm relative to mRojoA. These results can be partially ascribed to reversion of the V16T and I197Y mutations that are known to cause emission wavelength bathochromic shifts of 4 and 7 nm, respectively, when introduced into mCherry.16 These variants, which we call mSandys (Table 1 and Fig. 2, ESI Fig. 4†), all display quantum yield increases of 6–11-fold relative to mRojoA, representing absolute quantum yield increases of 0.11–0.24. The brightest variant, mSandy1, is as bright as mCherry with a quantum yield of 0.26 and a brightness of 21 mM−1 cm−1. mSandy1 also displays efficient red chromophore maturation, with no apparent green chromophore peak (λ = 510 nm, ESI Fig. 5†) in its absorption spectrum (Fig. 2), similar to what is observed for the efficiently-maturing mCherry. This is not the case for the other variants, which all display strong absorption peaks corresponding to the green chromophore (ESI Fig. 4†). These results indicate that aliphatic packing of the chromophore pocket can be detrimental to efficient chromophore maturation, possibly due to a trade-off between tight chromophore packing and maturation efficiency, since maturation requires sufficient flexibility for the chromophore-forming tripeptide to cyclize, oxidize, and dehydrate.27,28 Importantly, the comparable properties of mSandy1 and mCherry are not simply due to high sequence similarity between these two RFPs. As shown on Table 1, mSandy1 is as different from mCherry as it is from mRojoA, containing 6 mutations relative to either RFP. This result demonstrates how different sequence changes can produce similar functional effects in RFPs, and highlights the benefit of using CPD to identify improved sequences containing many mutations, which is an outcome that is difficult to achieve in a single round of directed evolution.Of the bright mSandy variants, mSandy1 is the only one that contains the Y197I mutation, which reverts the Tyr found at this position in mRojoA to the corresponding Ile found in mCherry. Since the single point mutation I197Y causes a large quantum yield decrease of 0.19 when introduced by itself into mCherry,16 we verified whether the Y197I reversion present in mSandy1 was primarily responsible for the large quantum yield increase observed in this variant. As shown on Table 1, the single point mutant mRojoA-Y197I displays an absolute quantum yield increase relative to mRojoA of approximately 0.05, less than a quarter of the increase obtained with mSandy1 (0.24). This result confirms that presence of the Y197I mutation alone is not sufficient to account for the high quantum yield of mSandy1, and that the other computationally-designed mutations contribute synergistically to enhance this property.
Evolution of mSandy2
Given that mSandy1 is as bright as mCherry but contains an amino acid at position 63 (Leu63) that has never been observed in natural fluorescent proteins and their engineered monomeric variants (ESI File 1†), we postulated that it could serve as a good starting template for the directed evolution of brighter monomeric RFPs by helping to escape the local fitness optimum of Discosoma-derived RFPs such as mCherry. We therefore subjected mSandy1 to random mutagenesis followed by high-throughput screening for enhanced brightness. After three rounds of directed evolution (Methods), we isolated mSandy2 (Table 1 and Fig. 2), a variant whose emission wavelength is not significantly altered but that displays an absolute quantum yield increase of 0.09 relative to mSandy1, making it approximately 33% brighter than its parent. mSandy2 is also 40% brighter than mCherry, an RFP that emits at a similar wavelength. However, chromophore maturation in mSandy2 is less efficient than in either mSandy1 or mCherry, as evidenced by the peaks at approximately 390 and 510 nm in its absorption spectrum (Fig. 2d) that correspond to neutral and anionic green chromophores (ESI Fig. 5†), respectively.27,28 Presence of these absorption peaks indicates a substantial amount of green chromophore-containing molecules in the RFP population in solution.mSandy2 contains six mutations from mSandy1, including two that occurred at designed positions (L163V and L199M). To evaluate whether these two mutations were sufficient to account for the large quantum yield increase in this variant, we generated the mSandy1-L163V/L199M double mutant. These two mutations together do not significantly improve the quantum yield of mSandy1 (Table 1), and are detrimental to chromophore maturation, as evidenced by the low overall absorbance of the red chromophore peak in the absorption spectrum (ESI Fig. 4†). The other four mutations are therefore responsible for increasing the quantum yield and compensating for the maturation deficiency caused by the L163V/L199M combination. Interestingly, three of the remaining four mutations occur on the RFP β-barrel surface, including a reversion of the H125R mutation that we introduced to favor monomer solubility. Nevertheless, these surface mutations do not cause oligomerization of mSandy2, which remains a monomer in solution (ESI Fig. 6†).
Crystal structure of mSandy2
We next solved the crystal structure of mSandy2 to evaluate the structural factors contributing to its high quantum yield. Crystals of mSandy2 were obtained at pH 8.5, and these diffracted at a resolution of 2.05 Å (Table 2). The unit cell corresponded to space group P212121 with eight molecules in the asymmetric unit. In all eight of these molecules, the p-hydroxybenzylidene moiety of the chromophore is sandwiched between the side chains of Leu63 and Ile197 (Fig. 3), a structural motif that has not been previously observed in fluorescent proteins. In these structures, the Leu63 side chain adopts two distinct rotamers (Fig. 1b). The mp rotamer (χ1 = gauche(−), χ2 = gauche(+)), which is found at this position in our computational model of mSandy1 (ESI Fig. 7†), fills the cavity above the p-hydroxybenzylidene moiety of the chromophore by packing tightly against it. In contrast, the alternate pt rotamer (χ1 = gauche(+), χ2 = trans), does not (ESI Fig. 1†). Interestingly, the mp rotamer of Leu63 is the minor conformer in all but two of the mSandy2 molecules in the crystal asymmetric unit, with occupancies between 34% and 53% (ESI Fig. 8†). Nevertheless, the Leu63 mp rotamer likely contributes to reduce non-radiative torsional motions within the chromophore p-hydroxybenzylidene moiety, leading to enhanced quantum yield.| mSandy2 | |
|---|---|
| a Highest resolution shell is shown in parentheses. | |
| PDB ID | 7RY2 |
| Crystallization conditions | 100 mM Tris buffer (pH 8.5) |
| 100 mM NaCl | |
| 20% PEG-3350 | |
| 34 mg mL−1 protein | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| Data collection | |
| Resolution (Å) | 80.31–2.05 |
| Space group | P212121 |
| Cell params. | |
| a, b, c (Å) | 59.68, 147.21, 240.94 |
| α, β, γ (°) | 90, 90, 90 |
| Molecules per asymm. unit | 8 |
| R pim | 0.052 (0.890) |
| CC1/2 | 0.999 (0.396) |
| I/σI | 11.9 (1.0) |
| Completeness (%) | 99.9 (97.3) |
| Multiplicity | 13.0 (10.9) |
| # unique reflections | 133![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 939 (6432) 939 (6432) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| Refinement | |
| R work/free | 0.1897/0.2227 |
| No. atoms | |
| Protein | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 901 901 |
| Chromophore | 184 |
| Water | 652 |
| RMSD | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.665 |
| Molprobity statistics | |
| All-atom clashscore | 1.47 |
| Ramachandran outliers (%) | 0.00 |
| Ramachandran allowed (%) | 2.02 |
| Ramachandran favored (%) | 97.98 |
| Rotamer outliers (%) | 0.13 |
To verify whether aliphatic packing mutations increase chromophore rigidity, we analyzed B-factors of individual chromophore atoms, which correspond to their average displacement in the crystal. Since both conformational flexibility and crystalline disorder can contribute to atomic B-factors, we calculated their Z-scores to account for variations between different crystal forms or different molecules in the asymmetric unit, and compared those of mSandy2 with those of mRojoA. B-Factor Z-scores for the chromophore p-hydroxybenzylidene moiety are lower in mSandy2 than in its dim parent mRojoA (Fig. 4, ESI Fig. 9†), confirming that this chromophore moiety is more rigid in mSandy2. This result confirms that the computationally-designed aliphatic packing mutations, in combination with those found by directed evolution, cause p-hydroxybenzylidene rigidification leading to quantum yield increases.
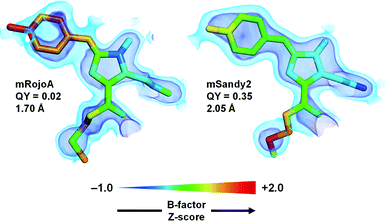 | ||
| Fig. 4 Chromophore rigidity. The red chromophores of mRojoA (chain A, PDB ID: 3NEZ) and mSandy2 (chain H, PDB ID: 7RY2) are shown as sticks with individual atoms colored according to their B-factor Z-score. The 2Fo − Fc map is shown in volume representation at two contour levels: 0.5 and 1.5 e Å−3 in light and dark blue, respectively. The crystal structure resolution and quantum yield (QY) are indicated for each protein. | ||
Discussion
In this work, we used CPD to optimize packing of the RFP chromophore with aliphatic residues to reduce its conformational flexibility and thereby enhance its quantum yield and overall brightness. In the brightest computationally designed variant, mSandy1, aliphatic packing of the chromophore causes an absolute quantum yield increase of 0.24 relative to the mRojoA parent without significantly affecting the molar extinction coefficient. This results in a brightness increase of approximately one order of magnitude. The quantum yield increase obtained by aliphatic packing described here is larger than those obtained by aromatic packing in triple-decker RFPs, mRojoA variants whose chromophore p-hydroxybenzylidene moiety is sandwiched between the side chains of an aromatic amino acid at position 63 (e.g., His, Phe, Tyr), and Tyr197.26 For example, the brightest triple-decker RFP, mRojo-VYGV, displays a modest absolute quantum yield increase of 0.03 and a 2-fold increase in brightness relative to mRojoA at pH 7.4.26 The approximately 5-fold higher quantum yield of mSandy1 (0.26) compared to mRojo-VYGV (0.05) suggests that aliphatic packing is a more efficient strategy for increasing RFP brightness than aromatic packing, possibly due to quenching by photoinduced electron transfer that could occur between the chromophore and neighboring aromatic residues in the triple-decker RFPs.29The aliphatic packing approach described here compares favorably with the computational core repacking procedure used to generate Azurite.20 Although the quantum yield increases obtained by these two CPD methods are similar (absolute quantum yield increases of 0.22 and 0.24 for Azurite and mSandy1, respectively), our aliphatic packing approach required the experimental screening of a total of 10 mutant sequences, a number several orders of magnitude smaller than those used to obtain Azurite (105 sequences). Furthermore, the V224R mutation that was largely responsible for the brightness increase in Azurite was not specified by the design procedure and was instead found due to the unintended incorporation of a more inclusive codon at position 224. We propose that our aliphatic packing procedure based on multistate design is more accurate than the single-state design protocol that yielded Azurite because it uses an ensemble of backbone templates to approximate conformational flexibility, which has been shown to result in greater prediction accuracy by reducing the number of false negative predictions.24,30 In addition, the library design algorithm used to create Azurite predicted an optimum degenerate codon at each designed residue position, but did not evaluate the energies of resulting individual sequences made from these combinations of amino acids. It is thus likely that many of these designed sequences were destabilized due to incompatible combinations of mutations that would be otherwise stabilizing in a different context. In contrast, we evaluated the top-ranked individual sequences to ensure that those mutation combinations were able to tightly pack the chromophore without destabilizing the protein fold. Importantly, our computational design approach also required the testing of a number of sequences that is many orders of magnitude lower than the number of sequences that are typically required to achieve comparable increases in quantum yield by directed evolution.4,5 These results highlight the advantage of CPD, which is to facilitate the identification of beneficial combinations of mutations that would have been difficult to predict from sequence comparison or to obtain through random mutagenesis.16
Directed evolution on the computationally designed mSandy1 yielded mSandy2, which is, to the best of our knowledge, the brightest Discosoma sp. derived monomeric RFP with an emission maximum above 600 nm reported to date. The quantum yield of mSandy2 (0.35) is comparable to those of mRuby (0.35) and mKate2 (0.40), two monomeric RFPs that have been used in many imaging applications.31,32 mRuby emits at the same wavelength as mSandy2 but is 40% brighter (39.2 mM−1 cm−1) due to a higher reported extinction coefficient. On the other hand, mKate2 is red-shifted by 27 nm but is approximately 10% less bright than mSandy2 (25 mM−1 cm−1). Interestingly, mRuby and mKate2 were engineered from natural RFPs isolated from the Entacmaea quadricolor sea anemone (eqFP611 and eqFP578, respectively), and both contain a Thr residue at position 63. In the crystal structures of these proteins (PDB ID: 3BXB33 and 3U0N34), the Thr63 side-chain methyl group packs against the chromophore p-hydroxybenzylidene moiety, likely contributing to chromophore rigidification. While several monomeric far-red fluorescent proteins (λem > 650 nm) with quantum yields ≥0.10 have been evolved from eqFP578 and eqFP611 (e.g., mKelly1,18 mMaroon1,35 and mGarnet36), to our knowledge, none have been successfully engineered from either DsRed23 or HcRed,37 and only relatively dim variants (quantum yield ≤ 0.10) with emission maxima <650 nm have been produced (e.g., mRouge,16 mPlum,38 mGinger1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 18). Presence of Thr at position 63 could partially explain why RFPs from Entacmaea quadricolor have been better templates for evolving far-red fluorescent proteins than those from Discosoma sp. (DsRed) or Heteractis crispa (HcRed), which instead contain a Pro residue at the corresponding position. Since RFP quantum yield tends to decrease as emission wavelength increases, we propose that the engineering of novel far-red fluorescent proteins could be initiated from a template where the chromophore p-hydroxybenzylidene moiety is already tightly packed. Our mSandy variants could be useful for this purpose.
18). Presence of Thr at position 63 could partially explain why RFPs from Entacmaea quadricolor have been better templates for evolving far-red fluorescent proteins than those from Discosoma sp. (DsRed) or Heteractis crispa (HcRed), which instead contain a Pro residue at the corresponding position. Since RFP quantum yield tends to decrease as emission wavelength increases, we propose that the engineering of novel far-red fluorescent proteins could be initiated from a template where the chromophore p-hydroxybenzylidene moiety is already tightly packed. Our mSandy variants could be useful for this purpose.
Although mSandy2 is bright, further improvements to its quantum yield should be possible. This could be achieved by using CPD to design second-shell mutations to stabilize the Leu63 side chain in the mp conformation that allows it to pack tightly against the chromophore p-hydroxybenzylidene. In addition to helping rigidify the chromophore, mutations introduced in the second shell should be less likely to affect the emission wavelength, as they are not in direct contact with the chromophore. However, increased chromophore rigidity could result in lower maturation efficiency, as is the case for mSandy2, but this undesirable property could be corrected by applying a negative screening procedure to eliminate bright RFPs that also emit high levels of green fluorescence.39 The computational aliphatic packing approach described here could also be applied to increase the brightness in other Pro63-containing RFPs such as mGinger1 (λem = 637 nm, quantum yield = 0.02)18 and mPlum (λem = 649 nm, quantum yield = 0.10),38 and in near-infrared fluorescent proteins derived from bacterial phytochromes40 by helping to improve packing of the biliverdin chromophore for increased rigidity. While many RFPs are now available, novel variants displaying high brightness in the far-red or near-infrared optical window are still desired to enable various types of applications. In this context, the CPD approach for enhancing quantum yield described here can expedite the creation of novel bright RFP templates for further engineering.
Methods
Computational protein design
All calculations were performed with the Triad protein design software (Protabit, Pasadena, CA, USA). The structure of mRojoA was obtained from the Protein Data Bank (PDB code: 3NEZ16). Following extraction of protein heavy-atom coordinates for the highest occupancy conformer, hydrogen atoms were added and the resulting structure was energy-minimized within Triad by performing 100 steps of conjugate gradient energy minimization in Cartesian space using the Phoenix energy function (see below) with added covalent terms from the Dreiding force field.41 Individual chains of the minimized mRojoA structure were extracted to generate an ensemble of 4 backbone templates for multistate design.Multistate design using a Monte Carlo with simulated annealing search algorithm for rotamer optimization and a fitness function corresponding to the Boltzmann weighted average energy (Temperature = 300 K) for sequence scoring was performed using the multi_design.py app within Triad. The 2002 backbone-dependent Dunbrack rotamer library42 with expansions of ±1 standard deviation around χ1 and χ2 was used to provide amino-acid side-chain conformations to be threaded onto each backbone template. Chromophore rotamers were generated by allowing all 36 combinations of the following dihedral angles around the indicated bonds (ESI Fig. 10†): CA2–CB2–CG2–CD1 (−15°, −10°, −5°, 0°, +5°, and +10°), N2–CA2–CB2–CG2 (−5°, 0°, +5°, +10°, +15°, and +20°). These angles were selected to encompass the range of values observed in the crystal structures of mCherry, mRojoA, and other related RFPs. Side-chain rotamers of first-shell residues surrounding the p-hydroxybenzylidene moiety of the chromophore (ESI Fig. 2a†) were optimized on each backbone template using the following amino acids: residue 63 (I, L, V), 143 (I, L, S, V), 161 (I, L, M, V), 163 (I, L, M, V), 197 (I, L, V), and 199 (I, L, M, V). Side-chain rotamers of second-shell residues S62, K70, M97, L165, V177, and E148 were also optimized, and the T16V mutation was introduced. The searched sequence space thus consisted of 2304 sequences.
Sequences were scored on each backbone template using the Phoenix energy function,16,43 which consists of a Lennard-Jones 12-6 van der Waals term from the Dreiding II force field41 with atomic radii scaled by 0.95, a direction-dependent hydrogen bond term with a well depth of 8.0 kcal mol−1 and an equilibrium donor–acceptor distance of 2.8 Å,44 an electrostatic energy term modelled using Coulomb's law with a distance-dependent dielectric of 10, an occlusion-based solvation potential with scale factors of 0.05 for nonpolar burial, 2.5 for nonpolar exposure, and 1.0 for polar burial,16 and a secondary structural propensity term with amino acid phi–psi propensities derived from the method of Shortle.45 The top 10 ranked sequences (ESI Fig. 2b†) were experimentally characterized.
Red fluorescent protein genes
Amino-acid sequences for all RFPs described here are listed on ESI Table 1.† His-tagged (N-terminus) genes for mRojoA and mCherry cloned into pET-11a (Novagen) were a gift from Stephen Mayo.16 Codon-optimized and his-tagged (N-terminus) genes for mRojoA variants cloned into the pET-29b(+) vector (Novagen) via NdeI and XhoI were obtained from Twist Bioscience. All plasmids were transformed into E. coli BL21-Gold (DE3) cells for protein expression. The mSandy2 gene was prepared as described below. All open-reading frames were verified by DNA sequencing.Random mutagenesis
Three rounds of random mutagenesis, starting from the mSandy1 gene and then on the pooled RFP genes from the top 1% cells obtained after cell sorting (see below), were performed using the protocol described by McCullum et al.46 Briefly, four cycles of error-prone polymerase chain reaction using Taq DNA polymerase (5 U) in 1× standard Taq buffer pH 8.3 (New England Biolabs) supplemented with MnCl2 (0.5 mM) and a mixture of deoxynucleotides (1 mM dTTP, 1 mM dCTP, 0.2 mM dATP, and 0.2 mM dGTP), were used to introduce approximately two mutations on the 732-bp RFP open reading frame. The resulting mutant library was cloned into pET-11a (Novagen) via NdeI and BamHI. Plasmids were transformed into E. cloni 10G Elite electrocompetent cells (Lucigen) and plated onto lysogeny broth (LB) agar supplemented with 100 μg mL−1 ampicillin. Following incubation at 37 °C overnight, all colonies on the agar plate were collected using 1 mL of LB supplemented with 100 μg mL−1 ampicillin, and the plasmid pool was harvested by DNA extraction (E.Z.N.A. Plasmid DNA Mini Kit, Omega Bio-tek). Purified plasmid DNA was then transformed into electrocompetent E. coli BL21-Gold (DE3) cells (Agilent) for protein expression.Fluorescence-activated cell sorting
BL21-Gold (DE3) cells containing the mSandy1 random library obtained as described above were used to inoculate 100 mL of LB supplemented with 100 μg mL−1 ampicillin. Cells were grown at 37 °C with shaking until they reached an optical density of 0.6 at 600 nm. At this point, cells expressing random mSandy1 mutants were harvested by centrifugation. Cells were washed twice with sterile phosphate buffer (20 mM sodium phosphate buffer, pH 7.4, 50 mM sodium chloride). Pellets were stored at 4 °C to allow for chromophore maturation. After two days, cells were resuspended in sterile phosphate buffer to a concentration of 107 cells per mL, and filtered using 40 μm cell strainers (Falcon).Cells were sorted (top 1%) using a MoFlo Astrios EQ cell sorter (Beckman Coulter) according to fluorescence intensity (λex = 561 nm, λem = 614 nm) into sterile flow cytometry test tubes (Beckman Coulter) containing LB supplemented with 100 μg mL−1 ampicillin. Sorted cells were washed twice with supplemented LB and grown overnight in 50 mL cultures at 37 °C with shaking. 5 mL of this culture was used for extraction of plasmid DNA. Two additional rounds of random mutagenesis and cell sorting were performed using the procedures described above. Cells collected from the third round of sorting were used to inoculate LB agar plates supplemented with 100 μg mL−1 ampicillin. Following overnight incubation at 37 °C, 96 individual colonies were transferred onto a Nunc MicroWell 96-well polypropylene plate (Thermo Scientific) containing 250 μL of LB supplemented with 100 μg mL−1 ampicillin. This plate was covered with a sterile breathable rayon film membrane (VWR) and incubated overnight at 37 °C with shaking. Cells included in this “mother” plate were used to inoculate cultures for protein expression and purification as described below.
Protein expression and purification in microplates
BL21-Gold (DE3) cells transformed with plasmids containing computationally-designed or evolved RFP variants were used to inoculate deep 24-well plates (Whatman) containing 5 mL of Overnight Express Instant TB medium (Novagen) supplemented with 100 μg mL−1 ampicillin (pET11-a) or kanamycin (pET29b(+)). These plates were sealed with sterile and pierced silicone sealing mats (Axygen) and incubated overnight at 37 °C with shaking. Cells were then harvested by centrifugation and washed twice with phosphate buffer pH 7.4. Pellets were stored at 4 °C to allow for chromophore maturation. After two days, cell pellets were resuspended in 400 μL of lysis buffer (50 mM sodium phosphate buffer, pH 8.0, 300 mM sodium chloride, 2.5 mM imidazole, 1× Bugbuster protein extraction reagent [Novagen], 1 mg mL−1 hen egg white lysozyme [Sigma], 10 U Benzonase nuclease [Novagen]) and plates were incubated at 25 °C with gentle shaking for 20 minutes. After incubation, clarified lysates were collected by centrifugation and proteins were purified using HisPur Ni-NTA spin plates (Thermo Scientific) according to the manufacturer's protocol. Protein purity was verified by SDS-PAGE. These proteins were used for initial spectroscopic characterization (see below).Protein expression and purification in large batches
RFP variants were expressed in E. coli BL21-Gold (DE3) cells (Agilent) using LB supplemented with 100 μg mL−1 ampicillin (pET11-a) or 50 μg mL−1 kanamycin (pET29b(+)). Cells were grown at 37 °C with shaking until they reached an optical density at 600 nm of 0.6. Protein expression was induced by adding 1 mM isopropyl β-D-1-thiogalactopyranoside (Thermo Scientific) and cultures were incubated overnight at 16 °C with shaking. Cells were then pelleted by centrifugation and resuspended in 8 mL lysis buffer (100 mM potassium phosphate buffer, pH 7.4, 5 mM imidazole, 1 mg mL−1 lysozyme, 50 U Benzonase nuclease [Novagen]). Cells were lysed using an EmulsiFlex-B15 cell disruptor (Avestin) and lysates were harvested by centrifugation. Proteins were purified by Ni-NTA affinity chromatography using Econo-Pac chromatography columns (Bio-Rad) according to the manufacturer's protocol. Eluted fractions were desalted using Macrosep Advance centrifugal devices (Pall) into phosphate buffer solution. Protein samples were stored at 4 °C for two days to allow for chromophore maturation prior to spectroscopic characterization. For crystallography experiments, an additional purification step consisting of gel filtration into 20 mM sodium phosphate buffer pH 7.4 was performed using an ÄKTA pure (GE Healthcare) fast protein liquid chromatography system equipped with a Superdex 75 (GE Healthcare) column. Protein purity was verified by SDS-PAGE.Spectroscopic characterization
Absorption, excitation and emission spectra were measured in phosphate-buffered saline (137 mM sodium chloride, 2.7 mM potassium chloride, 10 mM disodium phosphate, 1.8 mM monopotassium phosphate, pH 7.4) using an Infinite M1000 (Tecan) or Spectramax id3 (Molecular Devices) plate reader. Quantum yields were extrapolated by comparing the integrated fluorescence intensity of the mutant proteins with that of equally absorbing samples of mCherry and mRaspberry (quantum yields of 0.23 and 0.15, respectively) with excitation at 535 nm.Extinction coefficients were determined using the method described by Kredel and colleagues,47 which takes into account the presence of green chromophore in the population of RFP molecules, giving a more accurate estimate. Briefly, protein samples were diluted (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) into Britton–Robinson buffers48 pH 11–13 to allow measureable slow denaturation of the native red chromophore (peak at 568–605 nm, depending on the RFP) to the green form (452 nm). From the absorption spectrum recorded at different time intervals, a ratio of extinction coefficients of the native chromophore species to that of the denatured species (44
10) into Britton–Robinson buffers48 pH 11–13 to allow measureable slow denaturation of the native red chromophore (peak at 568–605 nm, depending on the RFP) to the green form (452 nm). From the absorption spectrum recorded at different time intervals, a ratio of extinction coefficients of the native chromophore species to that of the denatured species (44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1) was calculated, from which the value for the red chromophore was extrapolated.
000 M−1 cm−1) was calculated, from which the value for the red chromophore was extrapolated.
Protein crystallization
Purified mSandy2 was concentrated using Amicon Ultra-15 3K centrifugal devices (Millipore Sigma) to a concentration of 34 mg mL−1 in 200 mM sodium phosphate buffer pH 7.4. Crystallization drops were prepared by mixing 1 μL of protein solution with 1 μL of the mother liquor and sealed inside a reservoir containing an additional 500 μL of the mother liquor solution. Crystals used for X-ray data collection were obtained from mother liquor containing 0.1 M Tris buffer (pH 8.5), 0.1 M sodium chloride, and 20% PEG-3350 (Table 2).X-ray data collection and processing
Prior to X-ray data collection, crystals were harvested and cryoprotected by brief soaking in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of crystallization mother liquor and 4.0 M trimethylamine-N-oxide, then flash-cooled by rapid plunging into liquid nitrogen. We collected single-crystal X-ray diffraction data on beamline 8.3.1 at the Advanced Light Source. The beamline was equipped with a Pilatus3 S 6M detector (Dectris), the X-ray energy was set to 11
1 mixture of crystallization mother liquor and 4.0 M trimethylamine-N-oxide, then flash-cooled by rapid plunging into liquid nitrogen. We collected single-crystal X-ray diffraction data on beamline 8.3.1 at the Advanced Light Source. The beamline was equipped with a Pilatus3 S 6M detector (Dectris), the X-ray energy was set to 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 111 keV, and the crystals were maintained at a cryogenic temperature (100 K) throughout the course of data collection.
111 keV, and the crystals were maintained at a cryogenic temperature (100 K) throughout the course of data collection.
We processed the X-ray data using the Xia2 software,49 which performed indexing, integration, and scaling with XDS and XSCALE,50 followed by merging with Pointless.51 A resolution cutoff (2.05 Å) was taken where the signal-to-noise ratio of the individual reflection measurements fell to an average value of 1.0.
Structure determination
We obtained initial phase information for calculation of electron density maps by molecular replacement using the program Phaser,52 as implemented in v1.17.1.3660 of the PHENIX suite.53 We identified eight copies of the protein in the asymmetric unit using the coordinates from mCherry (PDB ID: 2H5Q21), consistent with an analysis of Matthews probabilities for the observed unit cell and molecular weight of the protein.54,55Next, we rebuilt the initial model using the electron density maps calculated from molecular replacement. We then performed additional, iterative refinement of atomic positions, individual atomic displacement parameters (B-factors), and occupancies using a riding hydrogen model and automatic weight optimization, until the model reached convergence. All model building was performed using Coot 0.8.9.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 56 and refinement steps were performed with phenix.refine (v1.17.1.3660) within the PHENIX suite.53,56 Restraints for the red and green chromophores were generated using phenix.elbow,57 starting from coordinates available in the Protein Data Bank58 (PDB ligand ID: NRQ or CH6 for red or green chromophore, respectively), and manually edited to ensure proper geometry of covalent bonds between the chromophore and rest of the protein (restraints available as ESI Files†). For unbiased comparison of B-factor Z-scores, a similar approach was used to re-refine the published mRojoA structure (PDB ID: 3NEZ), with the following modifications. We deleted the minor chromophore conformer, reset all isotropic atomic B-factors to a value of 20.0 Å2, applied random coordinate displacements (σ = 0.3 Å) to the atoms, and performed coordinate refinement against the structure factor data before proceeding to manual model building. The final mSandy2 model coordinates were deposited in the Protein Data Bank under accession code 7RY2. Further information regarding model building and refinement is presented in Table 2.
56 and refinement steps were performed with phenix.refine (v1.17.1.3660) within the PHENIX suite.53,56 Restraints for the red and green chromophores were generated using phenix.elbow,57 starting from coordinates available in the Protein Data Bank58 (PDB ligand ID: NRQ or CH6 for red or green chromophore, respectively), and manually edited to ensure proper geometry of covalent bonds between the chromophore and rest of the protein (restraints available as ESI Files†). For unbiased comparison of B-factor Z-scores, a similar approach was used to re-refine the published mRojoA structure (PDB ID: 3NEZ), with the following modifications. We deleted the minor chromophore conformer, reset all isotropic atomic B-factors to a value of 20.0 Å2, applied random coordinate displacements (σ = 0.3 Å) to the atoms, and performed coordinate refinement against the structure factor data before proceeding to manual model building. The final mSandy2 model coordinates were deposited in the Protein Data Bank under accession code 7RY2. Further information regarding model building and refinement is presented in Table 2.
Data availability
Structure coordinates for mSandy2 have been deposited in the RCSB Protein Data Bank with the following accession code: 7RY2 ([https://doi.org/10.2210/pdb7RY2/pdb]). The plasmid containing the mSandy2 gene is available from Addgene (Plasmid #177760).Author contributions
R. A. C. conceived the project. R. A. C. performed computational protein design experiments. S. L. performed directed evolution experiments. S. L. and D. P. H. performed spectral characterization of RFPs. S. L., M. G. E., and D. P. H. purified proteins. R. M. and M. C. T. crystallized proteins and performed X-ray diffraction experiments. R. A. C. performed model building and refinements. S. L. and R. A. C. wrote the manuscript. M. C. T. edited the manuscript.Conflicts of interest
The authors declare no competing interests.Acknowledgements
R. A. C. acknowledges grants from the Canada Foundation for Innovation (26503), Natural Sciences and Engineering Research Council of Canada (RGPIN-2016-04831), and the Human Frontier Science Program (RGP0041). S. L. is the recipient of an Ontario Graduate Scholarship and a postgraduate scholarship from the Natural Sciences and Engineering Research Council of Canada. The authors thank Shahrokh Ghobadloo for assistance with fluorescence-activated cell sorting, Noreen Ahmed for help with protein purification, John P. Pezacki for use of their fast protein liquid chromatography system and Spectramax plate reader, and Stephen L. Mayo for providing the mCherry and mRojoA expression vectors. Beamline 8.3.1 at the Advanced Light Source is operated by the University of California at San Francisco with grants from the National Institutes of Health (R01 GM124149 and P30 GM124169), Plexxikon Inc. and the Integrated Diffraction Analysis Technologies program of the US Department of Energy Office of Biological and Environmental Research. The Advanced Light Source (Berkeley, CA) is a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the US Department of Energy under contract number DE-AC02-05CH11231, Office of Basic Energy Sciences.References
- C. Ash, M. Dubec, K. Donne and T. Bashford, Effect of wavelength and beam width on penetration in light-tissue interaction using computational methods, Lasers Med. Sci., 2017, 32, 1909–1918 CrossRef PubMed.
- J. Livet, T. A. Weissman, H. Kang, R. W. Draft, J. Lu, R. A. Bennis, J. R. Sanes and J. W. Lichtman, Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system, Nature, 2007, 450, 56–62 CrossRef CAS PubMed.
- D. Cai, K. B. Cohen, T. Luo, J. W. Lichtman and J. R. Sanes, Improved tools for the Brainbow toolbox, Nat. Methods, 2013, 10, 540–547 CrossRef CAS PubMed.
- R. E. Campbell, O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias and R. Y. Tsien, A monomeric red fluorescent protein, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 7877–7882 CrossRef CAS PubMed.
- D. S. Bindels, L. Haarbosch, L. van Weeren, M. Postma, K. E. Wiese, M. Mastop, S. Aumonier, G. Gotthard, A. Royant, M. A. Hink and T. W. Gadella Jr, mScarlet: a bright monomeric red fluorescent protein for cellular imaging, Nat. Methods, 2017, 14, 53–56 CrossRef CAS PubMed.
- K. K. Yang, Z. Wu and F. H. Arnold, Machine-learning-guided directed evolution for protein engineering, Nat. Methods, 2019, 16, 687–694 CrossRef CAS PubMed.
- Y. Gumulya, J. Sanchis and M. T. Reetz, Many pathways in laboratory evolution can lead to improved enzymes: how to escape from local minima, ChemBioChem, 2012, 13, 1060–1066 CrossRef CAS PubMed.
- A. L. Antaris, H. Chen, S. Diao, Z. Ma, Z. Zhang, S. Zhu, J. Wang, A. X. Lozano, Q. Fan, L. Chew, M. Zhu, K. Cheng, X. Hong, H. Dai and Z. Cheng, A high quantum yield molecule–protein complex fluorophore for near-infrared II imaging, Nat. Commun., 2017, 8, 15269 CrossRef CAS PubMed.
- C. M. Megley, L. A. Dickson, S. L. Maddalo, G. J. Chandler and M. Zimmer, Photophysics and dihedral freedom of the chromophore in yellow, blue, and green fluorescent protein, J. Phys. Chem. B, 2009, 113, 302–308 CrossRef CAS PubMed.
- M. Drobizhev, R. S. Molina, P. R. Callis, J. N. Scott, G. G. Lambert, A. Salih, N. C. Shaner and T. E. Hughes, Local Electric Field Controls Fluorescence Quantum Yield of Red and Far-Red Fluorescent Proteins, Front. Mol. Biosci., 2021, 8, 633217 CrossRef CAS PubMed.
- Q. Sun, Z. Li, Z. Lan, C. Pfisterer, M. Doerr, S. Fischer, S. C. Smith and W. Thiel, Isomerization mechanism of the HcRed fluorescent protein chromophore, Phys. Chem. Chem. Phys., 2012, 14, 11413–11424 RSC.
- A. Broom, R. V. Rakotoharisoa, M. C. Thompson, N. Zarifi, E. Nguyen, N. Mukhametzhanov, L. Liu, J. S. Fraser and R. A. Chica, Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico, Nat. Commun., 2020, 11, 4808 CrossRef CAS PubMed.
- Y. Bouchiba, J. Cortes, T. Schiex and S. Barbe, Molecular flexibility in computational protein design: an algorithmic perspective, Protein Eng., Des. Sel., 2021, 34, gzab011 CrossRef PubMed.
- I. Samish, C. M. MacDermaid, J. M. Perez-Aguilar and J. G. Saven, Theoretical and computational protein design, Annu. Rev. Phys. Chem., 2011, 62, 129–149 CrossRef CAS PubMed.
- S. M. Malakauskas and S. L. Mayo, Design, structure and stability of a hyperthermophilic protein variant, Nat. Struct. Biol., 1998, 5, 470–475 CrossRef CAS PubMed.
- R. A. Chica, M. M. Moore, B. D. Allen and S. L. Mayo, Generation of longer emission wavelength red fluorescent proteins using computationally designed libraries, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 20257–20262 CrossRef CAS PubMed.
- T. M. Wannier, M. M. Moore, Y. Mou and S. L. Mayo, Computational Design of the β-Sheet Surface of a Red Fluorescent Protein Allows Control of Protein Oligomerization, PLoS One, 2015, 10, e0130582 CrossRef PubMed.
- T. M. Wannier, S. K. Gillespie, N. Hutchins, R. S. McIsaac, S. Y. Wu, Y. Shen, R. E. Campbell, K. S. Brown and S. L. Mayo, Monomerization of far-red fluorescent proteins, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E11294–E11301 CrossRef CAS PubMed.
- R. Heim and R. Y. Tsien, Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer, Curr. Biol., 1996, 6, 178–182 CrossRef CAS PubMed.
- M. A. Mena, T. P. Treynor, S. L. Mayo and P. S. Daugherty, Blue fluorescent proteins with enhanced brightness and photostability from a structurally targeted library, Nat. Biotechnol., 2006, 24, 1569–1571 CrossRef CAS PubMed.
- X. Shu, N. C. Shaner, C. A. Yarbrough, R. Y. Tsien and S. J. Remington, Novel chromophores and buried charges control color in mFruits, Biochemistry, 2006, 45, 9639–9647 CrossRef CAS PubMed.
- T. J. Lambert, FPbase: a community-editable fluorescent protein database, Nat. Methods, 2019, 16, 277–278 CrossRef CAS PubMed.
- M. V. Matz, A. F. Fradkov, Y. A. Labas, A. P. Savitsky, A. G. Zaraisky, M. L. Markelov and S. A. Lukyanov, Fluorescent proteins from nonbioluminescent Anthozoa species, Nat. Biotechnol., 1999, 17, 969–973 CrossRef CAS PubMed.
- J. A. Davey and R. A. Chica, Improving the accuracy of protein stability predictions with multistate design using a variety of backbone ensembles, Proteins, 2014, 82, 771–784 CrossRef CAS PubMed.
- M. Babor, D. J. Mandell and T. Kortemme, Assessment of flexible backbone protein design methods for sequence library prediction in the therapeutic antibody Herceptin–HER2 interface, Protein Sci., 2011, 20, 1082–1089 CrossRef CAS PubMed.
- A. T. Pandelieva, M. J. Baran, G. F. Calderini, J. L. McCann, V. Tremblay, S. Sarvan, J. A. Davey, J. F. Couture and R. A. Chica, Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif, ACS Chem. Biol., 2016, 11, 508–517 CrossRef CAS PubMed.
- M. M. Moore, S. K. Oteng-Pabi, A. T. Pandelieva, S. L. Mayo and R. A. Chica, Recovery of red fluorescent protein chromophore maturation deficiency through rational design, PLoS One, 2012, 7, e52463 CrossRef CAS PubMed.
- R. L. Strack, D. E. Strongin, L. Mets, B. S. Glick and R. J. Keenan, Chromophore formation in DsRed occurs by a branched pathway, J. Am. Chem. Soc., 2010, 132, 8496–8505 CrossRef CAS PubMed.
- A. M. Bogdanov, A. Acharya, A. V. Titelmayer, A. V. Mamontova, K. B. Bravaya, A. B. Kolomeisky, K. A. Lukyanov and A. I. Krylov, Turning On and Off Photoinduced Electron Transfer in Fluorescent Proteins by pi-Stacking, Halide Binding, and Tyr145 Mutations, J. Am. Chem. Soc., 2016, 138, 4807–4817 CrossRef CAS PubMed.
- J. A. Davey and R. A. Chica, Multistate Computational Protein Design with Backbone Ensembles, Methods Mol. Biol., 2017, 1529, 161–179 CrossRef CAS PubMed.
- S. Kredel, F. Oswald, K. Nienhaus, K. Deuschle, C. Rocker, M. Wolff, R. Heilker, G. U. Nienhaus and J. Wiedenmann, mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures, PLoS One, 2009, 4, e4391 CrossRef PubMed.
- D. Shcherbo, C. S. Murphy, G. V. Ermakova, E. A. Solovieva, T. V. Chepurnykh, A. S. Shcheglov, V. V. Verkhusha, V. Z. Pletnev, K. L. Hazelwood, P. M. Roche, S. Lukyanov, A. G. Zaraisky, M. W. Davidson and D. M. Chudakov, Far-red fluorescent tags for protein imaging in living tissues, Biochem. J., 2009, 418, 567–574 CrossRef CAS PubMed.
- S. Pletnev, D. Shcherbo, D. M. Chudakov, N. Pletneva, E. M. Merzlyak, A. Wlodawer, Z. Dauter and V. Pletnev, A crystallographic study of bright far-red fluorescent protein mKate reveals pH-induced cis–trans isomerization of the chromophore, J. Biol. Chem., 2008, 283, 28980–28987 CrossRef CAS PubMed.
- J. Akerboom, N. Carreras Calderon, L. Tian, S. Wabnig, M. Prigge, J. Tolo, A. Gordus, M. B. Orger, K. E. Severi, J. J. Macklin, R. Patel, S. R. Pulver, T. J. Wardill, E. Fischer, C. Schuler, T. W. Chen, K. S. Sarkisyan, J. S. Marvin, C. I. Bargmann, D. S. Kim, S. Kugler, L. Lagnado, P. Hegemann, A. Gottschalk, E. R. Schreiter and L. L. Looger, Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics, Front. Mol. Neurosci., 2013, 6, 2 CAS.
- B. T. Bajar, A. J. Lam, R. K. Badiee, Y. H. Oh, J. Chu, X. X. Zhou, N. Kim, B. B. Kim, M. Chung, A. L. Yablonovitch, B. F. Cruz, K. Kulalert, J. J. Tao, T. Meyer, X. D. Su and M. Z. Lin, Fluorescent indicators for simultaneous reporting of all four cell cycle phases, Nat. Methods, 2016, 13, 993–996 CrossRef CAS PubMed.
- A. Hense, B. Prunsche, P. Gao, Y. Ishitsuka, K. Nienhaus and G. U. Nienhaus, Monomeric Garnet, a far-red fluorescent protein for live-cell STED imaging, Sci. Rep., 2015, 5, 18006 CrossRef CAS PubMed.
- N. G. Gurskaya, A. F. Fradkov, A. Terskikh, M. V. Matz, Y. A. Labas, V. I. Martynov, Y. G. Yanushevich, K. A. Lukyanov and S. A. Lukyanov, GFP-like chromoproteins as a source of far-red fluorescent proteins, FEBS Lett., 2001, 507, 16–20 CrossRef CAS PubMed.
- L. Wang, W. C. Jackson, P. A. Steinbach and R. Y. Tsien, Evolution of new nonantibody proteins via iterative somatic hypermutation, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 16745–16749 CrossRef CAS PubMed.
- K. D. Piatkevich, J. Hulit, O. M. Subach, B. Wu, A. Abdulla, J. E. Segall and V. V. Verkhusha, Monomeric red fluorescent proteins with a large Stokes shift, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 5369–5374 CrossRef CAS PubMed.
- D. M. Shcherbakova, M. Baloban and V. V. Verkhusha, Near-infrared fluorescent proteins engineered from bacterial phytochromes, Curr. Opin. Chem. Biol., 2015, 27, 52–63 CrossRef CAS PubMed.
- S. L. Mayo, B. D. Olafson and W. A. Goddard, Dreiding – a Generic Force-Field for Molecular Simulations, J. Phys. Chem., 1990, 94, 8897–8909 CrossRef CAS.
- R. L. Dunbrack Jr, Rotamer libraries in the 21st century, Curr. Opin. Struct. Biol., 2002, 12, 431–440 CrossRef.
- H. K. Privett, G. Kiss, T. M. Lee, R. Blomberg, R. A. Chica, L. M. Thomas, D. Hilvert, K. N. Houk and S. L. Mayo, Iterative approach to computational enzyme design, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 3790–3795 CrossRef CAS PubMed.
- B. I. Dahiyat and S. L. Mayo, Probing the role of packing specificity in protein design, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 10172–10177 CrossRef CAS PubMed.
- D. Shortle, W. E. Stites and A. K. Meeker, Contributions of the large hydrophobic amino acids to the stability of staphylococcal nuclease, Biochemistry, 1990, 29, 8033–8041 CrossRef CAS PubMed.
- E. O. McCullum, B. A. Williams, J. Zhang and J. C. Chaput, Random mutagenesis by error-prone PCR, Methods Mol. Biol., 2010, 634, 103–109 CrossRef CAS PubMed.
- S. Kredel, K. Nienhaus, F. Oswald, M. Wolff, S. Ivanchenko, F. Cymer, A. Jeromin, F. J. Michel, K.-D. Spindler, R. Heilker, G. U. Nienhaus and J. Wiedenmann, Optimized and Far-Red-Emitting Variants of Fluorescent Protein eqFP611, Chem. Biol., 2008, 15, 224–233 CrossRef CAS PubMed.
- H. T. S. Britton and R. A. Robinson, CXCVIII.—Universal buffer solutions and the dissociation constant of veronal, J. Chem. Soc., 1931, 1456–1462 RSC.
- G. Winter, xia2: an expert system for macromolecular crystallography data reduction, J. Appl. Crystallogr., 2010, 43, 186–190 CrossRef CAS.
- W. Kabsch, XDS, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 125–132 CrossRef CAS PubMed.
- P. Evans, Scaling and assessment of data quality, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2006, 62, 72–82 CrossRef PubMed.
- A. J. McCoy, R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni and R. J. Read, Phaser crystallographic software, J. Appl. Crystallogr., 2007, 40, 658–674 CrossRef CAS PubMed.
- P. D. Adams, P. V. Afonine, G. Bunkoczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L. W. Hung, G. J. Kapral, R. W. Grosse-Kunstleve, A. J. McCoy, N. W. Moriarty, R. Oeffner, R. J. Read, D. C. Richardson, J. S. Richardson, T. C. Terwilliger and P. H. Zwart, PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 213–221 CrossRef CAS PubMed.
- K. A. Kantardjieff and B. Rupp, Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein–nucleic acid complex crystals, Protein Sci., 2003, 12, 1865–1871 CrossRef CAS PubMed.
- C. X. Weichenberger and B. Rupp, Ten years of probabilistic estimates of biocrystal solvent content: new insights via nonparametric kernel density estimate, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2014, 70, 1579–1588 CrossRef CAS PubMed.
- P. V. Afonine, R. W. Grosse-Kunstleve, N. Echols, J. J. Headd, N. W. Moriarty, M. Mustyakimov, T. C. Terwilliger, A. Urzhumtsev, P. H. Zwart and P. D. Adams, Towards automated crystallographic structure refinement with phenix.refine, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2012, 68, 352–367 CrossRef CAS PubMed.
- N. W. Moriarty, R. W. Grosse-Kunstleve and P. D. Adams, electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2009, 65, 1074–1080 CrossRef CAS PubMed.
- F. C. Bernstein, T. F. Koetzle, G. J. Williams, E. F. Meyer Jr, M. D. Brice, J. R. Rodgers, O. Kennard, T. Shimanouchi and M. Tasumi, The Protein Data Bank. A computer-based archival file for macromolecular structures, Eur. J. Biochem., 1977, 80, 319–324 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc05088e |
| This journal is © The Royal Society of Chemistry 2022 |