Open Access Article
Open Access ArticleSystematic synthesis of bisected N-glycans and unique recognitions by glycan-binding proteins†
Xuefeng Cao‡
a,
Shuaishuai Wang‡ a,
Madhusudhan Reddy Gadi‡
a,
Madhusudhan Reddy Gadi‡ a,
Ding Liua,
Peng G. Wanga,
Xiu-Feng Wanbcde,
Jian Zhangf,
Xi Chen
a,
Ding Liua,
Peng G. Wanga,
Xiu-Feng Wanbcde,
Jian Zhangf,
Xi Chen g,
Lauren E. Pepih,
Parastoo Azadih and
Lei Li
g,
Lauren E. Pepih,
Parastoo Azadih and
Lei Li *a
*a
aDepartment of Chemistry, Georgia State University, Atlanta, GA, USA. E-mail: lli22@gsu.edu
bMU Center for Research on Influenza Systems Biology (CRISB), University of Missouri, Columbia, MO, USA
cDepartment of Molecular Microbiology and Immunology, School of Medicine, University of Missouri, Columbia, MO, USA
dBond Life Sciences Center, University of Missouri, Columbia, MO, USA
eDepartment of Electrical Engineering & Computer Science, College of Engineering, University of Missouri, Columbia, MO, USA
fZ Biotech, LLC, Aurora, CO, USA
gDepartment of Chemistry, University of California, One Shields Avenue, Davis, CA, USA
hComplex Carbohydrate Research Center, University of Georgia, Athens, GA, USA
First published on 9th June 2022
Abstract
Bisected N-glycans represent a unique class of protein N-glycans that play critical roles in many biological processes. Herein, we describe the systematic synthesis of these structures. A bisected N-glycan hexasaccharide was chemically assembled with two orthogonal protecting groups attached at the C2 of the branching mannose residues, followed by sequential installation of GlcNAc and LacNAc building blocks to afford two asymmetric bisecting “cores”. Subsequent enzymatic modular extension of the “cores” yielded a comprehensive library of biantennary N-glycans containing the bisecting GlcNAc and presenting 6 common glycan determinants in a combinatorial fashion. These bisected N-glycans and their non-bisected counterparts were used to construct a distinctive glycan microarray to study their recognition by a wide variety of glycan-binding proteins (GBPs), including plant lectins, animal lectins, and influenza A virus hemagglutinins. Significantly, the bisecting GlcNAc could bestow (PHA-L, rDCIR2), enhance (PHA-E), or abolish (ConA, GNL, anti-CD15s antibody, etc.) N-glycan recognition of specific GBPs, and is tolerated by many others. In summary, synthesized compounds and the unique glycan microarray provide ideal standards and tools for glycoanalysis and functional glycomic studies. The microarray data provide new information regarding the fine details of N-glycan recognition by GBPs, and in turn improve their applications.
Introduction
The N-glycans on mammalian glycoproteins vary greatly in terms of structure, but the biological roles of these variations are largely unknown. Bisected N-glycans, which contain a β1-4 linked N-acetylglucosamine (GlcNAc) residue on the central β-mannoside, represent a unique class of N-glycans that play critical roles in many physiological and pathological processes, including cell adhesion and signaling, fertilization and fetal development, cell proliferation, tumor progression, neurogenesis, and immune responses.1–4 Importantly, bisected N-glycans may act as a suppressor of cancer metastasis. For example, upregulated bisected N-glycans on specific proteins, including E-cadherin, integrin, and epidermal growth factor receptor, could suppress tumor progression and migration.5 On the other hand, altered expression of bisected N-glycans were frequently observed in various cancers and other diseases.6 An increased level of bisecting GlcNAc was detected in the amyloid precursor proteins from the brains of Alzheimer's disease,7,8 which was suggested to have a protective role from additional β-amyloid production.7 Furthermore, the addition of a bisecting GlcNAc to the conserved N-glycans on the Fc of IgG antibody is also a common strategy to significantly enhance the antibody binding to the FcγRIIIa receptor, and consequently, the antibody-dependent cellular cytotoxicity (ADCC).9 These findings highlighted the critical functions of bisected N-glycans. However, the underlying mechanisms at the molecular level are largely unclear.One possible explanation is that the bisecting GlcNAc confers a dramatic conformational equilibrium shift of N-glycans, which may affect their recognition by glycan-binding proteins (GBPs) and thus influence corresponding biological processes.3 The most populated conformation of bisected N-glycans is termed “back-fold”, in which the α1-6 branch is flipped back towards the stem region as observed by NMR, FRET, and crystallography.10–12 It was reported that cell surface overexpression of bisected N-glycans profoundly affected the interaction of the cells with galactose-binding lectins (e.g., ricin, human galectins-1, -3, and -8), but enhanced the binding of lectin Phaseolus vulgaris Erythroagglutinin (PHA-E).3 It is reasonable to speculate that the bisecting GlcNAc may affect glycan recognition of other GBPs. However, the lack of structurally diverse bisected N-glycan probes has greatly impeded related studies.
The addition of the bisecting GlcNAc is catalyzed by β1-4-N-acetylglucosaminyltransferase III (MGAT3).13 MGAT3 acts on agalacto bi-, tri-, and tetra-antennary N-glycans as well as core-fucosylated ones. In contrast, bisected glycans do not undergo further branching catalyzed by MGAT2, MGAT4, MGAT5, and core-fucosylation by FUT8.13,14 Another study showed that the bisecting GlcNAc suppressed the addition of sialic acid, Lewis-type fucose, and generation of the HNK-1 epitope at N-glycan terminals, presumably due to general low activities of glycosyltransferases (GTs) towards bisected N-glycans in vivo.15 Glycosylation changes modulated by aberrant expression of MGAT3 may further affect glycan–protein interactions and thus influence particular biological processes. Therefore, it is of great significance to probe and quantify bisected N-glycans in complex biological samples. However, conventional approaches have been limited to PHA-E and Calsepa16 affinity,17 mass spectrometry (MS)-based glycomics analysis,18,19 or their combination.20,21 In addition, the glycan recognition of these lectins lacks selectivity and is not well defined. Other GBPs that are specific for bisected N-glycans, e.g., mouse dendritic cell inhibitory receptor 2 (mDCIR2),22 may also be applied, but a well elucidated binding profile is necessary. A comprehensive library of structurally defined bisected N-glycans is thus in high demand to (1) define recognition details of GBPs and promote their application, (2) be used as standards for MS-based glycoanalysis, (3) study the functions of individual bisected glycans in biological processes, and (4) understand the molecular details of their effects on N-glycan branching and extension.
Due to the intrinsic diversity and complexity in structures, separating individual N-glycans from natural sources in sufficient amounts remain a formidable challenge, particularly for bisected ones due to their general low abundance. As a result, numerous chemical23 and chemoenzymatic24 strategies have been developed over the past two decades to access complex N-glycans. However, owing to steric effects, the chemical synthesis of bisected N-glycans remains challenging, with only a few methodologies reported to generate non-extended structures.25–34 In a few other cases, recombinant GTs were used to extend specific bisected N-glycans by attaching Gal or Neu5Ac residues.26,32,35 Nevertheless, about 20 symmetric bisected N-glycans were prepared to date, whereas asymmetric ones have never been synthesized.
Given that the bisecting GlcNAc suppresses further actions on N-glycan by branching GTs and likely other GTs,14,15 we envisioned that a library of bisected biantennary N-glycans presenting short glycan determinants would cover a substantial portion of naturally existing bisected N-glycans. Herein, we describe the chemoenzymatic combinatorial synthesis of bisected N-glycans presenting 6 common glycan determinants, including GlcNAc, N-acetyl-lactosamine (LacNAc, Galβ1-4GlcNAc), 3-sialyl-LacNAc (3′SLN, Neu5Acα2-3LacNAc), 6-sialyl-LacNAc (6′SLN, Neu5Acα2-6LacNAc), Lewis X [LeX, Galβ1-4(Fucα1-3)GlcNAc], and sialyl-LeX [sLeX, Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAc]. A total of 36 bisected N-glycans are synthesized (Table 1), including 30 asymmetric structures (7–21 and 7i–21i, 15 pairs of positional isomers, and 9 pairs of Sia-linkage isomers) that have never been prepared before and 6 symmetric (1–6) ones. These bisected N-glycans are later fabricated together with their non-bisected counterparts as a unique glycan microarray to profile N-glycan recognition by a wide variety of GBPs.
Results
Divergent synthesis of bisected cores 7 and 7i
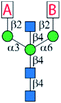
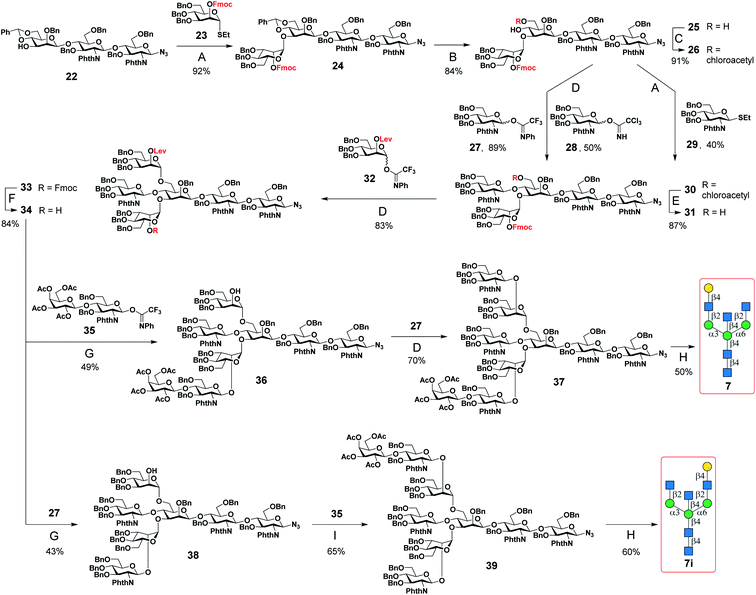
A major challenge in the synthesis of bisected N-glycans is to identify an optimal glycosylation sequence for adding the 3 monosaccharide residues (α1-3Man, α1-6Man, and β1-4GlcNAc) to the central β-mannoside of N-glycans. There are two commonly applied glycosylation sequences: (1) attaching the α1-3Man followed by the β1-4GlcNAc and finally the α1-6Man; (2) attaching the α1-3Man followed by the α1-6Man and finally the β1-4GlcNAc. With the consideration of the steric hindrance from the α1-3Man and α1-6Man, bisecting GlcNAc is typically introduced to the 4′′-OH of the central Man before the deprotection of its 6′′-OH and installation of the α1-6 branch (sequence 1).9,25,27–29,32,34,36 On the other hand, adding the bisecting GlcNAc to the 4′′-OH of the N-glycan acceptor containing both α1-3Man and α1-6Man would minimize protection and deprotection procedures of 6′′-OH. Some bisected N-glycans were prepared in this manner with good yields when optimal glycosylation conditions were applied.30,31,33 Here, we chose the first strategy to generate the bisected N-glycan cores as it consistently gave a better overall yield.Bisected “cores” 7 and 7i were divergently prepared from a common hexasaccharide 33 bearing two orthogonal protecting groups, fluorenylmethyloxycarbonyl (Fmoc) on C2 of the α1-3Man and levulinoyl (Lev) on C2 of the α1-6Man (Fig. 1). Briefly, the α1-3Man branch was first established by glycosylation of trisaccharide 22 (ref. 37) using thio donor 23,38 affording tetrasaccharide 24 in an excellent yield (92%). To further extend at 4′′ and 6′′-positions, the benzylidene acetal group of 24 was deprotected using ethanethiol and catalytic p-toluenesulfonic acid to provide 25. Regioselective esterification of the more reactive primary alcohol of 25 provided 26 with chloroacetyl protection, leaving the 4′′-OH for the continuous extension. Three glycosyl donors were tested for β1-4GlcNAcylation, in which the N-phenyl trifluoroacetimidate-based donor 27 was more efficient in providing 30 with a high yield of 89% than corresponding Schmidt (28) (ref. 36) and thioethyl donors (29).39 Subsequently, the chloroacetyl ester group was removed using thiocarbamide to obtain 31, which was glycosylated with glycosyl donor 32 to provide the key hexasaccharide 33 bearing Fmoc and Lev protecting groups. Chemoselective deprotection of either Fmoc or Lev would assist in generating two divergent asymmetric cores (Fig. 1). Initially, the Fmoc group of 33 was removed using triethylamine to produce 34. For the synthesis of core 7, 34 was first glycosylated with disaccharide donor 35 followed by Lev group deprotection using hydrazine acetate to provide 36 without purification. Compound 36 was further glycosylated with donor 27 to obtain the protected nonasaccharide 37. After 4 steps of global deprotection, including deprotection of phthaloyl (Phth) by ethylenediamine, acetylation of free amine, deacetylation, and hydrogenolysis under Pd(OH)2/C and H2 atm, as well as a P2 bio-gel chromatography cleanup and a final semi-preparative hydrophilic interaction chromatography (HILIC)-HPLC purification (Document S1†),40N-glycan 7 was obtained in a 50 mg scale with an overall yield of 50%. The synthetic route of Core 7i was similar to that of 7. Briefly, compound 34 was first glycosylated with donor 27 to obtain 38, which was used in the next step of Lev group deprotection without purification to provide 38. Finally, 38 was glycosylated with the disaccharide donor 35 to produce the protected nonasaccharide 39. After aforementioned global deprotection, cleanup, and purification, 53 mg of 7i was obtained with an overall yield of 60%.
Enzymatic modular extension to generate a comprehensive library of bisected biantennary N-glycans
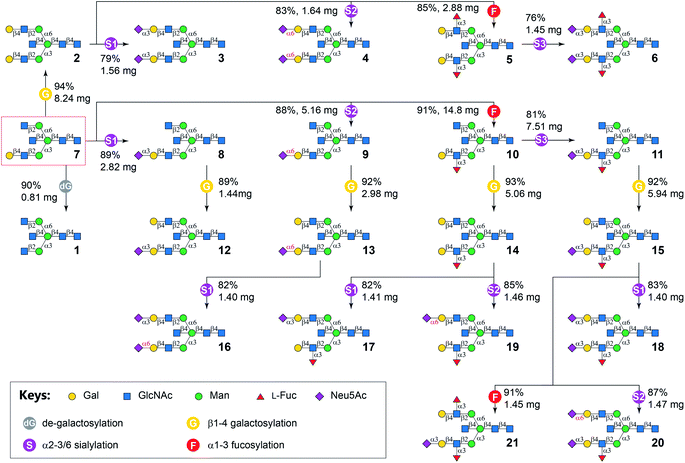
Six enzyme modules were selected to diversify cores 7 and 7i for the systematic synthesis of biantennary N-glycans containing the bisecting GlcNAc and presenting 6 common glycan determinants in a combinatorial fashion (Table 1). These include a de-galactosylation module (Module dG) catalyzed by Escherichia coli β-galactosidase (LacZ),41 a β1-4galactosylation module (Module G) catalyzed by bovine β1-4galactosyltransferase (b4GalT), which attaches a Gal residue specifically to a non-reducing end GlcNAc (except the bisecting GlcNAc) using uridine 5′-diphospho-α-galactose (UDP-Gal) as the donor. Since mammalian fucosyltransferases and sialyltransferases showed lower activity towards bisected N-glycans,15 bacterial GTs were used in the α1-3fucosylation module (Module F) and 3 sialylation modules (Module S1–S3). Module F, which adds an L-Fuc residue to GlcNAc within a LacNAc motif, includes the C-terminal 66 amino acid truncated Helicobacter pylori α1-3fucosyltransferase (Hp3FT)42 and guanosine 5′-diphospho-β-L-fucose (GDP-Fuc). Each of the sialylation modules contains one specific sialyltransferase together with Neisseria meningitidis CMP-sialic acid synthetase (NmCSS),43 cytidine diphosphate (CTP), and Neu5Ac for in situ generation of sugar donor CMP-Neu5Ac. Module S1 contains Pasteurella multocida α2-3sialyltransferase 1 E271F/R313Y mutant (PmST1-DM) which features a reduced sialidase activity to enhance the yields for α2-3sialylation of Gal residues,44 Module S2 contains Photobacterium damselae α2-6sialyltransferase (Pd26ST),45 and Module S3 contains PmST1 M144D mutant which was engineered to achieve efficient α2-3sialylation of the LeX epitope.46 All enzyme modules confer regio- and stereo-selective glycosylation of designated acceptors to achieve desired products, which have been validated by us and others in the preparation of N-glycans, O-mannosyl glycans, human milk oligosaccharides, and poly-LacNAc-containing glycans, etc.40,47–52As illustrated in Fig. 2, the synthesis of symmetric bisected N-glycans (1–6) was achieved through stepwise enzymatic modular extension of 7 following general procedures (Document S1†). The Core structure 7 was firstly converted to LacNAc-terminated N-glycan 2 using Module G in an excellent yield of 94% without galactosylation of the bisecting GlcNAc (Document S1,† MSn analysis). Unlike the forced reaction condition (19 mM acceptor, 4.2 equiv. donor, an excess amount of b4GalT, and a prolonged reaction time of 3–9 days) used to achieve galactosylation of the bisecting GlcNAc,53 the rather mild condition employed here eliminated the galactosylation on the bisecting GlcNAc in all Module G-involved reactions, which is consistent with results from previous reports.26,28,35 The product 2 was purified using HILIC-HPLC40 and used as an acceptor for further sialylation by Module S1 and Module S2 to produce Neu5Acα2-3/6-linked sialosides 3 and 4, as well as α1-3fucosylation by Module F to afford LeX-containing structure 5, respectively. HILIC-purified dodecasaccharide 5 was further extended by sialylation with Module S3 to yield sLeX-containing bisected N-glycan 6. Each modular reaction was allowed to proceed till complete conversion of the acceptor, which was monitored by HPLC with an evaporative light scattering detector (ELSD). Typically, glycosylations by Modules G, F, S2, and S3 could reach complete conversion within 6 hours, whereas reactions catalyzed by Modular S1 could be completed in half an hour. Prolonged reaction time for Modular S1 could lower the product yields due to residue sialidase activities of PmST1-DM.44 The agalacto bisected N-glycan 1 was prepared by LacZ-catalyzed de-galactosylation (Module dG) of 7 with 90% yield in 3 days. It could also be obtained from 7i at a much lower yield (33%) in 3 days. A similar preference of LacZ towards Gal on the α1-3Man branch was also observed for non-bisected N-glycans although with higher yields.41 All products were purified by HPLC before being used as substrates in the next step and characterized by mass spectrometry and 1H NMR spectroscopy (Document S1†).
The asymmetric bisected N-glycan targets (Table 1) were divided into two groups according to the number of monosaccharides on α1-3 and α1-6 branches. Synthetic targets in the first group (7–21) had more or the same number of monosaccharides on the α1-3 branch compared to that on the α1-6 branch. In comparison, the second group contained compounds 7i–21i (positional isomers of 7–21), where the glycan structures on the two branches of 7–21 are switched. The general strategy was to enzymatically assemble the α1-3 branch before the α1-6 branch for compounds 8–21, and assemble the α1-6 branch before the α1-3 branch for compounds 8i–21i. For example, the α1-3Man branch of 7 was extended by Modules S1, S2, F, and S3 (Fig. 2) similar to the procedures used for the synthesis of symmetric glycans 3–6 to form glycans 8–11, respectively. Subsequent β1-4galactosylation of purified 8–11 in parallel reactions using Module G produced 12–15 in good yields. N-Glycan 16 was derived from 13 by selective α2-3sialylation of the Gal residue on the α1-6 branch using Module S1, as the 6′SLN motif on the opposing branch is inert to PmST1-DM.54 The synthesis of 17 and 19, respectively, were achieved by regio-selective α2-3/6sialylation of the Gal residue on the α1-6 branch of 14 using Modules S1 and S2. The LeX motif is inert to both PmST1-DM in Module S1 (Document S1,† LC-HCD-MS/MS analysis of 17 and 17i showed the α2-3sialylation is only located on the non-fucosylated branch) and Pd26ST in Module S2.40 Finally, compound 18, 20 and 21 were synthesized from 15 in parallel reactions using Modules S1, S2, and F, respectively, in yields of 76–94% after HILIC-HPLC purification.
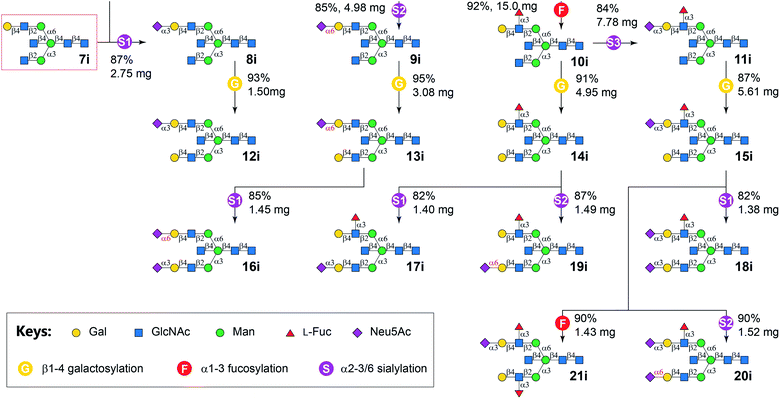
Similarly, N-glycans 8i–21i was synthesized from Core 7i with good yields (Fig. 3). It is worth noting that no apparent branch preference was observed for applied GTs. Furthermore, in contrast to the observation that bisecting GlcNAc suppress mammalian N-glycan extensions in a recent report,15 no significant difference in synthetic efficiency was observed for the synthesis of target bisected N-glycans compared to their non-bisected counterparts.40 This may be explained by a potential more relaxed substrate specificity of bacterial GTs used in this study than mammalian ones.
G, bovine β1-4galactosyltransferase (b4GalT) and UDP-Gal; F, C-terminal 66 amino acid truncated H. pylori α1-3fucosyltransferase (Hp3FT) and GDP-Fuc; S1, Pasteurella multocida α2-3sialyltransferase 1 E271F/R313Y mutant (PmST1-DM), Neisseria meningitidis CMP-sialic acid synthetase (NmCSS), CTP, and Neu5Ac; S2, Photobacterium damselae α2-6sialyltransferase (Pd26ST), NmCSS, CTP, and Neu5Ac; S3, PmST1-M144D, NmCSS, CTP, and Neu5Ac.
Glycan microarray fabrication and GBP profiling
The obtained structurally defined N-glycans are ideal standards for structure–function relationship studies. We sought to define the influence of the bisecting GlcNAc to N-glycan recognition by a wide variety of N-glycan-binding proteins. To this end, we constructed a glycan microarray presenting the 36 bisected N-glycans prepared in this study together with their non-bisected counterparts.40 Two linear glycans 3′SLNnT (L3, Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-4Glc) and 6′SLNnT (L6, Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4Glc) were used as controls. All 74 sequence-defined glycans contain a free reducing end and were derivatized using 2-amino-N-(2-aminoethyl)-benzamide (AEAB)55 for quantification and microarray fabrication (Document S1†).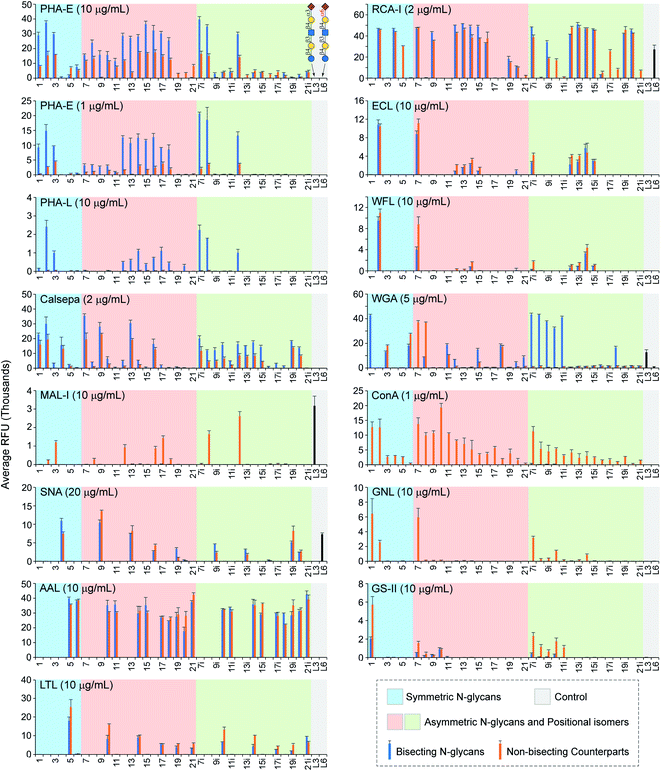 | ||
| Fig. 4 Microarray analysis of plant lectins using the N-glycan microarray with bisected N-glycans and their non-bisected counterparts. | ||
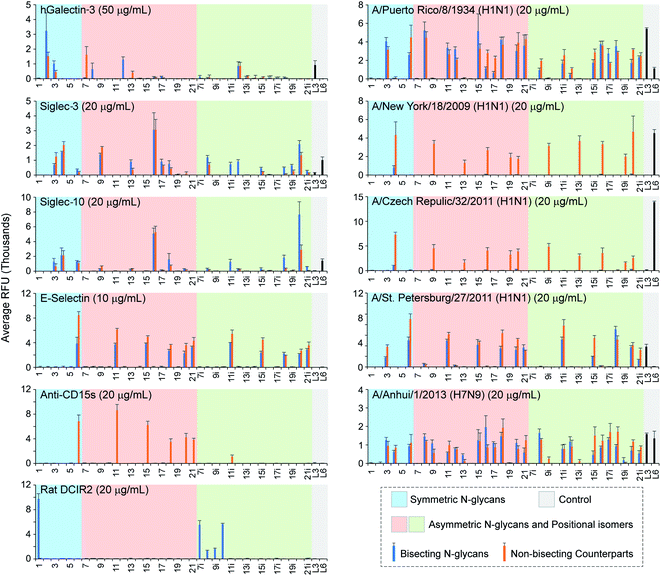
Phaseolus vulgaris erythroagglutinin (PHA-E) and its isolectin P. vulgaris leukoagglutinin (PHA-L) have distinct binding specificities towards N-glycans. PHA-E was reported to bind preferentially to biantennary Gal-terminated N-glycans containing the bisecting GlcNAc (compound 2) (ref. 59) but also bind to non-bisected complex N-glycans containing terminal Gal residues (e.g., 12–15) (Fig. 4).60 Our results revealed a clear binding preference of PHA-E to bisected N-glycans containing the terminal LacNAc determinant at both high (10 μg mL−1) and low (1 μg mL−1) PHA-E concentrations. Interestingly, PHA-E also strongly bound (RFU = 9197 at 1 μg mL−1) agalacto bisected glycan 1 which was not observed previously, even though lower than that to galacto bisected glycan 2 (RFU = 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 842). Additional α2-3sialylation was well tolerated (e.g., 3, 8, 8i), but α2-6sialylation or α1-3fucosylation are only tolerated on the α1-3Man branch (9–11, not 9i–11i). PHA-L is known to preferably bind to the β1-6GlcNAc branched multi-antennary N-glycans,59,60 which are not available in our array. Surprisingly, we observed relatively low (RFU < 3000 at 10 μg mL−1) but specific recognition of bisected N-glycans by PHA-L, with a binding pattern similar to that of PHA-E (1 μg mL−1). We also assayed Datura stramonium agglutinin (DSA) which is known to bind the GlcNAcβ1-4Manα1-3Man motif of multi-antennary N-glycans. No apparent binding was observed (Fig. S2†) as this array did not contain such structures.
842). Additional α2-3sialylation was well tolerated (e.g., 3, 8, 8i), but α2-6sialylation or α1-3fucosylation are only tolerated on the α1-3Man branch (9–11, not 9i–11i). PHA-L is known to preferably bind to the β1-6GlcNAc branched multi-antennary N-glycans,59,60 which are not available in our array. Surprisingly, we observed relatively low (RFU < 3000 at 10 μg mL−1) but specific recognition of bisected N-glycans by PHA-L, with a binding pattern similar to that of PHA-E (1 μg mL−1). We also assayed Datura stramonium agglutinin (DSA) which is known to bind the GlcNAcβ1-4Manα1-3Man motif of multi-antennary N-glycans. No apparent binding was observed (Fig. S2†) as this array did not contain such structures.
Calystegia sepium Lectin (Calsepa) had been used to detect bisected N-glycans,61 with a reported 5-fold stronger bindings to bisected N-glycans than non-bisected ones.16 Our results, however, showed high readouts (RFUs > 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) to both bisected and non-bisected glycans at high (10 μg mL−1, Fig. S2†) or low (2 μg mL−1, Fig. 4) Calsepa concentrations, although RFUs of some bisected glycans (e.g., 7i–15i) are 1 to 2 times higher than those of their non-bisected counterparts. Motif Finder analysis62 identified a minimal determinant of Galβ1-4GlcNAcβ1-2Manα1-3Man or GlcNAcβ1-2Manα1-3Man for Calsepa (Document S3†). Unlike PHA-E, Calsepa tolerated α2-6sialylation (4, 9, 13, 9i, 13i), but α2-3sialylation or α1-3fucosylation on the α1-3 branch could significantly suppress the binding (8, 10, 11).
000) to both bisected and non-bisected glycans at high (10 μg mL−1, Fig. S2†) or low (2 μg mL−1, Fig. 4) Calsepa concentrations, although RFUs of some bisected glycans (e.g., 7i–15i) are 1 to 2 times higher than those of their non-bisected counterparts. Motif Finder analysis62 identified a minimal determinant of Galβ1-4GlcNAcβ1-2Manα1-3Man or GlcNAcβ1-2Manα1-3Man for Calsepa (Document S3†). Unlike PHA-E, Calsepa tolerated α2-6sialylation (4, 9, 13, 9i, 13i), but α2-3sialylation or α1-3fucosylation on the α1-3 branch could significantly suppress the binding (8, 10, 11).
Results are shown as relative fluorescence units (RFUs) by averaging the background-subtracted fluorescence signals of 4 replicate spots, error bars represent the standard deviation (SD). Source data are provided as a Source Data file (Document S2†). L3, 3′SLNnT; L6, 6′SLNnT.
Maackia amurensis lectin I (MAL-I) and Sambucus nigra agglutinin (SNA) bound N-glycans presenting 3′SLN and 6′SLN, respectively, consistent with prior observations.60,63,64 Interestingly, while adding the bisecting GlcNAc completely blocked the binding of MAL-I to biantennary N-glycans, it had limited influence on SNA binding. Additionally, MAL-I preferably binds to 3′SLN on the α1-6Man branch (non-bisected counterparts of 8i and 12i) over the α1-3Man branch (non-bisected counterparts of 8 and 12), whereas SNA prefers to 6′SLN on the α1-3Man branch (9, 12 and their non-bisected counterparts), which are in agreement with our recent report.63
We assayed three Fuc-binding lectins including Aleuria aurantia lectin (AAL), Lotus tetragonolobus lectin (LTL), and Ulex europaeus agglutinin (UEA-I). AAL strongly bound (RFUs > 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) all fucosylated N-glycans, regardless of the presence of the bisecting GlcNAc or other modifications either on the Fuc-presenting branch or the other branch. On the other hand, LTL showed weak (RFU = 1855 for glycan 19i) to high (RFU = 17
000) all fucosylated N-glycans, regardless of the presence of the bisecting GlcNAc or other modifications either on the Fuc-presenting branch or the other branch. On the other hand, LTL showed weak (RFU = 1855 for glycan 19i) to high (RFU = 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 908 for glycan 5) binding signals only to LeX-presenting (bisected) N-glycans but not those with sLeX (6, 11, 15, 18, 20, 6i, 11i, 15i, 18i, 20i, and their non-bisected counterparts), indicating that an additional α2-3sialylation was not tolerated. No binding of UEA-I was observed as the glycan array did not contain its reported ligand, Fucα1-2Gal.65
908 for glycan 5) binding signals only to LeX-presenting (bisected) N-glycans but not those with sLeX (6, 11, 15, 18, 20, 6i, 11i, 15i, 18i, 20i, and their non-bisected counterparts), indicating that an additional α2-3sialylation was not tolerated. No binding of UEA-I was observed as the glycan array did not contain its reported ligand, Fucα1-2Gal.65
LacNAc-specific Ricinus communis agglutinin (RCA-I) strongly bound nearly all β1-4Gal terminated N-glycans or their α2-6sialylated derivatives (e.g., 2, 4, 7, 9, 7i, 9i), consistent with prior observations.60,63 In contrast, Erythrina cristagalli lectin (ECL) did not tolerate α2-6sialylation or other modifications on the LacNAc in the N-glycans (e.g., 3–6, 8–11, 8i–11i). Wisteria floribunda lectin (WFL) was reported to preferably bind to β-linked GalNAc on N-glycans, particularly LacdiNAc (GalNAcβ1-4GlcNAc), and to terminal Gal residues with lower avidity.66 We observed that WFL preferentially bound LacNAc on the α1-3 branch (7 and non-bisected counterpart) over the α1-6 branch (7i and its non-bisected counterpart). Like ECL, WFL did not accommodate additional sialylation or fucosylation. The impact of the bisecting GlcNAc to RCA–I binding is complicated, can be either tolerated (e.g.2, 4, 7, 9) or inhibited (5, 10i, 17i).65 On the other hand, the bisecting GlcNAc is tolerated by ECL and WFL.
Glycan recognition by wheat germ agglutinin (WGA) is complicated, with reported ligands of terminal 3′SLN, sLeX, LacNAc, and GlcNAc.50,65 In this array, WGA strongly bound non-bisected counterparts of 3, 5, 7, 8, 11, and 18 (Fig. 4, RFUs > 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000). The recognition of bisected N-glycans is strikingly different, with 1 and 7i–11i elicited highest binding signals, suggesting a determinant of the shared GlcNAcβ1-2Manα1-3(GlcNAcβ1-4)Man motif, same as that of mDCIR2.22
000). The recognition of bisected N-glycans is strikingly different, with 1 and 7i–11i elicited highest binding signals, suggesting a determinant of the shared GlcNAcβ1-2Manα1-3(GlcNAcβ1-4)Man motif, same as that of mDCIR2.22
The binding patterns of Man-specific lectins concanavalin A (ConA) and Galanthus nivalis lectin (GNL) towards non-bisected N-glycans are identical to our prior observation that they have a minimum glycan determinant of Manα1-3(Manα1-6)Man and Manα1-3Man, respectively.63 The addition of the bisecting GlcNAc completely abolished bindings of both lectins, which is expectable as this modification could cause a substantial conformational shift around the central Man residue.10–12 Interestingly, at a higher concentration of 10 mg mL−1, ConA showed strong bindings (RFUs > 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) to bisected N-glycans with at least one non-extended branch (1, 7–10, and 7i–10i) (Fig. S2†).
000) to bisected N-glycans with at least one non-extended branch (1, 7–10, and 7i–10i) (Fig. S2†).
As reported,67Griffonia simplicifolia lectin II (GS-II) only bound glycans with one or both GlcNAc-terminated branches (non-bisected counterparts of 1, 7–10, 7i–11i). Importantly, the binding can be diminished by the bisecting GlcNAc (1, 7, 7i–11i). It is also worth noting that the bisecting GlcNAc is not a ligand of GS-II (e.g., 2–6 and their non-bisected counterparts).
These results collectively revealed unique binding specificities of commonly used lectins to biantennary N-glycans, particularly the impact of the bisecting GlcNAc in N-glycan recognition.
Both human Siglec-3 and -10 are known to preferentially bind to Siaα2-6LacNAc trisaccharides.70 The binding pattern of them to asymmetric N-glycans is complicated (Fig. 5). Strikingly, bisected asymmetric N-glycan 16 and its non-bisected counterpart elicited highest binding signals for both Siglecs, whereas their positional isomers (16i and its non-bisected counterpart) showed no bindings. This result is consistent with our recent observation that Siglec-3 and -10 had a unique terminal epitope-dependent branch preference,71 where both branches of compound 16 presented preferred terminal-epitope (6′SLN on the α1-3 branch, a less preferred ligand 3′SLN on the α1-6 branch). Another observation is that the bisecting GlcNAc can be tolerated by both Siglecs.
Results are shown as relative fluorescence units (RFUs) by averaging the background-subtracted fluorescence signals of 4 replicate spots, error bars represent the standard deviation (SD). Source data are provided as a Source Data file (Document S2†). L3, 3′SLNnT; L6, 6′SLNnT.
Selectins are transmembrane glycoproteins containing C-type lectin domains that specifically bind to sLeX presenting glycans/glycoproteins. They are involved in chronic and acute inflammation processes and constitutive lymphocyte homing.72 We investigated the 3 human selectins on our microarray to explore their N-glycan recognition. P- and L-selectins did not bind to any N-glycans (data not shown) as their primary ligands are O-glycans and sulfated glycans, respectively.72 E-selectin bound all sLeX-containing N-glycans with or without the bisecting GlcNAc (Fig. 5). In contrast, the sLeX-specific monoclonal anti-CD15s antibody (clone CSLEX1) only bound non-bisected N-glycans with the ligand on the α1-3 branch (non-bisected counterparts of 6, 11, 15, 18, 20, 21, 11i), suggesting an intolerance of the bisecting GlcNAc and a strict α1-3 branch requirement.
The mouse dendritic cell inhibitory receptor 2 (mDCIR2) is the only bisected N-glycan-specific animal lectin discovered so far.22 The bisecting GlcNAc together with the non-extended β1-2GlcNAc on the α1-3 branch serves as the minimum glycan determinant of this C-type lectin.73 We found that rat dendritic cell inhibitory receptor (rDCIR2, Uniprot: Q5YIS1), which shares 68.5% sequence identity with mDCIR2, has the same specificity, as reflected by its strict preference to bisected N-glycans 1 and 7i–11i. It is worth noting that rDCIR2 tolerates α1-3fucosylation on the α1-6 branch (10i), but sialylation (8i, 9i, 11i) could significantly diminish the bindings.
Altogether, we identified protein-specific impacts of the bisecting GlcNAc on N-glycan recognition by animal lectins, and identified a second bisecting-specific animal lectin, rDCIR2.
Results showed that PR8 HA bound both α2-3 and α2-6 linked linear sialosides (3′SLNnT and 6′SLNnT) with a preference to 3′SLNnT (Fig. 5), consistent with prior observations.53 In contrast, a different specificity was observed towards N-glycans, where only α2-3sialylated N-glycans elicited binding signals disregarding the presence of α1-3fucosylation or the bisecting GlcNAc (e.g., 3, 8, 12 and their fucosylated counterparts 6, 11, 15). HAs of two 2009 H1N1 viruses (NY18 and CR32) had high binding preferences to α2-6sialylated glycans (Fig. 5).74 Interestingly, neither NY18 nor CR32 recognized bisected N-glycans encoded on the microarray with one exception of the symmetric glycan 4. We recently reported the recognition of α2-3-sialylated and sLeX-containing linear glycans by SP27 HA,47 which is further confirmed by this array. Surprisingly, SP27 HA showed specific bindings to sLeX-containing N-glycans (with or without the bisecting GlcNAc), suggesting a critical role of the α1-3fucose in the N-glycan recognition by SP27 HA. The only exceptions are compound 3 and its non-bisected counterpart that presenting 3′SLN on both branches. Different from the other 4 HA proteins, HA of A1 bound nearly all sialylated N-glycans with varied strength regardless of the presence of the bisecting GlcNAc (Fig. 5), indicating a broad specificity towards both α2-3sialosides and α2-6sialosides.75
Discussion
Systematic preparation of bisected N-glycans remains elusive, presumably due to steric effects in chemical synthesis. We tackled this challenge with a chemoenzymatic modular strategy. Firstly, two asymmetric bisected biantennary N-glycans (nonasaccharides 7 and 7i) were chemically synthesized with one branch galactosylated while the other agalactosylated. The uneven branches facilitated sequential extension by six dedicated enzyme modules. These judiciously selected GT modules featured robustness and regio/stereo-selectivity to achieve a highly efficient synthesis of 36 bisected N-glycans, which encompass variations in asymmetry, positional isomer, sialic acid linkage, and α1-3fucosylation.Our glycan microarray data provided new information regarding the influence of the bisecting GlcNAc on N-glycan recognition by GBPs. This was facilitated by the unique pairwise combination of bisected versus non-bisected N-glycans. As summarized in Table 2, many plant lectins could tolerate the bisecting GlcNAc, among which the bindings of PHA-E and Calsepa were enhanced. We observed that even at a low concentration (1 μg mL−1), PHA-E and Calsepa showed bindings to non-bisected N-glycans, which would pose a significant problem in their wide applications of bisected glycan identification and cancer biomarker discovery.76 Surprisingly, PHA-L exhibited specific recognition of bisected biantennary N-glycans, which could find promising implementation in probing such structures, e.g., in antibodies, where β1-6-branched glycans are absent. Unexpectedly, the bisecting GlcNAc barely impacted N-glycan recognition by assayed animal lectins, suggesting that the prior observation of decreased human lectin bindings to the cells that mainly presented bisected N-glycans3 may be caused by altered glycome rather than glycan recognition.
| GBP | Top N-glycan binder/ligand (gray) | Branch preference | Tolerance of bisecting | Tolerance of modifications | ||
|---|---|---|---|---|---|---|
| α2-3Sia | α2-6Sia | α1-3Fuc | ||||
| a Not applicable. b The bisecting GlcNAc enhanced binding. c The bisecting GlcNAc diminished binding. | ||||||
| PHA-E | 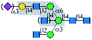 |
α1-6Man | Yes/+b | Yes | α1-3 Branch (yes); | α1-3 Branch (yes); |
| α1-6 Branch (no) | α1-6 Branch (no) | |||||
| PHA-L | 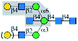 |
α1-6Man | Only to bisecting | Yes | —a | — |
| Calsepa | 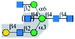 |
α1-3Man | Yes/+ | α1-6 Branch (yes); | Yes | α1-6 Branch (yes); |
| α1-3 Branch (suppress) | α1-3 Branch (suppress) | |||||
| MAL-I | 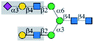 |
α1-6Man | No | — | — | No63 |
| SNA |  |
α1-3Man | Yes | — | — | — |
| RCA-I |  |
α1-3Man63 | Yes | No | Yes | No |
| ECL |  |
α1-3Man63 | Yes | No | No | No |
| WFL |  |
α1-3Man63 | Yes | No | No | No |
| AAL |  |
No | Yes | Yes | — | — |
| LTL |  |
No | Yes | No | — | — |
| ConA |  |
— | No | Yes | Yes | Yes |
| GNL | 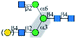 |
— | No | No | No | No |
| GS-II | 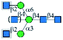 |
No | Yes/−c | No | No | No |
| Siglec-3 |  |
— | Yes | — | — | — |
| Siglec-10 |  |
— | Yes | — | — | — |
| E-selectin | 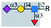 |
No | Yes | — | — | — |
| Anti-CD15s | 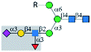 |
α1-3Man | No | — | — | — |
| rDCIR2 |  |
α1-3Man | Only to bisecting | — | — | — |
| PR8 (H1N1) |  |
No | Yes | — | — | Yes |
| NY18 (H1N1) |  |
No | No | — | — | — |
| CR32 (H1N1) |  |
No | No | — | — | — |
| SP27 (H1N1) |  |
No | Yes | — | — | — |
| A1 (H7N9) |  ; ;  |
No | Yes | — | — | Yes |
Furthermore, HA proteins from different flu strains exhibited disparate toleration of the bisecting GlcNAc modification. While 3′SLN- or sLeX-specific influenza viruses PR8 and SP27 could well tolerate this modification, the bisecting GlcNAc completely inhibited N-glycan recognition by 6′SLN-specific influenza viruses NY18 and CR32. The underlying mechanism is yet to be interpreted. Another interesting observation is the branch preference of the 4 bisecting-specific GBPs. PHA-E and PHA-L preferably bind to the α1-6Man branch in the presence of the bisecting GlcNAc, but Calsepa and rDCIR2 recognize the α1-3Man branch and the bisecting GlcNAc (Document S3†). Collectively, our results provide information on bisecting recognition and modulation, which could promote the application of these GBPs.
In summary, we achieved the systemic synthesis of (a)symmetric bisected N-glycans by combining chemical synthesis and enzymatic modular extension. The 30 asymmetric bisected glycans and the 6 symmetric ones are ideal standards for glycoanalysis and functional glycomics studies. In addition, the bisected versus non-bisected N-glycan microarray platform is a unique tool for investigating fine specificities of GBPs, including bisecting modulation and branch preferences.
Author contributions
L. L. conceived the project; X. Cao and M. R. G. performed chemical synthesis; L. L. and S. W. performed enzymatic synthesis; S. W. and D. L. performed mass spectrometry analysis; S. W. performed HPLC analysis; S. W. and M. R. G. analyzed NMR spectra; J. Z. fabricated the microarray; X. Chen provided sialyltransferase plasmids and revised the manuscript; X. W. provided flu HA proteins and revised the manuscript; L. P. and P. A performed and analyzed LC-HCD-MS/MS. L. L., S. W., and M. R. G. wrote the manuscript, which is edited an approved by all authors.Conflicts of interest
The authors declare no competing interests.Acknowledgements
The work was supported by United States National Institutes of Health (NIH) awards (U54HL142019 to LL, R44GM123820 to LL and JZ, and R21AI144433 to XFW). The work is partially supported by GlycoMIP, a National Science Foundation Materials Innovation Platform funded through Cooperative Agreement DMR1933525 and NIH R24GM137782. The glycan microarray analysis software MotifFinder was developed under the support of NIH SBIR award R43GM131430 to JZ.References
- Q. Chen, Z. Tan, F. Guan and Y. Ren, Front. Chem., 2020, 8, 511 CrossRef CAS PubMed.
- Y. Kizuka, S. Kitazume and N. Taniguchi, Biochim. Biophys. Acta, Gen. Subj., 2017, 1861, 2447–2454 CrossRef CAS PubMed.
- H. E. Miwa, Y. Song, R. Alvarez, R. D. Cummings and P. Stanley, Glycoconjugate J., 2012, 29, 609–618 CrossRef CAS PubMed.
- Y. Y. Zhao, M. Takahashi, J. G. Gu, E. Miyoshi, A. Matsumoto, S. Kitazume and N. Taniguchi, Cancer Sci., 2008, 99, 1304–1310 CrossRef CAS PubMed.
- N. Taniguchi, Y. Ohkawa, K. Maeda, Y. Harada, M. Nagae, Y. Kizuka, H. Ihara and Y. Ikeda, Mol. Aspects Med., 2020, 79, 100905 CrossRef PubMed.
- O. M. T. Pearce, Glycobiology, 2018, 28, 670–696 CrossRef CAS PubMed.
- K. Akasaka-Manya, H. Manya, Y. Sakurai, B. S. Wojczyk, Y. Kozutsumi, Y. Saito, N. Taniguchi, S. Murayama, S. L. Spitalnik and T. Endo, Glycobiology, 2010, 20, 99–106 CrossRef CAS PubMed.
- S. Schedin-Weiss, S. Gaunitz, P. Sui, Q. Chen, S. M. Haslam, K. Blennow, B. Winblad, A. Dell and L. O. Tjernberg, FEBS J., 2020, 287, 3221–3234 CrossRef CAS PubMed.
- G. Zou, H. Ochiai, W. Huang, Q. Yang, C. Li and L. X. Wang, J. Am. Chem. Soc., 2011, 133, 18975–18991 CrossRef CAS PubMed.
- S. W. Homans, R. A. Dwek and T. W. Rademacher, Biochem, 1987, 26, 6571–6578 CrossRef CAS PubMed.
- H. J. Stubbs, J. J. Lih, T. L. Gustafson and K. G. Rice, Biochem, 1996, 35, 937–947 CrossRef CAS PubMed.
- M. Nagae, M. Kanagawa, K. Morita-Matsumoto, S. Hanashima, Y. Kizuka, N. Taniguchi and Y. Yamaguchi, Sci. Rep., 2016, 6, 22973 CrossRef CAS PubMed.
- Y. Ikeda, H. Ihara, H. Tsukamoto, J. Gu and N. Taniguchi, in Handbook of Glycosyltransferases and Related Genes, Springer Japan, 2nd edn, 2014, pp. 209–222 Search PubMed.
- Handbook of Glycosyltransferases and Related Genes, ed. K. Ohtsubo, N. J. H. o. G. Taniguchi, R. Genes and N. Taniguchi, Honke, K., Fukuda, M., Narimatsu, H., Yamaguchi, Y. and Angata, T., Springer Japan, 2nd edn, 2014, pp. 223–232 Search PubMed.
- M. Nakano, S. K. Mishra, Y. Tokoro, K. Sato, K. Nakajima, Y. Yamaguchi, N. Taniguchi and Y. Kizuka, Mol. Cell. Proteomics, 2019, 18, 2044–2057 CrossRef PubMed.
- S. Nakamura-Tsuruta, N. Uchiyama, W. J. Peumans, E. J. Van Damme, K. Totani, Y. Ito and J. Hirabayashi, FEBS J., 2008, 275, 1227–1239 CrossRef CAS PubMed.
- J. P. Ribeiro and L. K. Mahal, Curr. Opin. Chem. Biol., 2013, 17, 827–831 CrossRef CAS PubMed.
- L. Dang, J. Shen, T. Zhao, F. Zhao, L. Jia, B. Zhu, C. Ma, D. Chen, Y. Zhao and S. Sun, Anal. Chem., 2019, 91, 5478–5482 CrossRef CAS PubMed.
- Q. Chen, Y. Zhang, K. Zhang, J. Liu, H. Pan, X. Wang, S. Li, D. Hu, Z. Lin, Y. Zhao, G. Hou, F. Guan, H. Li, S. Liu and Y. Ren, Genomics, Proteomics Bioinf., 2020 DOI:10.1016/j.gpb.2021.09.010.
- G. Lu and L. A. Holland, Anal. Chem., 2019, 91, 1375–1383 CrossRef CAS PubMed.
- H. Allam, K. Aoki, B. B. Benigno, J. F. McDonald, S. G. Mackintosh, M. Tiemeyer and K. L. Abbott, J. Proteome Res., 2015, 14, 434–446 CrossRef CAS PubMed.
- N. Kanazawa, T. Okazaki, H. Nishimura, K. Tashiro, K. Inaba and Y. Miyachi, J. Invest. Dermatol., 2002, 118, 261–266 CrossRef CAS PubMed.
- Y.-H. Lih, S. S. Shivatare and C.-Y. Wu, in Synthetic Glycomes, 2019, pp. 83–104 Search PubMed.
- L. Li, W. Guan, Z. Wu and P. G. Wang, in Synthetic Glycomes, 2019, pp. 105–124 Search PubMed.
- H. Paulsen, M. Heume and H. Nürnberger, Carbohydr. Res., 1990, 200, 127–166 CrossRef CAS PubMed.
- C. Unverzagt and J. Seifert, Tetrahedron Lett., 2000, 41, 4549–4553 CrossRef CAS.
- R. Schuberth and C. Unverzagt, Tetrahedron Lett., 2005, 46, 4201–4204 CrossRef CAS.
- S. Eller, R. Schuberth, G. Gundel, J. Seifert and C. Unverzagt, Angew. Chem., Int. Ed., 2007, 46, 4173–4175 CrossRef CAS PubMed.
- P. Wang, J. Zhu, Y. Yuan and S. J. Danishefsky, J. Am. Chem. Soc., 2009, 131, 16669–16671 CrossRef CAS PubMed.
- S. Eller, C. Raps, M. Niemietz and C. Unverzagt, Tetrahedron Lett., 2010, 51, 2648–2651 CrossRef CAS.
- M. Monnich, S. Eller, T. Karagiannis, L. Perkams, T. Luber, D. Ott, M. Niemietz, J. Hoffman, J. Walcher, L. Berger, M. Pischl, M. Weishaupt, C. Wirkner, R. G. Lichtenstein and C. Unverzagt, Angew. Chem., Int. Ed., 2016, 55, 10487–10492 CrossRef PubMed.
- K. Hammura, A. Ishikawa, V. H. R. Kumar, R. Miyoshi, Y. Yokoi, M. Tanaka, H. Hinou and S. I. Nishimura, ACS Med. Chem. Lett., 2018, 9, 889–894 CrossRef CAS PubMed.
- W. Yang, S. Ramadan, J. Orwenyo, T. Kakeshpour, T. Diaz, Y. Eken, M. Sanda, J. E. Jackson, A. K. Wilson and X. Huang, Chem. Sci., 2018, 9, 8194–8206 RSC.
- H. Weiss and C. Unverzagt, Angew. Chem., Int. Ed., 2003, 42, 4261–4263 CrossRef CAS PubMed.
- S. Andre, C. Unverzagt, S. Kojima, M. Frank, J. Seifert, C. Fink, K. Kayser, C. W. von der Lieth and H. J. Gabius, Eur. J. Biochem., 2004, 271, 118–134 CrossRef CAS PubMed.
- F. Yamazaki, T. Kitajima, T. Nukada, Y. Ito and T. Ogawa, Carbohydr. Res., 1990, 201, 15–30 CrossRef CAS PubMed.
- Y. Liu, Y. M. Chan, J. Wu, C. Chen, A. Benesi, J. Hu, Y. Wang and G. Chen, ChemBioChem, 2011, 12, 685–690 CrossRef CAS PubMed.
- M. Tersa, L. Raich, D. Albesa-Jové, B. Trastoy, J. Prandi, M. Gilleron, C. Rovira and M. E. Guerin, ACS Chem. Biol., 2018, 13, 131–140 CrossRef CAS PubMed.
- P. B. van Seeventer, J. Kerékgyártó, J. A. L. M. van Dorst, K. M. Halkes, J. P. Kamerling and J. F. G. Vliegenthart, Carbohydr. Res., 1997, 300, 127–138 CrossRef CAS PubMed.
- L. Li, Y. Liu, C. Ma, J. Qu, A. D. Calderon, B. Wu, N. Wei, X. Wang, Y. Guo, Z. Xiao, J. Song, G. Sugiarto, Y. Li, H. Yu, X. Chen and P. G. Wang, Chem. Sci., 2015, 6, 5652–5661 RSC.
- A. D. Calderon, J. Zhou, W. Guan, Z. Wu, Y. Guo, J. Bai, Q. Li, P. G. Wang, J. Fang and L. Li, Org. Biomol. Chem., 2017, 15, 7258–7262 RSC.
- S. W. Lin, T. M. Yuan, J. R. Li and C. H. Lin, Biochem, 2006, 45, 8108–8116 CrossRef CAS PubMed.
- Y. Li, H. Yu, H. Cao, S. Muthana and X. Chen, Appl. Microbiol. Biotechnol., 2012, 93, 2411–2423 CrossRef CAS PubMed.
- G. Sugiarto, K. Lau, Y. Li, Z. Khedri, H. Yu, D. T. Le and X. Chen, Mol. BioSyst., 2011, 7, 3021–3027 RSC.
- H. Yu, S. Huang, H. Chokhawala, M. Sun, H. Zheng and X. Chen, Angew. Chem., Int. Ed., 2006, 45, 3938–3944 CrossRef CAS PubMed.
- G. Sugiarto, K. Lau, J. Qu, Y. Li, S. Lim, S. Mu, J. B. Ames, A. J. Fisher and X. Chen, ACS Chem. Biol., 2012, 7, 1232–1240 CrossRef CAS PubMed.
- C. Chen, S. Wang, M. R. Gadi, H. Zhu, F. Liu, C.-C. Liu, L. Li, F. Wang, P. Ling and H. Cao, Chem. Commun., 2020, 56, 7549–7552 RSC.
- J. Ye, H. Xia, N. Sun, C.-C. Liu, A. Sheng, L. Chi, X.-W. Liu, G. Gu, S.-Q. Wang, J. Zhao, P. Wang, M. Xiao, F. Wang and H. Cao, Nat. Catal., 2019, 2, 514–522 CrossRef CAS.
- S. Wang, Q. Zhang, C. Chen, Y. Guo, M. R. Gadi, J. Yu, U. Westerlind, Y. Liu, X. Cao, P. G. Wang and L. Li, Angew. Chem., Int. Ed., 2018, 57, 9268–9273 CrossRef CAS PubMed.
- Z. Wu, Y. Liu, C. Ma, L. Li, J. Bai, L. Byrd-Leotis, Y. Lasanajak, Y. Guo, L. Wen, H. Zhu, J. Song, Y. Li, D. A. Steinhauer, D. F. Smith, B. Zhao, X. Chen, W. Guan and P. G. Wang, Org. Biomol. Chem., 2016, 14, 11106–11116 RSC.
- Z. Xiao, Y. Guo, Y. Liu, L. Li, Q. Zhang, L. Wen, X. Wang, S. M. Kondengaden, Z. Wu, J. Zhou, X. Cao, X. Li, C. Ma and P. G. Wang, J. Org. Chem., 2016, 81, 5851–5865 CrossRef CAS PubMed.
- S. Wang, C. Chen, M. R. Gadi, V. Saikam, D. Liu, H. Zhu, R. Bollag, K. Liu, X. Chen, F. Wang, P. G. Wang, P. Ling, W. Guan and L. Li, Nat. Commun., 2021, 12, 3573 CrossRef CAS PubMed.
- M. Weiss, D. Ott, T. Karagiannis, M. Weishaupt, M. Niemietz, S. Eller, M. Lott, M. Martínez-Orts, Á. Canales, N. Razi, J. C. Paulson and C. Unverzagt, ChemBioChem, 2020, 21, 3212–3215 CrossRef CAS PubMed.
- X. Meng, W. Yao, J. Cheng, X. Zhang, L. Jin, H. Yu, X. Chen, F. Wang and H. Cao, J. Am. Chem. Soc., 2014, 136, 5205–5208 CrossRef CAS PubMed.
- X. Song, B. Xia, S. R. Stowell, Y. Lasanajak, D. F. Smith and R. D. Cummings, Chem. Biol., 2009, 16, 36–47 CrossRef CAS PubMed.
- O. H. Hashim, J. J. Jayapalan and C. S. Lee, PeerJ, 2017, 5, e3784 CrossRef PubMed.
- D. C. Propheter, K. L. Hsu and L. K. Mahal, Methods Mol. Biol., 2011, 723, 67–77 CrossRef CAS PubMed.
- K. T. Pilobello, D. E. Slawek and L. K. Mahal, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 11534–11539 CrossRef CAS PubMed.
- R. D. Cummings and S. Kornfeld, J. Biol. Chem., 1982, 257, 11230–11234 CrossRef CAS PubMed.
- C. Gao, M. S. Hanes, L. A. Byrd-Leotis, M. Wei, N. Jia, R. J. Kardish, T. R. McKitrick, D. A. Steinhauer and R. D. Cummings, Cell Chem. Biol., 2019, 26, 535–547 CrossRef CAS PubMed.
- D. W. Heindel, S. Koppolu, Y. Zhang, B. Kasper, L. Meche, C. A. Vaiana, S. J. Bissel, C. E. Carter, A. A. Kelvin, M. Elaish, J. Lopez-Orozco, B. Zhang, B. Zhou, T. W. Chou, L. Lashua, T. C. Hobman, T. M. Ross, E. Ghedin and L. K. Mahal, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 26926–26935 CrossRef CAS PubMed.
- Z. Klamer, B. Staal, A. R. Prudden, L. Liu, D. F. Smith, G. J. Boons and B. Haab, Anal. Chem., 2017, 89, 12342–12350 CrossRef CAS PubMed.
- L. Li, W. Guan, G. Zhang, Z. Wu, H. Yu, X. Chen and P. G. Wang, Glycobiology, 2020, 30, 334–345 CrossRef CAS PubMed.
- X. Song, H. Yu, X. Chen, Y. Lasanajak, M. M. Tappert, G. M. Air, V. K. Tiwari, H. Cao, H. A. Chokhawala, H. Zheng, R. D. Cummings and D. F. Smith, J. Biol. Chem., 2011, 286, 31610–31622 CrossRef CAS PubMed.
- S. Narasimhan, J. C. Freed and H. Schachter, Carbohydr. Res., 1986, 149, 65–83 CrossRef CAS PubMed.
- O. Haji-Ghassemi, M. Gilbert, J. Spence, M. J. Schur, M. J. Parker, M. L. Jenkins, J. E. Burke, H. van Faassen, N. M. Young and S. V. Evans, J. Biol. Chem., 2016, 291, 24085–24095 CrossRef CAS PubMed.
- P. N. S. Iyer, K. D. Wilkinson and I. J. Goldstein, Arch. Biochem. Biophys., 1976, 177, 330–333 CrossRef CAS.
- L. G. Yu, N. Andrews, Q. Zhao, D. McKean, J. F. Williams, L. J. Connor, O. V. Gerasimenko, J. Hilkens, J. Hirabayashi, K. Kasai and J. M. Rhodes, J. Biol. Chem., 2007, 282, 773–781 CrossRef CAS PubMed.
- S. R. Stowell, C. M. Arthur, P. Mehta, K. A. Slanina, O. Blixt, H. Leffler, D. F. Smith and R. D. Cummings, J. Biol. Chem., 2008, 283, 10109–10123 CrossRef CAS PubMed.
- M. S. Macauley, P. R. Crocker and J. C. Paulson, Nat. Rev. Immunol., 2014, 14, 653–666 CrossRef CAS PubMed.
- S. Wang, C. Chen, M. Guan, D. Liu, X.-F. Wan and L. Li, Front. Mol. Biosci., 2021, 8, 645999 CrossRef CAS PubMed.
- R. D. Cummings and R. P. McEver, in Essentials of Glycobiology, ed. rd, A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart, M. Aebi, A. G. Darvill, T. Kinoshita, N. H. Packer, J. H. Prestegard, R. L. Schnaar and P. H. Seeberger, Cold Spring Harbor (NY), 2015, pp. 435–452, DOI:10.1101/glycobiology.3e.034.
- M. Nagae, K. Yamanaka, S. Hanashima, A. Ikeda, K. Morita-Matsumoto, T. Satoh, N. Matsumoto, K. Yamamoto and Y. Yamaguchi, J. Biol. Chem., 2013, 288, 33598–33610 CrossRef CAS PubMed.
- L. M. Chen, P. Rivailler, J. Hossain, P. Carney, A. Balish, I. Perry, C. T. Davis, R. Garten, B. Shu, X. Xu, A. Klimov, J. C. Paulson, N. J. Cox, S. Swenson, J. Stevens, A. Vincent, M. Gramer and R. O. Donis, Virology, 2011, 412, 401–410 CrossRef CAS PubMed.
- H. Zaraket, T. Baranovich, B. S. Kaplan, R. Carter, M. S. Song, J. C. Paulson, J. E. Rehg, J. Bahl, J. C. Crumpton, J. Seiler, M. Edmonson, G. Wu, E. Karlsson, T. Fabrizio, H. Zhu, Y. Guan, M. Husain, S. Schultz-Cherry, S. Krauss, R. McBride, R. G. Webster, E. A. Govorkova, J. Zhang, C. J. Russell and R. J. Webby, Nat. Commun., 2015, 6, 6553 CrossRef CAS PubMed.
- K. Dang, W. Zhang, S. Jiang, X. Lin and A. Qian, ChemistryOpen, 2020, 9, 285–300 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: The supporting information is available free of charge via the internet, including materials, experiment procedures, microarray fabrication and assay, HPLC, NMR, and MS analysis of synthesized compounds (Document S1), glycan microarray data (Document S2), and MotifFinder analysis results (Document S3). See https://doi.org/10.1039/d1sc05435j |
| ‡ Equal Contribution. |
| This journal is © The Royal Society of Chemistry 2022 |


![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S, MeOH
S, MeOH