Open Access Article
Open Access ArticleBiomimetic mimicry of formaldehyde-induced DNA–protein crosslinks in the confined space of a metal–organic framework†
Yu-Bai
Wei
 a,
Dong
Luo
a,
Dong
Luo
 a,
Xiao
Xiong
a,
Xiao
Xiong
 a,
Yong-Liang
Huang
a,
Yong-Liang
Huang
 b,
Mo
Xie
b,
Mo
Xie
 a,
Weigang
Lu
a,
Weigang
Lu
 *a and
Dan
Li
*a and
Dan
Li
 *a
*a
aCollege of Chemistry and Materials Science, Guangdong Provincial Key Laboratory of Functional Supramolecular Coordination Materials and Applications, Jinan University, Guangzhou 510632, P. R. China. E-mail: weiganglu@jnu.edu.cn; danli@jnu.edu.cn
bDepartment of Chemistry, Shantou University Medical College, Shantou, Guangdong 515041, P. R. China
First published on 21st March 2022
Abstract
DNA–protein crosslinks (DPCs) are highly toxic DNA lesions induced by crosslinking agents such as formaldehyde (HCHO). Building artificial models to simulate the crosslinking process would advance our understanding of the underlying mechanisms and therefore develop coping strategies accordingly. Herein we report the design and synthesis of a Zn-based metal–organic framework with mixed ligands of 2,6-diaminopurine and amine-functionalized dicarboxylate, representing DNA and protein residues, respectively. Combined characterization techniques allow us to demonstrate the unusual efficiency of HCHO-crosslinking within the confined space of the titled MOF. Particularly, in situ single-crystal X-ray diffraction studies reveal a sequential methylene-knitting process upon HCHO addition, along with strong fluorescence that was not interfered with by other metabolites, glycine, and Tris. This work has successfully constructed a purine-based metal–organic framework with unoccupied Watson–Crick sites, serving as a crystalline model for HCHO-induced DPCs by mimicking the confinement effect of protein/DNA interactions.
1. Introduction
DNA–protein interactions play significant roles in cellular functions such as transcriptional regulation, gene expression, cell division, and chromosomal organization.1 However, covalently linking proteins to DNA through crosslinking agents results in a specific type of DNA lesions known as DNA–protein crosslinks (DPCs). DPCs represent physical obstacles to the progression of DNA replication,2 which will ultimately threaten genomic integrity and cell viability.3 Thus, to gain a better understanding of DPCs is of great importance to advance our knowledge of their formation in biological systems and propose repair mechanisms accordingly. Yet no sophisticated models dedicated to mimicking DPCs have been reported to date.Numerous studies have demonstrated DPCs with formaldehyde (HCHO) as a crosslinker in vivo.4,5 The strikingly high crosslinking potency of HCHO made it a frequently used reagent for chemically binding DNA with proteins to study the impact of DPCs on DNA replication.6 HCHO-induced DPCs are reversible processes, which involve the formation of aminoacetal, a methylene knot bridging a wide range of DNA and proteins.7–9 The strikingly high potency of HCHO to crosslinking proteins to DNA has been exploited in various biochemical applications such as ChIP-Seq, which combines chromatin immunoprecipitation (ChIP) with DNA sequencing to identify the binding sites of DNA-associated proteins.10 Although many important biological insights have been revealed by ChIP-seq analysis, it suffers from tedious sample pre-treatments and notable inconsistencies between in vitro and in vivo; i.e., HCHO crosslinking is much less efficient in vitro than in vivo.11–21
All living organisms are compartmentalized into cells. A confined environment allows proteins to assemble and fold properly into active conformations. Researchers are increasingly realizing the impact of confinement on chemical reactivity. The past few decades have seen a surge of interest in the design of supramolecular assemblies for molecular recognition and mimicry of biological functions. Molecules in confined spaces could fundamentally change their chemical and physical properties. Confinement effects were observed not only in stabilizing reactive species, but also in accelerating chemical reactions and enhancing selectivity. For example, Fujita and co-workers explored supramolecular cages as reaction flasks to stack aromatic compounds and induce new intermolecular interactions and chemical phenomena.22–25
Metal–organic Frameworks (MOFs)26–33 are porous crystalline polymers formed by the assembly of metal clusters and organic linkers. Their nano-sized cavities inside can be viewed as uniformly arranged molecular flasks, which may exert a confinement effect on the guest molecules the same way as discrete molecular flasks, yet heterogeneously and collectively. Further, functional groups can be installed on the interior of the MOFs to simulate the interaction of enzymes and substrates in biological systems. For instance, Yaghi and his colleagues designed a multivariate MOF (MTV-IRMOF-74-III) with enzyme-like complexity and environment for multi-step oligopeptide synthesis.34 Similarly, the biologically relevant imidazole moieties and reactive copper-oxygen complexes were sequentially grafted within the framework of MOF-808 to mimic the particulate methane monooxygenase, realizing high selectivity of methane oxidation to methanol.35
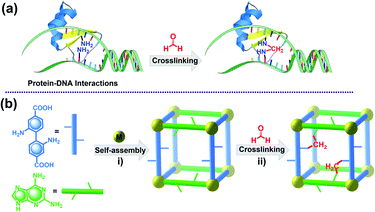
In this work, we report the design and synthesis of an amine-functionalized Zn-MOF (JNU-101) and its potential capacity as a crystalline model for mimicking HCHO-induced DPCs in vivo (Scheme 1). A mixed-ligand strategy was adopted to construct the framework, in which the two organic linkers, 2,6-diaminopurine (DAP) and 2,2′-diaminobiphenyl-4,4′-dicarboxylate (BPDC-2NH2), were judiciously selected to represent DNA and protein residues, respectively. Combined characterization techniques were applied to explore the HCHO-crosslinking efficiency within the confined space of JNU-101. Particularly, in situ single-crystal X-ray diffraction (SCXRD) studies reveal a sequential methylene-knitting process upon HCHO addition. The thus-amplified fluorescence was barely disturbed in the presence of other metabolites, Glycine (Gly), and tris(hydroxymethyl)aminomethane (Tris), the latter two are commonly used for quenching the HCHO-crosslinking activity. Isothermal titration calorimetry (ITC) studies confirm the sequential methylene knitting inside JNU-101 is a very favorable thermodynamic process, further attesting to the confinement effect on HCHO crosslinking inside JNU-101.
2. Results and discussion
2.1 Synthesis and structure
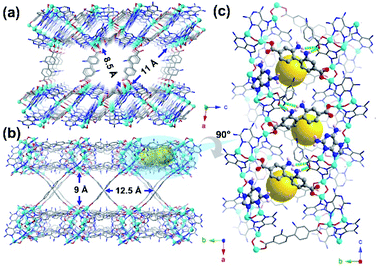
Solvothermal reaction of H2BPDC-2NH2, DAP, and Zn(NO3)2·6H2O in a solution of DMF/H2O/HNO3 at 135 °C for 72 h leads to the formation of yellow crystals of JNU-101 (Fig. 1). SCXRD reveals that JNU-101 crystallizes in a monoclinic crystal system with P2/c space group (Table S1†).36 Topological analysis unveils that JNU-101 features a 4,6-c network with the point symbol of {43.62.8}2{46.66.83}2 and transitivity of [2473] (Fig. S1†). The JNU-101 framework can be viewed as a pillar-layered structure with BPDC-2NH2 ligands as the pillars (Fig. 1a and b). There are three crystallographically independent Zn(II) atoms in the asymmetric unit, all of which adopt tetrahedral coordination geometry (Fig. S2a†).Among them, Zn1 and Zn2 form a dinuclear secondary building unit (SBU), Zn2O(DAP)2, with two chelating DAP at N3 and N9 as well as a bridging μ2-O2–, while the two axial positions are occupied by monodentated carboxylates of BPDC-2NH2 (Fig. S2b†). Zn3 is coordinated with two DAP at N7, a monodentated carboxylate of BPDC-2NH2, and a μ2-O2–, resulting in a mononuclear SBU, ZnO(DAP)2(η1-COO) (Fig. S2c†). The two SBUs are spaced by a DAP ligand, forming a grid-like substructure (Fig. S2d†), and these substructures are linked together through the μ2-O2– on the mononuclear SBUs into a one-dimensional (1D) chain-like structure along the c axis (Fig. S2e†). Adjacent 1D chains are further crosslinked through BPDC-2NH2 into a two-dimensional (2D) double-layered structure with intralayer cavities (Fig. 1c and S2f†). A closer look at the double-layered structure reveals that DAP and BPDC-2NH2 are located at the edge of the cavities in the proximity (highlighted in ball-and-stick models in Fig. 1c). Interestingly, the amino groups on DAP are orientated towards the cavities, while the amino groups on BPDC-2NH2 are orientated away from the cavities, held by hydrogen bonding with adjacent BPDC-2NH2 (O⋯H, 2.184 Å) and DAP (N⋯H, 2.548 Å) (Fig. 1c).
2.2 Sequential methylene knitting
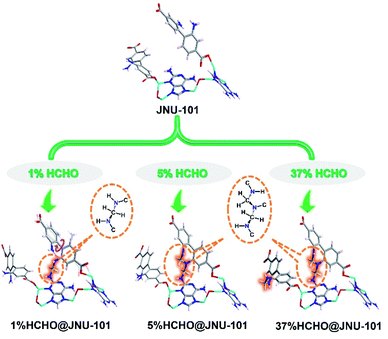
Inspired by the proximity of DAP and BPDC-2NH2 inside JNU-101 as well as the efficiency of HCHO-induced DPCs in vivo, the freshly prepared single crystals of JNU-101 were immersed in 1% HCHO solution for 1 day, and the SCXRD data were collected for the HCHO-loaded crystals (termed 1%HCHO@JNU-101) at 100 K. Although gas adsorption measurements were not successful (Fig. S4†), indicating JNU-101 losing its crystalline upon solvent removal, single crystal to single crystal transformation in HCHO solution is self-evidence of its hydrolytic stability. Further, HCHO is not freely adsorbed in the channels of JNU-101via physisorption, but chemically linked to the amino groups on DAP and BPDC-2NH2 by forming a methylene knot (Fig. S5†), which bears many similarities to the HCHO-induced DPCs in vivo.37 In our case, the seemingly simple methylene knitting involves a conformational change of BPDC-2NH2. Specifically, the amino group on BPDC-2NH2, originally orientated away due to hydrogen bonding (Fig. 1c), has to flip around to the same side of the amino group on DAP to form the methylene knot. Some resemblance can be seen in the structural and molecular mechanism of the HCHO-responsive transcription factor HxIR,38 in which the conformational change and resulting intrahelical methylene knot formation between the side-chains of Cys11 and Lys13 on HxIR's helix α1 (Cys–CH2–Lys) may allosterically regulate its DNA-binding domain.39Surprisingly, three methylene knots were observed in the crystal structure of 37%HCHO@JNU-101 (JNU-101 soaked in 37% HCHO solution for 1 day), prompting interest in their formation sequence. Therefore, single-crystal to single-crystal (SC–SC) transformation studies of JNU-101 in 5%, 10%, and 15% HCHO solutions were also performed, and their SCXRD data were collected at 100 K (termed 5%HCHO@JNU-101, 10%HCHO@JNU-101, 15%HCHO@JNU-101, respectively). As shown in Fig. 2, the first methylene knitting occurs between the amino group on DAP and the amino group on the intralayered BPDC-2NH2 in 1% HCHO as described above; the second tandem methylene knitting further bridges the two amino groups on the intralayered BPDC-2NH2 upon increasing the HCHO concentration to 5%; the third methylene knitting takes place between the two amino groups on the interlayered BPDC-2NH2 upon increasing the HCHO concentration to 37%. It appears that the 2-amino group on DAP is rather reactive and it can trigger the “N-terminal flipping” of BPDC-2NH2 and form the first methylene knot even in 1% HCHO solution. The amino groups on the intralayered BPDC-2NH2 were observed more prone to methylene knitting compared to the amino groups on the interlayered ones. We suspect that one of the amino groups on the intralayered BPDC-2NH2 was involved in the first methylene knitting, and its conformation constraint might facilitate the formation of the second knot in 5% HCHO solution. Yet neither of the amino groups on the interlayered BPDC-2NH2 was constrained and therefore did not form the third knot until they were soaked in 37% HCHO solution.
2.3 Thermodynamic studies
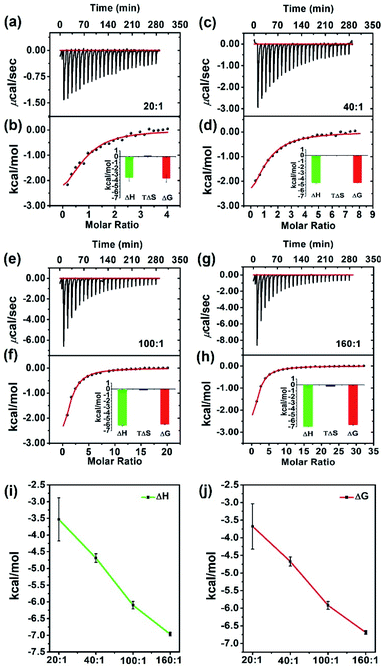
The binding constant (ka), enthalpy change (ΔH), entropy change (ΔS), and Gibbs free energy change (ΔG) of a chemical process can be directly quantified by Isothermal Titration Calorimetry (ITC),40–42 and this technique has already been frequently used to study the binding energy of substrate at the active site in homogeneous biological systems.43,44 Only recently has it been applied to measure host–guest interactions in heterogeneous systems, e.g. porous materials.45–50 To further explore the high efficiency of HCHO crosslinking in the confined space of JNU-101, we conducted ITC studies to characterize the liquid-phase thermodynamics of HCHO in JNU-101 (Fig. 3). As shown in Fig. 3a and b, a JNU-101 suspension (0.50 mM, 1.8 mL) was titrated with an HCHO solution (10 mM, 300 μL), and the corresponding thermogram (termed 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, molarity ratio of HCHO titrant to JNU-101 suspension) was obtained. The HCHO binding to JNU-101 is thermodynamically favored as evidenced by the calculated Gibbs free energy change (ΔG = −3.6782 ± 0.6455 kcal mol−1) and enthalpy change (ΔH), indicating the methylene knot formation is energetically favored with significant heat evolution.
1, molarity ratio of HCHO titrant to JNU-101 suspension) was obtained. The HCHO binding to JNU-101 is thermodynamically favored as evidenced by the calculated Gibbs free energy change (ΔG = −3.6782 ± 0.6455 kcal mol−1) and enthalpy change (ΔH), indicating the methylene knot formation is energetically favored with significant heat evolution.
The titration of JNU-101 suspension (0.50 mM, 1.8 mL) was then carried out with an increased HCHO concentration (20 mM, 300 μL), and the obtained thermogram (termed 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) exhibits a more negative ΔG (−4.6870 ± 0.1293 kcal mol−1) (Fig. 3c and d). A further increase of HCHO concentration (termed 100
1) exhibits a more negative ΔG (−4.6870 ± 0.1293 kcal mol−1) (Fig. 3c and d). A further increase of HCHO concentration (termed 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 160
1 and 160![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) results in even more negative ΔG (−5.9216 ± 0.1128 kcal mol−1 and −6.6875 ± 0.0535 kcal mol−1) (Fig. 3e–h). The negative correlations between ΔH and HCHO concentration as well as ΔG and HCHO concentration (Fig. 3i and j) confirm the HCHO crosslinking inside JNU-101 is a thermodynamically very favorable process.
1) results in even more negative ΔG (−5.9216 ± 0.1128 kcal mol−1 and −6.6875 ± 0.0535 kcal mol−1) (Fig. 3e–h). The negative correlations between ΔH and HCHO concentration as well as ΔG and HCHO concentration (Fig. 3i and j) confirm the HCHO crosslinking inside JNU-101 is a thermodynamically very favorable process.
To prove the confinement effect on HCHO crosslinking inside JNU-101, control experiments were carried out and ITC thermograms were obtained from the titration of a suspension of Zn(NO3)2·6H2O, DAP, and H2BPDC-2NH2 with HCHO titrants of different concentrations. As shown in Fig. S19,† the ΔG and ΔH values are rather small compared to the titration of JNU-101 suspension, indicating the HCHO crosslinking of free DAP and BPDC-2NH2 is substantially less favored. To further quantify if there is any crosslinking, high-resolution electrospray ionization time-of-flight mass spectrometry (HR ESI-TOF MS) was performed. As can be seen in Fig. S20,† DAP and H2BPDC-2NH2 molecular ions can be identified, yet no signals of any crosslinking products can be found. This is in stark contrast to the high efficiency of HCHO crosslinking inside JNU-101. On the other hand, 13C solid-state nuclear magnetic resonance spectroscopy (13C SSNMR) studies corroborate the methylene knot formation in 1%HCHO@JNU-101, 5%HCHO@JNU-101, and 37%HCHO@JNU-101, as evidenced by the appearance of carbon signals of aminoacetals in the region of 50–80 ppm (Fig. S21†).
2.4 Exclusive HCHO recognition
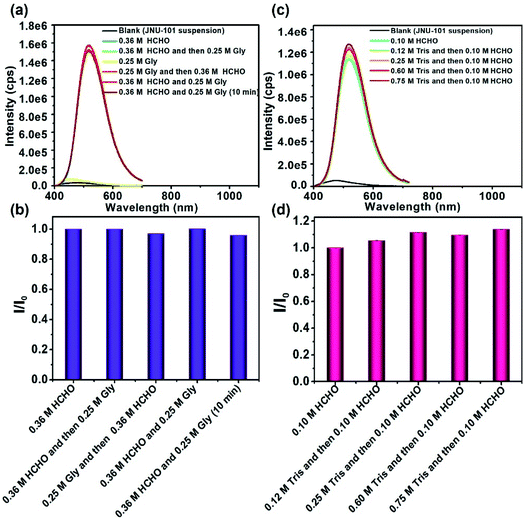
Significant fluorescence enhancement has been observed for the JNU-101 suspension upon HCHO addition, which could partially be attributed to the ligand-to-ligand charge transfer (LLCT) through bridging methylene knots.51 Human metabolism may produce a variety of biologically relevant species, including formaldehyde, formic acid, acetone, ethanol, lactic acid, urea, amino acids, and so on.52–57 Enlightened by the efficiency of HCHO crosslinking in JNU-101, we decided to look into if these metabolic products would interfere with the HCHO-crosslinking-induced fluorescence. Fig. S23† depicts the emission signals of JNU-101 suspension upon addition of different metabolites. Remarkably, the emission intensity of JNU-101 was increased by 19 times upon addition of HCHO, while it was barely changed in the presence of other metabolites. In addition, amino acids hardly affect the fluorescence response of JNU-101 toward HCHO (Fig. S24†), and such fluorescence response was observed at the extremely low concentration of HCHO, further proving the high efficiency of HCHO crosslinking in JNU-101 (Fig. S25†). These results demonstrate an exclusive recognition of HCHO without the interference of other metabolites, suggesting the potential of JNU-101 for accurately quantifying the HCHO level in vivo.Gly and Tris are two commonly used agents for quenching HCHO crosslinking in ChIP analysis.58–67 Hence, we decided to investigate whether the HCHO crosslinking in JNU-101 can be quenched by Gly and Tris. As shown in Fig. 4a and b, the addition of Gly, whether before or after HCHO addition, barely affects the emission intensity, indicating no quenching effect of Gly on HCHO crosslinking in JNU-101. Tris is supposed to be more effective in quenching the HCHO crosslinking activity,68 yet the emission intensity was hardly disturbed upon addition of Tris of different concentrations (Fig. 4c and d), implying also no quenching effect of Tris on HCHO crosslinking in JNU-101. These results further demonstrate the confinement effect of JNU-101, in which the amino groups on the ligands were restricted in close vicinity, facilitating an efficient aminoacetal formation without having to pay entropy penalty, as would for free amino groups.
2.5 Potential clinical applications
Physiological HCHO levels were reported to be around 0.1 mM in human blood.69–71 Clinical data show elevated HCHO concentrations for patients suffering from cancers or Alzheimer's disease.70,72,73 For example, the HCHO concentrations were found 0.756 ± 0.12 mM in breast cancer tissues and 0.726 ± 0.06 mM in the lung cancer tissues, much higher than that in the normal tissues adjacent to the cancer sites (0.196 ± 0.06 mM).74 The excess production of HCHO by tumor tissues renders it a biomarker of certain cancerization, osseous metastasis, and neurodegenerative diseases. Inspired by the ultralow detection limit of JNU-101 for HCHO in aqueous solutions, we further investigated whether it could produce an effective fluorescence response in vitro within the above-mentioned HCHO concentrations. Fluorescence optical imaging of JNU-101 shows no obvious fluorescence response after being exposed to a cell culture (DMEM + 2% FBS) containing less than 0.20 mM of HCHO (Fig. S28†), yet a distinctive greenish fluorescence response when HCHO concentration was increased to 0.40 mM (Fig. 5c), suggesting the prospect of being developed into an auxiliary diagnostic tool. Powder X-ray diffraction (PXRD) measurements were also carried out to confirm the framework integrity of JNU-101 in above cell culture media (Fig. S29†). Clinical data also show elevated HCHO concentrations in the exhaled air of lung cancer patients.75,76 We thus explored its potential of sensing HCHO vapor in a closed environment. As depicted in Fig. S30,†JNU-101 exhibits a quick fluorescence response to an HCHO vapor of ca. 3.8 ppm (the saturated HCHO vapor concentration over 1% HCHO aqueous solution), suggesting the prospect of being integrated into health care systems for monitoring HCHO in exhalation.3. Conclusions
In summary, we report a zinc-based biological metal–organic framework (JNU-101) constructed with mixed ligands of 2,6-diaminopurine and 2,2′-diaminobiphenyl-4,4′-dicarboxylate. The integration of structural characteristics of DNA and protein residues renders JNU-101 a crystalline model for HCHO-induced DNA-protein crosslinks. In situ single-crystal X-ray diffraction reveals a sequential methylene-knitting process upon HCHO addition. Isothermal titration calorimetry assays unveil large negative Gibbs free energy change for the methylene-knitting process. Fluorescence studies in aqueous solutions demonstrate an exclusive HCHO recognition of JNU-101 that was not interfered with by other metabolites, Gly, and Tris, the latter two are commonly used reagents for quenching the HCHO-crosslinking activity in ChIP analysis. Overall, we have successfully constructed an amine-functionalized metal–organic framework that could potentially serve as a crystalline model for HCHO-induced DNA-protein crosslinks by mimicking the confinement effect of protein/DNA interactions. The crystalline nature of MOF materials may be utilized to acquire structural evolution information in the process of crosslinking by X-ray diffraction technology. The detailed structural elucidation of HCHO crosslinking in JNU-101 is of great significance for the in-depth understanding of the HCHO-induced DNA-protein cross-links in biological systems, which may be conducive to the study of DNA damages derived from DPCs and possible repair mechanisms.Data availability
All data are available in the main text and ESI.†Author contributions
The manuscript was written through contributions of all authors. W. Lu, and D. Li conceived the research ideas. Y.-B. Wei, D. Luo, Y.-L. Huang and M. Xie performed experiments including the fabrication and modification of MOFs. Y.-B. Wei and X. Xiong carried out the experiments and collected the data; Y.-B. Wei, W. Lu, and D. Luo analyzed and processed the experimental data. Y.-B. Wei, W. Lu, and D. Li. wrote the paper. All authors have approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 21731002, 21975104, 22150004, 22101099, and 21801095), the Guangdong Major Project of Basic and Applied Research (2019B030302009), the Guangdong Basic and Applied Basic Research Foundation (No. 2020A1515011005), and the Special Fund Project for Science and Technology of Guangdong (No. STKJ2021172). We thank Prof. W. Wang and S.-Y. Ding (Lanzhou University) for solid–state 13C NMR measurements and their helpful discussions.Notes and references
- M. L. Bulyk, Curr. Opin. Biotechnol., 2006, 17, 422–430 CrossRef CAS PubMed
.
- B. Vaz, M. Popovic and K. Ramadan, Trends Biochem. Sci., 2017, 42, 483–495 CrossRef CAS PubMed
.
- J. Stingele, M. S. Schwarz, N. Bloemeke, P. G. Wolf and S. Jentsch, Cell, 2014, 158, 327–338 CrossRef CAS PubMed
.
- S. J. Zanton and B. F. Pugh, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 16843–16848 CrossRef CAS PubMed
.
- M. J. Solomon, P. L. Larsen and A. Varshavsky, Cell, 1988, 53, 937–947 CrossRef CAS PubMed
.
- S. Pfuhler and H. Uwe Wolf, Environ. Mol. Mutagen., 1996, 27, 196–201 CrossRef CAS PubMed
.
- K. Lu, W. Ye, L. Zhou, L. B. Collins, X. Chen, A. Gold, L. M. Ball and J. A. Swenberg, J. Am. Chem. Soc., 2010, 132, 3388–3399 CrossRef CAS PubMed
.
- J. Kennedy-Darling and L. M. Smith, Anal. Chem., 2014, 86, 5678–5681 CrossRef CAS PubMed
.
- G. Quievryn and A. Zhitkovich, Carcinogenesis, 2000, 21, 1573–1580 CrossRef CAS PubMed
.
- T. S. Furey, Nat. Rev. Genet., 2012, 13, 840–852 CrossRef CAS PubMed
.
- D. S. Gilmour and J. T. Lis, Proc. Natl. Acad. Sci. U. S. A., 1984, 81, 4275–4279 CrossRef CAS PubMed
.
- D. S. Gilmour and J. T. Lis, Mol. Cell. Biol., 1985, 5, 2009–2018 CAS
.
- P. J. Farnham, Nat. Rev. Genet., 2009, 10, 605–616 CrossRef CAS PubMed
.
- L. Teytelman, D. M. Thurtle, J. Rine and A. van Oudenaarden, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 18602–18607 CrossRef CAS PubMed
.
- D. Park, Y. Lee, G. Bhupindersingh and V. R. Iyer, PloS One, 2013, 8, e83506 CrossRef PubMed
.
- D. Jain, S. Baldi, A. Zabel, T. Straub and P. B. Becker, Nucleic Acids Res., 2015, 43, 6959–6968 CrossRef CAS PubMed
.
- W. C. Gasper, G. K. Marinov, F. Pauli-Behn, M. T. Scott, K. Newberry, G. DeSalvo, S. Ou, R. M. Myers, J. Vielmetter and B. J. Wold, Sci. Rep., 2014, 4, 5152 CrossRef CAS PubMed
.
- D. M. Thurtle and J. Rine, Genes Dev., 2014, 28, 245–258 CrossRef CAS PubMed
.
- L. Teytelman, B. Özaydın, O. Zill, P. Lefrançois, M. Snyder, J. Rine and M. B. Eisen, PloS One, 2009, 4, e6700 CrossRef PubMed
.
- A. K. Nagaich, D. A. Walker, R. Wolford and G. L. Hager, Mol. cell, 2004, 14, 163–174 CrossRef CAS PubMed
.
- L. Schmiedeberg, P. Skene, A. Deaton and A. Bird, PLoS One, 2009, 4, e4636 CrossRef PubMed
.
- M. Yoshizawa, J. Nakagawa, K. Kumazawa, M. Nagao, M. Kawano, T. Ozeki and M. Fujita, Angew. Chem., Int. Ed., 2005, 44, 1810–1813 CrossRef CAS PubMed
.
- M. Yoshizawa, M. Nagao, K. Kumazawa and M. Fujita, J. Organomet. Chem., 2005, 690, 5383–5388 CrossRef CAS
.
- Y. Yamauchi, M. Yoshizawa and M. Fujita, J. Am. Chem. Soc., 2008, 130, 5832–5833 CrossRef CAS PubMed
.
- J. Rebek, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 10423–10424 CrossRef CAS PubMed
.
- A. Kirchon, L. Feng, H. F. Drake, E. A. Joseph and H.-C. Zhou, Chem. Soc. Rev., 2018, 47, 8611–8638 RSC
.
- S. Furukawa, J. Reboul, S. Diring, K. Sumida and S. Kitagawa, Chem. Soc. Rev., 2014, 43, 5700–5734 RSC
.
- P. Deria, J. E. Mondloch, O. Karagiaridi, W. Bury, J. T. Hupp and O. K. Farha, Chem. Soc. Rev., 2014, 43, 5896–5912 RSC
.
- X. Lian, Y. Fang, E. Joseph, Q. Wang, J. Li, S. Banerjee, C. Lollar, X. Wang and H.-C. Zhou, Chem. Soc. Rev., 2017, 46, 3386–3401 RSC
.
- W. Lu, Z. Wei, Z. Y. Gu, T. F. Liu, J. Park, J. Park, J. Tian, M. Zhang, Q. Zhang, T. Gentle, IIIrd, M. Bosch and H.-C. Zhou, Chem. Soc. Rev., 2014, 43, 5561–5593 RSC
.
- O. M. Yaghi, M. O'Keeffe, N. W. Ockwig, H. K. Chae, M. Eddaoudi and J. Kim, Nature, 2003, 423, 705–714 CrossRef CAS PubMed
.
- H. Furukawa, K. E. Cordova, M. O'Keeffe and O. M. Yaghi, Science, 2013, 341, 1230444 CrossRef PubMed
.
- H. Cai, L. Xu, M. Li and D. Li, Chin. Sci. Bull., 2016, 61, 1762–1773 CrossRef
.
- A. M. Fracaroli, P. Siman, D. A. Nagib, M. Suzuki, H. Furukawa, F. D. Toste and O. M. Yaghi, J. Am. Chem. Soc., 2016, 138, 8352–8355 CrossRef CAS PubMed
.
- J. Baek, B. Rungtaweevoranit, X. Pei, M. Park, S. C. Fakra, Y. S. Liu, R. Matheu, S. A. Alshmimri, S. Alshehri, C. A. Trickett, G. A. Somorjai and O. M. Yaghi, J. Am. Chem. Soc., 2018, 140, 18208–18216 CrossRef CAS PubMed
.
- Y.-B. Wei, CCDC 2041867, CSD Communications, 2020, DOI:10.5517/CCDC.CSD.CC26JQP5.
- E. A. Hoffman, B. L. Frey, L. M. Smith and D. T. Auble, J. Biol. Chem., 2015, 290, 26404–26411 CrossRef CAS PubMed
.
- H. Yurimoto, R. Hirai, N. Matsuno, H. Yasueda, N. Kato and Y. Sakai, a Member of the Duf24 Protein Family, is a DNA-Binding Protein That Acts as a Positive Regulator of the Formaldehyde-Inducible Hxlab Operon in Bacillus Subtilis, Mol. Microbiol., 2005, 57, 511–519 CrossRef CAS PubMed
.
- R. Zhu, G. Zhang, M. Jing, Y. Han, J. Li, J. Zhao, Y. Li and P. R. Chen, Nat. Commun., 2021, 12, 581 CrossRef CAS PubMed
.
- O. L. M. Ernesto Freire and M. Straume, Anal. Chem., 1990, 18, 950A–959A CrossRef
.
- M. W. Freyer and E. A. Lewis, Methods Cell Biol., 2008, 84, 79–113 CAS
.
- A. Velazquez-Campoy and E. Freire, Nat. Protoc., 2006, 1, 186–191 CrossRef CAS PubMed
.
- S. L. a. E. Freire, Curr. Opin. Struct. Biol., 2001, 5, 560–566 Search PubMed
.
- M. M. Pierce, C. S. Raman and B. T. Nall, Methods, 1999, 2, 213–221 CrossRef PubMed
.
- V. Agostoni, R. Anand, S. Monti, S. Hall, G. Maurin, P. Horcajada, C. Serre, K. Bouchemal and R. Gref, J. Mater. Chem. B, 2013, 1, 4231–4242 RSC
.
- A. Aykac, M. Noiray, M. Malanga, V. Agostoni, J. M. Casas-Solvas, E. Fenyvesi, R. Gref and A. Vargas-Berenguel, Biochim. Biophys. Acta. Gen. Subj., 2017, 1861, 1606–1616 CrossRef CAS PubMed
.
- S. Kato, R. J. Drout and O. K. Farha, Cell Rep. Phys. Sci., 2020, 1, 100006 CrossRef
.
- J. C. Moreno-Pirajan and L. Giraldo, Rev. Sci. Instrum., 2012, 83, 015117 CrossRef PubMed
.
- C. J. Penn, J. M. Gonzalez and I. Chagas, Front. Chem., 2018, 6, 307 CrossRef PubMed
.
- R. J. Drout, S. Kato, H. Chen, F. A. Son, K. I. Otake, T. Islamoglu, R. Q. Snurr and O. K. Farha, J. Am. Chem. Soc., 2020, 142, 12357–12366 CrossRef CAS PubMed
.
- Y.-B. Wei, M.-J. Wang, D. Luo, Y.-L. Huang, M. Xie, W. Lu, X. Shu and D. Li, Mater. Chem. Front., 2021, 5, 2416–2424 RSC
.
- S. Bouatra, F. Aziat, R. Mandal, A. C. Guo, M. R. Wilson, C. Knox, T. C. Bjorndahl, R. Krishnamurthy, F. Saleem, P. Liu, Z. T. Dame, J. Poelzer, J. Huynh, F. S. Yallou, N. Psychogios, E. Dong, R. Bogumil, C. Roehring and D. S. Wishart, PloS One, 2013, 8, e73076 CrossRef CAS PubMed
.
- W. Jumpathong, W. Chan, K. Taghizadeh, I. R. Babu and P. C. Dedon, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E4845–E4853 CrossRef CAS PubMed
.
- N. Psychogios, D. D. Hau, J. Peng, A. C. Guo, R. Mandal, S. Bouatra, I. Sinelnikov, R. Krishnamurthy, R. Eisner, B. Gautam, N. Young, J. Xia, C. Knox, E. Dong, P. Huang, Z. Hollander, T. L. Pedersen, S. R. Smith, F. Bamforth, R. Greiner, B. McManus, J. W. Newman, T. Goodfriend and D. S. Wishart, PloS One, 2011, 6, e16957 CrossRef CAS PubMed
.
- S. Becker, L. Kortz, C. Helmschrodt, J. Thiery and U. Ceglarek, J. Chromatogr. B, 2012, 883–884, 68–75 CrossRef CAS PubMed
.
- K. Oliphant and E. Allen-Vercoe, Microbiome, 2019, 7, 91 CrossRef PubMed
.
- W. Lin, L. P. Conway, A. Block, G. Sommi, M. Vujasinovic, J. M. Lohr and D. Globisch, Analyst, 2020, 145, 3822–3831 RSC
.
- C. Klockenbusch and J. Kast, J. Biomed. Biotechnol., 2010, 2010, 927585 Search PubMed
.
- C. Klockenbusch, J. E. O'Hara and J. Kast, Anal. Bioanal. Chem., 2012, 404, 1057–1067 CrossRef CAS PubMed
.
- Y. Lai, R. Yu, H. J. Hartwell, B. C. Moeller, W. M. Bodnar and J. A. Swenberg, Cancer Res., 2016, 76, 2652–2661 CrossRef CAS PubMed
.
- J. Z. Rappoport, L. Schmiedeberg, P. Skene, A. Deaton and A. Bird, PLoS One, 2009, 4, e4636 CrossRef PubMed
.
- Q. You, A. Y. Cheng, X. Gu, B. T. Harada, M. Yu, T. Wu, B. Ren, Z. Ouyang and C. He, Nat. Biotechnol., 2021, 39, 225–235 CrossRef CAS PubMed
.
- H. Zaidi, E. A. Hoffman, S. J. Shetty, S. Bekiranov and D. T. Auble, J. Biol. Chem., 2017, 292, 19338–19355 CrossRef CAS PubMed
.
- P. Y. Zeng, C. R. Vakoc, Z. C. Chen, G. A. Blobel and S. L. Berger, Biotechniques, 2006, 41, 694–698 CrossRef CAS PubMed
.
- S. M. N. Brett, A. Kaufman, R. L. Hallberg, C. A. Slaughter, P. S. Perlman and R. A. Butow, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 7772–7777 CrossRef PubMed
.
- J. Wells and P. J. Farnham, Methods, 2002, 26, 48–56 CrossRef CAS PubMed
.
- L. Valášek, B. Szamecz, A. G. Hinnebusch and K. H. Nielsen, Methods Enzymol., 2007, 429, 163–183 Search PubMed
.
- C. H. Wu, S. Chen, M. R. Shortreed, G. M. Kreitinger, Y. Yuan, B. L. Frey, Y. Zhang, S. Mirza, L. A. Cirillo, M. Olivier and L. M. Smith, PLoS One, 2011, 6, e26217 CrossRef CAS PubMed
.
- H. Heck and M. Casanova, Regul. Toxicol. Pharmacol., 2004, 40, 92–106 CrossRef CAS PubMed
.
- Z. Tong, J. Zhang, W. Luo, W. Wang, F. Li, H. Li, H. Luo, J. Lu, J. Zhou, Y. Wan and R. He, Neurobiol. Aging, 2011, 32, 31–41 CrossRef CAS PubMed
.
- Z. Tong, C. Han, W. Luo, X. Wang, H. Li, H. Luo, J. Zhou, J. Qi and R. He, Age, 2013, 35, 583–596 CrossRef PubMed
.
- S. E. Ebeler, A. J. Clifford and T. Shibamoto, J. Chromatogr. B, 1997, 702, 211 CrossRef CAS
.
- L. Trézl, I. Rusznák, E. Tyihák, T. Szarvas and B. Szende, Biochem. J., 1983, 214, 289 CrossRef PubMed
.
- Z. Tong, W. Luo, Y. Wang, F. Yang, Y. Han, H. Li, H. Luo, B. Duan, T. Xu, Q. Maoying, H. Tan, J. Wang, H. Zhao, F. Liu and Y. Wan, PloS One, 2010, 5, e10234 CrossRef PubMed
.
- M. Hakim, Y. Y. Broza, O. Barash, N. Peled, M. Phillips, A. Amann and H. Haick, Chem. Rev., 2012, 112, 5949–5966 CrossRef CAS PubMed
.
- A. Wehinger, A. Schmid, S. Mechtcheriakov, M. Ledochowski, C. Grabmer, G. A. Gastl and A. Amann, Int. J. Mass Spectrom., 2007, 265, 49–59 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available: physical measurements, and other characterizations. CCDC 2081103–2081107. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d2sc00188h |
| This journal is © The Royal Society of Chemistry 2022 |