Open Access Article
Open Access ArticleA reversible plasmonic nanoprobe for dynamic imaging of intracellular pH during endocytosis†
Jin
Wang
 a,
Qiao
Yu
a,
Xiang-Ling
Li
a,
Qiao
Yu
a,
Xiang-Ling
Li
 *b,
Xue-Li
Zhao
c,
Hong-Yuan
Chen
a and
Jing-Juan
Xu
*b,
Xue-Li
Zhao
c,
Hong-Yuan
Chen
a and
Jing-Juan
Xu
 *a
*a
aState Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210023, China. E-mail: xujj@nju.edu.cn; xlli@njtech.edu.cn
bCollege of Life Science and Pharmaceutical Engineering, Nanjing Tech University, Nanjing 211816, China
cCollege of Chemistry and Molecular Engineering, Zhengzhou University, Zhengzhou 450001, China
First published on 5th April 2022
Abstract
Understanding the pH evolution during endocytosis is essential for our comprehension of the fundamental processes of biology as well as effective nanotherapeutic design. Herein, we constructed a plasmonic Au@PANI core–shell nanoprobe, which possessed significantly different scattering properties under acidic and basic conditions. Encouragingly, the scattering signal of Au@PANI nanoprobes displayed a positive linear correlation with the pH value not only in PBS but also in nigericin-treated cells. Ultimately, benefiting from the excellent response properties as well as the excellent biocompatibility and stability, the Au@PANI nanoprobes have successfully enabled a dynamic assessment of the evolving pH in the endosomal package as the endosome matured from early to late, and eventually to the lysosome, by reporting scattering signal changes.
1 Introduction
The intracellular pH, an essential physiological parameter, has been proved to be crucial for maintaining the function of various organelles inside cells,1,2 while abnormal pH evolution could induce the dysfunction of cells, which is related to various diseases such as cancer, Alzheimer's, etc.3–5 Moreover, the trafficking and release processes of cells after the uptake of external substances are closely associated with fluctuations of pH in the cellular compartment.6 Specifically, the cellular internalization of biomacromolecules or nanoparticles triggers a series of activities, including early endosome formation, evolution, and maturation, and subsequently targeted trafficking, culminating in the arrival of lysosomes.7,8 Owing to the great importance for the development of pathologies as well as therapeutics such as drug delivery, the endosomal maturation and acidification progress has currently attracted extensive interest from researchers.9–11 To date, diverse forms of intracellular pH-responsive probes have been developed, including fluorescence,12 electrochemistry,13 SERS,14,15etc. Moreover, organelle-specific targeting can be further achieved by ingenious targeting design. For instance, Guo's group reported a new fluorescein/cyanine hybrid sensor, realizing ratiometric detection of pH fluctuation in mitochondria.16 Over recent decades, nanotechnology has unveiled significant prospects for the development of pH-responsive nanoparticles.17 For example, numerous nanoparticle-based nanomaterials have been used to fabricate nanoprobes capable of monitoring pH fluctuations during important physiological processes such as the cell cycle or cellular autophagy,10,18 as well as tracking the maturation process of endosomes.19,20 Despite the great progress, the development of nanoprobes for real-time tracking of pH changes still suffers from several limitations. The first limitation is the stability of the probes; the commonly used probes based on fluorescent dyes or fluorescent proteins21,22 are still not immune to the effects of photobleaching and cellular autofluorescence,23 and some organic small molecule dyes have a tendency to rapid leak from cells. The second one is the structural and functional integrity of the cell; for example, some SERS and electrochemical-based sensing devices could cause irreversible damage to cells,15,24 while polymer nanoparticle-based probes may also cause damage to the organelle due to the use of large amounts of block copolymers.25 These largely hampered the long-term, continuous monitoring of pH-related biological processes occurring inside living cells. Accordingly, the development of nanoprobes with excellent photostability, biocompatibility and signal-to-noise ratio for dynamic monitoring of intracellular pH evolution remains a pressing issue.Polyaniline (PANI), a kind of conductive polymer with conjugated molecular structures, has received significant attention in biosensing and biomedicine, due to its good biocompatibility, low cost, and mild synthesis conditions.26–28 In general, PANI exists in six different states, namely pernigraniline salt (PS), pernigraniline base (PB), emeraldine salt (ES), emeraldine base (EB), leucoemeraldin salt (LS), and leucoemeraldin base (LB).29 The switch between all these states can be realized by protonation/deprotonation and applying electrochemical potential.30 Wang's group systematically investigated the plasmonic switching behavior of (Au nanorod core)/(PANI shell) nanostructures, and found that the plasmon resonance of the Au core can be modulated reversibly by varying the dielectric function of the PANI shell through proton doping.31 Moreover, it has been proved that integrating a transition metal into the PANI matrix could increase the electron-delivery efficiency and thus decrease the transition pH value of PANI from the EB to the ES state, which facilitates its application in cells and in vivo. For example, Qu's group reported an Au core/PANI shell nanostructure (Au@PANI) as an intelligent tumor acidic microenvironment-activated photothermal agent, which showed a much higher photothermal effect at pH 6.5 owing to the increased electron-delivery efficiency, greatly improving the effectiveness of tumor ablation.32 Beyond photothermal therapy (PTT), the distinctive optical response properties of Au@PANI modulated by proton doping have led to a wide range of applications in surface-enhanced Raman scattering (SERS),33 electrochemistry sensing,34 and other fields as well.35,36
It is noteworthy that, as a promising technology, dark-field microscope (DFM)-based single-particle imaging has inspired extensive research on biosensing and cellular imaging recently.37–39 The scattering signal is generated by the local surface plasmon resonance (LSPR) of nanoparticles, which offers many advantages over other integral average-based measurement methods, such as low sample consumption, simple preparation, and low background interference.40,41 Furthermore, the excellent photostability of plasmonic nanoparticles enables long-term tracking of chemical reactions during biochemistry processes.42,43 Hence, combined with the unique optical properties of PANI, we believe that the utilization of Au@PANI for dynamic single-particle tracking of intracellular pH fluctuations based on the changes of light scattering will be of great potential interest. Herein, we synthesized a pH-sensitive Au@PANI core–shell nanoprobe, in which the PANI shell could respond to the pH variation of the circumstances, and the Au core would give feedback on the proton-doping levels of PANI by the fluctuations of single-particle Rayleigh scattering under DFM. Benefiting from the stability and biocompatibility of PANI, this nanoprobe was able to maintain good dispersion and responsiveness both in PBS and in living cells. In light of this, it was eventually applied for real-time quantitative monitoring of pH evolution during intracellular endosome maturation and provided a dynamic assessment of the evolution of endosomal pH as the endosome matures from early to late, and finally to the lysosome, by reporting the scattering signal changes of the intracellular Au@PANI nanoparticle (NP) populations over time (Scheme 1).
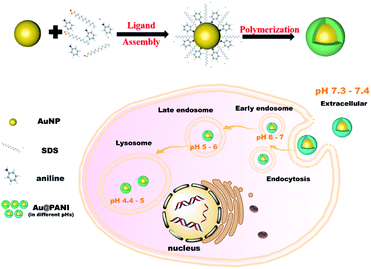 | ||
| Scheme 1 Synthetic process of the Au@PANI nanoprobe and schematic representations of the endosomal pathway for nanoparticle cellular uptake. | ||
2 Results and discussion
2.1 Synthesis and characterization of the pH-responsive nanoprobes
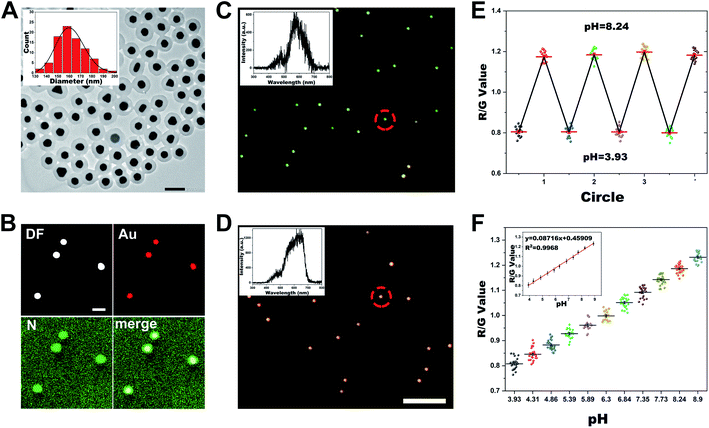
The synthesis process of Au@PANI core–shell NPs was according to previously reported work with some modifications.43 Firstly, colloidal Au nanoparticles (AuNPs) with an average diameter of ca. 80 nm were employed as the cores, which could produce brighter dark-field scattered light due to their relatively strong LSPR, enabling effective differentiation of the nanoprobe's signal from the cellular background. Thereafter, the PANI shell was constructed on the Au core through surfactant-assisted chemical oxidative polymerization of aniline in a strongly acidic environment. According to a previous report,44 during the synthesis process, the PANI chains grew in the protonated pernigraniline form by adding anilinium cations in para positions, and at the end of oxidation, they would be reduced by excess aniline to the protonated ES form. As illustrated by the TEM images of AuNPs before and after the PANI coating (Fig. S1A† and 1A), the average diameter of AuNPs was 80.4 nm and the thickness of PANI shells was measured to be about 40 nm. Elemental mappings in Fig. 1B of Au and N elements further confirmed the formation of the PANI shell. As shown in Fig. S1B,† compared with the UV-vis spectrum of AuNPs, two new absorption peaks at 425 and 800 nm emerged on that of PANI coated AuNPs, accompanied by the plasmonic resonance of AuNPs shifted to 565 nm. Accordingly, the color of the suspension changed from pink to fuchsia, and finally purple during the synthesis process. The zeta potential of Au@PANI NPs dispersed in PBS (pH ca. 7.3) was measured to be −17.1 mV (Fig. S2B†), owing to the outer coating of SDS as a protective agent. From this, it can be assumed that the nanoprobe was able to immobilize on the positively charged glass slide for subsequent single-particle studies. As displayed in Fig. S1C,† the Au@PANI NPs exhibited a uniform and monodisperse orange color under the DFM in PBS of pH ca. 7.3. The characteristic Rayleigh scattering peak of Au was located at ca. 630 nm, while the shoulder peak at ca. 460 nm was the scattering peak of PANI.29 Therefore, the above results proved the potential of the Au@PANI nanoprobe for single-particle studies based on Rayleigh scattering under DFM.2.2 The pH-responsive behavior of the plasmonic nanoprobes
As is known, by changing the acid–base properties of the environment, emeraldine PANI can switch between ES or EB states by proton doping and dedoping. Each state has different optical properties, such as dielectric constant, which further influence the plasmon resonance state of the Au core. Initially, we examined the feasibility of the Au@PANI NPs to respond to changes in the pH value. The UV-vis spectra in Fig. S2A† revealed that the characteristic absorption peak of Au@PANI NPs was red-shifted from 561 to 625 nm when the pH of solution changed from 1 to 12, owing to EB possessing a higher refractive index than ES. Additionally, we found that the zeta potential of Au@PANI NPs in an alkaline environment (pH 12) was about −22 mV, but in an acidic environment (pH 1), it actually decreased to −2 mV (Fig. S2B†), which may also result from the EB–ES form transition of PANI by proton doping. As for the single-particle measurement, as depicted in Fig. 1C and D, the Au@PANI NPs displayed a green color under the DFM in a pH 3.93 environment and changed to orange-red at a pH of 8.90. The obtained scattering spectra of single Au@PANI NP revealed that when pH changed from 8.90 to 3.93, the scattering peak shifted from 640 nm to 575 nm accompanied by a significant decrease in intensity. To better visualize the overall performance of the Au@PANI nanoprobes, an RGB analysis of the scattered light was conducted by using Image J software. Specifically, the R (red), G (green), and B (blue) chrominance information of the nanoprobe from the dark-field image could be obtained by splitting color channels, from which the R/G ratio was then derived.45 As displayed in Fig. S2C,† the average R/G value was ca. 0.81 under acid conditions (pH 3.93) and increased to 1.23 at a pH of 8.90. The above results demonstrated that Au@PANI NPs could respond sensitively to pH variation.To realize real-time monitoring of pH changes, we then examined the reversible response behavior of the Au@PANI nanoprobe. The nearly completely reversible oscillation of the plasmonic responsiveness of Au@PANI NPs could be observed during four-cycle pH switching when the pH was repeatedly varied between 3.93 and 8.90 (Fig. 1E and S3†). Thereafter, the plasmonic response kinetics during the acidic/basic switching process of the Au@PANI NPs were assessed. As shown in Fig. S4,† the response time of these Au@PANI NPs was about 0.75 s as their environment was switched between the basic state and acidic state. With the above results, we further investigated the plasmonic performance of Au@PANI NPs in response to a series of pH changes from 3.93 to 8.90, which covered the so-called physiological pH range.12 As illustrated in Fig. S5,† Au@PANI NPs displayed a gradual evolution of the scattering color with increasing pH, changing from green to yellow, and finally orange-red, specifically. The single-particle scattering spectra exhibited a gradual red-shift accompanied by an obvious increase in the scattered intensity (Fig. S6†). Meanwhile, the calculated R/G value displayed a positive relationship with increasing pH value, and a linear correlation between the R/G value and the pH value was obtained as shown in Fig. 1F.
Besides, the stability and anti-interference capability of Au@PANI NPs were investigated for a better application in living cells. As illustrated in Fig. S7,† we found that after 24 h of dispersion in a cell culture medium (DMEM+10% FBS) and cell lysate at pH ∼7.4, the negative potential of Au@PANI NPs became smaller and the diameter measured by dynamic light scattering (DLS) increased, which may be caused by the adsorption of peptides or protein macromolecules. However, the scattering color and R/G value of the Au@PANI NPs in both the cell culture medium and cell lysates hardly changed after over 24 h, indicating the good monodispersity and stability of Au@PANI NPs in the complex cellular environment. In addition, the effects of cations (K+, Mg2+, Na+, and Ca2+), anions (Cl−, SO42−, and CO32−), and small molecules (glutathione, glucose, cysteine, and H2O2), which were common in the intracellular environment, were studied. As shown in Fig. S8,† these potentially interfering substances had negligible influence on the scattering properties of Au@PANI NPs. All the results validated the excellent stability and selectivity of the established nanoprobe for pH determination, which could be further used for the measurement of pH in complex matrices.
2.3 Intracellular pH calibration and internalization mechanism of Au@PANI NPs
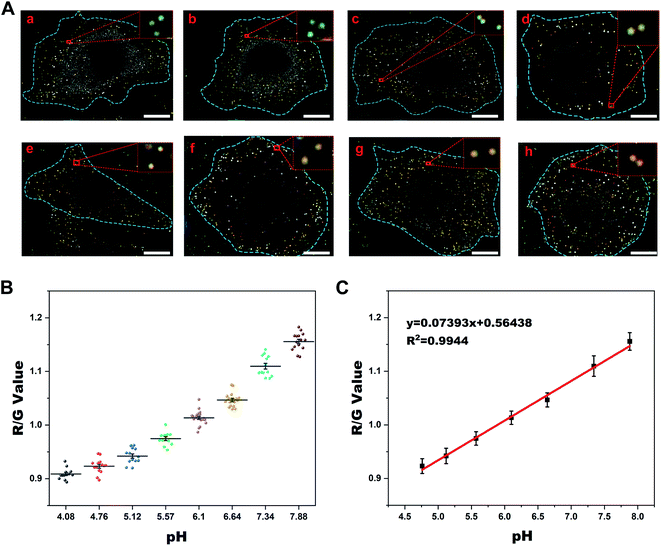
The excellent plasmonic properties of Au@PANI NPs for pH sensing inspired us to apply them in intracellular pH imaging. A standard CCK-8 assay was carried out and the result demonstrated that cell viability was hardly changed with the concentration of Au@PANI NPs less than 1.6 pM after incubation for 48 h (Fig. S9†). Considering that only a concentration of 0.16 pM was employed in the subsequent optical imaging experiments, the biocompatibility of Au@PANI NPs under experimental conditions could be ensured. To employ our Au@PANI nanoprobes to accurately quantify intracellular pH values, an intracellular pH calibration curve was established. For this purpose, HeLa cells treated with nanoprobes were incubated with high K+-buffers containing 10 μM nigericin (a K+/H+ antiporter commonly used for the modulation of intracellular pH)46 of different pH values varying from 4.08 to 7.88, which was employed for equalizing the pH inside the cells and the pH of the surrounding medium. The results indicated that Au@PANI NPs in HeLa cells with lower intracellular pH displayed a green scattering color under DFM, while in cells with higher intracellular pH they exhibited a red-shifted scattering color (Fig. 2A). By collecting and analyzing the scattering signal of the endocytosed Au@PANI NPs in each group, a standard curve of R/G value versus intracellular pH can be established (Fig. 2B and C). Apparently, there was a difference between the standard curve established intracellularly and that in PBS, which may be due to the complex physical environment inside the cell that leads to the doping of oxidative species or adsorption of biomolecules and thus affects the proton exchange properties of PANI.28,47Before the endocytic pathway imaging of Au@PANI NPs, we also investigated their endocytic mechanism with reference to previous studies.48,49 Fig. S10† shows time-dependent dark-field images of the internalization process. As illustrated in Fig. S11,† an obvious decrease in cell uptake was observed when cells were incubated at 4 °C, indicating that the endocytosis was energy-dependent. Besides, by treating cells with different inhibitors, it was revealed that the internalization of Au@PANI NPs seems to follow multiple pathways including caveolae- and clathrin-mediated endocytosis, as proposed in published work.50 As is known, in these reported endocytic pathways, endosomes were always the first intracellular compartments encountered by internalized nanoparticles.51
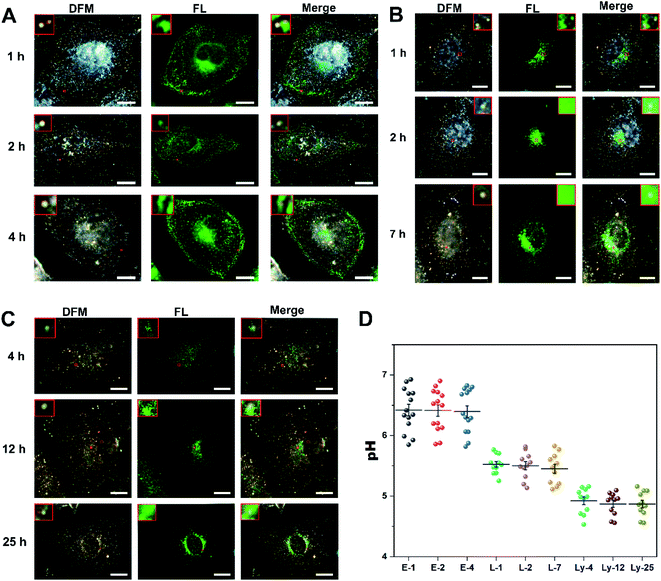
Furthermore, to better clarify the intracellular transportation of the endocytosed Au@PANI NPs, co-localization experiments were conducted. HeLa cells were transfected with a green fluorescent protein (GFP)-fused Rab5a (early endosome membrane protein) or Rab7a (late endosome membrane protein) biomarker, which could specifically identify early endosomes and late endosomes, respectively. Then the cells were incubated with Au@PANI NPs for different times. Additionally, for the co-localization with the lysosome, the cells were stained with lyso-tracker green dye for 40 min before imaging. Fluorescence and dark-field correlation imaging results and the acquired signals of Au@PANI nanoprobes are presented in Fig. 3 and S12.† The co-localization of Au@PANI NPs with early endosomes was observed to be close to the cellular membrane (Fig. 3A), and the calculated average pH value of this category of NPs was ca. 6.4 (Fig. 3D). As shown in Fig. 3B, the Au@PANI NPs co-localized with the late endosomes were mostly distributed near the nucleus, and their scattering signals displayed a yellow or yellow-green color with an average pH value of ca. 5.5. Likewise, the co-localization of Au@PANI NPs with lysosomes mainly took place around the perinuclear region, and the monodispersed nanoprobes exhibited a green scattering signal with an average pH value of ca. 4.9 (Fig. 3C). The above results demonstrated that Au@PANI nanoprobes were observed in all components of the endolysosomal pathway after endocytosis into cells, and the obtained pH values of nanoprobes captured in each component were consistent with those reported in previous work as well.52 Consistent with these co-localization studies, TEM images (Fig. S13†) showed the corresponding Au@PANI NP-containing vesicles characteristic of early endosomes (ca. 200 nm in diameter), late endosomes (ca. 500 nm in diameter), and lysosomes (multi-layer spherical vesicles).49 Notably, Au@PANI NPs were mainly distributed near the cellular plasmonic membrane as individual particles when incubated with cells for 1 h. Afterward, Au@PANI NPs were gradually transported to the perinuclear region and eventually localized around the nucleus. Although occasionally two NPs were located in the same vesicle simultaneously, most of them remained monodisperse. The average diameter of the endocytosed nanoparticles was calculated to be ca. 160 nm (Fig. S13F†), demonstrating that the probe maintained structural stability after endocytosis into cells.
2.4 Dynamic monitoring of pH evolution during the endosomal maturation process in living cells
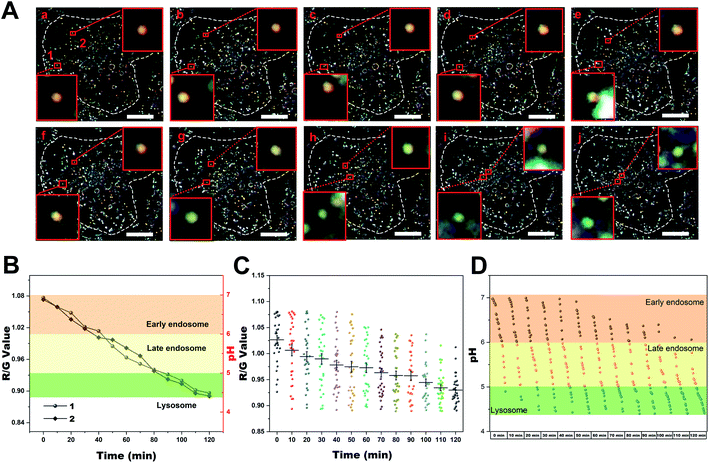
Encouraged by the above results, the capability of Au@PANI NPs for dynamic imaging was investigated. They were employed to real-time track the dynamic pH evolution during the endosomal mature process by the scattering signal changes extracted from the living-cell DFM images over time. Typically, after incubation with Au@PANI NPs (0.16 pM) for 1 h, the adhered cells were washed with PBS to remove the uninternalized nanoparticles and further cultured with phenol red-free DMEM for real-time live-cell imaging. This step was performed to ensure that continuous uptake events do not confound the observation of the vesicle transport kinetics of nanoparticles and the efficient reporting of intracellular pH by Au@PANI NPs. From the time-series images in Fig. 4A, the real-time transport process of nanoprobes packaged in endosomes within the cell could be observed. We selected two representative nanoparticles and analyzed the acidification process as well as the intracellular transport processes of the endosomes where they were located. It was found that the two Au@PANI NPs possessed similar acidification trends. Concretely, the endocytosed nanoprobes were first distributed adjacent to the cell membrane when the cells were incubated with the nanoprobes for 1 h without further incubation, showing orange scattered light, and then gradually transported around the nucleus, during which the scattering signal changed from orange to yellow gradually. Finally, the endocytosed nanoprobes located around the nucleus and the scattered signal evolved to yellow-green or green. As seen in the R/G value & pH versus time profile of Fig. 4B, the two nanoparticles stayed in the early endosomes within the first 30 min, and then evolved to the lysosomes after 80 min by further acidification. Furthermore, we performed a statistical analysis of a large number of probes endocytosed by the cells to comprehensively explore the endosomal maturation process in which they were packaged. As shown in Fig. 4A and C, the scattering signal of the intracellular Au@PANI NPs displayed a large distribution range initially, illustrating the heterogeneity of the intracellular pH. The pH evolution profile derived from the standard curve is demonstrated in Fig. 4D, from which the intracellular transport of the endosomal packaged nanoprobes in various compartments within 2 h could be inferred. It was evident that the endosomes encapsulating Au@PANI NPs were mainly early endosomes at the beginning of the observation, with most of them exhibiting a pH of 6.0–7.0, followed by gradual acidification, and finally almost all of them converted into more acidic late endosomes or lysosomes after 2 h. This indicated that the traffic trajectories and acidification trends of individual particles were consistent with the overall changes.As a further clarification of the intracellular vesicle transport and acidification process of the Au@PANI NPs after endocytosis, cells were divided into eight groups, and each group was further incubated for different times after 1 h incubation with the nanoprobes. Then the scattering signal of the nanoprobes in each group of cells was collected separately. As demonstrated in Fig. S14,† in the group of HeLa cells that were further cultured for 0–2 h, respectively, the intracellular nanoprobes were gradually moved to the area close to the nucleus with a progressive blue-shift of the scattered signal. The evolution of pH over time clearly showed the gradual acidification of the endosomes where the probes were packaged, which was consistent with the results obtained from single-cell real-time tracking experiments. Subsequently, with the incubation time prolonged, further acidification of the endosomes occurred. After 6 h of incubation, the endocytosed nanoprobes were all distributed in the perinuclear area and exhibited a green scattering signal with a pH below 5.0, implying that the nanoprobes eventually all entered the lysosome (Fig. S14B†). All these results suggested that the Au@PANI nanoprobes followed a typical endocytosis pathway upon vesicle transportation, in which they first encountered early endosomes (pH 6.0–7.0), and then transported to late endosomes (pH 5.0–6.0), and finally trafficked into the acidic lysosome (pH 4.4–5.0), which was concordant with the reported pH evolution during endosome maturation.53
3 Conclusion
In brief, we have synthesized a simple and robust plasmonic core–shell nanoprobe that could reversibly respond to pH variation. The plasmonic resonance of the Au core could be regulated reversely by changing the dielectric properties of the PANI shell through proton doping and dedoping. Notably, a significant scattering peak shift of 65 nm has been achieved for the plasmonic modulation. With the excellent photostability and biocompatibility, the plasmonic resonance changes of the Au@PANI NPs exhibited high sensitivity and selectivity towards various pH values within the range of intracellular pH. By incorporating a fluorescent labeling strategy for co-localization, the intracellular vesicular traffic process of the Au@PANI NPs was accurately captured and the pH variations in each period were quantitatively measured. Therefore, the pH-responsive nanoprobes were employed to track the dynamic pH changes during the endocytosis process, which were observed to proceed from near-neutral pH (7.0–6.0) at early time points toward more classical late endosome pH values of 6.0 to 5.0, and finally mature to the lysosome (pH 5.0–4.4). This study not only provides an in-depth insight into the intracellular internalization and transportation process of nanoparticles but also provides an accurate mapping of the pH evolution during the vesicular transport that nanoparticles undergo in endocytosis, which will be enlightened for the design of efficient drug delivery techniques and controlled-release nanoprobes in diagnosis and treatment of diseases.4 Experimental
4.1 Synthesis of Au@PANI core–shell nanoprobes
The AuNPs with a diameter of about 80 nm (16 pM) were purchased from TED PELLA, INC. Then the Au@PANI core–shell NPs were prepared as follows. 1.5 mL 80 nm-AuNPs were concentrated at 5000 rpm for 6 min, followed by redispersion in 0.5 mL of SDS (40 mM). After adding 2 μL aniline (10%) to the solution, the obtained mixture was shaken for 1 min. Afterward, the solution was quickly mixed with 1 mL (NH4)2S2O8 solution (2 mM in 10 mM HCl) and shaken for 30 s. Finally, after being incubated at room temperature for 5 h, the mixture was concentrated at 5000 rpm for 5 min and redispersed in pH ∼7.3 PBS.4.2 The pH-responsive properties of Au@PANI core–shell NPs
First, the core–shell nanostructures were adsorbed on a positively charged glass slide by electrostatic adsorption. Then, PBS buffer with different pH values was cast on the glass slide. The solution was kept on the slide for 10 min to ensure that the ion exchange between the PANI shell of the nanostructure and the solution was in balance, and then the images and scattering spectra of the nanoparticles were recorded under a DFM. For single-particle dark-field scattering measurements, after several Au@PANI NPs were located under the DFM, ∼100 μL of the solution at a specific pH value was added dropwise on the glass slide, and images were captured 10 min later. Then the solution was drawn away, PBS with another pH value was gently added, and pictures were taken again after 10 min to record the changes of these nanoparticles. This procedure was repeated to obtain optical signal changes of the nanoprobe at different pH values.4.3 Cell culture and DFM imaging
HeLa cells were cultured in high-glucose DMEM with 10% fetal bovine serum (FBS) in a cell incubator supplied with 5% CO2 at 37 °C. For dark-field imaging experiments, 105 cells were seeded in 60 mm dishes with a 30 mm glass-bottom (Corning) and cultured overnight before use. For the establishment of intracellular pH standard curves, high K+ buffer solutions with different pH values containing 120 mM KCl, 1 mM CaCl2, 30 mM NaCl, 20 mM HEPES, 0.5 mM MgSO4, 5 mM glucose, 20 mM NaOAc, 1 mM NaH2PO4, and 10.0 μM nigericin was used for changing intracellular pH values. For the tracking of intracellular transport of Au@PANI NPs, cells were incubated with 0.16 pM Au@PANI NPs for 1 h, washed with PBS, and further incubated in high-glucose DMEM with 10% FBS. After that, the dark-field images of the individual cell were captured every 10 min.4.4 Colocalization experiments
HeLa cells seeded in 60 mm-dishes were pretreated with early endosomes-GFP and late endosomes-GFP for 16 h and then incubated with the Au@PANI nanoprobes for another 1 h, 2 h, 4 h, or 7 h. For colocalization with the lysosome, the cells were stained with lyso-tracker green dyes for 40 min after being incubated with nanoprobes for 4 h, 12 h, or 25 h. The obtained images were analyzed using Image J software.4.5 Visualization of intracellular Au@PANI NPs using TEM
First, cells were treated with 0.16 pM Au@PANI NPs for 1 h and washed with PBS and then further incubated in high-glucose DMEM with 10% FBS for 0 h, 1 h, 2 h, 4 h, and 6 h, respectively. After that, the cells were fixed with 2.5% glutaraldehyde and stained with 1% OsO4 at 4 °C. Images of cell slices were taken with an FEI Tecnai Spirit G2 BioTWIN TEM using a beam voltage of 100 kV.Data availability
All experimental or computational data associated with this article have been provided in the ESI.†Author contributions
J. W., X. L. and J. X. conceived the idea. J. W. conducted most of the experiments. Q. Y. and X. Z. performed some characterization and performance tests. J. W., X. L. and J. X. wrote the paper. All the authors discussed the results and commented on the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the National Key R&D Program of China (Grant No. 2021YFA0910003), the National Natural Science Foundation (Grants 22034003 and 21991080) of China, the Excellent Research Program of Nanjing University (ZYJH004) and the Natural Science Research Project of Jiangsu Higher Education (20KJB150020).Notes and references
- J. Llopis, J. M. McCaffery, A. Miyawaki, M. G. Farquhar and R. Y. Tsien, Proc. Natl. Acad. Sci. U.S.A., 1998, 95, 6803–6808 CrossRef CAS PubMed.
- J. R. Casey, S. Grinstein and J. Orlowski, Nat. Rev. Mol. Cell Biol., 2010, 11, 50–61 CrossRef CAS PubMed.
- B. Fang, D. Wang, M. Huang, G. Yu and H. J. I. J. o. N. Li, Int. J. Neurosci., 2010, 120, 591–595 CrossRef CAS PubMed.
- R. A. Gatenby, E. T. Gawlinski, A. F. Gmitro, B. Kaylor and R. J. Gillies, Cancer Res., 2006, 66, 5216–5623 CrossRef CAS PubMed.
- B. A. Webb, M. Chimenti, M. P. Jacobson and D. L. Barber, Nat. Rev. Cancer, 2011, 11, 671–677 CrossRef CAS PubMed.
- Q. Mu, G. Jiang, L. Chen, H. Zhou, D. Fourches, A. Tropsha and B. Yan, Chem. Rev., 2014, 114, 7740–7781 CrossRef CAS PubMed.
- P. Foroozandeh and A. A. Aziz, Nanoscale Res. Lett., 2018, 13, 339 CrossRef PubMed.
- S. Lee, Z. Zhang and Y. Yu, Angew. Chem., Int. Ed., 2021, 60, 26734–26739 CrossRef CAS PubMed.
- X. Wang, Y. Wang, W. Pan, J. Wang and X. Sun, ACS Sustain. Chem. Eng., 2021, 9, 3718–3726 CrossRef CAS.
- X. S. Zheng, C. Zong, X. Wang and B. Ren, Anal. Chem., 2019, 91, 8383–8389 CrossRef CAS PubMed.
- E. Persi, M. Duran-Frigola, M. Damaghi, W. R. Roush, P. Aloy, J. L. Cleveland, R. J. Gillies and E. Ruppin, Nat. Commun., 2018, 9, 2997 CrossRef PubMed.
- Y. Ma, H. Liang, Y. Zeng, H. Yang, C. L. Ho, W. Xu, Q. Zhao, W. Huang and W. Y. Wong, Chem. Sci., 2016, 7, 3338–3346 RSC.
- L. Ning, X. Li, D. Yang, P. Miao, Z. Ye and G. Li, Anal. Chem., 2014, 86, 8042–8047 CrossRef CAS PubMed.
- A. Jaworska, K. Malek and A. Kudelski, Spectrochim. Acta, Part A, 2021, 251, 119410 CrossRef CAS PubMed.
- Y. Bi, H. Di, E. Zeng, Q. Li, W. Li, J. Yang and D. Liu, Anal. Chem., 2020, 92, 9574–9582 CrossRef CAS PubMed.
- Y. Chen, C. Zhu, J. Cen, Y. Bai, W. He and Z. Guo, Chem. Sci., 2015, 6, 3187–3194 RSC.
- C. He, K. Lu and W. Lin, J. Am. Chem. Soc., 2014, 136, 12253–12256 CrossRef CAS PubMed.
- S. S. Li, M. Zhang, J. H. Wang, F. Yang, B. Kang, J. J. Xu and H. Y. Chen, Anal. Chem., 2019, 91, 8398–8405 CrossRef CAS PubMed.
- R. L. Zhang, F. W. Pratiwi, B. C. Chen, P. Chen, S. H. Wu and C. Y. Mou, ACS Appl. Mater. Interfaces, 2020, 12, 42472–42484 CrossRef CAS PubMed.
- K. J. F. Carnevale, R. A. Riskowski and G. F. Strouse, ACS Nano, 2018, 12, 5956–5968 CrossRef CAS PubMed.
- K. K. Yu, K. Li, H. H. Qin, Q. Zhou, C. H. Qian, Y. H. Liu and X. Q. Yu, ACS Appl. Mater. Interfaces, 2016, 8, 22839–22848 CrossRef CAS PubMed.
- M. Na, Y. Han, Y. Chen, S. Ma, J. Liu and X. Chen, Anal. Chem., 2021, 93, 5185–5193 CrossRef CAS PubMed.
- X. Hu, Q. Zhang, X. Dai, J. Sun and F. Gao, ACS Appl. Bio Mater., 2021, 4, 7663–7672 CrossRef CAS PubMed.
- M. Xu, X. Ma, T. Wei, Z. X. Lu and B. Ren, Anal. Chem., 2018, 90, 13922–13928 CrossRef CAS PubMed.
- C. Wang, Y. Wang, Y. Li, B. Bodemann, T. Zhao, X. Ma, G. Huang, Z. Hu, R. J. DeBerardinis, M. A. White and J. Gao, Nat. Commun., 2015, 6, 8524 CrossRef CAS PubMed.
- Q. Hao, Q. Xu, S. Niu, C. Ding and X. Luo, Anal. Chem., 2021, 93, 10679–10687 CrossRef CAS PubMed.
- E. B. Choi, J. Choi, S. R. Bae, H.-O. Kim, E. Jang, B. Kang, M.-H. Kim, B. Kim, J.-S. Suh, K. Lee, Y.-M. Huh and S. Haam, Nano Res., 2014, 8, 1169–1179 CrossRef.
- J. Yang, J. Choi, D. Bang, E. Kim, E. K. Lim, H. Park, J. S. Suh, K. Lee, K. H. Yoo, E. K. Kim, Y. M. Huh and S. Haam, Angew. Chem., Int. Ed., 2011, 50, 441–444 CrossRef CAS PubMed.
- Y. Lu, S. H. Lam, W. Lu, L. Shao, T. H. Chow and J. Wang, Nano Lett., 2022, 22, 1406–1414 CrossRef CAS PubMed.
- W. Lu, N. Jiang and J. Wang, Adv. Mater., 2017, 29, 1604862 CrossRef PubMed.
- N. Jiang, L. Shao and J. Wang, Adv. Mater., 2014, 26, 3282–3289 CrossRef CAS PubMed.
- E. Ju, K. Dong, Z. Liu, F. Pu, J. Ren and X. Qu, Adv. Funct. Mater., 2015, 25, 1574–1580 CrossRef CAS.
- H. Chen, Z. Liu, S. Li, C. Su, X. Qiu, H. Zhong and Z. Guo, Theranostics, 2016, 6, 1096–1104 CrossRef CAS PubMed.
- N. Hui, X. Sun, S. Niu and X. Luo, ACS Appl. Mater. Interfaces, 2017, 9, 2914–2923 CrossRef CAS PubMed.
- H. Liu, P. Huang, F. Y. Wu and L. Ma, Mikrochim. Acta, 2021, 188, 155 CrossRef CAS PubMed.
- P. Tang, X. Jiang, Y. Wang, H. Chen, Y. S. Zhang, P. Gao, H. Wang, X. Li and J. Zhou, Anal. Chem., 2017, 89, 9758–9766 CrossRef CAS PubMed.
- J. Wang, X. L. Li, H. Y. Chen and J. J. Xu, Anal. Chem., 2020, 92, 15647–15654 CrossRef CAS PubMed.
- M. X. Li, C. H. Xu, N. Zhang, G. S. Qian, W. Zhao, J. J. Xu and H. Y. Chen, ACS Nano, 2018, 12, 3341–3350 CrossRef CAS PubMed.
- X. L. Li, Z. L. Zhang, W. Zhao, X. H. Xia, J. J. Xu and H. Y. Chen, Chem. Sci., 2016, 7, 3256–3263 RSC.
- J. Wang, T. Zhao, X. L. Li, X. L. Luo, H. Y. Chen and J. J. Xu, ACS Appl. Nano Mater., 2021, 4, 7319–7329 CrossRef CAS.
- Z. Ye, L. Wei, X. Zeng, R. Weng, X. Shi, N. Wang, L. Chen and L. Xiao, Anal. Chem., 2018, 90, 1177–1185 CrossRef CAS PubMed.
- Y. Ling, D. Zhang, X. Cui, M. Wei, T. Zhang, J. Wang, L. Xiao and Y. Xia, Angew. Chem., Int. Ed., 2019, 58, 10542–10546 CrossRef CAS PubMed.
- L. Zhang, Y. Li, D. W. Li, C. Jing, X. Chen, M. Lv, Q. Huang, Y. T. Long and I. Willner, Angew. Chem., Int. Ed., 2011, 50, 6789–6792 CrossRef CAS PubMed.
- I. Sapurina and J. Stejskal, Polym. Int., 2008, 57, 1295–1325 CrossRef CAS.
- T. Xie, C. Jing and Y. T. Long, Analyst, 2017, 142, 409–420 RSC.
- P. Anees, K. V. Sudheesh, P. Jayamurthy, A. R. Chandrika, R. V. Omkumar and A. Ajayaghosh, Chem. Sci., 2016, 7, 6808–6814 RSC.
- Y. Zhang, Y. Wang, X. Yang, Q. Yang, J. Li and W. Tan, Small, 2020, 16, e2001177 CrossRef PubMed.
- L. Ding, C. Yao, X. Yin, C. Li, Y. Huang, M. Wu, B. Wang, X. Guo, Y. Wang and M. Wu, Small, 2018, 14, e1801451 CrossRef PubMed.
- M. Liu, Q. Li, L. Liang, J. Li, K. Wang, J. Li, M. Lv, N. Chen, H. Song, J. Lee, J. Shi, L. Wang, R. Lal and C. Fan, Nat. Commun., 2017, 8, 15646 CrossRef CAS PubMed.
- D. Manzanares and V. Cena, Pharmaceutics, 2020, 12, 371 CrossRef CAS PubMed.
- M. Podinovskaia, C. Prescianotto-Baschong, D. P. Buser and A. Spang, Elife, 2021, 10, e70982 CrossRef PubMed.
- B. Yameen, W. I. Choi, C. Vilos, A. Swami, J. Shi and O. C. Farokhzad, J. Controlled Release, 2014, 190, 485–499 CrossRef CAS PubMed.
- N. Oh and J. H. Park, Int. J. Nanomed., 2014, 9, 51–63 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2sc01069k |
| This journal is © The Royal Society of Chemistry 2022 |