Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Topologically switchable and gated transcription machinery†
Pu
Zhang‡
a,
Amit
Fischer‡
 a,
Yu
Ouyang
a,
Yang Sung
Sohn
b,
Rachel
Nechushtai
b,
Junji
Zhang
a,
Yu
Ouyang
a,
Yang Sung
Sohn
b,
Rachel
Nechushtai
b,
Junji
Zhang
 c,
He
Tian
c,
He
Tian
 c,
Chunhai
Fan
c,
Chunhai
Fan
 d and
Itamar
Willner
d and
Itamar
Willner
 *a
*a
aInstitute of Chemistry, Center for Nanoscience and Nanotechnology, The Hebrew University of Jerusalem, Jerusalem 91904, Israel. E-mail: itamar.willner@mail.huji.ac.il
bInstitute of Life Science, The Hebrew University of Jerusalem, Jerusalem 91904, Israel
cKey Laboratory for Advanced Materials, School of Chemistry and Molecular Engineering, East China University of Science and Technology, Shanghai, China
dSchool of Chemistry and Chemical Engineering, Frontiers Science Center for Transformative Molecules, National Center for Translational Medicine, Shanghai Jiao Tong University, 200240, Shanghai, China
First published on 18th August 2022
Abstract
Topological barriers control in nature the transcription machinery, thereby perturbing gene expression. Here we introduce synthetically designed DNA templates that include built-in topological barriers for switchable, triggered-controlled transcription of RNA aptamers. This is exemplified with the design of transcription templates that include reversible and switchable topological barriers consisting of a Sr2+-ion-stabilized G-quadruplex and its separation by kryptofix [2.2.2], KP, for the switchable transcription of the malachite green (MG) RNA aptamer, the T-A·T triplex barrier being separated by a fuel-strand for the cyclic triggered transcription of the 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI)-binding aptamer, and the use of a photoactivated cis/trans azobenzene-modified nucleic acid barrier for the switchable “ON”/“OFF” transcription of the MG RNA aptamer. By applying a mixture of topologically triggered templates consisting of the photoresponsive barrier and the T-A·T triplex barrier, the gated transcription of the MG aptamer or the DFHBI-binding aptamer is demonstrated. In addition, a Sr2+-ion/KP topologically triggered DNA tetrahedra promoter-transcription scaffold, for the replication of the MG RNA aptamer, and T7 RNA polymerase are integrated into DNA-based carboxymethyl cellulose hydrogel microcapsules acting as cell-like assemblies. The switchable, reversible transcription of the MG RNA aptamer in a cell-like containment is introduced.
Introduction
Topological barriers associated with condensed chromatin regulate the transcription of mRNA through the suppression of the accessibility of transcription factors or RNA polymerase to the transcription machinery.1–5 The development of in vitro programmed reversible topologically regulated transcription machinery could lead to triggered, dose-controlled gene expression for medical applications.6 The base sequence encoded in nucleic acids provides a means to reversibly modulate and switch the topologies of DNA nanostructures by auxiliary triggers. For example, ion-induced formation of G-quadruplexes and their separation in the presence of crown ethers,7,8 the stabilization of T–T and C–C mismatched duplexes by metal ions, such as Hg2+ or Ag+, and their separation by metal-ion binding ligands,9–11 the reversible pH-stimulated formation of i-motif structures,12 the formation of T-A·T triplex structures and their disassembly by fuel strand-driven displacement processes,13 and the photochemical stabilization/destabilization of DNA duplexes by photoisomerizable intercalators, such as azobenzene units,14,15 provide a rich “tool-box” of reconfigurable DNA topologies. Indeed, these switchable DNA structures were applied to assemble DNA switches16 and DNA machines,17–19 such as tweezers,20,21 walkers22–25 or nanocarriers,26–28 to design “smart” materials acting as gated drug carriers for controlled drug release, such as SiO2 nanoparticles,29–31 metal–organic framework nanoparticles32–34 or microcapsules,35,36 and to prepare stimuli-responsive hydrogels exhibiting switchable stiffness properties for shape-memory,37,38 self-healing,39,40 controlled drug-release41 and mechanical actuating applications.42The reconfigurable DNA structures allow, in principle, the engineering of DNA templates that include topological barriers for operating reversible and cyclic transcription machineries. Here, we report on the design of switchable transcription machineries driven by Sr2+-ion stabilized G-quadruplex/kryptofix [2.2.2] (KP), T-A·T triplex displacement strands, and photoresponsive trans/cis azobenzene intercalator units. By mixing two scaffolds of topologically reconfigurable transcription machineries composed of trans-azobenzene-duplex bridged blocker topologies and the T-A·T triplex, the programmed and reversible gated operation of transcription machineries synthesizing target RNAs is demonstrated. It should be noted that in a recent report,43 the triggered operation of a topologically hindered transcription machinery was reported. In this system, the DNA template was caged in a DNA tetrahedral nanostructure. The separation of the tetrahedral nanostructure by a fuel strand yielded an active template that facilitated the RNA polymerase/NTPs replication of RNAs. This system is, however, not reversible and could not be cycled across “ON”–“OFF” states. In addition, we demonstrate the Sr2+-ions/KP switchable transcription machinery in hydrogel microcapsule cell-like containments as a model system for operating a triggered transcription process in a cell-like microenvironment.44–46
Results and discussion
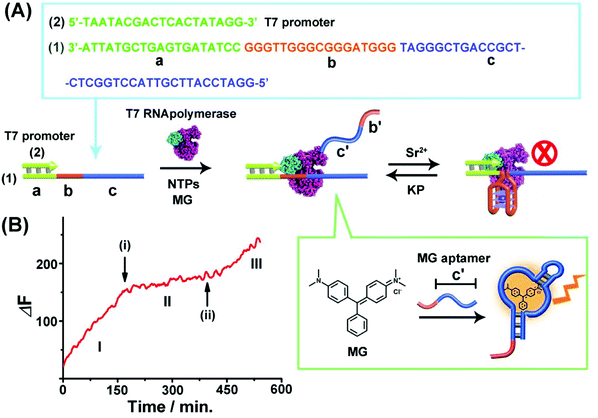
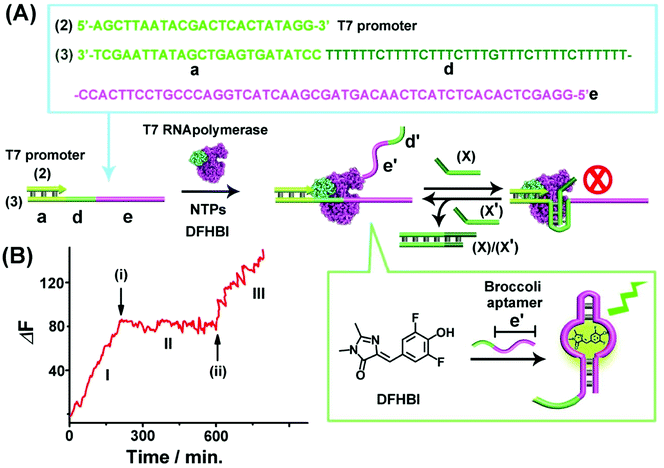
Fig. 1(A) depicts the reversible Sr2+-ion stabilized G-quadruplex/kryptofix [2.2.2] (KP)-induced switchable transcription of the malachite green (MG) RNA aptamer. The association of MG with its aptamer switches-ON the fluorescence of MG (λem = 665 nm) and, thus, the transcription process is followed by the temporal fluorescence changes of the transcribed MG/aptamer complex. The transcription module includes a DNA duplex template consisting of scaffold (1) and the T7 promoter (2). The strand (1) includes three domains: (a) the T7 promoter binding sequence, (b) the guanosine (G)-rich sequence that self-assembles under appropriate conditions to the G-quadruplex unit, and the domain (c) that is complementary to the MG aptamer sequence. Subjecting the template (1)/(2) to T7 RNA polymerase and the NTPs mixture, in the presence of MG, results in the triggered transcription machinery that replicates the domains (b) and (c) to form the RNA strand (b')/(c'), where (c') corresponds to the MG aptamer. The generation of the MG aptamer is followed by the time-dependent fluorescence changes of the formation of the MG/aptamer complex.47Fig. 1(B), part I depicts the time-dependent fluorescence changes upon the transcription of the MG aptamer. The addition of Sr2+ ions to the reaction machinery, time marked (i), reconfigures domain (b) into the Sr2+-ion stabilized G-quadruplex structure. The formation of the G-quadruplex topological barrier blocks the transcription process and inhibits the formation of the aptamer, Fig. 1(A) and (B), part II. At time marked (ii), KP is added to the system, resulting in the separation of the Sr2+-ion stabilized G-quadruplex blocker units, and the reactivation of the transcription of the MG aptamer, Fig. 1(A) and (B), part III. For further experiments supporting the formation of the Sr2+-ion stabilized G-quadruplex, see ESI, Fig. S1 and S2† and accompanying discussion. It should be noted that in contrast to the common application of a promoter/duplex template configuration of the transcription machinery, we apply throughout the study promoter/single strand templates as transcription machineries. We find that the promoter/duplex template assemblies lead to lower (incomplete) switching efficiencies upon the triggered topological reconfiguration of the template strand, as compared to the switching efficiency of the promoter/single strand configuration (see Fig. S3† ESI and accompanying discussion). This is due to the fact that the triggered topological reconfiguration of the promoter/double stranded configuration includes always a residual content of a non-reconfigured double-stranded template that generates the transcription product. In addition, we note that for the G-quadruplex/KP reversible modulated ON/OFF switchable synthesis of the MG aptamer, we employed the Sr2+-ion stabilized G-quadruplex (rather than the common K+-ion stabilized/crown ether switching system). This is due to the fact that the K+-ion stabilized G-quadruplex/crown ether system was found to quench the fluorescence of the MG/aptamer complex, thus perturbing the readout signal of the respective transcription machinery.The second switchable transcription process included the T-A·T triplex as the topological blocker unit. This is exemplified in Fig. 2(A) with the switchable transcription of the Broccoli aptamer that binds 3,5-difluoro-4-hydroxybenzylidene imidazolinone, DFHBI, to yield a fluorescent DFHBI/aptamer complex (λem = 500 nm).48 The transcription machinery is composed of the template (2)/(3) where the scaffold (3) includes the domains (a), (d) and (e). Domain (a) corresponds to the T7 promoter (2) binding sites, domain (d) consists of a thymine (T)-rich sequence, being capable of forming a T-A·T triplex structure in the presence of an appropriate trigger, and domain (e) is complementary to the broccoli aptamer sequence. Subjecting the template (2)/(3) to a T7 RNA polymerase and NTPs mixture, in the presence of the DFHBI ligand, activates the transcription machinery and the synthesis of the broccoli RNA aptamer that binds DFHBI. The fluorescence changes of the resulting DFHBI/aptamer complex transduce the transcription process. Fig. 2(B), part I, shows the time-dependent fluorescence changes upon the formation of the DFHBI/aptamer complex. At time marked (i), the strand (X) is added to the transcription machinery. The strand (X) reconfigures domain (d) of scaffold (3) into a T-A·T triplex blocker that switches-OFF the transcription process, Fig. 2(A) and (B), part II. The blocker strand is engineered, however, to include a toehold tether, that upon further addition of the strand (X′) displaces the blocker unit, to yield the duplex (X)/(X′), resulting in the uncaging of the topological barrier and the reactivation of the transcription process generating the DFHBI binding aptamer, Fig. 2(B), part III, time marked (ii). Moreover, an alternative template (2)/(4) consisting of a T-A·T triplex, switchable transcription machinery for synthesizing the MG aptamer was engineered and the results of this system are presented in Fig. S4.† In this system, the scaffold (4) includes the domains (a), (d) and (c), where (a) provides the promoter binding site, (d) corresponds to the T-rich sequence that forms, in the presence of the triggering strand (X), the T-A·T triplex, and the domain (c) is complementary to the MG aptamer sequence. Fig. S4(A) and (B)† demonstrate the T-A·T triplex-based switchable transcription of the MG aptamer. That is, the reversible topological T-A·T triplex blocker allows the switchable transcription of the DFHBI-binding RNA aptamer or the MG RNA aptamer. It should be noted, however, that in contrast to the triplex blocker that allows the transcription of the DFHBI or the MG aptamer by appropriate design of the template machinery, the Sr2+-ion triggered G-quadruplex blocker unit could not be applied to design a DFHBI-binding aptamer transcription machinery. The DFHBI-binding aptamer includes a G-quadruplex domain that associates with the Sr2+ ions and this perturbs the fluorescence features of DFHBI, thus disturbing the readout of the potentially G-quadruplex switching of the DFHBI transcription machinery.
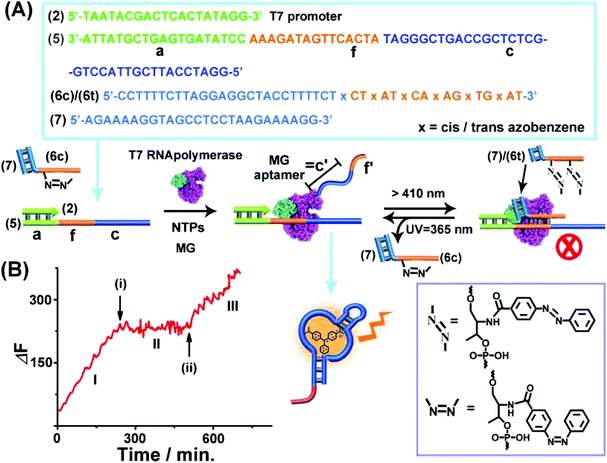
The third topologically triggered transcription machinery involved light-induced control over the transcription machinery using trans/cis azobenzene photoisomerizable blocker units. The trans azobenzene units modifying the nucleic acid strand intercalate into duplex nucleic acid structures, thereby cooperatively stabilizing the duplex DNA structure, whereas the photoisomerized cis-azobenzene tether forms a steric hindrance between the duplex nucleic acid structure thereby destabilizing the duplex structures. Thus, by the appropriate design of a photoisomerizable auxiliary azobenzene-functionalized strand, the light-induced switchable transcription machinery is realized. Fig. 3(A) depicts the principle to reversibly modulate the switchable transcription machinery by light. The template composed of the duplex (2)/(5) and the auxiliary duplex consisting of (7) and the photoresponsive cis-azobenzene functionalized strand (6c) acts, in the presence of T7 RNA polymerase and NTPs, as the photo-triggered transcription machinery. (The strand (6c) is composed of two domains, one domain hybridizes with (7), and the other domain contains the photoisomerized cis-azobenzene units). The strand (5) being a part of the transcription machinery includes the domains (a), (f) and (c). Domain (a) provides the T7 promoter with a binding site, the domain (f) is engineered to interact with the photoresponsive blocker unit (6)/(7), and domain (c) is complementary to the MG aptamer sequence. The cis-azobenzene (6c)/(7) auxiliary unit does not interact with domain (f) of the transcription machinery; thus subjecting the transcription machinery to a T7 RNA polymerase and NTPs mixture, in the presence of MG, triggers-ON the transcription of the MG aptamer, (c′), which is followed by the fluorescence changes of the MG/aptamer complex. Fig. 3(B), part I shows the time-dependent fluorescence changes, resulting in the formation of the fluorescent MG/aptamer complex upon the transcription of the MG aptamer. At the time marked with an arrow (i), the system is illuminated at λ > 410 nm, leading to the photoisomerization of the cis-azobenzene-functionalized module (6c)/(7) to the trans-state, (6t)/(7). This results in the hybridization of the trans-azobenzene topological unit to the domain (f) of template (5), forming a topological barrier that switches-OFF the transcription process, Fig. 3(A), and the switched-OFF transcription process is demonstrated in Fig. 3(B), part II. Further irradiation of the system, λ = 365 nm, photoisomerizes the trans-azobenzene blocker element into the cis-state (6c)/(7), resulting in the separation of the cis-azobenzene (6c)/(7) unit from the template and the reactivation of the transcription process, Fig. 3(A) and (B), arrow (ii), part III. It should be noted that the use of the duplex blocker unit (6t)/(7) is important to switch-OFF the transcription process. Applying (6t) as a single strand yielding the duplex between (6t) and domain (f) of (5) and the single toehold tether linked to (6t) still allows the T7 RNA polymerase to proceed. Presumably,49,50 the rigid duplex structure of (6t)/(7) introduces a bulky topological barrier that perturbs and inhibits the transcription of the scaffold.
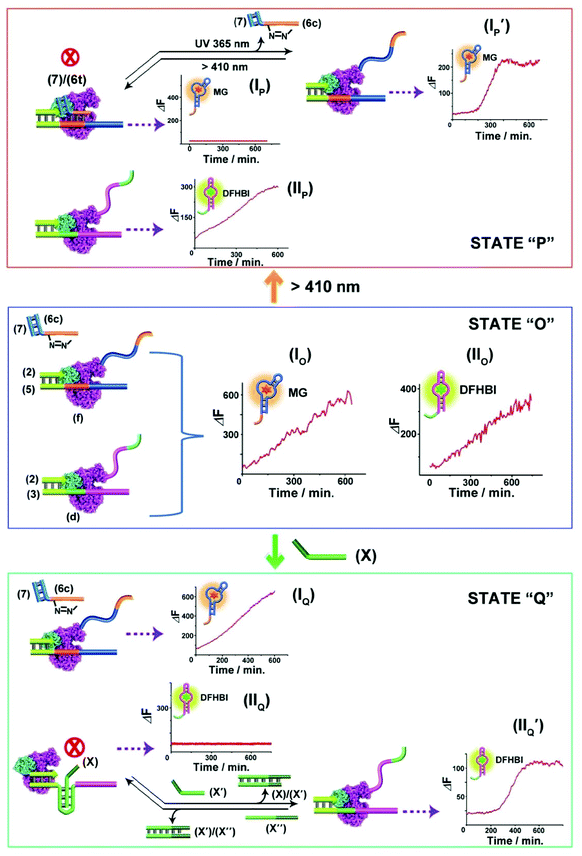
The successful design of topologically switchable transcription machineries using different triggers to yield two different aptamers (the MG and DFHBI-binding aptamer) suggested that gated transcription machinery guiding the selective synthesis of target RNAs could be designed. Fig. 4 presents the gated operation of two scaffolds of transcription machineries synthesizing the MG and DFHBI-binding aptamers, using the photoresponsive trans-azobenzene blocker and the T-A·T triplex blocker as gating units. The system consists of a mixture of the two scaffolds of transcription machineries, state “O” composed of the (2)/(5) and (6c)/(7) photoresponsive machinery that transcribes the MG aptamer and the T-A·T triplex-responsive template (2)/(3) that transcribes the DFHBI-binding aptamer. Subjecting the mixture to a T7 RNA polymerase and NTPs mixture, in the presence of MG and DFHBI, triggers the two scaffolds of transcription machineries to yield the MG aptamer and DFHBI-binding aptamer, panels IO and IIO, respectively. The irradiation of state “O”, λ > 410 nm, transforms the (6c)/(7) module into the trans-azobenzene module (6t)/(7) that hybridizes with domain (f) of template (5), resulting in the blocking of the photoresponsive machinery, state “P”, panel IP, while the triplex responsive machinery is unaffected, panel IIP. That is, the photochemical transition of state “O” to state “P” gates the transcription machinery by blocking the transcription of the MG aptamer, panel IP, while allowing the transcription of the DFHBI-binding aptamer, Fig. 4, panel IIP. The caged inactive MG aptamer transcription machinery can be, however, switched between “ON” and “OFF” states by the cyclic photoinduced isomerization of the photoresponsive units between the (6c)/(7) (λ = 365 nm) and the (6t)/(7) (λ > 410 nm) units, respectively, panel IP′. Similarly, state “O” can be triggered by the strand (X) that transforms state “O” into state “Q”, where the domain (d) of strand (3) is reconfigured into the topological T-A·T triplex blocked the transcription machinery. That is, subjecting strand (X) to state “O” results in the transcription of the MG aptamer, by the photoresponsive machinery panel IQ, while the transcription of the DFHBI-binding aptamer is blocked, panel IIQ. The T-A·T triplex topologically blocked machinery can be, however, reversibly triggered between “ON” and “OFF” states. The treatment of the system with the anti-blocker strand, (X′), and the subsequent re-treatment of the switched-ON transcription machinery of the DFHBI-binding aptamer with the strand (X′′) result in the cyclic activation/deactivation of the DFHBI-binding aptamer formation, panel IIQ′.
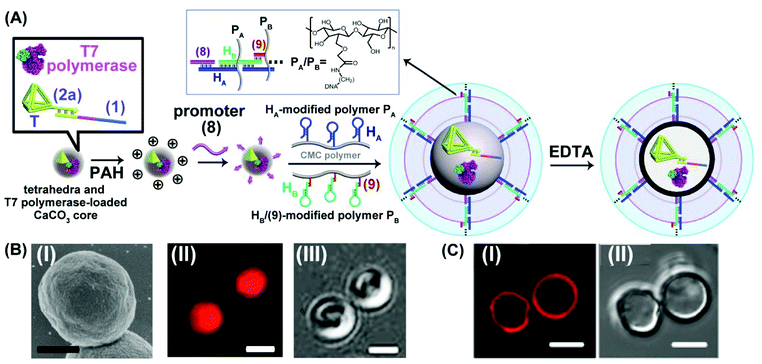
The different scaffolds of topologically switchable transcription machineries are operated in a homogeneous buffer solution. Their operation in a cell-like containment could provide a further complex design of transcription processes mimicking native systems. Different cell-like containments acting as cell-like containments were developed in the past few years. These include liposomes,51,52 polymersomes,53–55 proteinosomes,56–59 colloidosomes60 and coacervate microdroplets.61 Our laboratory developed in the past few years a versatile method to design nucleic acid-based hydrogel microcapsules, and particularly stimuli-responsive microcapsules.62 These microcapsules were loaded with drugs, nanoparticles or proteins and used as functional carriers for the release of the loads. The preparation of the microcapsules involves the impregnation of CaCO3 microparticles with the loads, the functionalization of the particles with promoter-nucleic acid tethers, and the interaction of the modified microparticles with two polymer chains functionalized with two types of pre-engineered hairpins HA and HB. Subjecting the promoter modified microparticles to the hairpin-functionalized polymer chains triggered the inter-polymer hybridization chain reaction (HCR) resulting in a hydrogel coating crosslinked by the inter-hybridized HA/HB duplexes. The subsequent etching of the CaCO3 core templates with ethylenediaminetetraacetic acid, EDTA, yielded substrate-loaded microcapsules. We find that besides using microcapsules as drug carriers, they can be applied as cell-mimicking containments. By appropriate engineering of the hydrogel coating, molecular entities with a molecular weight ca. ≥5 kDa are retained in the cell-like containment with no leakage to the bulk solution, yet a molecular or ionic substrate of a lower molecular weight, ≤5 kDa, can diffuse in-and-out across the microcapsular boundaries.35 We find that the nucleic acid-based hydrogel microcapsules reveal beneficial properties as compared to other protocell containments. The microcapsules revealed high-loading capacities, rapid exchange of low-molecular-weight agents, allowing effective communication with the bulk surrounding solution, low leakage properties towards constituents exhibiting molecular weight ≤ ca. 5 kD, and relatively high stabilities.63 Accordingly, we designed a cell-like containment in which the topologically triggered switchable transcription of the MG aptamer is demonstrated, Fig. 5. The assembly of the microcapsules loaded with MG aptamer transcription machinery is depicted in Fig. 5(A). The duplex consisting of the template scaffold (1) hybridized with the nucleic acid tetrahedra (T)-functionalized T7 promoter (2a) was deposited together with T7 RNA polymerase on CaCO3 microparticles. (The tetrahedral units were tethered to the T7 promoter to ensure the non-leakable containers of the transcription template to the resulting microcapsules). The loaded CaCO3 particles were, then, coated with polyallylamine hydrochloride, PAH, modified with the nucleic acid promoter (8), and the particles were subjected to two polymer chains PA and PB consisting of carboxymethyl cellulose, CMC, functionalized with the hairpins HA/HB. The hairpin HA was tethered to polymer PA, and hairpin HB was anchored through hybridization with a short tether (9) to polymer PB. (The specific anchoring of HB with (9) is essential for directionality reasons dictating the inter-hybridization of HA/HB). Under these conditions, the promoter strands (8), functionalizing the particles, trigger the HCR process, where (8) opens hairpin HA, and opened HA, opens hairpin HB, and vica versa, resulting in a CMC hydrogel coating of the polymer chains crosslinked by the duplexes comprising inter-cross opened hairpins HA/HB. The subsequent etching of the CaCO3 cores with EDTA resulted in hydrogel-stabilized microcapsules loaded with the tetrahedra-modified (1)/(2a) template, (1)/T-(2a), and T7 RNA polymerase. Fig. 5(B), panel I, shows the SEM image of the T7 RNA polymerase and (1)/T-(2a) loaded CaCO3 microparticles coated with the CMC hydrogel before etching. For comparison, the bright field and fluorescence confocal microscopy images of the hydrogel coated CaCO3 particles loaded with T7 RNA polymerase and (1)/T-(2a) prior to etching are provided in panel II and panel III. The fluorescence and bright-field images of the EDTA etched microcapsules are shown in Fig. 5(C) panels I and II. It should be noted that a fluorophore (Cy5) was used to label (1)/T-(2a) for the fluorescence characterization of the microcapsules. Spherical microcapsules with ca. 2.5 μm diameter are formed. The loadings of the microcapsules with the Cy5-labeled (1)/T-(2a) scaffold was estimated to be ca. 2.4 × 10−10 moles in the overall volume of capsules (or 2.6 × 10−16 moles per capsule) and of the T7 RNA polymerase 1.11 × 10−11 moles in the overall mixture of capsules (or 1.23 × 10−17 moles per capsule). The loadings of the microcapsules with fluorescent (1)/T-(2a) was evaluated by following the fluorescence intensities of the resulting microcapsules and concomitantly evaluating the residual fluorescent (1)/T-(2a) in the washing solutions upon the preparation of the microcapsules. The loadings of T7 RNA polymerase were estimated by following the absorbance feature of the protein. (For details, see ESI, Fig. S5†). The resulting microcapsules were stable for at least three days, and no leakage from the microcapsules was detected within this time-interval, Fig. S6.† The intact structure of the tetrahedra-modified strand (2a), T-(2a), was confirmed by electrophoretic characterization, Fig. S7,† and the intact structure of (1)/T-(2a), under the conditions applied to prepare the microcapsules (Fig. S8†), and the activity of the (1)/T-(2a) and T7 RNA polymerase, subjected to the conditions to prepare the microcapsule (Fig. S9†) were evaluated by independent experiments. While it is impossible to monitor either the structure of the (1)/T-(2a) in the microcapsules or the activity of T7 RNA polymerase in the microcapsules, we subjected CaCO3 microparticles to independent impregnation with the (1)/T-(2a) template or with the T7 RNA polymerase. The impregnated microparticles were then subjected to the EDTA etching process under identical conditions used to prepare the microcapsules, and the resulting (1)/T-(2a) solution was analyzed by electrophoresis and compared to a non-etched (1)/T-(2a) solution, Fig. S8† and accompanying discussion. The results demonstrate that the deposition of (1)/T-(2a) on the CaCO3 core and the EDTA etching had no effect on the intact structure of (1)/T-(2a). In addition, the activity of the T7 RNA polymerase subjected to the deposition onto the CaCO3 particles and the subsequent etching by EDTA was compared to the parent T7 RNA polymerase activity by comparing the transcription rates of the MG RNA aptamer by the two enzyme systems, Fig. S9.† We find that the T7 RNA polymerase retained 70% of the activity of the parent enzyme after the CaCO3 impregnation/EDTA etching.
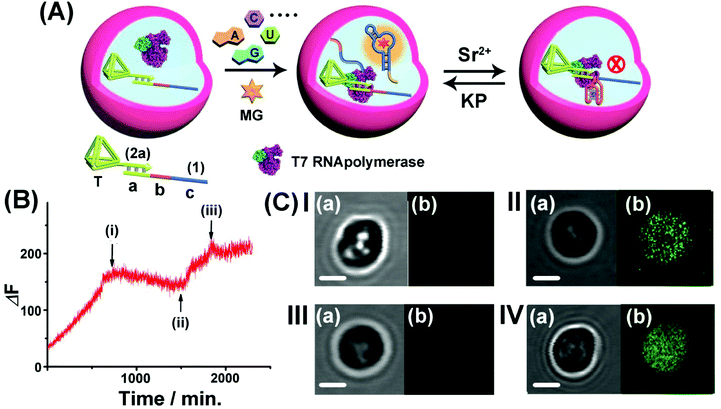
Realizing that the T7 RNA polymerase preserves ca. 70% of its activity and the template (1)/T-(2a) retains its intact structure in the microcapsule, cell-like containment and the permeability features of NTPs, MG, Sr2+ ions and KP through the microcapsule boundaries, we applied the system as a functional topologically switchable transcription machinery of the MG RNA aptamer in a cell-like containment assembly, Fig. 6. (For the operation of the Sr2+-ion/KP-switchable tetrahedra (2a)/(1) transcription machinery in a homogeneous buffer solution, see Fig. S10† and accompanying discussion). The scaffold (1) consists of the three domains (a), (b) and (c), where the sequence (a) corresponds to the T7 promoter binding site, domain (b) includes a guanosine (G)-rich sequence that reveals, in the presence of Sr2+ ions and KP, reversible reconfigurable formation and separation of the G-quadruplex structure, and domain (c) consists of a complementary sequence to the MG RNA aptamer. The treatment of the microcapsules with NTPs and MG triggers-ON the intracapsular transcription of the MG RNA aptamer. Subjecting the microcapsules to Sr2+ ions is, then, anticipated to reconfigure domain (b) of (1) into the Sr2+-ion stabilized G-quadruplex unit that acts as a topological barrier for the transcription process. Under these conditions, the transcription of the MG aptamer is blocked. Further addition of KP to the system separates the G-quadruplex, resulting in the reactivation of the transcription process in the microcapsule. Thus, switchable intramicrocapsule operation of the topologically triggered transcription proceeds in the system. The switchable intramicrocapsule operation of Sr2+-ions/KP-stimulated “ON”/“OFF” transcription process is followed by the time-dependent fluorescence changes of the MG/aptamer complex in the presence of NTPs and MG, and demonstrated in Fig. 6(B). At the time (i), marked with an arrow, Sr2+ ions are added to the system. This reconfigures domain (b), of scaffold (1), into a Sr2+-ion stabilized G-quadruplex barrier that blocks the transcription process. At time-interval marked (ii), KP is added to the system. The permeation of KP into the microcapsule separates the Sr2+-ion stabilized G-quadruplex, resulting in the reactivation of the transcription process. At time-interval marked (iii), Sr2+ ions were re-added to the system, and re-blocked the transcription process in microcapsules. It should be noted that the switchable fluorescence changes observed in the microcapsules show noisy fluorescence intensities, as compared to the fluorescence data recorded for an analogous homogeneous buffer system. This is attributed to the fact that microcapsular hydrogel containments represent a microheterogeneous environment that reveals light-scattering that perturbs the fluorescence intensities. Furthermore, the relatively low number of microcapsules used in the total reaction volume, required the operation of the fluorescence spectrophotometer at enhanced sensitivities that resulted in further background noise. The switchable intracapsular transcription of the MG aptamer was further supported by bright field and confocal fluorescence microscopy experiments, Fig. 6(C). Panel I presents the bright field (a) and confocal fluorescence microscopy (b) images of the same domain of the microcapsules loaded with the (1)/T-(2a) template and T7 RNA polymerase in the presence of auxiliary NTPs and MG, at t = 0. Only the bright field image of the microcapsule is visible. Panel II depicts the bright field (a) and confocal fluorescence microscopy (b) images of the microcapsules loaded with the (1)/T-(2a) template and T7 RNA polymerase, after a time interval of 24 hours. The green fluorescence of the transcribed MG aptamer loaded microcapsule is observed, indicating the successful transcription of the MG aptamer. Panel III depicts the experimental results corresponding to microcapsules loaded with the (1)/T-(2a) template and T7 RNA polymerase subjected to NTPs, MG and Sr2+ ions, for a time interval of 24 hours. While the bright field image of the capsule is visible (a), no fluorescence of the MG aptamer loaded capsules is observed (b), consistent with the Sr2+-ion stabilized G-quadruplex topological inhibition of the transcription process. Panel IV depicts the images generated upon treatment of the switched-OFF microcapsule system shown in panel III with KP for a time interval of 24 hours. In addition to the bright field image of the microcapsule (a), the green fluorescence of the MG/aptamer complex formed in the microcapsule as a result of switched-ON transcription of the MG aptamer is observed (b). The result is consistent with the KP stimulated separation of the Sr2+-ion stabilized G-quadruplex topological barrier that allows the activation of the transcription process. It should be noted that all fluorescence changes of the MG/aptamer complex are confined to the microcapsule containments. Centrifuged precipitation of the microcapsules does not lead to any detectable fluorescence of the MG/aptamer complex in the surrounding buffer solution, and the loaded precipitated microcapsules can be resuspended by gentle shaking. Specifically, the introduction of a means to gate the transcription machinery, in bulk buffer solutions, suggests that the incorporation of gated transcription/translation machinery into protocell containments could enable the triggered programmed synthesis of target proteins in analogy to natural processes.
Conclusions
The study introduced an artificial topological means to switch the transcription of RNA. Three different topological barriers to switch the transcription reaction were introduced including Sr2+-ion stabilized G-quadruplex units, T-A·T triplex structures and photoisomerizable azobenzene-nucleic acid blockers. Other topologically triggered reconfigurable nucleic acid structures, such as redox-triggered ligand/aptamer complexes, enzyme-triggered blocker units consisting of aptamer–ligand complexes or enzyme/DNAzyme nicked duplex blockers could provide a means to switch the transcription processes. In addition, in order to mimic native regulation transcription processes, we introduce synthetic transcription machineries and gated transcription machineries that could lead to dose-controlled and programmed gene expression. The artificial model systems mimicking native topologically modulated transcription machinery introduce a dynamic means to yield programmed and modulated RNAs for possible therapeutic applications. Furthermore, primary attempts to integrate the topologically triggered transcription machinery into cell-mimicking containment microcapsules were provided. While the machinery advances systems chemistry by establishing control over gene expression using supramolecular complexes, important challenges are ahead of us. In particular, the significance of the present results rests on the incorporation of the switchable transcription machinery into artificial cell containments. The co-addition of dynamic networks activating the transcription machinery in cell-like containments or the coupling of the transcription machinery to co-added translation machinery in cell-like containments could be important future steps to design synthetic cell-like containments mimicking native systems.Author contributions
P. Z., A. F. and I. W. formulated the concepts and methodology of the study. The manuscript was written through the contributions of all authors. All authors participated in the experimental work and have given approval regarding the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the Israel Science Foundation and the Minerva Center for Biohybrid Complex Systems.Notes and references
- S. L. Berger, Nature, 2007, 447, 407–412 CrossRef CAS PubMed.
- M. Fuxreiter, P. Tompa, I. Simon, V. N. Uversky, J. C. Hansen and F. J. Asturias, Nat. Chem. Biol., 2008, 4, 728–737 CrossRef CAS PubMed.
- D. A. Rodionov, Chem. Rev., 2007, 107, 3467–3497 CrossRef CAS PubMed.
- K. Agaronyan, Y. I. Morozov, M. Anikin and D. Temiakov, Science, 2015, 347, 548–551 CrossRef CAS PubMed.
- A. Dauba, F. Z. Braikia, C. Oudinet and A. A. Khamlichi, Cell. Mol. Immunol., 2021, 18, 776–778 CrossRef CAS PubMed.
- T. I. Lee and R. A. Young, Cell, 2013, 152, 1237–1251 CrossRef CAS PubMed.
- J. S. Kahn, A. Trifonov, A. Cecconello, W. Guo, C. Fan and I. Willner, Nano Lett., 2015, 15, 7773–7778 CrossRef CAS PubMed.
- S. Lilienthal, A. Fischer, W. C. Liao, R. Cazelles and I. Willner, ACS Appl. Mater. Interfaces, 2020, 12, 31124–31136 CrossRef CAS PubMed.
- R. Freeman, T. Finder and I. Willner, Angew. Chem., 2009, 121, 7958–7961 CrossRef.
- Y. Miyake, H. Togashi, M. Tashiro, H. Yamaguchi, S. Oda, M. Kudo, Y. Tanaka, Y. Kondo, R. Sawa and T. Fujimoto, J. Am. Chem. Soc., 2006, 128, 2172–2173 CrossRef CAS PubMed.
- A. Ono, S. Cao, H. Togashi, M. Tashiro, T. Fujimoto, T. Machinami, S. Oda, Y. Miyake, I. Okamoto and Y. Tanaka, Chem. Commun., 2008, 4825–4827, 10.1039/b808686a.
- K. Gehring, J.-L. Leroy and M. Gueron, Nature, 1993, 363, 561–565 CrossRef CAS PubMed.
- Y. Hu, A. Cecconello, A. Idili, F. Ricci and I. Willner, Angew. Chem., Int. Ed. Engl., 2017, 56, 15210–15233 CrossRef CAS PubMed.
- A. S. Lubbe, W. Szymanski and B. L. Feringa, Chem. Soc. Rev., 2017, 46, 1052–1079 RSC.
- C. Wang, M. P. O’Hagan, Z. Li, J. Zhang, X. Ma, H. Tian and I. Willner, Chem. Soc. Rev., 2022, 51, 720–760 RSC.
- F. Wang, X. Liu and I. Willner, Angew. Chem., Int. Ed. Engl., 2015, 54, 1098–1129 CrossRef CAS PubMed.
- J. Bath and A. J. Turberfield, Nanoscience and Technology: A Collection of Reviews from Nature Journals, 2010, pp. 124–133 Search PubMed.
- P. O'Neill, P. W. Rothemund, A. Kumar and D. K. Fygenson, Nano Lett., 2006, 6, 1379–1383 CrossRef PubMed.
- C. Teller and I. Willner, Curr. Opin. Biotechnol., 2010, 21, 376–391 CrossRef CAS PubMed.
- J. Elbaz, Z.-G. Wang, R. Orbach and I. Willner, Nano Lett., 2009, 9, 4510–4514 CrossRef CAS PubMed.
- B. Yurke, A. J. Turberfield, A. P. Mills, F. C. Simmel and J. L. Neumann, Nature, 2000, 406, 605–608 CrossRef CAS PubMed.
- T. Omabegho, R. Sha and N. C. Seeman, Science, 2009, 324, 67–71 CrossRef CAS PubMed.
- Z. G. Wang, J. Elbaz and I. Willner, Nano Lett., 2011, 11, 304–309 CrossRef CAS PubMed.
- P. Yin, H. Yan, X. G. Daniell, A. J. Turberfield and J. H. Reif, Angew. Chem., 2004, 116, 5014–5019 CrossRef.
- M. You, Y. Chen, X. Zhang, H. Liu, R. Wang, K. Wang, K. R. Williams and W. Tan, Angew. Chem., Int. Ed. Engl., 2012, 51, 2457–2460 CrossRef CAS PubMed.
- R. A. Muscat, J. Bath and A. J. Turberfield, Nano Lett., 2011, 11, 982–987 CrossRef CAS PubMed.
- J. Wang, L. Yue, Z. Li, J. Zhang, H. Tian and I. Willner, Nat. Commun., 2019, 10, 4963 CrossRef PubMed.
- Z. G. Wang, J. Elbaz and I. Willner, Angew. Chem., Int. Ed. Engl., 2012, 51, 4322–4326 CrossRef CAS PubMed.
- C. H. Lu and I. Willner, Angew. Chem., Int. Ed., 2015, 54, 12212–12235 CrossRef CAS PubMed.
- C.-H. Lu and I. Willner, Angew. Chem., 2015, 127, 12380–12405 CrossRef.
- M. Vazquez-Gonzalez and I. Willner, Langmuir, 2018, 34, 14692–14710 CrossRef CAS PubMed.
- W. H. Chen, G. F. Luo, M. Vazquez-Gonzalez, R. Cazelles, Y. S. Sohn, R. Nechushtai, Y. Mandel and I. Willner, ACS Nano, 2018, 12, 7538–7545 CrossRef CAS PubMed.
- P. Zhang, A. Fischer, Y. Ouyang, J. Wang, Y. S. Sohn, R. Nechushtai, E. Pikarsky, C. Fan and I. Willner, Chem. Sci., 2021, 12, 14473–14483 RSC.
- Z. Zhou, M. Vazquez-Gonzalez and I. Willner, Chem. Soc. Rev., 2021, 50, 4541–4563 RSC.
- A. Fischer, S. Lilienthal, M. Vazquez-Gonzalez, M. Fadeev, Y. S. Sohn, R. Nechushtai and I. Willner, J. Am. Chem. Soc., 2020, 142, 4223–4234 CrossRef CAS PubMed.
- P. Zhang, L. Yue, M. Vazquez-Gonzalez, Z. Zhou, W. H. Chen, Y. S. Sohn, R. Nechushtai and I. Willner, ACS Nano, 2020, 14, 1482–1491 CrossRef CAS PubMed.
- J. S. Kahn, Y. Hu and I. Willner, Acc. Chem. Res., 2017, 50, 680–690 CrossRef CAS PubMed.
- M. Vazquez-Gonzalez and I. Willner, Angew. Chem., Int. Ed. Engl., 2020, 59, 15342–15377 CrossRef CAS PubMed.
- X. Liu, J. Zhang, M. Fadeev, Z. Li, V. Wulf, H. Tian and I. Willner, Chem. Sci., 2019, 10, 1008–1016 RSC.
- C. Wang, M. Fadeev, M. Vázquez-González and I. Willner, Adv. Funct. Mater., 2018, 28, 1803111 CrossRef.
- C. Wang, X. Liu, V. Wulf, M. Vazquez-Gonzalez, M. Fadeev and I. Willner, ACS Nano, 2019, 13, 3424–3433 CrossRef CAS PubMed.
- Z. Li, G. Davidson-Rozenfeld, M. Vazquez-Gonzalez, M. Fadeev, J. Zhang, H. Tian and I. Willner, J. Am. Chem. Soc., 2018, 140, 17691–17701 CrossRef CAS PubMed.
- K. Jiao, B. Zhu, L. Guo, H. Zhou, F. Wang, X. Zhang, J. Shi, Q. Li, L. Wang, J. Li and C. Fan, J. Am. Chem. Soc., 2020, 142, 10739–10746 CrossRef CAS PubMed.
- B. C. Buddingh and J. C. M. van Hest, Acc. Chem. Res., 2017, 50, 769–777 CrossRef CAS PubMed.
- M. Marguet, C. Bonduelle and S. Lecommandoux, Chem. Soc. Rev., 2013, 42, 512–529 RSC.
- Y. Tu, F. Peng, A. Adawy, Y. Men, L. K. Abdelmohsen and D. A. Wilson, Chem. Rev., 2016, 116, 2023–2078 CrossRef CAS PubMed.
- J. R. Babendure, S. R. Adams and R. Y. Tsien, J. Am. Chem. Soc., 2003, 125, 14716–14717 CrossRef CAS PubMed.
- J. C. Savage, P. Shinde, Y. Yao, M. A. Davare and U. Shinde, Chem. Commun., 2021, 57, 1344–1347 RSC.
- K. Adelman and J. T. Lis, Nat. Rev. Genet., 2012, 13, 720–731 CrossRef CAS PubMed.
- A. J. Neil, B. P. Belotserkovskii and P. C. Hanawalt, Biochemistry, 2012, 51, 8964–8970 CrossRef CAS PubMed.
- Y. Elani, R. V. Law and O. Ces, Nat. Commun., 2014, 5, 5305 CrossRef CAS PubMed.
- Z. Nourian and C. Danelon, ACS Synth. Biol., 2013, 2, 186–193 CrossRef CAS PubMed.
- H. Che, S. Cao and J. C. M. van Hest, J. Am. Chem. Soc., 2018, 140, 5356–5359 CrossRef CAS PubMed.
- R. J. R. W. Peters, I. Louzao and J. C. M. van Hest, Chem. Sci., 2012, 3, 335–342 RSC.
- R. S. Rikken, H. Engelkamp, R. J. Nolte, J. C. Maan, J. C. van Hest, D. A. Wilson and P. C. Christianen, Nat. Commun., 2016, 7, 12606 CrossRef CAS PubMed.
- R. Booth, Y. Qiao, M. Li and S. Mann, Angew. Chem., Int. Ed. Engl., 2019, 58, 9120–9124 CrossRef CAS PubMed.
- X. Huang, M. Li, D. C. Green, D. S. Williams, A. J. Patil and S. Mann, Nat. Commun., 2013, 4, 2239 CrossRef PubMed.
- X. Huang, A. J. Patil, M. Li and S. Mann, J. Am. Chem. Soc., 2014, 136, 9225–9234 CrossRef CAS PubMed.
- Y. Qiao, M. Li, R. Booth and S. Mann, Nat. Chem., 2017, 9, 110–119 CrossRef CAS PubMed.
- Z. Wang, M. C. van Oers, F. P. Rutjes and J. C. van Hest, Angew. Chem., Int. Ed. Engl., 2012, 51, 10746–10750 CrossRef CAS PubMed.
- M. Li, R. L. Harbron, J. V. Weaver, B. P. Binks and S. Mann, Nat. Chem., 2013, 5, 529–536 CrossRef CAS PubMed.
- W.-C. Liao and I. Willner, Adv. Funct. Mater., 2017, 27, 1702732 CrossRef.
- P. Zhang, A. Fischer, Y. Ouyang, J. Wang, Y. S. Sohn, O. Karmi, R. Nechushtai and I. Willner, Chem. Sci., 2022, 13, 7437–7448 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2sc01599d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |