Open Access Article
Open Access ArticleEngineering living cells with cucurbit[7]uril-based supramolecular polymer chemistry: from cell surface engineering to manipulation of subcellular organelles†
Fang
Huang
ab,
Jiaxiong
Liu
ab and
Yiliu
Liu
 *ab
*ab
aSouth China Advanced Institute for Soft Matter Science and Technology, School of Emergent Soft Matter, South China University of Technology, Guangzhou 510640, China. E-mail: liuyiliu@scut.edu.cn
bGuangdong Provincial Key Laboratory of Functional and Intelligent Hybrid Materials and Devices, South China University of Technology, Guangzhou 510640, China
First published on 6th July 2022
Abstract
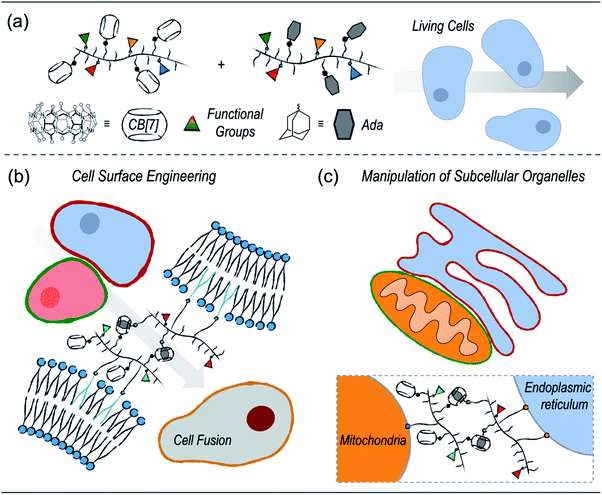
Supramolecular polymer chemistry, which closely integrates noncovalent interactions with polymeric structures, is a promising toolbox for living cell engineering. Here, we report our recent progress in exploring the applications of cucurbit[7]uril (CB[7])-based supramolecular polymer chemistry for engineering living cells. First, a modular polymer-analogous approach was established to prepare multifunctional polymers that contain CB[7]-based supramolecular recognition motifs. The supramolecular polymeric systems were successfully applied to cell surface engineering and subcellular organelle manipulation. By anchoring polymers on the cell membranes, cell–cell interactions were established by CB[7]-based host–guest recognition, which further facilitated heterogeneous cell fusion. In addition to cell surface engineering, placing the multifunctional polymers on specific subcellular organelles, including the mitochondria and endoplasmic reticulum, has led to enhanced physical contact between subcellular organelles. It is highly anticipated that the CB[7]-based supramolecular polymer chemistry will provide a new strategy for living cell engineering to advance the development of cell-based therapeutic materials.
Introduction
Engineering living cells with synthetic materials is emerging as a powerful strategy for biotechnology innovation.1–12 Engineered cells with modulated or augmented cellular functions have applications in various fields, ranging from therapeutical agents to living soft materials.13–19 For example, cell surface engineering modulating communications between the cell and its surrounding environment has led to unique applications such as selective cell-based therapy,20–22 targeting drug delivery systems,23–26 universal blood,27,28 and cell cryopreservation.29,30 In addition to molecular engineering of cell surfaces, substantial progress has also been made in manipulating the subcellular organelles to regulate cellular functions. Strategies for organelle-targeted intracellular synthesis and in situ self-assembly have been established to controllably interfere with cellular behaviors, including cell migration, proliferation, and apoptosis.31–35Synthetic polymers are one of the most popular artificial materials used in living cell engineering, mainly because of their accessibility and structural diversity.36,37 Nevertheless, the current demand for precision, modularity, and complexity creates further challenges in developing advanced polymeric systems. The emerging application of DNA technology in living cell engineering is an example.38–42 With programmable sequencing and precise base pairing, delicate DNA nanostructures with high modularity can be controllably constructed inside the cell or on cell surfaces. With the success of DNA systems from a polymer perspective, the integration of specific noncovalent recognition with a structure-controlled polymer is key. As a result, supramolecular polymer chemistry that integrates synthetic polymers with noncovalent interactions may provide new methods for living cell engineering.43–48
Cucurbituril-based host–guest recognition has been widely used in constructing supramolecular architectures.49–52 In particular, cucurbit[7]uril (CB[7]), which has an ultra-strong binding affinity (Ka > 109) toward specific guests, such as adamantanes or ferrocenes in aqueous solution, retains its high fidelity of guest recognition within complex biological environments.53–59 Recently, Wang and coworkers demonstrated that CB[7]-based recognition can be used to anchor drug-loading liposomes on macrophages for cell-hitchhiking delivery.60 In a separate study, they also showed that a multivalent CB[7]-grafted hyaluronic acid can induce mitochondrial aggregation and fusion via host–guest coupling.61 These pioneering works demonstrate that the CB[7]-based host–guest interactions hold great promise in engineering living cells.
Herein, we report our recent progress in exploring the applications of CB[7]-based supramolecular polymer chemistry in living cell engineering (Scheme 1). First, considering that the current CB[7]-decorated polymers are mostly limited to simple polyethene glycols (PEGs) or naturally derived polymers, we established a polymer-analogous synthetic approach to modularly expand the CB[7]-based supramolecular polymer system. As a result, different polymers with desired functional motifs, including CB[7] or its counterpart guest groups, organelle-targeting ligands, as well as fluorescence probes, were simply synthesized. These polymers were further applied to different applications of living cell engineering. On one hand, polymers with cell membrane anchoring moieties can effectively modify the surfaces of cancer cells and dendritic cells. Cell–cell interactions between dendritic cells and cancer cells were established by the CB[7]-based noncovalent recognition, which further led to heterogeneous cell fusion. On the other hand, specifically targeting subcellular organelles, including the endoplasmic reticulum (ER) and mitochondria (MITO), was achieved by introducing corresponding targeting ligands onto the polymers. Enhanced contact between the ER and MITO was successfully realized by the host–guest recognition between the polymers.
Results and discussion
Polymer design and modular synthesis
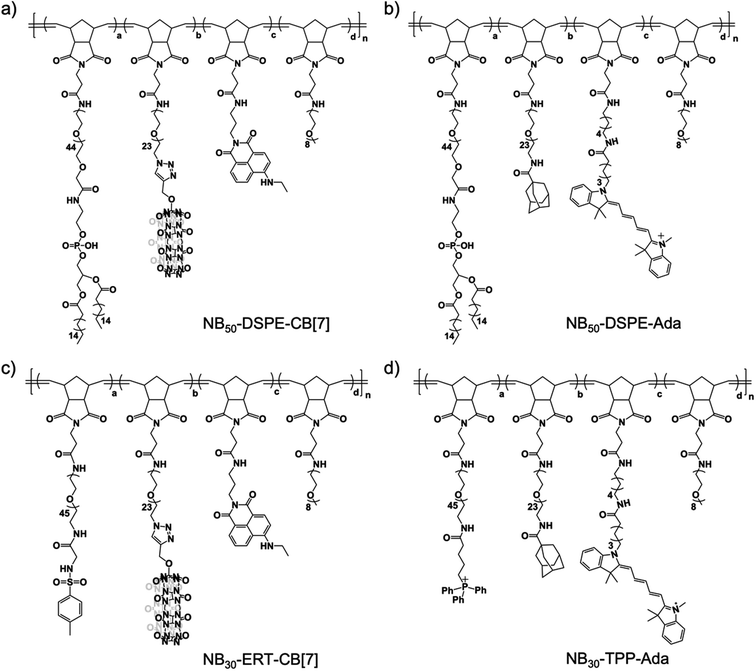
In our design, the desired polymers were decorated with CB[7] or its counterpart guest group. Adamantane was chosen as the guest group because of its high binding affinity to CB[7]. In addition to the supramolecular recognition pair, targeting ligands were introduced to the polymers for anchoring the polymers at specific cellular locations. For cell surface anchoring, polymeric side chains that contain double hydrophobic alkyl chains (DSPE) were introduced. The hydrophobic alkyl chains can insert into the lipid bilayer of the cell membrane, fixing the polymers to the cell surfaces.62 For organelle anchoring, two widely used targeting ligands, a p-toluenesulfonyl (Tosyl) group and a triphenylphosphonium (TPP) group, were chosen for specific ER and MITO locating, respectively.63,64 In addition, naphthalimide-based fluorophore (NTI) with green emission and cyanine 5 (Cy5) with red emission were introduced as fluorescence probes to track the location of the polymers using confocal laser scanning microscopy (CLSM). The structures of the designed polymers are shown in Fig. 1.A polymer-analogous approach was used to synthesize the designed polymer modularly.65,66 First, precursor polymers containing active ester moieties were synthesized via ring-opening metathesis polymerization (ROMP). Precursor polymers with a degree of polymerization of 30 (NB30-PF5) and 50 (NB50-PF5) were obtained with a standard ROMP protocol using a norbornene monomer with a pentafluorophenyl ester group (Fig. S4 and S5†).67 Because of the high reactivity of the pentafluorophenyl ester groups toward amines, the desired multifunctional polymers were prepared by directly reacting the precursor polymers with varied functional molecules with amino groups. One more step with a copper-catalyzed azide–alkyne cyclo-addition reaction (CuAAC) was required to obtain the CB[7]-containing polymers. As shown in Fig. 2, reacting the precursor polymer NB50-PF5 with the amino-containing molecules 1–4 produces the azide-containing polymer NB50-DSPE-N3. NB50-DSPE-CB[7] was readily obtained with the CuAAC reaction between the azide groups with the alkynyl functionalized CB[7].68 The polymers were dialyzed and then characterized by 1H NMR and gel permeation chromatography (GPC). The detailed synthetic procedure and characterization information are in the ESI (Fig. S14–S17 and Table S1†). Specifically, NB50-DSPE-CB[7] and NB50-DSPE-Ada, which contain double alkyl chains for cell membrane insertion, were prepared for cell surface engineering. NB30-ERT-CB[7] and NB30-TPP-Ada, as well as their fluorophore-free analogue polymers, NB30-ERT-CB[7]-2 and NB30-TPP-Ada-2, were used for the manipulation of subcellular organelles (Fig. 1 and Table 1).
 | ||
| Fig. 2 Polymer-analogous reaction. The synthesis of CB[7]-containing polymer NB50-DSPE-CB[7] is used as an example. | ||
| Polymer | Ligand | S-Motif | Dye | a | b | c | d | n |
|---|---|---|---|---|---|---|---|---|
| a Ligand: targeting ligands; S-motif: supramolecular motif; Dye: fluorophore. | ||||||||
| NB50-DSPE-CB[7] | DSPE | CB[7] | NTI | 0.10 | 0.10 | 0.06 | 0.74 | 50 |
| NB50-DSPE-Ada | DSPE | Ada | Cy5 | 0.10 | 0.10 | 0.02 | 0.78 | 50 |
| NB30-ERT-CB[7] | Tosyl | CB[7] | NTI | 0.10 | 0.10 | 0.06 | 0.74 | 30 |
| NB30-ERT-CB[7]-2 | Tosyl | CB[7] | — | 0.10 | 0.10 | — | 0.80 | 30 |
| NB30-TPP-Ada | TPP | Ada | Cy5 | 0.10 | 0.10 | 0.02 | 0.78 | 30 |
| NB30-TPP-Ada-2 | TPP | Ada | — | 0.10 | 0.10 | — | 0.80 | 30 |
Cell surface engineering toward heterogeneous cell fusion
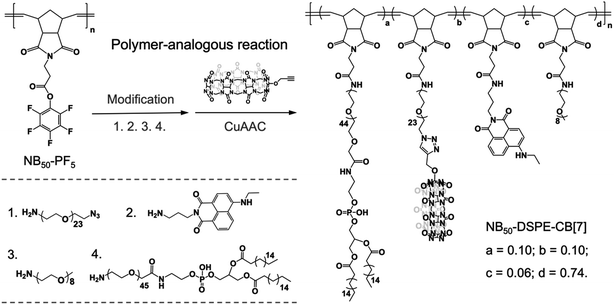
Dendritic cells (DCs) are the most potent and crucial antigen-presenting cells in the body that can directly capture and process antigens of cancer cells, then present these cancer-specific antigens to immune cells to elicit an immune response against the cancer epitopes.69 DC fusion vaccines have shown great potential for clinical efficacy. Fusing DCs with inactivated tumor cells enable DCs to obtain all of the antigen information of tumor cells and preserve their native antigen presentation properties.70 However, homogeneously and heterogeneously fused cells were randomly obtained in the process of fusion, and selectively inducing heterogeneous cell fusion is of great concern when preparing efficient a DCs/tumor fusion vaccine. Selective cell–cell attachment is important for heterogeneous cell fusion.71 Recently, Teramura and coworkers modified cell membranes with two poly(ethylene glycol)-lipid (PEG-lipid) derivatives carrying a pair of recognizable oligopeptides to induce cell–cell attachment, which greatly reduced the non-specific fusion between cells and enhanced the efficiency of cell fusion.72 On the basis of the high guest specificity and strong binding affinity of CB[7]-based host–guest recognition, CB[7]-based supramolecular polymer chemistry may provide a new method to promote heterogeneous cell fusion.In this study, human prostate cancer cells (PC-3) and dendritic cells (DC 2.4) were used as model cell lines, and NB50-DSPE-CB[7] and NB50-DSPE-Ada were chosen as supramolecular polymer systems. Prior to cell surface anchoring, the cytotoxicity of NB50-DSPE-CB[7] and NB50-DSPE-Ada were evaluated by a CCK-8 assay. Both of the polymers exhibited no obvious toxicity toward PC-3 or DC 2.4 cells after 24 h of incubation over a range of concentrations (Fig. S22†). The anchoring ability of the polymers on cell surfaces was then investigated. PC-3 and DC 2.4 cells were modified with NB50-DSPE-CB[7] and NB50-DSPE-Ada, respectively, at a concentration of 500 μg mL−1. The polymers with the fluorophores were tracked using CLSM. As shown in Fig. 3, after an incubation of 25 min, NB50-DSPE-CB[7] (green) and NB50-DSPE-Ada (red) were successfully labeled on the cell surfaces with minimal internalization. The retention of the polymers on the cell surfaces was also investigated. As confirmed by CLSM, NB50-DSPE-CB[7] remains on the surface of the PC-3 cells even after 24 h, albeit with less fluorescence intensity. In contrast, NB50-DSPE-Ada was gradually internalized into the DC 2.4 cells in approximately 3 h (Fig. S18†), which was likely attributed to the hydrophobic and positively charged moieties on the polymers, such as Ada and Cy5, facilitating the cellular internalization.73
 | ||
| Fig. 3 CLSM images of PC-3 and DC 2.4 cells modified with NB50-DSPE-CB[7] (green) and NB50-DSPE-Ada (red), respectively. | ||
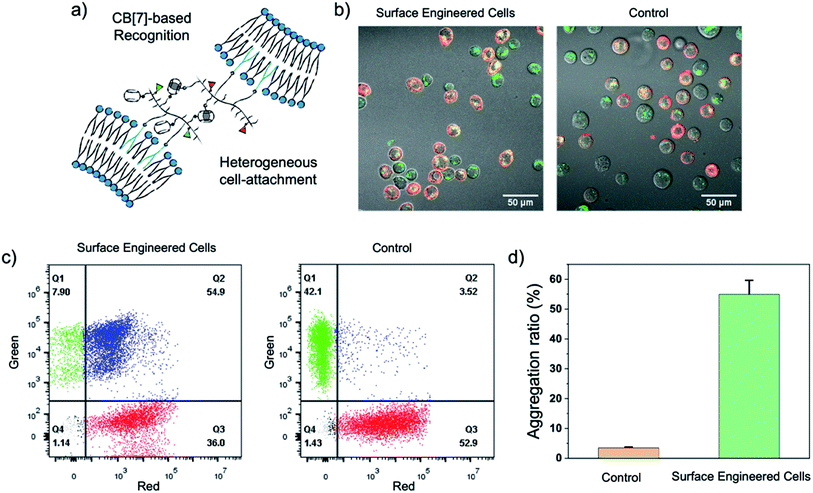
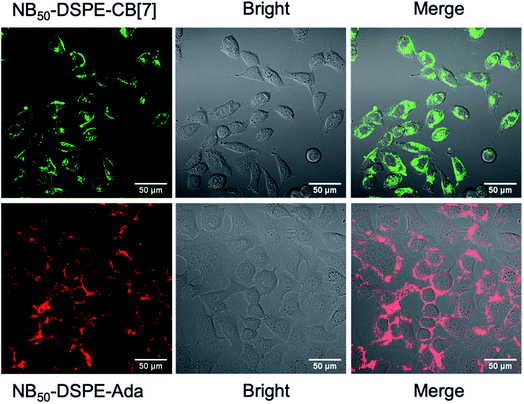
Fresh surface-engineered PC-3 and DC 2.4 cells were mixed and shaken at 300 rpm for 30 min. The interaction of the two types of cells was evaluated by CLSM and flow cytometry. A representative CLSM image is shown in Fig. 4b, and apparent heterogeneous cell aggregation was observed. As a control, PC-3 cells modified with NB50-DSPE-N3 and DC 2.4 cells modified with NB50-DSPE-Ada were mixed and shaken. In the control, no CB[7] was on the surface of the PC-3 cells, which means no host–guest recognition existed. As shown in Fig. 4b, the cells were individually distributed in the confocal dish with little aggregation. The cell aggregation was mostly attributed to the CB[7]-based host–guest recognition. The dynamic action of cell aggregation was also carefully studied by time-dependent scanning of the cells after the shaking procedure (Fig. S19†). Most of the cells were firmly bound together and not moving, which means these cells were already attached to each other via interfacial polymer assembly that occurred during shaking. Interestingly, there were still a few stray cells that moved around and gradually attached to the other type of cells. The process appears to be purposeful rather than a random action, which is attributed to the CB[7]-based recognition. In addition to the CLSM imaging, a quantitative evaluation of the cell aggregation was performed using flow cytometry. As shown in Fig. 4c, no obvious aggregation was detected in the control sample; an aggregation degree of only 3.52% was expressed in the Q2 quadrant of the scatter plot. In contrast, the aggregation ratio increased significantly to 54.9% for the cells modified with NB50-DSPE-CB[7] and NB50-DSPE-Ada. Overall, the results confirmed that the cell–cell interactions could be established by applying the CB[7]-based supramolecular polymer systems, which provides a biocompatible, specific, and efficient approach to realize heterogeneous cell assembly.
With the successful realization of heterogeneous cell attachments, inducing cell fusion to create hybrid cells was investigated. The conventional PEG approach was applied to induce cell fusion. Briefly, PC-3 cells modified by NB50-DSPE-CB[7] were mixed with equal number of DC 2.4 cells modified by NB50-DSPE-Ada. The mixture was shaken for 30 min to allow for sufficient heterogeneous cell attachment. The mixture was then treated with a 50 wt% PEG solution to induce cell fusion.72 After discarding the PEG solution by centrifugation, the cells were resuspended in culture medium and observed by confocal microscopy.
As shown in Fig. 5, distinct assembled cells were observed at the very beginning if the cells were priority modified with NB50-DSPE-CB[7] and NB50-DSPE-Ada. Mixed red and green fluorescence were distributed both on the cell surfaces and intracellularly (zoom image), indicating successful heterogeneous cell fusion via selective cell–cell attachment. In the control sample in which the cells were not modified beforehand by cell surface engineering, cells were mono-dispersed in the solution. In addition, it is mainly manifested as the fusion of homogeneous cells because only red fluorescence was observed on the surface and inside the fused cells. After 24 h of incubation, for the cells modified with polymers, the green fluorescence was uniformly labeled around the periphery of the fused cell membrane, and overlapping red and green fluorescence partially merged to yellow inside the cytoplasm. This phenomenon was consistent with the results of the retention of polymers on the cell surface, where NB50-DSPE-CB[7] remained anchored to the cell surfaces even after 24 h. However, most of the NB50-DSPE-Ada was internalized into cells within 3 h. In the control sample, only internalized red fluorescence was observed in fused cells. In general, the above results demonstrated that CB[7]-based supramolecular polymer chemistry can effectively induce heterogeneous cell–cell attachment, which offers a feasible approach for the preparation of DC-based fusion cells.v
 | ||
| Fig. 5 CLSM merged images of heterogeneous cell fusion taken at 0 h and 24 h. PC-3 cells (modified with NB50-DSPE-CB[7], green) and DC 2.4 cells (modified with NB50-DSPE-Ada, red). | ||
Enhancing the contact between the mitochondria and endoplasmic reticulum
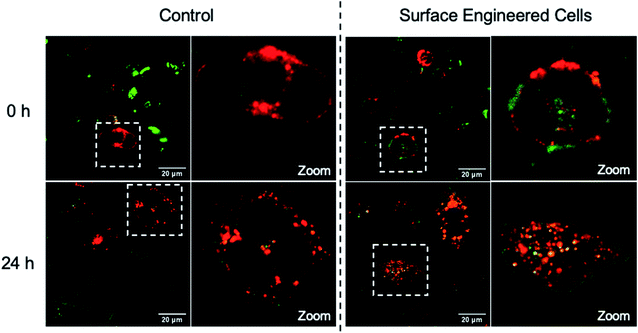
The MITO and ER are subcellular organelles that are crucial to maintaining the normal metabolic and biological functions of eukaryotic cells. The communication between the MITO and ER is strongly associated with the special physical contact sites between the two organelles, namely MITO-associated membranes (MAMs). Such intimate contact allows for the transfer of lipids and ions, as well as cell signaling, between the two subcellular organelles.74 Disruption of MAMs could directly affect the cellular homoeostatic functions, including lipid metabolism, calcium homoeostasis, unfolded protein response, ER stress, and MITO bioenergetics, which have been implicated in neurodegenerative diseases,75 liver diseases,76 and cancer.77 The MITO–ER tethering protein-based approach is currently the only means to artificially manipulate MITO–ER contacts. The physical tethering between MITO–ER could be realized by CB[7]-based host–guest recognition. In this study, we explored the possibility of applying CB[7]-based supramolecular polymer chemistry in enhancing the contact of the two subcellular organelles.To this end, NB30-ERT-CB[7] and NB30-TPP-Ada, which contain the MITO targeting TPP ligand and ER targeting Tosyl ligand, were used. First, the biocompatibility of polymers were confirmed that no obvious cytotoxicity was detected in PC-3 cells after treatment with NB30-ERT-CB[7] or NB30-TPP-Ada for 24 h (Fig. S22†). The targeting ability of the polymers was then evaluated. After incubation of NB30-ERT-CB[7] (green) with PC-3 cells for 8 h, the cells were stained by commercially available ER Tracker Red, Mito-Tracker Red CMXRos and Lyso-Tracker Red dyes. Pearson's colocalization coefficient (PCC) was used to quantify the degree of colocalization. As shown in Fig. 6, the fluorescence of NB30-ERT-CB[7] was well matched with that of ER Tracker Red with a PCC of 0.73. In contrast, non-overlapping fluorescence was observed between NB30-ERT-CB[7] and Mito-Tracker Red, which displays a negative correlation PCC of −0.29. The fluorescence of NB30-ERT-CB[7] was partially overlapped with that of Lyso-Tracker Red and displayed a rather low PCC of 0.45 (Fig. S20†). Based on these results, NB30-ERT-CB[7] exhibited good ER-targeting capability, although some of the polymers were trapped in lysosomes. Similarly, the MITO targeting ability of the Cy5 labeled NB30-TPP-Ada (red) in PC-3 cells was demonstrated by co-staining with Mito-Tracker Green, Lyso-Tracker Green, and ER-Tracker Green. A high colocalization between NB30-TPP-Ada and MITO was observed, producing a high PCC of 0.80. As expected, there was no clear colocalization fluorescence in the lysosome or ER. The PCC was −0.1 (Fig. S20†) and 0.16 for the lysosome and ER, respectively; these low values may be attributed to the lysosome escape capacity of TPP ligands. Overall, by introducing targeting ligands, the polymers show effective and orthogonal locating properties.
 | ||
| Fig. 6 Colocalization images of NB30-ERT-CB[7] and NB30-TPP-Ada with MITO and ER. Pearson's colocalization coefficient (PCC) was used to quantify the degree of colocalization. | ||
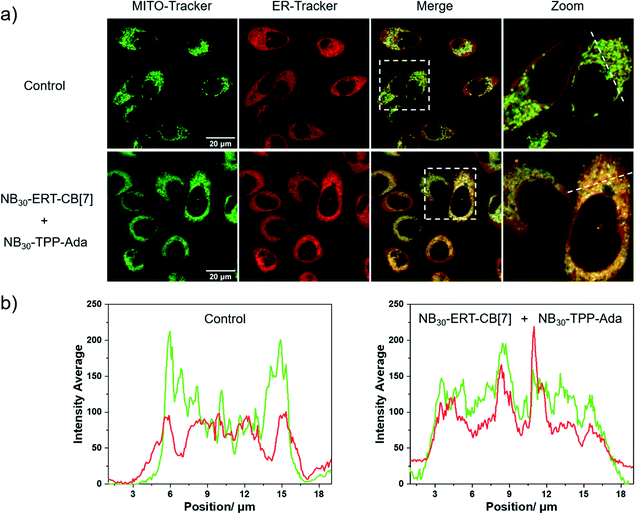
Having demonstrated the successful subcellular organelle targeting of the polymers, we further applied the polymers in promoting the physical contact between the ER and MITO. Instead of using fluorophores on the polymers, ER Tracker Red and Mito-Tracker Green staining were used for visualizing the construct distribution of the ER and MITO for better fluorescence tracking of the subcellular organelles. Accordingly, the fluorophore-free polymers NB30-ERT-CB[7]-2 and NB30-ERT-CB[7]-2 were used to treat the cells to avoid interference. The PC-3 cells were pretreated with 200 μg mL−1 of NB30-ERT-CB[7]-2 for 12 h, and subsequently treated with the same concentration of NB30-TPP-Ada-2 for another 12 h. As shown in Fig. 7, the ER and MITO fluorescence were distributed separately in the control cells that were not treated with the polymers. However, for the NB30-ERT-CB[7]-2 and NB30-TPP-Ada-2 treated cells, the green fluorescence of the MITO highly overlapped with the ER red fluorescence. The fluorescence intensity profile of regions of interest (ROI) across cells (Fig. 7b, white dotted lines indicated in the CLSM images) show that green and red peaks fluctuated consistently. Further evidence of the enhanced contact between MITO and ER was provided by transmission electron microscopy (TEM). As shown in Fig. 8, the ER was more tightly twined around the MITO within the polymer-treated cells. The average distance between the outer mitochondrial membrane and ER is approximately 60 nm in the untreated PC-3 cells, while the physical linkage length between the ER and MITO was reduced to less than 15 nm after treatment with the supramolecular polymer system.
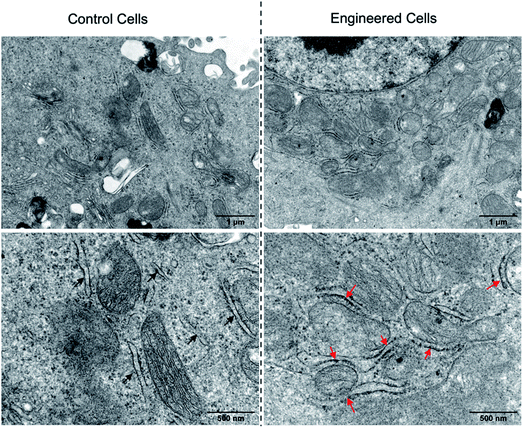 | ||
| Fig. 8 TEM images of PC-3 cells without (left) or with (right) the treatment of NB30-ERT-CB[7]-2 and NB30-TPP-Ada-2. The red arrows reveal the increased contact between the ER and MITO. | ||
Conclusions
This work demonstrates the feasibility and potential of CB[7]-based supramolecular polymer chemistry in living cell engineering. Through a polymer-analogous approach, multifunctional polymers that contain CB[7]-based supramolecular recognition motifs were simply synthesized, greatly expanding the types of CB[7]-based supramolecular polymer systems, providing a modular synthetic platform for polymers toward living cell engineering. Manipulation of the interaction between cells or subcellular organelles was successfully realized with the polymers containing CB[7] or an adamantane group, targeting ligands, PEG side chains, and fluorophores. The modulation helps to solve the problem of non-specific fusion in the preparation of tumor vaccines and also provides a promising new treatment paradigm to address diseases associated with dysfunction of the ER–mitochondria contact such as neurodegenerative diseases. We anticipate that this line of research will provide a new strategy of living cell engineering to advance the development of cell-based therapeutic materials.Data availability
All experimental data is available within the article and the ESI.†Author contributions
F. Huang, J. Liu and Y. Liu conceived and designed the study. F. Huang and J. Liu performed the experiments. F. Huang, J. Liu and Y. Liu analyzed the data. Y. Liu supervised the work. All authors contributed to the finalization of the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We gratefully acknowledge the financial support from National Natural Science Foundation of China (21901077), Natural Science Foundation of Guangdong (2016ZT06C322), the Research Fund Program of Guangdong Provincial Key Laboratory of Functional and Intelligent Hybrid Materials and Devices (2019B121203003), and open project of Key Lab of Organic Optoelectronics & Molecular Engineering.References
- E. Saxon and C. R. Bertozzi, Science, 2000, 287, 2007–2010 CrossRef CAS PubMed.
- C. M. Csizmar, J. R. Petersburg and C. R. Wagner, Cell Chem. Biol., 2018, 25, 931–940 CrossRef CAS PubMed.
- Z. Liu, X. Xu and R. Tang, Adv. Funct. Mater., 2016, 26, 1862–1880 CrossRef CAS.
- C. W. Shields IV, L. L.-W. Wang, M. A. Evans and S. Mitragotri, Adv. Mater., 2020, 32, 1901633 CrossRef PubMed.
- B. J. Kim, H. Cho, J. H. Park, J. F. Mano and I. S. Choi, Adv. Mater., 2018, 30, 1706063 CrossRef PubMed.
- D. A. Uhlenheuer, K. Petkau and L. Brunsveld, Chem. Soc. Rev., 2010, 39, 2817–2826 RSC.
- J. Brinkmann, E. Cavatorta, S. Sankaran, B. Schmidt, J. van Weerd and P. Jonkheijm, Chem. Soc. Rev., 2014, 43, 4449–4469 RSC.
- K. Petkau and L. Brunsveld, Org. Biomol. Chem., 2013, 11, 219–232 RSC.
- J. Park, B. Andrade, Y. Seo, M.-J. Kim, S. C. Zimmerman and H. Kong, Chem. Rev., 2018, 118, 1664–1690 CrossRef CAS PubMed.
- J. Cao, O. T. Zaremba, Q. Lei, E. Ploetz, S. Wuttke and W. Zhu, ACS Nano, 2021, 15, 3900–3926 CrossRef CAS PubMed.
- M. M. Stevens and J. H. George, Science, 2005, 310, 1135–1138 CrossRef CAS PubMed.
- A. J. R. Amaral and G. Pasparakis, Acta Biomater., 2019, 90, 21–36 CrossRef CAS PubMed.
- Q. Wang, H. Cheng, H. Peng, H. Zhou, P. Y. Li and R. Langer, Adv. Drug Delivery Rev., 2015, 91, 125–140 CrossRef CAS PubMed.
- J. Lee, I. S. Choi, T. I. Oh and E. Lee, Chem.–Eur. J., 2018, 24, 15725–15743 CrossRef CAS PubMed.
- S. Abbina, E. M. J. Siren, H. Moon and J. N. Kizhakkedathu, ACS Biomater. Sci. Eng., 2018, 4, 3658–3677 CrossRef CAS PubMed.
- P. Q. Nguyen, N.-M. D. Courchesne, A. Duraj-Thatte, P. Praveschotinunt and N. S. Joshi, Adv. Mater., 2018, 30, 1704847 CrossRef PubMed.
- L. Sun, Z. Xiong, F. Shen, Z. Wang and Z. Liu, Sci. China: Chem., 2021, 64, 719–733 CrossRef CAS.
- X. Liu, M. E. Inda, Y. Lai, T. K. Lu and X. Zhao, Adv. Mater., 2022, 2201326 CrossRef CAS PubMed.
- H. Jia, Y. Zhu, Q. Duan and F. Wu, Chem. Soc. Rev., 2021, 50, 6240–6277 RSC.
- X. Xiong, H. Liu, Z. Zhao, M. B. Altman, D. Lopez-Colon, C. Yang, L. J. Chang, C. Liu and W. Tan, Angew. Chem., Int. Ed., 2013, 52, 1472–1476 CrossRef CAS PubMed.
- D. Zhang, Y. Zheng, Z. Lin, X. Liu, J. Li, H. Yang and W. Tan, Angew. Chem., Int. Ed., 2020, 59, 12022–12028 CrossRef CAS PubMed.
- C. Plumet, A. S. Mohamed, T. Vendeuvre, B. Renoux, J. Clarhaut and S. Papot, Chem. Sci., 2021, 12, 9017–9021 RSC.
- X. Huang, F. Zhang, H. Wang, G. Niu, K. Y. Choi, M. Swierczewska, G. Zhang, H. Gao, Z. Wang, L. Zhu, H. S. Choi, S. Lee and X. Chen, Biomaterials, 2013, 34, 1772–1780 CrossRef CAS PubMed.
- T. Liu, C. Gao, D. Gu and H. Tang, Drug Delivery Transl. Res., 2022 DOI:10.1007/s13346-022-01149-y.
- D. Y. Lee, B.-H. Cha, M. Jung, A. S. Kim, D. A. Bull and Y.-W. Won, J. Biol. Eng., 2018, 12, 28 CrossRef CAS PubMed.
- C. Gao, Q. Cheng, J. Wei, C. Sun, S. Lu, C. H. T. Kwong, S. M. Y. Lee, Z. Zhong and R. Wang, Mater. Today, 2020, 40, 9–17 CrossRef CAS.
- B. Wang, G. Wang, B. Zhao, J. Chen, X. Zhang and R. Tang, Chem. Sci., 2014, 5, 3463–3468 RSC.
- Y. Zhao, M. Fan, Y. Chen, Z. Liu, C. Shao, B. Jin, X. Wang, L. Hul, S. Wang, Z. Liao, D. Ling, R. Tang and B. Wang, Sci. Adv., 2020, 6, eaaw9679 CrossRef CAS PubMed.
- J. Huang, J. Guo, L. Zhou, G. Zheng, J. Cao, Z. Li, Z. Zhou, Q. Lei, C. J. Brinker and W. Zhu, ACS Appl. Bio Mater., 2021, 4, 2996–3014 CrossRef CAS PubMed.
- W. Zhu, J. Guo, J. O. Agola, J. G. Croissant, Z. Wang, J. Shang, E. Coker, B. Motevalli, A. Zimpel, S. Wuttke and C. J. Brinker, J. Am. Chem. Soc., 2019, 141, 7789–7796 CrossRef CAS PubMed.
- Q. Zhang, W. Tan and B. Xu, ChemPlusChem, 2022, 87, e202200060 CAS.
- X. Sun, Y. Dong, Y. Liu, N. Song, F. Li and D. Yang, Sci. China: Chem., 2021, 65, 31–47 CrossRef.
- A. N. Shy, B. J. Kim and B. Xu, Matter, 2019, 1, 1127–1147 CrossRef PubMed.
- J. Liu and B. Liu, Prog. Polym. Sci., 2022, 129, 101545 CrossRef CAS.
- Y. Dai, T. Li, Z. Zhang, Y. Tan, S. Pan, L. Zhang and H. Xu, J. Am. Chem. Soc., 2021, 143, 10709–10717 CrossRef CAS PubMed.
- Y. Teramura and H. Iwata, Soft Matter, 2010, 6, 1081–1091 RSC.
- M. C. Arno, Macromol. Rapid Commun., 2020, 41, 2000302 CrossRef CAS PubMed.
- P. Shi and Y. Wang, Angew. Chem., Int. Ed., 2021, 60, 11580–11591 CrossRef CAS PubMed.
- Z. He, Q. Chen, F. Chen, J. Zhang, H. Li and J.-M. Lin, Chem. Sci., 2016, 7, 5448–5452 RSC.
- Z. Guo, L. Zhang, Q. Yang, R. Peng, X. Yuan, L. Xu, Z. Wang, F. Chen, H. Huang, Q. Liu and W. Tan, Angew. Chem., Int. Ed., 2022, 61, e202111151 CAS.
- F. Li, Y. Liu, Y. Dong, Y. Chu, N. Song and D. Yang, J. Am. Chem. Soc., 2022, 144, 4667–4677 CrossRef CAS PubMed.
- X. Guo, F. Li, C. Liu, Y. Zhu, N. Xiao, Z. Gu, D. Luo, J. Jiang and D. Yang, Angew. Chem., Int. Ed., 2020, 59, 20651–20658 CrossRef CAS PubMed.
- T. Aida, E. W. Meijer and S. I. Stupp, Science, 2012, 335, 813–817 CrossRef CAS PubMed.
- K. Petkau-Milroy, M. H. Sonntag and L. Brunsveld, Chem.–Eur. J., 2013, 19, 10786–10793 CrossRef CAS PubMed.
- B. Qin, Z. Yin, X. Tang, S. Zhang, Y. Wu, J. Xu and X. Zhang, Prog. Polym. Sci., 2020, 100, 101167 CrossRef CAS.
- P. Shi, E. Ju, J. Wang, Z. Yan, J. Ren and X. Qu, Mater. Today, 2017, 20, 16–21 CrossRef CAS.
- Y. Gu, B. Liu, Q. Liu, Y. Hang, L. Wang, J. L. Brash, G. Chen and H. Chen, ACS Appl. Mater. Interfaces, 2019, 11, 47720–47729 CrossRef CAS PubMed.
- G. Morgese, B. F. M. de Waal, S. V. Aramburu, A. R. A. palmans, L. Albertazzi and E. W. Meijer, Angew. Chem., Int. Ed., 2020, 59, 17229–17233 CrossRef CAS PubMed.
- J. Lagona, P. Mukhopadhyay, S. Chakrabarti and L. Isaacs, Angew. Chem., Int. Ed., 2005, 44, 4844–4870 CrossRef CAS PubMed.
- K. Kim, N. Selvapalam, Y. H. Ko, K. M. Park, D. Kim and J. Kim, Chem. Soc. Rev., 2007, 36, 267–279 RSC.
- E. Masson, X. Ling, R. Joseph, L. K. Mensah and X. Lu, RSC Adv., 2012, 2, 1213–1247 RSC.
- S. J. Barrow, S. Kasera, M. J. Rowland, J. del Barrio and O. A. Scherman, Chem. Rev., 2015, 115, 12320–12406 CrossRef CAS PubMed.
- Y. Liu, Y. Zhang, H. Yu and Y. Liu, Angew. Chem., Int. Ed., 2021, 60, 3870–3880 CrossRef CAS PubMed.
- K. M. Park, J. Murray and K. Kim, Acc. Chem. Res., 2017, 50, 644–646 CrossRef CAS PubMed.
- P. Neirynck, J. Brinkmann, Q. An, D. W. J. van der Schaft, L.-G. Milroy, P. Jonkheijm and L. Brunsveld, Chem. Commun., 2013, 49, 3679–3681 RSC.
- K. L. Kim, G. Sung, J. Sim, J. Murray, M. Li, A. Lee, A. Shrinidhi, K. M. Park and K. Kim, Nat. Commun., 2018, 9, 1712 CrossRef PubMed.
- H. Wang, Y. Yan, Y. Yi, Z. Wei, H. Chen, J. Xu, H. Wang, Y. Zhao and X. Zhang, CCS Chem., 2020, 2, 739–748 CrossRef CAS.
- H. Wang, H. Wu, Y. Yi, K. Xue, J. Xu, H. Wang, Y. Zhao and X. Zhang, CCS Chem., 2021, 3, 1413–1425 CrossRef CAS.
- H. Wu, H. Chen, B. Tang, Y. Kang, J. Xu and X. Zhang, Langmuir, 2020, 36, 1235–1240 CrossRef CAS PubMed.
- C. Gao, Q. Cheng, J. Li, J. Chen, Q. Wang, J. Wei, Q. Huang, S. M. Lee, D. Gu and R. Wang, Adv. Funct. Mater., 2021, 31, 2102440 CrossRef CAS.
- C. Sun, Z. Wang, L. Yue, Q. Huang, Q. Cheng and R. Wang, J. Am. Chem. Soc., 2020, 142, 16523–16527 CrossRef CAS PubMed.
- D. C. Church and J. K. Pokorski, Angew. Chem., Int. Ed., 2020, 59, 11379–11383 CrossRef CAS PubMed.
- J. Zielonka, J. Joseph, A. Sikora, M. Hardy, O. Ouari, J. Vasquez-Vivar, G. Cheng, M. Lopez and B. Kalyanaraman, Chem. Rev., 2017, 117, 10043–10120 CrossRef CAS PubMed.
- H. Deng, Z. Zhou, W. Yang, L.-s. Lin, S. Wang, G. Niu, J. Song and X. Chen, Nano Lett., 2020, 20, 1928–1933 CrossRef CAS PubMed.
- A. Das and P. Theato, Chem. Rev., 2016, 116, 1434–1495 CrossRef CAS PubMed.
- X. Yang, Q. Ji, J. Liu and Y. Liu, CCS Chem., 2022 DOI:10.31635/ccschem.022.202201936.
- K. Zhang, M. A. Lackey, Y. Wu and G. N. Tew, J. Am. Chem. Soc., 2011, 133, 6906–6909 CrossRef CAS PubMed.
- H. Chen, Z. Huang, H. Wu, J. Xu and X. Zhang, Angew. Chem., Int. Ed., 2017, 56, 16575–16578 CrossRef CAS PubMed.
- S. K. Wculek, F. J. Cueto, A. M. Mujal, I. Melero, M. F. Krummel and D. Sancho, Nat. Rev. Immunol., 2020, 20, 7–24 CrossRef CAS PubMed.
- D. Galati and S. Zanotta, Cytotherapy, 2018, 20, 1309–1323 CrossRef CAS PubMed.
- A. Bahadori, L. B. Oddershede and P. M. Bendix, Nano Res., 2017, 10, 2034–2045 CrossRef CAS.
- A. Yoshihara, S. Watanabe, I. Goel, K. Ishihara, K. N. Ekdahl, B. Nilsson and Y. Teramura, Biomaterials, 2020, 253, 120113 CrossRef CAS PubMed.
- T. Osaka, T. Nakanishi, S. Shanmugam, S. Takahama and H. Zhang, Colloids Surf., B, 2009, 71, 325–330 CrossRef CAS PubMed.
- M. A. Carpio, R. E. Means, A. L. Brill, A. Sainz, B. E. Ehrlich and S. G. Katz, Cell Rep., 2021, 34, 108827 CrossRef CAS PubMed.
- S. Lee, W. Wang, J. Hwang, U. Namgung and K.-T. Min, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 16074–16079 CrossRef CAS PubMed.
- M. I. Hernández-Alvarez, D. Sebastián, S. Vives, S. Ivanova, P. Bartoccioni, P. Kakimoto, N. Plana, S. R. Veiga, V. Hernández and N. Vasconcelos, Cell, 2019, 177, 881–895 CrossRef PubMed , e817..
- M. Xia, Y. Zhang, K. Jin, Z. Lu, Z. Zeng and W. Xiong, Cell Biosci., 2019, 9, 1–19 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2sc02797f |
| This journal is © The Royal Society of Chemistry 2022 |