Open Access Article
Open Access ArticleMonitoring and modulating a catalytic hybridization circuit for self-adaptive bioorthogonal DNA assembly†
Xue
Gong‡
a,
Shizhen
He‡
a,
Ruomeng
Li‡
a,
Yingying
Chen
a,
Kaiyue
Tan
a,
Yeqing
Wan
a,
Xiaoqing
Liu
 a and
Fuan
Wang
a and
Fuan
Wang
 *ab
*ab
aCollege of Chemistry and Molecular Sciences, Wuhan University, 430072 Wuhan, P. R. China. E-mail: fuanwang@whu.edu.cn
bWuhan Research Center for Infectious Diseases and Cancer, Chinese Academy of Medical Sciences, Wuhan 430072, China
First published on 10th August 2022
Abstract
Constructing artificial domino nanoarchitectures, especially dynamic DNA circuits associated with the actuation of biological functions inside live cells, represents a versatile and powerful strategy to regulate the behaviors and fate of various living entities. However, the stepwise operation of conventional DNA circuits always relies on freely diffusing reactants, which substantially slows down their operation rate and efficiency. Herein, a self-adaptive localized catalytic circuit (LCC) is developed to execute the self-sustained bioorthogonal assembly of DNA nanosponges within a crowded intracellular environment. The LCC-generated DNA scaffolds are utilized as versatile templates for realizing the proximity confinement of LCC reactants. Single-molecule-detecting fluorescence correlation spectroscopy (FCS) is used to explore the reaction acceleration of the catalytic circuit. This self-adaptive DNA circuit facilitates the bioorthogonal assembly of highly branched DNA networks for robust and accurate monitoring of miRNA targets. Based on its intriguing and modular design, the LCC system provides a pivotal molecular toolbox for future applications in early disease diagnosis.
Introduction
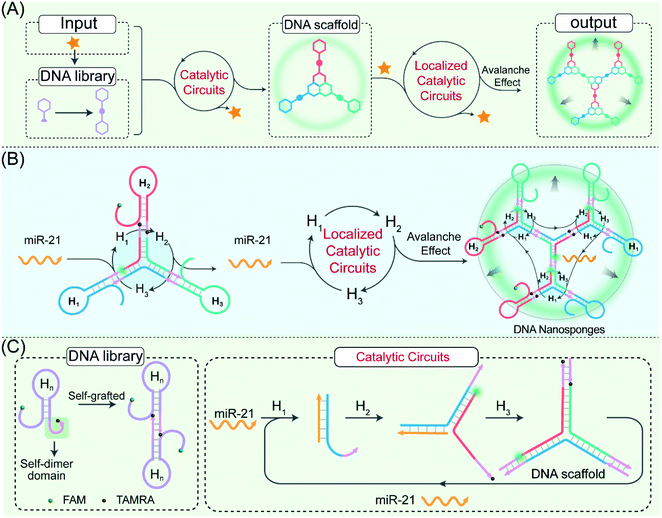
Biological systems routinely reconfigure their growth, self-repair, and transportation via the assembly of various domino nanoarchitectures.1,2 For instance, the cytoskeleton, consisting of various dynamic protein assemblies, can form intricate signal networks for ensuring cellular stability, motility, and division.3,4 For mimicking these naturally occurring structures, the integration of synthetic self-assembled nanostructures with the actuation of biological behaviors showed great potential for clinical diagnosis and therapy as well as drug delivery.5–8 The assembly of synthetic circuitry with cellular function needs the self-assembly of biocompatible architectures within crowded and complex biological environments. However, in practice, synthetic circuits have so far relied on freely diffusing reactants, which substantially slows down their operation rate and efficiency.9–12 The colocalization of synthetic circuitry reactants to enhance their effective concentration represents one charming way to realize non-natural assemblies in living cells.With its highly predictable base-pairing, low cost, ease of synthesis, and high biocompatibility, DNA has been recognized as a highly promising biomolecule for designing localized molecular biocomputing circuits for different aims.13–17 A wide range of localized DNA circuits has been implemented on DNA origami, molecular scaffolds, and spherical nucleic acids (SNAs).18–22 Such a spatially confined arrangement plays a pivotal role in increasing the effective concentration of reactants through proximity confinement and simultaneously achieves reduced crosstalk between these spatially separated reaction processes.23–26 However, the proximal immobilization of DNA circuitry elements on DNA origami requires time-consuming and disturbing optimization procedures to facilitate the efficient execution of various biocomputing events. Meanwhile, these circuits, immobilized on these as-synthetic or -assembled molecular scaffolds, are sensitive to the scaffold defects where these non- or mis-incorporated circuitry components could easily terminate the propagation of cascaded signal transactions along these different scaffolds. Besides, the complicated adjustment and optimization of these circuitry reactants and scaffolds are always confronted with a lack of scalability and modularity for the flexible design and efficient execution of these biocomputing circuits. It is expected that the circuitry-generated DNA scaffolds could be utilized as versatile self-confined proximity-guided templates to realize the facile and reliable arrangement of circuit reactants for achieving precisely localized biocomputing circuits. Thus, it remains a huge challenge for designing a self-templated and localized DNA circuit that exquisitely integrates the accelerating cross-catalytic reaction within the self-supplemented molecular matrix.27–31 The self-sustained proximity-accelerated catalytic reaction could facilitate the robust monitoring of biomolecule targets, thus making the self-adaptive localized catalytic circuits highly desirable for clinical diagnostics, yet they are still unexplored.
Herein, by using endogenous microRNA (miRNA) as a model promoter, we construct a self-adaptive localized catalytic circuit (LCC) that allows the biorthogonal assembly of DNA nanosponges for efficient amplified imaging of miRNA. In the LCC system, the key concept is to introduce structural symmetry in DNA sequences to achieve interconnecting catenated DNA reactants with self-confined hybridization capability. These engineered self-supplemented catenated DNA reactants are packaged into the folate-(FA)-modified poly(D,L-lactic-co-glycolic acid) (PLGA) nanovesicles, which facilitate their efficient and selective accumulation/endocytosis in target tumour cells. Then, the endogenous miRNA intiated an autonomous and successive catalytic DNA assembly in a self-sustaining and self-adapting manner, leading to the bioorthogonal assembly of highly branched DNA nanosponges. In the LCC system, the promoter could be recycled for multiple rounds of catalytic hybridization without being consumed, thus resulting in a remarkably amplified transduction of the corresponding promoter. The circuit modules and mechanisms are extensively investigated in vitro and in vivo by using the single-molecule-detecting fluorescence correlation spectroscopy (FCS) technique. Based on a systematic investigation, the catenated DNA probes with symmetrical fragments could increase the local concentration of circuitry reactants for accelerating the localized catalytic reaction and could also reduce the cellular diffusibility of circuitry products for achieving high-contrast imaging of cellular biomarkers. Specifically, the intracellular location of biomarkers is scarcely affected during the self-adaptive hierarchical assembly of the compact DNA nanosponges, thus facilitating the fast and precise positioning of intracellular analytes with high spatial resolution. As a versatile “plug-and-play” approach, our proposed LCC system shows great potential for constructing complex cascading DNA circuits in live cells, which reveals high potential applications in early disease diagnosis and drug-delivery.
Results and discussion
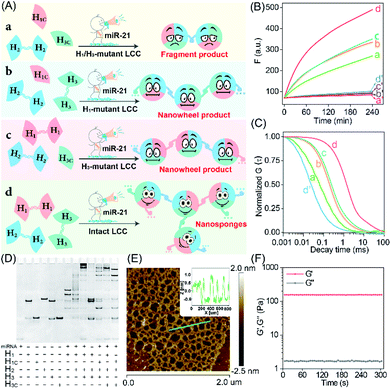
The self-adaptive bioorthogonal regulation of the LCC is realized via the spatially confined catalytic hybridization reaction of three elaborately designed DNA reactants (Fig. 1, for details, see Fig. S1†). Each of the DNA reactant units, H1, H2, and H3, is grafted with a symmetrical sequence at its 3′-end to form the catenated structure, and therefore is prone to assembling into Y-shaped DNA units with three self-grafted building blocks, which then lead to the ultimate formation of the advanced DNA architectures by the localized catalytic hybridization reaction (Fig. 1A). MiR-21 (T), a new oncogene biomarker, is selected as the model promoter to demonstrate the stimulus-responsive self-sustained DNA assembly (Fig. 1B). The promoter T opens the catenated H1 through the strand-displacement mechanism. The activated H1 immediately hybridizes with the toehold of the catenated H2, generating the transition-state intermediate T-H1·H2 triplex that was attached to the other two metastable hairpin building blocks. As a result, the opened H2 hybridizes and opens the catenated H3 to re-generate the promoter and produce the intermediate Y-shaped H1·H2·H3 triplex tethered with three metastable hairpin building blocks in the self-confined configuration. The released promoter is able to hybridize with the nearby tethered H1 of the as-assembled Y-shaped H1·H2·H3 triplex for substantially promoting the localized catalytic hybridization reaction of LCC reactants, thus realizing the self-sustained hierarchical assembly of DNA nanosponges (Fig. 1C). Noteworthily, a large number of Y-shaped DNAs are thus cross-linked, via the interconnecting catenated DNA reactants, for assembling into the DNA nanosponges. In order to monitor the self-assembly process, H2 is labeled with a FAM fluorophore and TAMRA quencher at its 5′- and 3′-end, respectively. Therefore, each promoter could initiate an “avalanche” reaction acceleration process in the LCC system, leading to the sustained assembly of DNA nanosponges for generating the amplified turn-on fluorescence transduction of the promoter.To investigate the detailed reaction procedure of our LCC system, a primary fluorescence experiment was performed. The catenated DNA reactant mixture showed no obvious fluorescence change without the promoter, while a significantly increased fluorescence signal was obtained upon its introduction with the promotor (Fig. S2†), confirming that miR-21 could effectively catalyze the assembly of DNA nanostructures. To investigate whether the developed LCC system could accelerate the reaction kinetics, a non-localized hybridization procedure was established by altering the symmetrical domain. For consistency, here the symmetrical domain of H1 and H3 was substituted with a random sequence to generate H1C and H3C that could not form catenated conformation (Fig. 2A, for a detailed reaction process, see Fig. S3A†). The kinetic characterization of the LCC system under different conditions was investigated. As shown in Fig. 2B, the introduction of miR-21 into the LCC system induced a rapid fluorescence increase (curve d), while a relatively slow and weak fluorescence emission enhancement was obtained in the defective localized system (curves a, b and c, for details, see Fig. S3B and C†), indicating that the constrain of DNA reactants into self-adaptive structures could enhance the overall reaction performance. The higher reaction rate of the LCC system was ascribed to the self-adaptive reactant-to-template design and the accompanying successive localization of reactants, which maintains structural integrity and increases the relative concentration of the self-constraint DNA reactants.
As a powerful single-molecule-detecting technique, FCS allows the measurement of the fluorescence fluctuations caused by the Brownian motion of a single molecule.32,33 Therefore, FCS was further utilized to explore the performance of the LCC system at the single-molecule level. An extremely fast molecular diffusion was observed in the catenated H1/H3-mutant system while comparatively slower signal attenuation emerged in the catenated H1 or H3-mutant system (Fig. 2C). In contrast, the slowest molecular diffusion signal appeared in the intact LCC system, indicating that a substantial amount of polymer nanostructure was achieved to constrain the single molecular diffusion. The characteristic diffusion coefficient (D) assay showed that the H1/H3-mutant LCC system exhibited 14.23-fold faster molecular diffusion than the intact LCC system, 1.25-fold faster fluorescence fluctuation than the mere H1-mutant LCC system, or 3.56-fold higher molecular diffusion than the mere H3-mutant LCC system (Fig. S4†). These single molecular detection results demonstrated that the self-supplementing assembly of the localized catalytic circuit was equipped with faster reaction profiles. The native polyacrylamide gel electrophoresis (PAGE) analysis also confirmed that the miR-21-initiated LCC resulted in the formation of high-molecular-weight products (Fig. 2D). The morphological characterization of the LCC system was performed by atomic force microscopy (AFM). As expected, the crosslinked hydrogel networks with a height of ∼1.5 nm were observed (Fig. 2E), suggesting that the localized catalytic hybridization reaction proceeded as anticipated. Compared to the tiny spots of the LCC system without the promotor, the H1-mutant LCC system generated copolymer dsDNA nanowires (Fig. S5†), further confirming the correctness of our localized catalytic hybridization reaction mechanism. These results indicated that interconnecting catenated DNA reactants with self-confined capability could enhance the reaction rate for guaranteeing effective signal amplification.
Then, we examined whether the promoter-responsive self-adaptive reactant-to-template hybridization reaction could form a macroscopic DNA hydrogel at high concentrations. After overnight incubation, a small amount of promoter (50 nM) could induce the obvious sol–gel transition. Even after a three-day incubation at room temperature, the formation of gelation solution was stable enough without fluidity (Fig. S6†). The rheology measurement was further performed to characterize the mechanical strength (Fig. 2F). Even with 2.0 wt% DNA, the storage modulus (G′) was significantly higher than the loss modulus (G′′), demonstrating the solid properties of the DNA hydrogel sample. These results confirmed that the self-confined LCC system is encoded with self-accelerated characterization and a substantially higher signal amplification performance, thus providing a useful platform in bioanalytical fields.
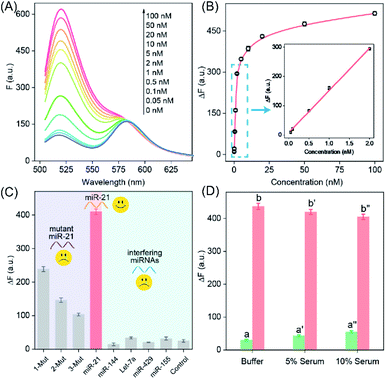
The highly crosslinked self-assembly of the DNA hydrogel and the efficient amplification features of the LCC system encouraged us to further apply this strategy for detecting miRNA (miR-21). Obviously, the fluorescence intensity increased with the gradually elevated concentration of miR-21 (Fig. 3A), indicating that the promoter-responsive LCC system was highly dependent on the dosage of the analyte. The limit of detection (LOD) was acquired to be 17.5 pM according to the conventional 3σ calculation method (Fig. 3B). The selectivity of the LCC system was then evaluated by testing its capacity for discrimination toward these mutant miR-21 sequences. According to Fig. 3C, only intact miR-21 could generate a remarkable fluorescence signal, while the other control counterparts showed a comparably lower fluorescence readout, indicating the remarkable specificity of the present LCC system. In addition, the sensing performance could be maintained even in DMEM with 10% serum (Fig. 3D), suggesting high stability and acceptable accuracy of the present LCC system in complex biological fluids.
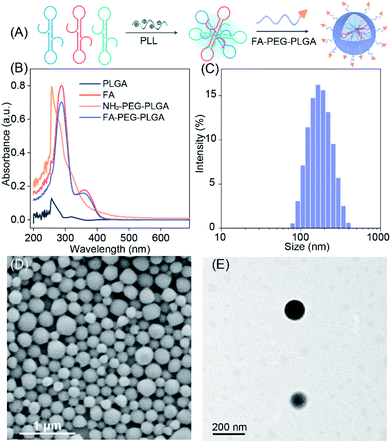
To efficiently deliver the self-templated DNA reactants, the FA coupled PLGA copolymers (FA–PEG–PLGA) were synthesized for encapsulating these LCC reactants (Fig. 4A).34 The synthesis and conjugation of these copolymers were demonstrated by 1H-NMR spectra (Fig. S7 and S8†) and UV-vis absorption spectra (Fig. 4B). The DNA-loaded FA–PEG–PLGA nanovesicles (FA-nanovesicles) were synthesized using the double emulsion-solvent evaporation method.35 The DNA reactants were encapsulated into the nanovesicles by complexing the DNA with cationic poly L-lysine (PLL), and the encapsulation efficiency was determined to be 45%. The average diameter of the as-achieved FA-nanovesicles was 172 nm with a polydispersity index (PDI) of 0.15 (Fig. 4C). The external surface morphology of the FA-nanovesicles was analyzed by scanning electron microscopy (SEM) and transmission electron microscopy (TEM) (Fig. 4D and E, respectively), which showed a spherical shape with a smooth surface. The as-prepared FA-nanovesicles could be redispersed in DMEM even with 10% serum without obvious aggradation (Fig. S9†), suggesting the high stability of our FA-nanovesicles. The in vitro cytotoxicity evaluation and hemolysis assays demonstrated their favorable biocompatibility for further bio-applications without notable side effects (Fig. S10†).
To identify whether these FA-nanovesicles could effectively target specific cancer cells, their cellular uptake was evaluated by confocal laser scanning microscopy (CLSM) and flow cytometry. With different expressions of the folate receptor (FR) on the cell membrane, human breast cancer (MCF-7) and normal MCF-10A cells were used as models to investigate the living-cell imaging.36–39 Noteworthily, here the nanovesicles only contained random FAM-modified DNA to evaluate the FR-mediated endocytosis. As shown in Fig. S11,† FA-nanovesicles exhibited an enhanced fluorescence signal as compared to the unmodified nanovesicles after 3 h of incubation in MCF-7 cells. These enhancement phenomena were abrogated by a free FA competition inhibition assay for validating the specific interaction between FA and the FR. In contrast, CLSM and flow cytometry analysis revealed that the cellular uptake of FA-nanovesicles by MCF-10A cells was not affected by the competition of free FA (Fig. S12†). The specific interaction between FA and the FR was further demonstrated using a competitive inhibition assay in FR-positive human cervical cancer cells (HeLa) (Fig. S13†),40 thus confirming the specifically FA-promoted receptor-mediated endocytosis into FR-positive cells. The endocytosis inhibition experiments in MCF-7 cells showed that the FA-decorated nanovesicle internalization was predominant via the clathrin-mediated endocytosis and micropinocytosis processes (Fig. S14†).
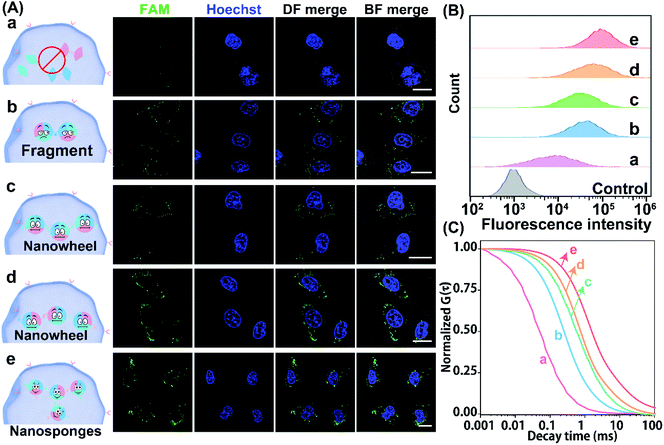
Having demonstrated the reliable receptor-mediated endocytosis, we then explored the LCC system for imaging low-abundance miRNA. Herein, the miR-21-overexpressed MCF-7 cell was chosen as a model to verify the LCC amplifier. At the optimized incubation time (Fig. S15†), a bright fluorescence signal was obtained in MCF-7 cells that were incubated with the intact LCC-packaged FA-nanovesicle (sample e in Fig. 5A), suggesting that the stimulus-responsive catalytic hybridization reaction could proceed in the complicated intracellular environment for the sensitive monitoring of low-abundance miR-21. By contrast, the H1/H3-mutant FA-nanovesicle showed a relatively weaker fluorescence signal (sample b), which was consistent with the quantitative flow cytometry assay (Fig. 5B and S16†). A remarkably enhanced fluorescence signal was observed in the intact LCC amplifier as compared to that in the H1 or H3-mutant LCC amplifier (samples c and d). This enhanced fluorescence signal of the intact LCC system originated from the localized hybridization with increased DNA concentration, thus providing accelerated reaction kinetics. Furthermore, a substantially decreased fluorescence signal was observed in MCF-7 cells that were pre-treated with the anti-miR-21 inhibitor (sample a), confirming that the miR-21 molecule was indeed present in MCF-7 cells for initiating the self-adaptive catalytic assembly of DNA.
Moreover, the accuracy and robustness of the designed LCC system were further evaluated using FCS to differentiate the varied expressions of miR-21 in single MCF-7 cells. As shown in Fig. 5C, the most intense signal attenuation was observed in MCF-7 cells that were treated with the H1/H3-mutant system, followed by the H1 or H3-mutant system, while a minimal diffusion signal was exhibited in MCF-7 cells treated with an intact LCC system, implying that constraining interactions of spatially proximal circuit elements could enhance the reaction kinetics in the complex intracellular environment. As expected, a remarkable diffusion signal appeared in anti-miR-21 inhibitor pre-treated cells, implying that the specific miR-21-activated the LCC system. Quantitative analysis showed that the H1/H3-mutant LCC displayed 2.40- and 31.39-fold higher fluorescence diffusion than the H3-mutant and intact LCC amplifier (Fig. S17†), respectively. The depressed diffusion of fluorescence of the intact LCC system was attributed to the self-adaptive reactant-to-template design and the accompanying successive localization of reactants that accelerated the entire reaction process and constrained the single molecular diffusion. Furthermore, the diffusion coefficient of the miR-21-suppressed LCC amplifier was 45.28-fold higher than that of the non-pretreated LCC system, suggesting that the specific miR-21-activated the LCC amplifier in single live cells. These results further manifested that our LCC-guided cellular assembly of DNA nanosponges could reliably differentiate varied expressions of miR-21.
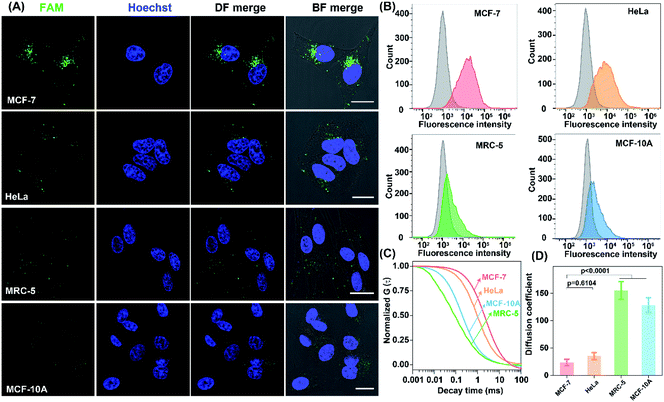
The precise in situ localization of miR-21 in the LCC system encouraged us to implement our system for discriminating different cell types with distinct miR-21 expressions. As shown in Fig. 6A, an extremely intense fluorescence signal was exhibited in MCF-7 cells while a comparatively weaker fluorescence activation emerged in HeLa cells. In contrast, nearly no detectable fluorescence response appeared in normal human lung fibroblast (MRC-5) and MCF-10A cells, implying a relatively higher expression of miR-21 in tumour cells than that in normal cells, which was consistent with previous research.41–45 Meanwhile, the performance of our designed LCC system to distinguish the varied miR-21 expression levels in different cells was further verified by a quantitative flow cytometry assay (Fig. 6B). In addition, the shortest characteristic diffusion time was exhibited in MCF-10A and MRC-5 cells, followed by HeLa cells, while the slowest molecular diffusion signal was shown in MCF-7 cells (Fig. 6C). The characteristic diffusion coefficient assay indicated that the normal MCF-10A cells achieved 6.45-fold higher signal diffusion than the tumour MCF-7 cells, and 4.37-fold higher fluorescence diffusion than the tumour HeLa cells (Fig. 6D). Meanwhile, the noncancerous MRC-5 cells showed 5.42-fold faster fluorescence fluctuation than the MCF-7 cells and 3.62-fold faster molecular diffusion in HeLa cells. These results demonstrated that the designed LCC system could reliably differentiate the varied expression levels of miR-21 in different cells, suggesting a great potential application in single-cell analysis.
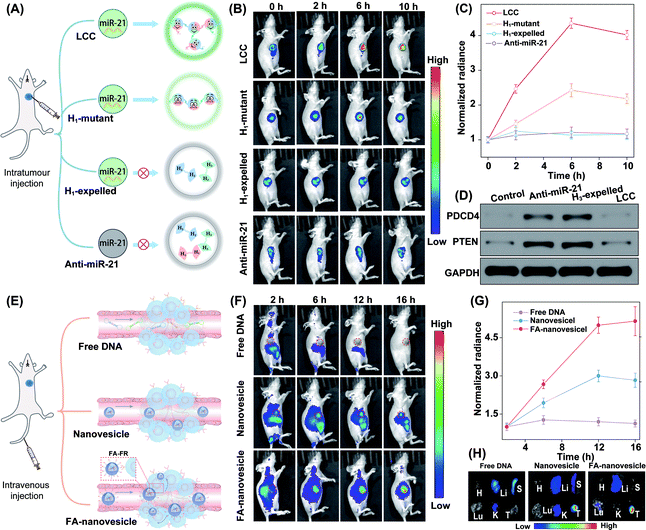
The satisfactory intracellular stimulus-responsive assembly performance of the LCC amplifier was confirmed in vitro, which inspired us to explore a more challenging in vivo application of the intelligent DNA self-assembly. Mice bearing orthotropic MCF-7 breast tumours were chosen as a model to evaluate the stimulus-responsive performance of our LCC system. To achieve a reliable and accurate comparison of the miR-21-guided assembly of DNA nanostructures, these different LCC systems were first intratumourally injected to ensure comparable and constant DNA reactants (Fig. 7A). For the enhanced light penetration ability, H2 was functionalized with a Cy5/quencher pair for in vivo bioimaging. As shown in Fig. 7B, the intact LCC system presented a remarkably enhanced fluorescence signal in the tumour region compared to the H1-mutant system, indicating the localized hybridization characteristics of the LCC that accelerated the entire self-assembly procedure and simultaneously promoted the generation of an enhanced fluorescence signal. As a control, the H1-expelled tumour showed a negligible change in intratumoural fluorescence with prolonged incubation time. Quantitative analysis showed that intact LCC-treated mice exhibited a 1.8-fold higher intratumoural fluorescence signal than those treated with the H1-mutant LCC system at 6 h (Fig. 7C). In addition, nearly no enhanced Cy5 signal was observed in the anti-miR-21-pretreated tumour. These results demonstrated that the miR-21-responsive assembly could indeed proceed in vivo. In our design, the activation of the LCC was based on the displacement of the promoter, in which the content of the initiator would not be influenced as well as the function of the living cells. According to the western blot analysis (Fig. 7D), the designed LCC system showed little effect on the expressions of downstream PDCD4 and PTEN proteins in mice bearing MCF-7 tumours, demonstrating the bioorthogonal assembly of DNA nanosponges in vivo.
In addition, the miR-21-activated LCC-amplified bioorthogonal assembly of DNA nanosponges was further evaluated in mice bearing orthotopic MCF-7 tumours via intravenous administration (Fig. 7E). As shown in Fig. 7F, an intravenously injected free LCC system was quickly eliminated without fluorescence in the tumour site. By contrast, the LCC-packaged nanovesicles could gradually accumulate in the tumour site. Quantitative analysis showed that the FA-nanovesicles exhibited a 1.7-fold higher intratumoural fluorescence signal than bare nanovesicles at 12 h post-injection (Fig. 7G). The enhanced Cy5 signal of FA-nanovesicles in the tumour site was attributed to the FA-favoured cellular uptake that promoted the miR-21-catalytic assembly performance. In addition, the fluorescence signal of harvested tumours and major organs at 16 h after injection was evaluated. The intratumoural Cy5 signal in FA-nanovesicle-treated mice was 1.5-fold higher than that of the bare nanovesicle-treated mouse group (Fig. 7H and S18†). These results significantly demonstrated FA-promoted tumour-addressable characters and miR-21-responsive DNA assembly. Furthermore, hematoxylin and eosin (H&E) staining (Fig. S19†) and haematology assay (Fig. S20†) demonstrated the satisfactory systemic biocompatibility of these functional nanovesicles.
Conclusions
In summary, a stimulus-responsive localized catalytic circuitry system was facilely engineered using the rationally designed interconnecting catenated DNA reactants for self-adaptive catalytic DNA assembly. The autonomous and cascade bioorthogonal assembly was realized for in situ generation of the three-dimensional hyper-branched DNA architectures on the nanoscale. By accurately monitoring the assembly of DNA reactants and products in a self-confined environment, the crucial miRNA biomarker was robustly detected in complex intracellular environments and even under in vivo conditions. The real-time assembly of DNA nanosponges can be monitored by the single-molecule-detecting FCS technique. The key concept of the bioorthogonal regulated circuits is to utilize the sequence/structural symmetry of DNA probes to achieve the catenated geometry conformation for realizing the self-adaptive catalytic acceleration hybridization. The promoter could continue to initiate the self-supplemented acceleration reaction to assemble DNA nanosponges, thus enabling a more reliable in situ localization of miRNAs. The self-adaptive catalytic circuit paves a new way for developing high-performance DNA computing circuits and nanorobots, thus exhibiting promising potential in clinical diagnosis and therapeutic evaluation.Ethical statement
All animal experimental protocols were approved by the Institutional Animal Care and Use Committee of the Animal Experiment Center of Wuhan University (Wuhan, China). Four to six week-old female BALB/c nude mice were purchased from Charles River Company and raised in a specific pathogen-free grade laboratory according to the guidelines for laboratory animals established by the Wuhan University Center for Animal Experiment/A3-Lab. All animals were housed with a 12 h light/dark cycle at 22 °C, 40% relative humidity, and food and water ad libitum.Data availability
All relevant data is presented in the paper and ESI.†Author contributions
X. G. and F. W. conceived and designed the experiments. X. G., S. H., and R. L. performed the main experiments and analysed the data. Y. C. and K. T. synthesized FA-PLGA. Y. W. collected fluorescence images. X. G., X. L. and F. W. wrote the manuscript. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (22074112 and 21874103), the Fundamental Research Funds for the Central Universities (2042022kf1175), and the Central Funds Guiding the Local Science and Technology Development of Shenzhen (2021Szvup101).Notes and references
- M. C. Good, J. G. Zalatan and W. A. Lim, Science, 2011, 332, 680–686 CrossRef CAS.
- C. Agapakis, P. Boyle and P. Silver, Nat. Chem. Biol., 2012, 8, 527–535 CrossRef CAS PubMed.
- Y. Tu, F. Peng, A. Adawy, Y. Men, L. K. Abdelmohsen and D. A. Wilson, Chem. Rev., 2016, 116, 2023–2078 CrossRef CAS PubMed.
- P. K. Mattila and P. Lappalainen, Nat. Rev. Mol. Cell Biol., 2008, 9, 446–454 CrossRef CAS PubMed.
- M. Vazquez-González, C. Wang and I. Willner, Nat. Catal., 2020, 3, 256–273 CrossRef.
- M. Liu, Q. Zhang, D. Chang, J. Gu, J. D. Brennan and Y. Li, Angew. Chem., Int. Ed., 2017, 56, 6142–6146 CrossRef CAS PubMed.
- J. Liu, T. Wu, X. Lu, X. Wu, S. Liu, S. Zhao, X. Xu and B. Ding, J. Am. Chem. Soc., 2019, 141, 19032–19037 CrossRef CAS PubMed.
- B. Liu and J. Liu, Matter, 2019, 1, 825–847 CrossRef.
- S. Bi, S. Yue and S. Zhang, Chem. Soc. Rev., 2017, 46, 4281–4298 RSC.
- X. Gong, J. Wei, J. Liu, R. Li, X. Liu and F. Wang, Chem. Sci., 2019, 10, 2989–2997 RSC.
- J. Wei, H. Wang, X. Gong, Q. Wang, H. Wang, Y. Zhou and F. Wang, Nucleic Acids Res., 2020, 48, e60 CrossRef CAS PubMed.
- Q. Wang, K. Tan, H. Wang, J. Shang, Y. Wan, X. Liu, X. Weng and F. Wang, J. Am. Chem. Soc., 2021, 143, 6895–6904 CrossRef CAS PubMed.
- S. Yue, Y. Li, Z. Qiao, W. Song and S. Bi, Trends Biotechnol., 2021, 39, 1160–1172 CrossRef CAS PubMed.
- M. Oishi and K. Saito, ACS Nano, 2020, 14, 3477–3489 CrossRef CAS PubMed.
- C. Hong, Q. Wang, Y. Chen, Y. Gao, J. Shang, X. Weng, X. Liu and F. Wang, Chem. Sci., 2021, 12, 15339–15346 RSC.
- H. Wang, Y. He, J. Wei, H. Wang, K. Ma, Y. Zhou, X. Liu, X. Zhou and F. Wang, Angew. Chem., Int. Ed., 2022, e202115489 CAS.
- K. Shi, S. Y. Xie, R. Y. Tian, S. Wang, Q. Lu, D. H. Gao, C. Y. Lei, H. Z. Zhu and Z. Nie, Sci. Adv., 2021, 7, eabc7802 CrossRef CAS PubMed.
- G. Chatterjee, N. Dalchau, R. A. Muscat, A. Phillips and G. A. Seelig, Nat. Nanotechnol., 2017, 12, 920–927 CrossRef CAS PubMed.
- H. Bui, S. Shah, R. Mokhtar, T. Song, S. Garg and J. Reif, ACS Nano, 2018, 12, 1146–1155 CrossRef CAS PubMed.
- T. Q. Song, S. Shah, H. Bui, S. Garg, A. Eshra, D. Fu, M. Yang, R. Mokhtar and J. Reif, J. Am. Chem. Soc., 2019, 141, 16539–16543 CrossRef CAS PubMed.
- B. Li, Y. Liu, Y. Liu, T. Tian, B. Yang, X. Huang, J. Liu and B. Liu, ACS Nano, 2020, 14, 8116–8125 CrossRef CAS PubMed.
- H. Peng, X. F. Li, H. Zhang and X. C. Le, Nat. Commun., 2017, 8, 14378 CrossRef PubMed.
- F. Yang, Y. R. Cheng, Y. Cao, Y. Y. Zhang, H. F. Dong, H. T. Lu and X. J. Zhang, Anal. Chem., 2019, 91, 9828–9835 CrossRef CAS PubMed.
- C. Jung, P. B. Allen and A. D. Ellington, ACS Nano, 2017, 11, 8047–8054 CrossRef CAS PubMed.
- W. Engelen, S. P. W. Wijnands and M. Merkx, J. Am. Chem. Soc., 2018, 140, 9758–9767 CrossRef CAS PubMed.
- B. Wang, C. Thachuk, A. D. Ellington, E. Winfree and D. Soloveichik, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E12182–E12191 CrossRef CAS PubMed.
- F. Wang, C. H. Lu and I. Willner, Chem. Rev., 2014, 114, 2881–2941 CrossRef CAS PubMed.
- Z. Zhu, R. Wu and B. Li, Chem. Sci., 2019, 10, 1953–1961 RSC.
- L. Qi, M. Yang, D. Chang, W. Zhao, S. Zhang, Y. Du and Y. Li, Angew. Chem., Int. Ed., 2021, 60, 24823–24827 CrossRef CAS PubMed.
- M. Xiao, W. Lai, H. Yu, Z. Yu, L. Li, C. Fan and H. Pei, J. Am. Chem. Soc., 2021, 143, 3448–3454 CrossRef CAS PubMed.
- T. Song, A. Eshra, S. Shah, H. Bui, D. Fu, M. Yang, R. Mokhtar and J. Reif, Nat. Nanotechnol., 2019, 14, 1075–1081 CrossRef CAS PubMed.
- S. A. Kim, K. G. Heinze and P. Schwille, Nat. Methods, 2007, 4, 963–973 CrossRef CAS PubMed.
- H. Liu, C. Dong and J. Ren, J. Am. Chem. Soc., 2014, 136, 2775–2785 CrossRef CAS PubMed.
- S. Jain, V. V. Rathi, A. K. Jain, M. Das and C. Godugu, Nanomedicine, 2012, 7, 1311–1337 CrossRef CAS PubMed.
- X. Gong, R. Li, J. Wang, J. Wei, K. Ma, X. Liu and F. Wang, Angew. Chem., Int. Ed., 2020, 59, 21648–21655 CrossRef CAS PubMed.
- J. Feng, Z. Xu, F. Liu, Y. Zhao, W. Yu, M. Pan, F. Wang and X. Liu, ACS Nano, 2018, 12, 12888–12901 CrossRef CAS PubMed.
- Y. Shao, J. Zhao, J. Yuan, Y. Zhao and L. Li, Angew. Chem., Int. Ed., 2021, 60, 8923–8931 CrossRef CAS.
- Y. Chen, X. Gong, Y. Gao, Y. Shang, J. Shang, S. Yu, R. Li, S. He, X. Liu and F. Wang, Chem. Sci., 2021, 12, 15710–15718 RSC.
- J. Wei, H. Wang, Q. Wu, X. Gong, K. Ma, X. Liu and F. Wang, Angew. Chem., Int. Ed., 2020, 59, 5965–5971 CrossRef CAS PubMed.
- W. Zhang, Z. L. Yu, M. Wu, J. G. Ren, H. F. Xia, G. L. Sa, J. Y. Zhu, D. W. Pang, Y. F. Zhao and G. Chen, ACS Nano, 2017, 11, 277–290 CrossRef CAS PubMed.
- J. Wei, X. Gong, Q. Wang, M. Pan, X. Liu, J. Liu, F. Xia and F. Wang, Chem. Sci., 2018, 9, 52–61 RSC.
- R. Deng, K. Zhang and J. Li, Acc. Chem. Res., 2017, 50, 1059–1068 CrossRef CAS.
- J. Wang, S. Yu, Q. Wu, X. Gong, S. He, J. Shang, X. Liu and F. Wang, Angew. Chem., Int. Ed., 2021, 60, 10766–10744 CrossRef CAS PubMed.
- A. F. J. Jou, Y. T. Chou, I. Willner and J. A. A. Ho, Angew. Chem., Int. Ed., 2021, 133, 21841–21846 CrossRef.
- Y. Wan, G. Li, L. Zou, H. Wang, Q. Wang, K. Tan, X. Liu and F. Wang, Anal. Chem., 2021, 93, 11052–11059 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedure, DNA sequences, AFM characterization studies, cell imaging, flow cytometric analysis, and additional figures, schemes and tables. See https://doi.org/10.1039/d2sc03757b |
| ‡ X. Gong, S. He and R. Li contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |