Open Access Article
Open Access ArticleDiscovery of pentaene polyols by the activation of an enediyne gene cluster: biosynthetic implications for 9-membered enediyne core structures†
Jian
Pan
 a,
Qingwen
Tan
a,
Saibin
Zhu
a,
Xiaohui
Yan
d,
Yu
Li
a,
Zhoukang
Zhuang
a,
Xiangcheng
Zhu
ac,
Yanwen
Duan
abc and
Yong
Huang
a,
Qingwen
Tan
a,
Saibin
Zhu
a,
Xiaohui
Yan
d,
Yu
Li
a,
Zhoukang
Zhuang
a,
Xiangcheng
Zhu
ac,
Yanwen
Duan
abc and
Yong
Huang
 *ab
*ab
aXiangya International Academy of Translational Medicine, Central South University, Changsha, Hunan 410013, China. E-mail: jonghuang@csu.edu.cn
bHunan Engineering Research Center of Combinatorial Biosynthesis and Natural Product Drug Discovery, Changsha, Hunan 410205, China
cNational Engineering Research Center of Combinatorial Biosynthesis for Drug Discovery, Changsha, Hunan 410205, China
dState Key Laboratory of Natural and Biomimetic Drugs, Peking University, Beijing, China, State Key Laboratory of Component-Based Chinese Medicine, Tianjin University of Traditional Chinese Medicine, Tianjin, China
First published on 31st October 2022
Abstract
The identification and characterization of enediyne polyketide synthases (PKSEs) revealed that PKSE-bound polyene is a common intermediate, while its subsequent tailoring steps to enediyne cores remain obscure. Herein, we report pentaene polyols 5–7 and cinnamic acid derivatives 8 and 9 biosynthesized from an activated enediyne biosynthetic gene cluster in Streptomyces sp. CB02130. The C-1027 pksE could partially complement production of these polyene polyols in a CB02130 mutant where the native pksE is inactivated. The yields of 5–7 were improved by increasing the cellular pool of L-Phe through either gene inactivation of a prephenate dehydrogenase WlsPDH or supplementation of L-Phe. A flexible ammonia lyase WlsC4 is responsible for biosynthesis of 8 and 9 from L-Phe. The co-localization of wlsPDH and PKSE gene cassette supports their close evolutionary relationships and an enediyne genome mining strategy using WlsPDH. These findings not only provide a facile approach to activate silent enediyne BGCs, but suggest that a polyene epoxide intermediate may be formed for construction of 9-membered enediyne macrocycles.
Introduction
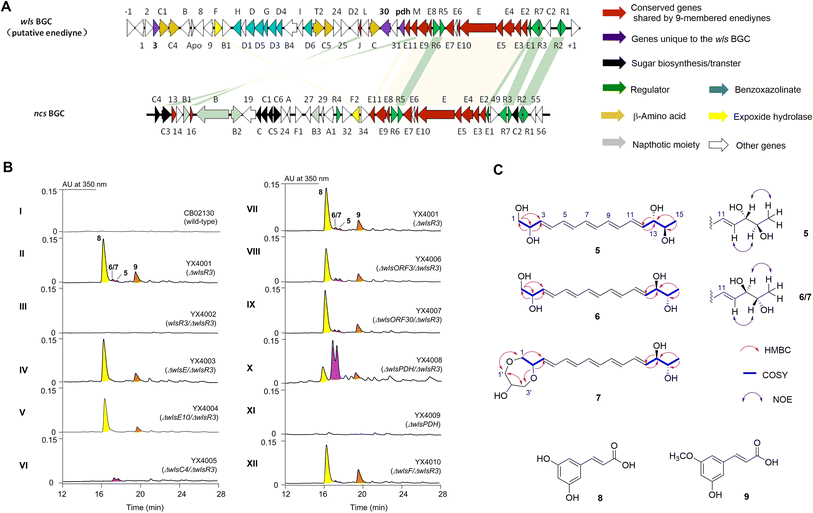
Enediyne natural products are characterized by the unique conjugation of two acetylenic groups to a double bond within a 9- or 10-membered carbocycle (Fig. 1 and S1†).1–4 The last two decades have witnessed deciphering the unique biosynthetic logic of enediyne natural products, including the identification of their biosynthetic gene clusters (BGCs),5–12 the discovery and characterization of highly reducing iterative polyketide synthases (PKSEs) for enediyne assembly,13–20 as well as novel enzymology to construct peripheral moieties into enediyne architectures.21–26 However, the biosynthesis of enediyne cores has been a topic of great interest and remains largely a mystery. The carbon skeleton of enediyne cores is formed from iterative condensation of malonyl-CoA catalyzed by PKSEs, leading to the isolation of several polyenes 1–4,13–16,18–20 among which β-hydroxy acid 4 or its PKSE-bound analog was considered as the common intermediate for 9- or 10-enediyne core biosynthesis.13,16,20 However, further tailoring steps for enediyne construction and how the 9- or 10-membered rings diverge remain unknown. Some underlying reasons may include that (1) the functions of many conserved genes for enediyne biosynthesis are unknown and (2) only a few metabolites, e.g.1–4, related to enediyne core processing were discovered.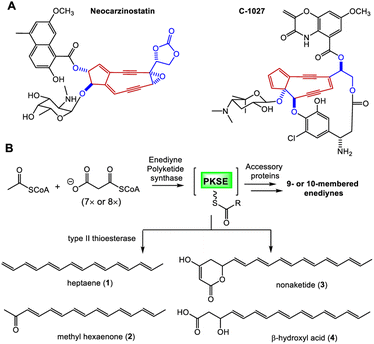 | ||
| Fig. 1 Proposed biosynthesis of enediyne cores. (A) Representative 9-membered enediynes. (B) Biosynthesis of PKSE-catalyzed formation of polyenes from acetate. | ||
We and our colleagues recently reported PKSE gene cassette-based genome mining approaches to discover enediyne producers, leading to the isolation of new anthraquinone-fused enediynes and several high-yield producers of C-1027.27–30 These studies not only show wide-spread enediyne BGCs and their huge biosynthetic potential, but highlight significant challenges to activate silent enediyne BGCs and harness novel chemistry, including intermediate steps towards enediyne core biosynthesis.
Here, we report the activation of a putative Wuliangshan (wls) enediyne BGC in Streptomyces sp. CB02130 and the discovery of three novel pentaene polyols 5–7 and cinnamic acid derivatives 8 and 9. Their biosynthetic study suggests that 5–7 are likely shunt metabolites during the construction of 9-membered enediynes, which support epoxidation of the PKSE-tethered polyene intermediate during the biosynthesis of 9-membered enediynes.
Results and discussion
Bioinformatics analysis revealed a subclass of putative enediyne BGCs
Bioinformatic analysis revealed a sub-cluster of putative enediyne BGCs with a conserved regulatory network with neocarzinostatin (Fig. 2 and S2†).7S. sp. CB02130 was prioritized as a prototype for this sub-family since it could produce the heptaene metabolite 1, while no enediyne natural products could be identified. Heterologous expression of WlsPKSE and a type II thioesterase WlsE10 in Escherichia coli also accumulated 1, which further supports the operation of previously reported enediyne PKS chemistry in CB02130 (Fig. S3 and Table S2†).Discovery of 5–9 from an activated wls BGC
Since manipulation of pathway-situated regulators could improve the yield of C-1027,32,33 we systematically inactivated putative negative regulators in wls BGC by gene replacement of wlsR2 and wlsR3 using a thiostrepton-resistance gene. An extra copy of putative positive regulators, including wlsR1, wlsR5, wlsR6, and wlsR7, was also individually introduced into CB02130 through site-specific integration (Fig. S4†).34 When we inactivated wlsR3 encoding a TetR regulator, a series of metabolites 5–9 were produced, while ΔwlsR2 and mutants overexpressing wlsR1, wlsR5, wlsR6, and wlsR7 had no obvious metabolite overproduction (Fig. 2B, panel II; S5, S9†). Introduction of wlsR3 to ΔwlsR3 mutant restored its phenotype to the wild-type strain (Fig. 2B, panel III). Interestingly, deletion of wlsE and wlsE10 in ΔwlsR3 only abolished the production of 5–7. Inactivation of wlsC4 encoding a 4-methylideneimidazole-5-one-dependent ammonia lyase abolished the production of 8 and 9 (Fig. 2B, panels VI). The above data suggest the activation of wls BGC in ΔwlsR3.Compounds 5–7 are likely related to enediyne core biosynthesis, but the low titer hindered their isolation. Further analysis of wls BGC revealed two potential chorismate-related pathways: WlsD and WlsG share high sequence identity with SgcD and SgcG for the biosynthesis of benzoxazolinate from chorismate in C-1027,31 while wlsOrf3 and wlsPDH encode putative chorismatase and prephenate dehydrogenase (PDH), respectively. WlsOrf3 and wlsPDH, along with wlsOrf30 predicted to encode a 2-hydroxyacyl-CoA lyase, are not present in the known enediyne BGCs (Fig. S2†). We therefore deleted wlsOrf3, wlsOrf30, and wlsPDH in ΔwlsR3, respectively, to investigate their roles in 5–9 biosynthesis (Fig. 2B, panels VII–X). No new metabolites and obvious yield change of 5–9 were observed in ΔwlsORF3/ΔwlsR3 and ΔwlsORF30/ΔwlsR3; in contrast, the yields of 5–7 increased by about 8-fold in ΔwlsPDH/ΔwlsR3, while gene replacement of wlsPDH in CB02130 wild-type had no obvious changes (Fig. 2B, panel XI). Gene inactivation of wlsR3 or wlsR3 and wlsPDH also led to significant overproduction of heptaene 1, while further overexpression of putative positive regulators wlsR1 and wlsR6 under the control of ermEp* in ΔwlsR3 had no effects towards the production of 1 and 5–7 (Fig. S14 and S15†).
Scaled-up fermentation of ΔwlsR3 or ΔwlsPDH/ΔwlsR3 mutants thus led to the isolation of 8 and 9, or 5–7, respectively. Compound 5 was isolated as a colorless powder that was analyzed for the molecular formula C15H22NaO4, by HR-ESI-MS (m/z 289.1403 [M + Na]+, calcd 289.1416) (Fig. S6†). It contains five degrees of unsaturation, consistent with a conjugated pentaene structure with characteristic UV/Vis absorption at λmax 312.2, 330.2, and 348.2 nm. Analyses of 1D and 2D NMR spectra of 5 revealed that it contains one terminal methyl as a doublet at λ 1.1 (J = 6.6 Hz, H15) and one terminal hydroxylmethyl at λ 3.49 (2H, m, H1), three oxymethine protons [3.62 (1H, t, J = 6.6), H14; 3.90 (1H, m), H13; 4.17 (1H, m), H2], two trans olefinic protons [5.74 (1H, dd, J = 15, 3.0 Hz), H12; 5.73 (1H, dd, J = 15, 2.4 Hz), H3], and eight overlapping olefinic protons at λ 6.27–6.33 (m, H5–H10) (Table S4 and Fig. S16–S26†). HSQC and HMBC experiments showed that these proton signals correlated with their corresponding 13C NMR bands and established their connections. The geometry of the remaining three double bonds is likely in the E configuration, since (1) 5 is the iterative acting PKSE product and (2) the overlapping signals of H5–H10 would suggest a pseudo-centrosymmetric structure, similar to all trans configurations of 1–4. Therefore, the planar structure of 5 was identified as (3E, 5E, 7E, 9E, 11E)-pentadeca-3, 5, 7, 9, 11-pentaene-1, 2, 13, 14-tetraol.
Compound 6 was likely a diastereoisomer of 5, since both compounds had identical HR-ESI-MS and similar NMR data (Table S4 and Fig. S27–S39†). Compound 7 was isolated as a colorless powder with an almost identical UV/Vis spectrum to 5, suggesting a similar molecular structure. The difference of 7 is that it contains two extra hydroxylmethyl groups [3.59 (2H, br m), H1′; 3.48 (2H, m), H3′] and one oxymethine proton at λ 3.65 (1H, m, H2′), corresponding to a three-carbon glycerol unit (Fig. S40–S53†). HMBC correlations from H-1 to C-1′, H-2 to C-3′, as well as H-1′ to C-3′ and H-3′ to C-1′ established the connection of the glycerol unit to the pentaene skeleton of 7.
The 13,14-diol in 5/6/7 was assigned with an anti-configuration, due to the relatively large 3JH,H constants (6.6 Hz) and the presence of NOE between H-15/H-13 and H-12/H-14 and the absence of NOE between H-12/H-15.39 The characteristic electronic circular dichroism of 5 and 6 also suggested that they are diastereoisomers (Fig. S7†). However, 5 had no optical rotation when measured in methanol, suggesting a pseudo mesomeric structure. Several highly symmetric polyene polyols with a tetraene unit flanked by diols were isolated, while their optical rotation ranged from −2 to −15.40 In contrast, 6 exhibits an optical rotation of [α]25D +149 (c 0.04, MeOH) and less signal overlapping in both its 1H and 13C spectra, indicating rather different stereochemistry. Taken together, the relative stereochemistry of 5–7 is shown in Fig. 2B. The structures of 8 and 9 were determined by their corresponding 1D and 2D NMR and HR-ESI-MS, which are the known cinnamic acid derivatives (Fig. S6 and S54–S61†).
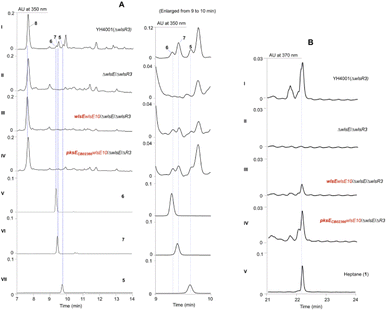
Since PKSE genes belonging to 9-membered enediynes could cross-complement C-1027 or neocarzinostatin production when the native pksE gene was inactivated,13 we therefore overexpressed wlsE10 and pksE for C-1027 biosynthesis from Streptomyces sp. CB02366 in the ΔwlsE/ΔwlsR3 mutant (Fig. 3).30 Both mutants overexpressing pksECB02366 and wlsE could produce 1 and partially restored the production of 5–7 in the wlsE null mutant, suggesting the operation of enediyne PKS chemistry and interaction of C-1027 PKSE with accessory enzymes in CB02130.
Since the 1, 2-diol functionality is present in many known 9-membered enediyne natural products, epoxide hydrolases including SgcF and NcsF2 from C-1027 or neocarzinostatin BGCs were proposed for the hydrolysis of epoxide intermediates.35–37 To study the role of WlsF, a close homolog of SgcF and NcsF2 for the formation of vicinal diols in 5–7, it was inactivated in ΔwlsR3. The production of 5–7 was unchanged in the resulting mutant (Fig. 2B, panel XII). We next inactivated wlsORF8 and wlsORF24 encoding putative monooxygenases with high sequence similarity to SpoT4 and SpoORF18 in sporolides BGC,10 as well as putative oxidoreductase genes wlsL and wlsE9, and cytochrome P450 monooxygenase gene wlsE7 in the ΔwlsR3 mutant, while the production of 5–7 seemed unchanged in the resulting mutant strains ΔwlsORF8/ΔwlsR3, ΔwlsORF24/ΔwlsR3, ΔwlsL/ΔwlsR3, ΔwlsE7/ΔwlsR3, and ΔwlsE9/ΔwlsR3 (Fig. S13†).
Characterization of WlsC4 as an amino lysase
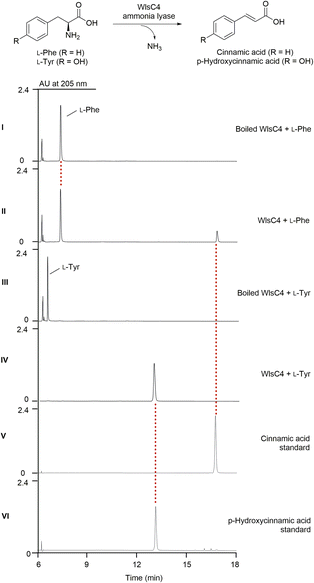
Both 8 and 9 disappeared when wlsC4 was inactivated in ΔwlsR3 (Fig. 2B, panel VI). This suggests that 9 was likely a product of WlsC4, while further O-methylation of 8 may yield 9. Bioinformatics analysis showed that WlsC4 shares high sequence similarity to several L-Tyr or L-Phe ammonia mutases or lyases, including SgcC4 (80.45% amino acid sequence identity) known as amino mutase that converts L-Tyr into (S)-β-tyrosine.41–43 However, in vitro assay confirmed that purified WlsC4 from heterologous expression in E. coli utilized both L-Tyr or L-Phe as substrates to produce 4-hydroxyl cinnamic acid or cinnamic acid, while it prefers L-Tyr (Fig. 4). To decipher whether L-Tyr or L-Phe was the substrate for WlsC4 in vivo, both compounds were individually added to the production medium of the ΔwlsR3 mutant as a sole nitrogen source (Fig. S11†). Surprisingly, 5–9 could only be produced with the exogenous L-Phe, while the production of heptaene 1 was comparable when either L-Tyr or L-Phe was used. These data suggest that L-Phe may be the genuine substrate for biosynthesis of cinnamic acid derivatives 8 and 9, while the cellular concentration of L-Phe is important to regulate the production of pentaene polyols 5–7. In addition, supplementation of L-Phe (6 mM) to the original production medium significantly increased the yields of 5–7 in ΔwlsR3, even comparable to those in ΔwlsPDH/ΔwlsR3 (Fig. S12†). WlsPDH shares 30.69% sequence identity with characterized prephenate dehydrogenase MtPDH from Mycobacterium tuberculosis H37Rv and it can oxidatively decarboxylate prephenate derived from chorismate to 4-hydroxyphenylpyruvate, followed by its conversion into L-Tyr by tyrosine aminotransferase. Gene inactivation of wlsPDH likely increased the cellular L-Phe concentration by removing the branch point to L-Tyr from chorismate, since it can oxidatively decarboxylate prephenate derived from chorismate, which thus led to enhanced pentaene polyol production (Fig. 2B, panel X).In silico genome mining of enediyne BGCs containing wlsPDH homologs
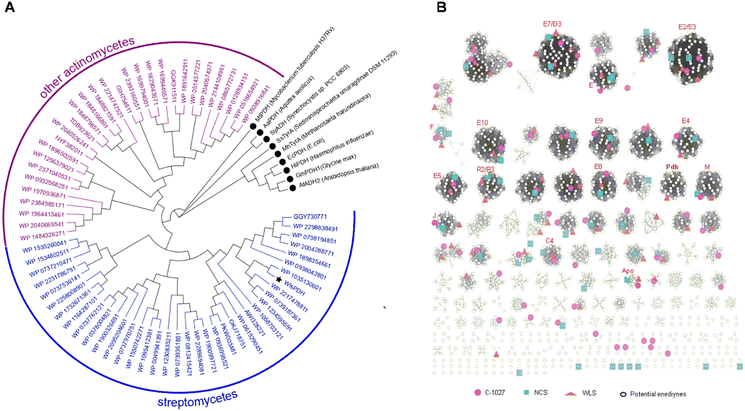
We next studied the distribution of WlsPDH homologs in GenBank and its evolutionary relationship with enediyne BGCs (Fig. 5). Using WlsPDH as an in silico probe, a GenBank BlastP search resulted in 67 homologous PDH proteins clustered with putative enediyne BGCs (Table S5†). Phylogenetic analysis revealed that WlsPDH and homologous proteins are from actinomycetes, and they are clustered in one clade clearly separated from the known PDHs or arogenate dehydrogenases. These 67 putative enediyne BGCs were then winnowed into 31 representative BGCs by a PKSE similarity threshold of 93.86%, which resulted in an enediyne genome neighborhood network (Tables S6–S36†). The newly discovered enediyne BGCs likely all belong to the 9-membered enediyne family, since they contain the 9-membered enediyne conserved genes, including E2, E7–E9, E11, as well as M and J, while all of them contain one PDH.Proposed pathway for putative enediyne natural product biosynthesis in wls BGC
Gene inactivation of wlsR3 led to activation of wls BGC in S. sp. CB02130 and the production of polyene polyol 5–7, suggesting a common polyene epoxide precursor for the 9-membered enediyne core (Fig. 6): a hydrogen atom may be abstracted from the terminal methyl group of the polyene intermediate 10 to yield resonance and generate 11,19,20,38 followed by epoxidation of C15–C16 to produce 12 and then ring closure of C4–C15 in a site-specific fashion and cleavage of oxygenated C16. In contrast, quenching of the allylic radical 12 by a hydroxyl radical, followed by nucleophilic addition of H2O or glycerol to its terminal epoxide and hydrolysis mediated by WlsE10 and subsequent decarboxylation would afford 5–7. Compared to 1–4, the production of these pentaene polyols suggests accessory enzyme-mediated oxidation for 9-membered enediyne core biosynthesis. Alternative mechanisms for the construction of the enediyne core without the epoxide formation, including carbocation or electrocyclic chemistry, may also be possible. These data support the proposal that 9- and 10-membered enediyne biosynthesis diverges in post-PKS steps.17 We also envision the converged biosynthesis of a putative 9-membered enediyne natural product from the enediyne core and a cinnamic acid derivative via chorismate metabolism in CB02130. In addition, the co-localization of homologous PDHs in these 31 BGCs suggests it as a convenient probe to discover potential enediyne BGCs, while their role in coordinating chorismate metabolism and enediyne biosynthesis needs further study.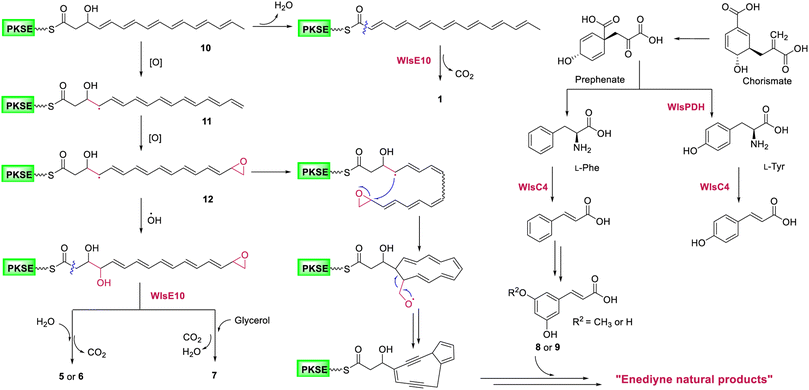 | ||
| Fig. 6 A proposed biosynthetic mechanism for potential 9-membered enediyne natural products in S. sp. CB02130. | ||
Conclusion
Biosynthetic study of enediyne natural products continuingly provides new natural products and new enzymology.3,4,22 Here, we have reported the isolation and structure elucidation of 5–9 from the activated wls enediyne BGC by gene inactivation of a negative transcriptional regulator wlsR3. The correlation of the polyene polyols 5–7 with enediyne core biosynthesis was established by genetic complementation of a wlsE/wlsR3 mutant using PKSE from the C-1027 biosynthetic pathway. We further discovered that an ammonia lyase WlsC4 is involved in the formation of 8 and 9, and proposed that WlsPDH could be used as an in silico probe to discover enediyne BGCs. Although we were unable to obtain genuine enediynes from S. sp. CB02130, the isolation and initial biosynthetic studies of 5–9 is one step forward to understanding how nature builds 9-membered enediyne chromophores, especially in this specific sub-class.Data availability
Experimental procedures and characterization for all new compounds are available in the ESI.†Author contributions
Conceptualization, Y. H., Y. D. and J. P.; methodology, J. P., Q. T., S. Z., and Y. L; formal analysis, J. P., Q. T., and Y. H.; resources, X. Y., Z. Z., X. Z.; writing – original draft, Y. H. and J. P.; writing – review and editing, Y. H. and J. P. with input from all authors. Funding acquisition, Y. H., Y. D., J. P., X. Y.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in parts by NSFC grants 82173688 (Y. H.) and 81872779 (X. Y.), the Science and Technology Innovation Program of Hunan Province (2021RC4067), and Hunan Provincial Natural Science Foundation of China (2021JJ30791) (Y. H.), the Chinese Ministry of Education 111 Project (BP0820034) (to Y. D.), the State Key Laboratory of Natural and Biomimetic Drugs (No. K202116 to X. Y.), the Hunan Provincial Innovation Foundation for Postgraduate and the Fundamental Research Funds for the Central Universities of Central South University (CSU) 2020zzts247 (to J. P.). We thank the Center for Advanced Research in CSU for NMR experiments.References
- K. C. Nicolaou, A. L. Smith and E. W. Yue, Proc. Natl. Acad. Sci., 1993, 90, 5881–5888 CrossRef CAS PubMed.
- U. Galm, M. H. Hager, S. G. Van Lanen, J. Ju, J. S. Thorson and B. Shen, Chem. Biol., 2005, 105, 739–758 CAS.
- B. Shen, Hindra, X. Yan, T. Huang, H. Ge, D. Yang, Q. Teng, J. D. Rudolf and J. R. Lohman, Bioorg. Med. Chem. Lett., 2015, 25, 9–15 CrossRef CAS PubMed.
- X. Yan, Nat. Prod. Rep., 2022, 39, 703–728 RSC.
- W. Liu, S. D. Christenson, S. Standage and B. Shen, Science, 2002, 297, 1170–1173 CrossRef CAS.
- J. Ahlert, E. Shepard, N. Lomovskaya, E. Zazopoulos, A. Staffa, B. O. Bachmann, K. Huang, L. Fonstein, A. Czisny, R. E. Whitwam, C. M. Farnet and J. S. Thorson, Science, 2002, 297, 1173–1176 CrossRef CAS.
- W. Liu, K. Nonaka, L. Nie, J. Zhang, S. D. Christenson, J. Bae, S. G. Van Lanen, E. Zazopoulos, C. M. Farnet, C. F. Yang and B. Shen, Chem. Biol., 2005, 12, 293–302 CrossRef CAS PubMed.
- S. G. Van Lanen, T. J. Oh, W. Liu, E. Wendt-Pienkowski and B. Shen, J. Am. Chem. Soc., 2007, 129, 13082–13094 CrossRef CAS PubMed.
- Q. Gao and J. S. Thorson, FEMS Microbiol. Lett., 2008, 282, 105–114 CrossRef CAS PubMed.
- A. L. Lane, S. J. Nam, T. Fukuda, K. Yamanaka, C. A. Kauffman, P. R. Jensen, W. Fenical and B. S. Moore, J. Am. Chem. Soc., 2013, 135, 4171–4174 CrossRef CAS PubMed.
- S. Y. Ma, Y. S. Xiao, B. Zhang, F. L. Shao, Z. K. Guo, J. J. Zhang, R. H. Jiao, Y. Sun, Q. Xu and R. X. Tan, Org. Lett., 2017, 19, 6208–6211 CrossRef CAS.
- J. R. Lohman, S. X. Huang, G. P. Horsman, P. E. Dilfer, T. Huang, Y. Chen, E. Wendt-Pienkowski and B. Shen, Mol. Biosyst., 2013, 9, 478–491 RSC.
- J. Zhang, S. G. Van Lanen, J. Ju, W. Liu, P. C. Dorrestein, W. Li, N. L. Kelleher and B. Shen, Proc. Natl. Acad. Sci., 2008, 105, 1460–1465 CrossRef CAS.
- R. Kong, L. P. Goh, C. W. Liew, Q. S. Ho, E. Murugan, B. Li, K. Tang and Z. X. Liang, J. Am. Chem. Soc., 2008, 130, 8142–8143 CrossRef CAS PubMed.
- H. Sun, R. Kong, D. Zhu, M. Lu, Q. Ji, C. W. Liew, J. Lescar, G. Zhong and Z. X. Liang, Chem. Commun., 2009, 7399–7401 RSC.
- K. Belecki, J. M. Crawford and C. A. Townsend, J. Am. Chem. Soc., 2009, 131, 12564–12566 CrossRef CAS.
- G. P. Horsman, Y. Chen, J. S. Thorson and B. Shen, Proc. Natl. Acad. Sci., 2010, 107, 11331–11335 CrossRef CAS PubMed.
- X. Chen, Z. F. Guo, P. M. Lai, K. H. Sze and Z. Guo, Angew Chem. Int. Ed., 2010, 49, 7926–7928 CrossRef CAS.
- K. Belecki and C. A. Townsend, Angew. Chem., Int. Ed., 2012, 51, 11316–11319 CrossRef CAS PubMed.
- K. Belecki and C. A. Townsend, J. Am. Chem. Soc., 2013, 135, 14339–14348 CrossRef CAS.
- S. G. Van Lanen and B. Shen, Curr. Top. Med. Chem., 2008, 8, 448–459 CrossRef CAS.
- Z. X. Liang, Nat. Prod. Rep., 2010, 27, 499–528 RSC.
- X. Yan, J. J. Chen, A. Adhikari, C. N. Teijaro, H. Ge, I. Crnovcic, C. Y. Chang, T. Annaval, D. Yang, C. Rader and B. Shen, Org. Lett., 2018, 20, 5918–5921 CrossRef CAS.
- D. R. Cohen and C. A. Townsend, Nat. Chem., 2018, 10, 231–236 CrossRef CAS.
- D. R. Cohen and C. A. Townsend, Angew. Chem. Int. Ed., 2018, 57, 5650–5654 CrossRef CAS.
- G. L. Ma, H. T. Tran, Z. J. Low, H. Candra, L. M. Pang, Q. W. Cheang, M. Fang and Z. X. Liang, J. Am. Chem. Soc., 2021, 143, 11500–11509 CrossRef CAS.
- X. Yan, H. Ge, T. Huang, Hindra, D. Yang, Q. Teng, I. Crnovčić, X. Li, J. D. Rudolf, J. R. Lohman, Y. Gansemans, X. Zhu, Y. Huang, L. X. Zhao, Y. Jiang, F. Van Nieuwerburgh, C. Rader, Y. Duan and B. Shen, mBio, 2016, 7, e02104–e02116 CrossRef CAS.
- X. Yan, J. J. Adhikari A, D. Yang, I. Crnovcic, N. Wang, C. Y. Chang, C. Rader and S. B. Chen, Org. Lett., 2017, 19, 6192–6195 CrossRef CAS PubMed.
- Z. Wang, Z. Wen, L. Liu, X. Zhu, B. Shen, X. Yan, Y. Duan and Y. Huang, J. Nat. Prod., 2019, 82, 2483–2488 CrossRef CAS PubMed.
- X. Yan, Hindra, H. Ge, D. Yang, T. Huang, I. Crnovcic, C. Y. Chang, S. M. Fang, T. Annaval, X. Zhu, Y. Huang, L. X. Zhao, Y. Jiang, Y. Duan and B. Shen, J. Nat. Prod., 2018, 81, 594–599 CrossRef CAS PubMed.
- S. G. Van Lanen, S. Lin and B. Shen, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 494–499 CrossRef CAS PubMed.
- Y. Chen, M. Yin, G. P. Horsman and B. Shen, J. Nat. Prod., 2011, 74, 420–424 CrossRef CAS PubMed.
- Y. Chen, M. Yin, G. P. Horsman, S. Huang and B. Shen, J. Antibiot., 2010, 63, 482–485 CrossRef CAS.
- M. Bierman, R. Logan, K. O'Brien, E. T. Seno, R. N. Rao and B. E. Schoner, Gene, 1992, 116, 43–49 CrossRef CAS.
- S. Lin, G. P. Horsman, Y. Chen, W. Li and B. Shen, J. Am. Chem. Soc., 2009, 131, 16410–16417 CrossRef CAS PubMed.
- S. Lin, G. P. Horsman and B. Shen, Org. Lett., 2010, 12, 3816–3819 CrossRef CAS PubMed.
- G. P. Horsman, A. Lechner, Y. Ohnishi, B. S. Moore and B. Shen, Biochemistry, 2013, 52, 5217–5224 CrossRef CAS PubMed.
- H. M. Ge, T. Huang, J. D. Rudolf, J. R. Lohman, S. X. Huang, X. Guo and B. Shen, Org. Lett., 2014, 16, 3958–3961 CrossRef CAS PubMed.
- N. Matsumori, D. Kaneno, M. Murata, H. Nakamura and K. Tachibana, J. Org. Chem., 1999, 64, 866–876 CrossRef CAS PubMed.
- M. Bae, H. Kim, Y. Shin, B. Y. Kim, S. K. Lee, K. B. Oh, J. Shin and D. C. Oh, Mar. drugs, 2013, 11, 2882–2893 CrossRef PubMed.
- L. Xiang and B. S. Moore, J. Biol. Chem., 2002, 277, 32505–32509 CrossRef CAS PubMed.
- S. D. Christenson, W. Liu, M. D. Toney and B. Shen, J. Am. Chem. Soc., 2003, 125, 6062–6063 CrossRef CAS PubMed.
- S. D. Christenson, W. Wu, M. A. Spies, B. Shen and M. D. Toney, Biochemistry, 2003, 42, 12708–12718 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Full experimental details and procedures, Fig. S1–S61; Tables S1–S36. See DOI: https://doi.org/10.1039/d2sc04379c |
| This journal is © The Royal Society of Chemistry 2022 |