Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Correction: Ultrasensitive recognition of AP sites in DNA at the single-cell level: one molecular rotor sequentially self-regulated to form multiple different stable conformations
Beidou
Feng
a,
Kui
Wang
a,
Yonggang
Yang
a,
Ge
Wang
b,
Hua
Zhang
*a,
Yufang
Liu
a and
Kai
Jiang
a
aHenan Key Laboratory of Green Chemical Media and Reactions, Ministry of Education, Key Laboratory of Green Chemical Media and Reactions, Collaborative Innovation Center of Henan Province for Green Manufacturing of Fine Chemicals, Henan Key Laboratory of Organic Functional Molecules and Drug Innovation, School of Chemistry and Chemical Engineering, School of Environment, College of Physics and Materials Science, Henan Normal University, Xinxiang 453007, China. E-mail: zhanghua1106@163.com
bXinxiang Medical University, Xinxiang 453000, P. R. China
First published on 29th April 2022
Abstract
Correction for ‘Ultrasensitive recognition of AP sites in DNA at the single-cell level: one molecular rotor sequentially self-regulated to form multiple different stable conformations’ by Beidou Feng et al., Chem. Sci., 2019, 10, 10373–10380, DOI: 10.1039/C9SC04140K.
The authors regret that the descriptions of the DNA sequences in the original legend of Fig. 2 were inappropriate. The correct descriptions in the legend of Fig. 2 are presented below, and the detailed DNA sequences were added in page S2 of the revised ESI file associated with the original article (DOI: 10.1039/C9SC04140K) and are copied below.
Detailed DNA sequences (also included in the revised ESI):
DNA sequence: TTCTAGGCTCCTAGGACCCC TTCTAGGCTCCTAGGACCCC TTCTAGGCTCCTAGGACCCC;
2 AP sites in DNA sequence: TTCTAGG(RDG)(RDG)CCTAGGACCCC TTCTAGGCTCCTAGGACCCC TTCTAGGCTCCTAGGACCCC;
4 AP sites in DNA sequence: TTCTAG(RDG)(RDG)(RDG)(RDG)CTAGGACCCC TTCTAGGCTCCTAGGACCCC TTCTAGGCTCCTAGGACCCC;
6 AP sites in DNA sequence: TTCTA(RDG)(RDG)(RDG)(RDG)(RDG)(RDG)TAGGACCCC TTCTAGGCTCCTAGGACCCC TTCTAGGCTCCTAGGACCCC;
8 AP sites in DNA sequence: TTCT(RDG)(RDG)(RDG)(RDG)(RDG)(RDG)(RDG)(RDG)AGGACCCC TTCTAGGCTCCTAGGACCCC TTCTAGGCTCCTAGGACCCC;
10 AP sites in DNA sequence: TTC(RDG)(RDG)(RDG)(RDG)(RDG)(RDG)(RDG)(RDG)(RDG) (RDG)GGACCCC TTCTAGGCTCCTAGGACCCC TTCTAGGCTCCTAGGACCCC;
14 AP sites in DNA sequence: TTC(RDG)(RDG)(RDG)(RDG)(RDG)(RDG)(RDG)(RDG)(RDG)(RDG) (RDG)(RDG)(RDG)(RDG)CCC TTCTAGGCTCCTAGGACCCC TTCTAGGCTCCTAGGACCCC
In addition, the authors regret that the abbreviation and the naming of compounds were incorrect in the original article and ESI. The correct name for the compound abbreviated HDM in Scheme S1 should be (R)-3,5-dimethoxy-4-oxopentanal. The correct version of Scheme S1 is presented below.
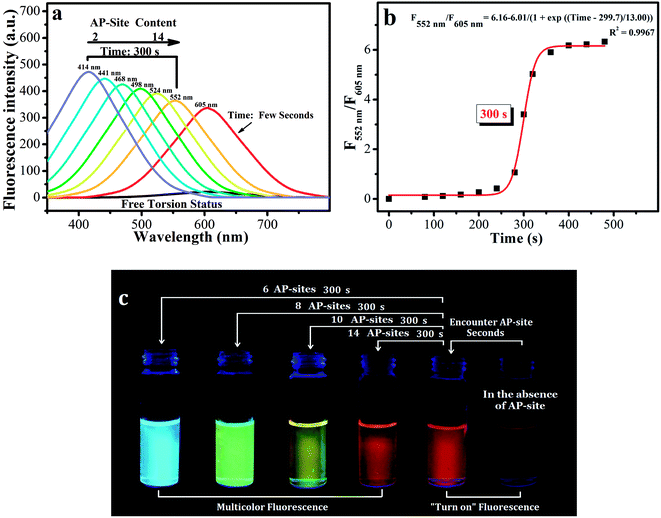
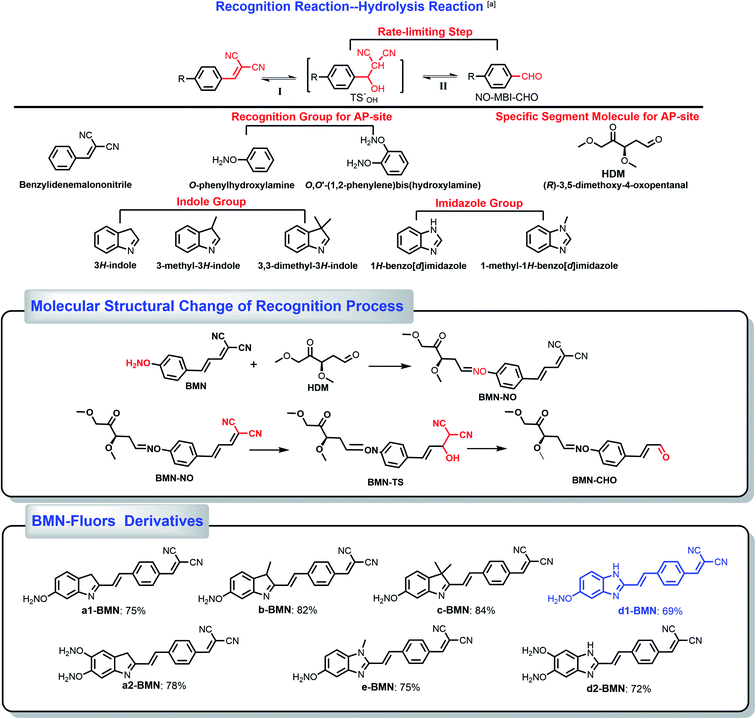 | ||
| Scheme 1 Derivative molecules. The hydrolysis transformation of 2-(4-vinylbenzylidene)malononitrile derivatives. | ||
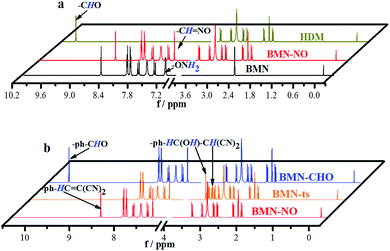
(E)-2-(3-(4-(aminooxy)phenyl)allylidene)malononitrile (Fig. 3a,b and Scheme S1) was used as the simplest molecular fragment to better explain the structural changes during the recognition process. Its correct abbreviation is BMN, which has been corrected in Fig. 3a, b and Scheme S1. The caption of Fig. 3a has also been updated. In addition, the corresponding 1H NMR titration spectra have been included in the revised ESI file associated with the original article (DOI: 10.1039/C9SC04140K).
The Royal Society of Chemistry apologises for these errors and any consequent inconvenience to authors and readers.
| This journal is © The Royal Society of Chemistry 2022 |