Open Access Article
Open Access ArticleCombining hyperspectral imaging and electrochemical sensing for detection of Pseudomonas aeruginosa through pyocyanin production†
R. David
Dunphy‡
 a,
Perrine
Lasserre‡
a,
Perrine
Lasserre‡
 b,
Lily
Riordan
c,
Katherine R.
Duncan
b,
Lily
Riordan
c,
Katherine R.
Duncan
 c,
Christopher
McCormick
b,
Paul
Murray
*a and
Damion K.
Corrigan
*bd
c,
Christopher
McCormick
b,
Paul
Murray
*a and
Damion K.
Corrigan
*bd
aDepartment of Electronic and Electrical Engineering, University of Strathclyde, Glasgow G1 1XW, UK. E-mail: paul.murray@strath.ac.uk
bDepartment of Biomedical Engineering, University of Strathclyde, Glasgow G1 1QE, UK. E-mail: damion.corrigan@strath.ac.uk
cStrathclyde Institute of Pharmacy and Biomedical Sciences, University of Strathclyde, Glasgow G4 0RE, UK
dDepartment of Chemistry, University of Strathclyde, Glasgow G1 1BX, UK
First published on 10th June 2022
Abstract
Despite bacterial biofilms representing a common form of infection, notably on medical devices post implantation, their detection and characterisation with existing methods is not sufficient to inform clinicians about biofilm presence or treatment response in affected patients. This study reports the development and use of a combined hyperspectral imaging (HSI) and electrochemical platform to monitor biofilm formation optically and electrochemically. Firstly, production of pyocyanin, a common pigmented and redox active secondary metabolite produced by P. aeruginosa, is monitored by combined HSI and square-wave voltammetry. Secondly, P. aeruginosa biofilm formation is characterised directly using electrochemical impedance spectroscopy. This suite of optical and electrochemical measurements allows for combined monitoring of secondary metabolite/virulence factor production along with direct monitoring of biofilm formation on the sensor surface. Crucially, the easy to deploy and low-cost nature of the selected sensing technologies means the approach can be developed for enhanced study of biofilms and/or at the point of care.
Introduction
The National Biofilms Innovation Centre estimated the worldwide annual cost of biofilms to human health at $387 billion USD, representing five percent of global health expenditure.1 Microorganisms in the biofilm form are reported up to 1000 times more antibiotic resistant than planktonic specimens of the same species.2–4 In many cases, the concentrations of antibiotics required to treat biofilm infections cannot be administered as it would lead to systemic toxicity and adverse side effects for patients.5 A particular problem arises with the use of implantable medical devices, as they account for over half of hospital-acquired infections,6 often a result of biofilm formation.7 Some methods to lower infection include, enhanced sterilisation procedures, for instance, low-intensity intermittent ultrasonication-induced bursting of microbubbles or photodynamic therapy. Yet, these strategies in addition to device modifications, such as coatings (metal, anti-microbial or hydrogel), have not proved sufficient to lower infection risks and rates.8,9 Furthermore, biofilm-related device infections are rarely detected before they become systemic often leading to device failure. One of the most effective strategies to tackle biofilm-related infections is the invasive procedure of removing and replacing the infected implant.7 For these reasons, it is clear that biofilm control and treatments are not yet optimal. This is mainly due to the lack of reliable detection and diagnostic methods.2,3Biofilm presence is commonly confirmed by optical systems using microscopy or staining approaches such as crystal violet.10–15 Culture methods are routinely used to grow and identify pathogens but involve invasive sampling and destructive end-point characterisation.12,13 Furthermore, slow-growing biofilm microorganisms do not always grow in vitro.3,9 An alternative method to culture is to use molecular techniques for pathogen identification, but these do not discriminate between biofilm-constituting organisms and extracellular DNA or planktonic bacteria contained in a sample.2,3,16 Recent fluorescence-based techniques and next-generation sequencing have not yet found their place in the clinic due to cost and expertise requirements,3 and no consensus has been reached over standardisation or quantitative strategies.10,17 Furthermore, low replicability both within and between laboratories does not facilitate comparisons and development of universal models, detection, treatments or point-of-care solutions.12,18 Non-invasive and reliable systems are therefore needed for biofilm characterisation and quantification. Downscaling from known macro systems, such as Calgary devices, modified Robbin's devices or CDC biofilm reactors, to micro techniques could decrease resource costs and analysis time while improving real-time monitoring.12
These are features of electrochemical sensors, which enable selective, sensitive and rapid analyte detection, with possibilities for higher throughput and miniaturisation.19 Electrochemically active metabolites can be directly detected through amperometry or voltammetry. The chemical signal from oxidation or reduction processes is converted into a measurable electrical signal, an application of which is square-wave voltammetry (SWV), a fast and sensitive technique, where peak amplitude is directly related to analyte concentration.20 Using an alternating current instead, electrochemical impedance spectroscopy (EIS) assesses surface characteristics from fitting electrical circuits modelling physiochemical properties of the studied system.21–23 Faradaic impedance is produced upon occurrence of redox reactions close to the electrode surface, which can be exploited for quantitative analysis.24 EIS has previously proven to be a successful strategy in the detection of biofilms.11,25
Pseudomonas aeruginosa is a World Health Organization priority pathogen,26 and one of the most commonly associated pathogens in hospital-acquired infections.7,8,27,28 It is responsible for acute infections in healthy subjects, its pathogenicity in immuno-compromised patients often results in chronic infections because of its resistance to antibiotics and to the host immune response.7 Pyocyanin is ubiquitously produced by P. aeruginosa and one of its main virulence factors, characteristically produced by this bacterial species in a biofilm phenotype, its most efficient survival strategy.27,29–31 As a well-characterised compound across the electrochemical literature, pyocyanin has led to a wide range of sensing strategies through amperometric, voltammetric, or impedance-based detection.20,29,30,32
Hyperspectral imaging (HSI) is a photographic imaging technique that captures detailed spectral information for each pixel of an image. The resulting hyperspectral datacube can be thought of as a collection of greyscale images corresponding to narrow bands of the electromagnetic spectrum. This effectively allows a complete reflectance spectrum to be constructed for each pixel of an image, enabling computer-assisted analysis of processes that are detectable in the visible or infrared spectrum. HSI enables the detection of specific substances by matching absorption bands to known spectral signatures. HSI has the advantage over UV spectroscopy and other analytical techniques that it allows identified compounds to be localised over the area of an affected sample.
Biomedical applications of HSI range from in vivo identification of cancer cells33 to the classification of Shiga toxin-producing Escherichia coli.34 The use of HSI has previously been proposed for the detection of environmental biofilm through colour changes within the visible to near-IR (VNIR) range, resulting from biofilm formation or interaction with the bacteria-substrate.35 While hyperspectral microscopy has frequently been used for microbiological applications such as the detection of E. coli36 or single-cell identification of food-borne pathogens,37 macroscopic applications are of increasing interest.38 Hyperspectral techniques applied to biofilms include hyperspectral fluorescence imaging, which has been used to assess E. coli and Salmonella biofilms on stainless steel,39 and Raman chemical imaging spectroscopy for identifying a range of pathogens.40 Due to their non-destructive nature, hyperspectral methods are particularly well suited for time series studies, and have been used to monitor the effect of antibiotics on biofilm growth.41
In this study, we propose a robust and multifaceted combined HSI and electrochemical sensing approach to the detection of the P. aeruginosa virulence factor pyocyanin. The reported approach will enhance the ability to study and therefore improve understanding of biofilm formation and maturation on medical implant materials. Additionally, because of the two technologies involved, the system has potential for future point-of-care applications. Pyocyanin's blue pigmentation serves to quantify P. aeruginosa growth using VNIR-HSI for the first time. The trends observed using VNIR-HSI are supported using SWV to evaluate the quantity of pyocyanin in samples of two strains of P. aeruginosa over 48 hours of growth and supplemented by EIS for biofilm assessment. We show that the electrochemical detection of pyocyanin can be correlated with its spectral signature and demonstrate, using time series data, that the growth and spatial distribution of the bacteria can be documented.
Materials & methods
Custom-built test cell design
A 3D-printed custom-built test cell was designed and manufactured in-house to facilitate the image capture and electrochemical monitoring of P. aeruginosa pyocyanin production and biofilm formation. The design allows for samples to be grown, imaged from above before electrochemical measurements are taken using the potentiostat without disruption. The plate consists of a baseplate with four slots to position screen-printed electrodes, which is bolted to a clear acrylic enclosure with wells around the electrode measuring area. Acrylic cylinders were inserted into the enclosure wells and sealed with O-rings to reproduce the shape of a 96-well microtiter plate, as this is the most common format for biofilm measurements.25 The baseplate and the enclosure are bolted together after sliding electrodes into the designated slots. The custom-built test cell was cleaned with 70% ethanol in dH2O (v/v). Fig. 1 shows close-ups of a well of the custom-built test cell containing lysogeny broth (LB) medium (Fig. 1b) and P. aeruginosa biofilm (Fig. 1c).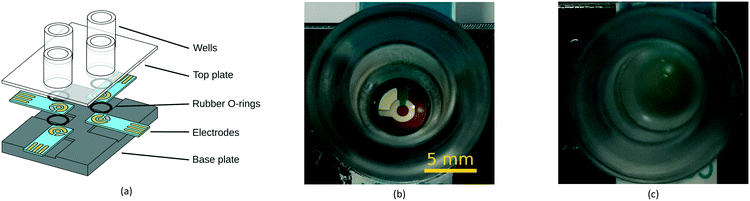 | ||
| Fig. 1 Biofilm formation in the custom-built test cell. (a) Exploded view of the test cell, (b) P. aeruginosa in LB medium at 0 h and (c) after 6 h of incubation. | ||
Pyocyanin calibration
Pyocyanin standard (P0046) and LB medium (L3522) were purchased from Sigma. Pyocyanin was dissolved in 100% ethanol at a concentration of 10 mg mL−1. Further dilutions were prepared in triplicates, in LB medium at the following concentrations: 10 mM, 5 mM, 2.5 mM, 1 mM, 0.50 mM, 0.25 mM, 0.10 mM, 0.05 mM, 0.025 mM and 0.01 mM.Screen-printed gold electrodes (C223-BT DropSens, Oviedo, Spain) with a 1.6 mm diameter working electrode were used to conduct experiments. 50 μL of culture or media blank was deposited onto electrodes. The custom-built test cell was placed on a lateral translation stage located under the VNIR hyperspectral camera, and illuminated by four 60 W halogen lamps. The plate was positioned to align each well with the lens of the camera as it was being imaged, minimising differences in illumination that might otherwise have been caused by the relative position of the well, camera and lamps. To avoid inconsistent illumination resulting from the edges of the plate, the outermost wells were not used. For each sample, an image was first captured, and electrochemical measurements followed directly after.
Hyperspectral images were captured using a Hamamatsu CCD/Specim C8484-05G VNIR pushbroom hyperspectral camera system, with 2 × 2 pixel binning used to compensate for sensor noise and an exposure time of 0.04 s. 512 wavelength bands in the range 386–1006 nm were measured, of which 440 bands in the range 379.6–909.9 nm were retained for analysis, as bands outside this range are negatively impacted by sensor noise. The spectral reflectance was obtained from the raw hyperspectral data using one-point calibration for each image, according to the formula:
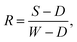 | (1) |
Square wave voltammograms were acquired following settings previously described,42 with a 5 A current, a 3 mV step potential, and a 15 Hz frequency. The potential ranged from −0.7 V to 0.0 V vs. Ag/AgCl. For EIS data generation, frequencies were scanned between 0.1 Hz and 100 kHz at a 0.01 V AC potential with 11 frequencies per decade at 67 frequencies. EIS spectra were measured against OCP. EIS and SWV data were acquired with a PalmSens potentiostat and analysed with Origin.
P. aeruginosa monitoring in a 96-well plate format
Wells were inoculated with an OD600 value of 0.5 or 1 from an overnight P. aeruginosa culture of either PA14 or LESB58 respectively. Distilled water was added to any empty well, ensuring sufficient ambient water availability to avoid sample concentration. Images of the plate were taken first, and sampling of each well occurred in sterile conditions where well contents were transferred to 0.5 mL sterile microcentrifuge tubes and stored on ice until SWV and EIS acquisition. 50 μL were pipetted onto the electrode area of gold screen printed electrodes and tested with SWV with a wider potential window from −0.7 to 1.0 V and EIS.Hyperspectral images were normalised using the standard normal variate (SNV) approach, and adjusted relative to the LB baseline, with the formula:
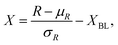 | (2) |
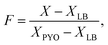 | (3) |
P. aeruginosa biofilm monitoring in the custom-built test cell
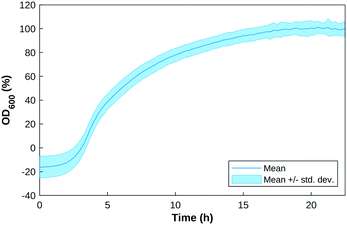
The custom-built test cell was assembled with the electrodes in the slots. An overnight P. aeruginosa (PA14 and LESB58) culture was inoculated in 200 μL of LB medium in each well, at an initial OD600 value of 0.5 or 1. OD600 measurements were taken every 15 minutes for 24 hours as a growth check. Fig. 2 shows the growth curve of P. aeruginosa samples over a 24-hour period.The operation was repeated in triplicate following the same image capture and SWV and EIS settings established in the above section at 0, 24 hours and 48 hours.
Mean reflectance spectra were calculated from a region of each hyperspectral image corresponding to the location of the reference electrode, which had the most Lambertian spectrum of any portion of the samples.
The spectra were normalised to a mean of μ = 0 as in eqn (2). The mean reflectance for the corresponding area of a control sample containing only LB was used as the baseline reflectance spectrum and subtracted from each of the sample spectra.
Results and discussion
SWV and HSI standard pyocyanin calibration
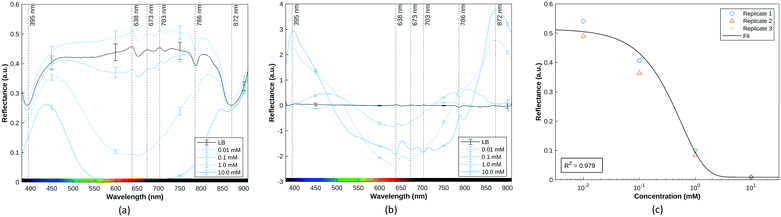
A pyocyanin standard calibration curve was first established with SWV and HSI with 50 μL deposits on screen-printed electrodes contained in the custom-built test cell. Images of increasing pyocyanin concentrations were captured and immediately followed with the acquisition of a square wave voltammogram.43 As different oxidised and reduced forms of pyocyanin itself have different pigmentations,44 HSI was always performed prior SWV to capture images before any sample modification could occur at that specific time point. Since SWV took place at each time point as well, pyocyanin produced could have undergone oxidation or reduction. In that case, HSI was able to measure newly formed pyocyanin more accurately whereas SWV assesses the overall production diffused in the culture medium.As extensive work has been conducted on pyocyanin electrochemical detection, its purpose here serves as a basis for interpretation of hyperspectral observations. Additionally, its peak potential is distinct from possible common confounding compounds,12,13,45 and is also distinguishably detected in polymicrobial environments.46 The average SWV current responses obtained for one order of magnitude are depicted in Fig. 4a for trend visualisation and readability. SWV responses for each concentration tested were extrapolated into a dose response curve. An approximately linear relationship was observed between pyocyanin concentration and SWV peak current (Fig. 4b). One trend noticed with calibration is the shift in potential at which the pyocyanin peak occurs. This can be attributed to pH dependent changes as demonstrated previously43 or use of a quasi Ag/AgCl reference electrode rather than a fritted system.47,48
The calibration curve obtained for the HSI data, shown in Fig. 3, reveals local maxima in the difference between the responses at the highest and lowest concentration at the wavelengths 395 nm, 638 nm, 703 nm, and 872 nm, with the 638 nm band corresponding to the centre of the main red absorption band. These curves are derived from the mean reflectance of known pyocyanin concentrations imaged in a 96-well plate placed on top of a Spectralon tile. Fig. 3b shows the SNV with the mean LB reflectance subtracted. On this plot, an ideal LB sample should be represented by a line with a constant adjusted reflectance of 0. Deviations from this at the edges of the spectrum are caused by sensor noise and variation in the samples. Fig. 3c shows the response at 638 nm as a function of concentration. An exponential decay curve of the form y = ae−bx+c + d was fitted to this data. The exponential curve was chosen to account for the diminishing change in the spectrum as the solution approaches the saturation point, in line with the Beer–Lambert law, which states that the transmittance of a sample at a given wavelength is a function of the exponential of its concentration.49 The constant term d accounts for the reflectance. This curve fits the data with R2 value of 0.98 (Fig. 3c and S1†). The trend obtained for the SWV response is in line with HSI results for each sample.
 | ||
| Fig. 3 (a) Mean reflectance spectra of pyocyanin in LB at four concentrations. Error bars representing the standard deviation have been included at 150 nm intervals, and a plot of the individual replicates can be found in Fig. S3.† (b) The same spectra normalised using SNV and baseline subtraction. (c) Exponential curve fit to calibration data for 638 nm. | ||
 | ||
| Fig. 4 (a) Average SWV current amplitude for pyocyanin solutions from 0.5 mM to 10 mM (n = 3) (b) SWV peak amplitude response to increasing pyocyanin concentrations between [−0.35: −0.15] V. | ||
SWV and HSI pyocyanin detection from P. aeruginosa inoculum in a 96-well plate format
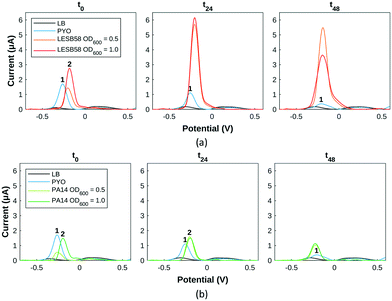
After defining pyocyanin detection profiles with SWV and HSI, the growth of two strains of P. aeruginosa, PA14 and LESB58, was monitored with both approaches to detect and assess pyocyanin production. A transparent 96-well plate format was used to optimise bacterial culture and sample repeatability while allowing for higher throughput. Hyperspectral images were captured from the 96-well plate and wells were then sampled in sterile conditions for transfer onto gold screen-printed electrodes for SWV. Time points were chosen to ensure sufficient time for bacterial growth, pyocyanin production and biofilm formation.14,15,50,51 LESB58, known to be the more virulent strain of the two tested,52 produced on average more pyocyanin than PA14,53 when assessed at two initial seeding concentrations.Pyocyanin standards and Pseudomonas spp. cultures were in a background of LB medium. As reported,43 LB is a complex culture medium which shows an intense oxidation peak at 0.85 V. This peak is not reproducible across samples and time, meaning the potential window of detection considered will be between −0.7 V and 0.6 V since pyocyanin and other signature metabolites are detected below 0.6 V.
For LESB58 samples (Fig. 5a), two overlapping peaks (peak 2) are initially present at −0.25 V, which do not exactly overlap the pyocyanin standard (peak 1). Their amplitude is proportional to the initial bacterial inoculation concentration, the less concentrated samples at the level of the control and the more concentrated at double the amount of the control. After 24 hours, both LESB58 culture concentrations have produced a culture medium containing >2 mM pyocyanin as evidenced by the two peaks at −0.25 V overlapping the pyocyanin standard curve. After 48 hours, pyocyanin quantities have slightly decreased for the initially less concentrated LESB58 samples and dropped at the higher concentrations (Table S1†).
Concerning PA14 samples (Fig. 5a), the less concentrated samples show the presence of pyocyanin at the same potential whereas PA14 samples at a higher concentration exhibit a slightly shifted peak (numbered 2) in the same potential region. After 24 hours, all PA14 samples contain approximately 0.5 mM pyocyanin (peak 1). The less concentrated samples have produced pyocyanin while the more concentrated one has converted the most likely precursor (phenazine-1-carboxylic acid) into pyocyanin. Pyocyanin levels decreased after 24 hours.
Other possible peaks present (Fig. 5) can be attributed to 5-methylphenazine-1-carboxylic acid (5-MCA) around −0.1 V, a direct precursor of pyocyanin within the Pseudomonas quorum sensing pathway: its presence testifies of bacterial growth and communication between bacteria.54,55 For PA14 samples, the Pseudomonas quinolone signal (PQS) is detected around +0.2 V (Fig. 5).43,54,55 From square wave voltammogram monitoring extrapolated into pyocyanin estimations, PA14 and LESB58 show different behaviours in terms of pyocyanin production: LESB58 samples showing more pyocyanin production than PA14 (Table S1†), confirming earlier reports.53 Furthermore, pyocyanin production by both strains reflects previous reports of higher production within the first hours of growth, followed by either a plateau or decrease in pyocyanin production.43 This has been linked to P. aeruginosa exoproduct synthesis being heightened to establish infection during acute phases,31 further evolving towards a decrease at chronic stages.56 Overall, pyocyanin production trends were in agreement with previous reports.48,50,57
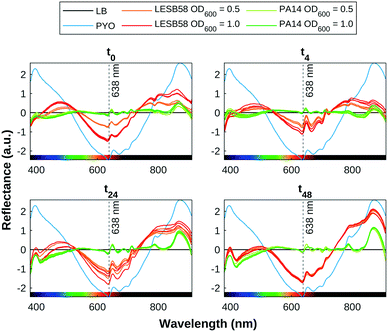
The presence of pyocyanin detected by SWV correlates with pigmentation observed in the HSI data, as shown in Fig. 6. The pyocyanin absorption band centred around 638 nm can be clearly seen in the mean spectra for the positive control samples, which is increasingly approached by the spectra of the LESB58 samples over time. Two smaller absorption bands at 395 nm and 872 nm that are present in all samples are produced by LB. The 395 nm band overlaps with an absorbance band in the UV range that increased over time in the bacterial samples, and may be indicative of biofilm growth. Further work is required to confirm the source of this feature. Table 1 shows an estimation of the quantity of pyocyanin in representative samples based on the fit found for 638 nm of y = 0.49e−1.78x+0.39 + 0.0089 to the 96-well plate calibration data, as shown in Fig. 3. Estimated values for PA14 before 48 h were lower than the error margin of the method. Negative values were adjusted to 0.
| t = 0 | t = 4 h | t = 24 h | t = 48 h | |
|---|---|---|---|---|
| LESB58 OD600 0.5 | 60.6 | 80.0 | 225.3 | 524.5 |
| LESB58 OD600 1.0 | 160.8 | 153.6 | 283.5 | 544.2 |
| PA14 OD600 0.5 | 0 | 0 | 16.7 | 45.9 |
| PA14 OD600 1.0 | 0 | 0 | 28.3 | 48.3 |
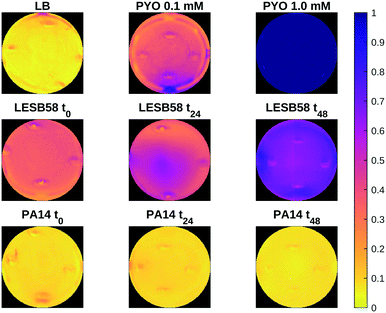
Fig. 7 shows a heat map of the differential feature map F for the 638 nm absorption band as calculated by eqn (3), showing an increase in the amount of pyocyanin over time for the LESB58 samples, with initially greater production in the centre of the well. Pyocyanin production in the PA14 samples was below the detection limit of this method. Four small artefacts are visible around the centre of each well, which are caused by specular reflection from the surface of the liquid. These areas were omitted from the analysis.
The combined detection of pyocyanin through electrochemical methods along with HSI has enabled the reliable detection of P. aeruginosa presence through its electroactive and pigmented biomarker. Furthermore, pyocyanin quantities were detected within the clinically relevant range.31 However, differences in trends and estimations were observed between both approaches, which could result from the data acquisition angle and nature of tested sample. It has been reported that biofilms form layers and gradients.58 Imaging occurred from the top of the plate, where the biofilm forms and the most oxygenated form of pyocyanin, in its blue form, is the most concentrated.59 Contrastingly, SWV measurements were taken after pipetting samples on screen-printed sensors, deposits being a mix of biofilm and diffused pyocyanin in the medium, in its transparent form as the growing biofilm gradually shields the liquid medium from optimal oxygenation, which might explain the diverging trends.
The detection of pyocyanin relies only on the relative absorption of light over a few representative wavelengths, opening up the possibility of developing a low-cost sensor that leverages the transmittance at those specific wavelengths. Such a sensor would be limited to settings where the presence of confounding pigments can be ruled out, whereas HSI has the ability to distinguish between spectrally distinct pigments.
Integrated P. aeruginosa biofilm detection in a custom-built test cell
After calibrating with pyocyanin standards and confirming detection in bacterial inoculum, EIS was used to demonstrate the presence of biofilm formation when bacterial samples were cultured in the custom-built test cell. Being able to grow biofilms directly above the sensor and simultaneously imaging the interface maintained the coherence of the bacterial structure as much as possible for detection. Limiting the number of samples per support minimised movement and temperature disturbances.EIS measurements were analysed but not presented at the previous stage since biofilm integrity was disrupted by sample pipetting from wells onto screen-printed electrodes.58 SWV results shown in the ESI† showed more variability between replicates and the likely presence of other metabolic products than pyocyanin alone, with higher peak amplitudes, suggesting that other metabolic pathways were favoured over pyocyanin production. Pyocyanin metabolic precursors are characterised by diverse and distinct pigmentation profiles.60 This could lead to an overall alteration of sample spectra, however a response from the chosen detection wavelengths from pyocyanin confirmed with electrochemical detection is a confident validation. As the hyperspectral data focused on pyocyanin and almost none was detected in the custom-built test cell (Fig. S2†), the focus here was to detect biofilm formation with EIS, as the direct correlation between these two virulence factors still remains to be demonstrated.61
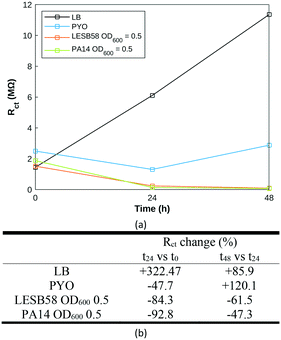
Nyquist plots (Fig. S4†) were fitted to a modified Randles equivalent circuit.62 The evolution of LB across time was very distinct from pyocyanin and P. aeruginosa samples (Fig. 8a), which might result from a slow evaporation process occurring over time. The pyocyanin standard exhibits a decrease in charge transfer resistance (Rct) over the first 24 hours followed by an increase over the next 24 hours, closer to the behaviour of LB alone. Both bacterial strains display identical Rct trends: a steep decrease within the first 24 hours maintained at a slower rate over the next 24 hours (Fig. 8). It appears that the initial Rct value and its change over the first time period could be specific to the P. aeruginosa strain, reflecting the synthesis of electroactive secondary metabolites, attributable to phenazine compounds or Pseudomonas quinolone signal. A decrease in Rct over time has previously proven to be indicative of biofilm extent.58 The biofilm is formed at the air liquid interface, more distant from the electrode measuring area in this experiment than in previous demonstrations, which likely explains the fewer observed changes. Using the custom-built test cell to grow and characterise P. aeruginosa biofilm, it was possible to monitor biofilm formation in both strains using impedance measurements. Biofilm formation occurred within the timeframe of previously published work.15 Further work is required to establish whether a hyperspectral approach can be used to differentiate between P. aeruginosa in biofilm and in planktonic form.
EIS was chosen to enable surface assessment of the electrode, useful for further transfer of the method to bacterial biofilms where no redox mediators are produced, having only tested our system with one of the ESKAPE pathogens; P. aeruginosa, which is an outlier in this respect. Further research is required to establish whether the presented method will work with multi-species biofilms, which are more reflective of clinical or even environmental cases,63 for testing in later clinical samples.
In contrast to other spectroscopic methods that do not have a spatial component, such as UV spectroscopy, HSI can be used to gain an understanding of the spatial distribution of metabolites produced by pathogens. Due to diffusion through the liquid culture in relatively small wells the differences seen in Fig. 7 are not pronounced, but more distinctive distribution patterns may be found in other settings. While of limited use in a clinical setting, this approach may allow for further research to increase understanding of how bacterial pathogens interact with their environments and disperse over time, which may be of interest in areas such as biofilm characterisation, antibiotic susceptibility testing, or medical device infections.
While HSI relies on large and expensive equipment, identifying wavelengths that correspond to diagnostic spectral features can allow the development of cheaper filter-based MSI sensors that only capture the relevant wavelengths. These sensors could be simplified and miniaturised as electrochemical equipment is also prone to integration and miniaturisation, outlining the perspectives of detection and identification of specific microorganisms that are frequent causes of infection and implementation at the point of care.64 Additionally, the strategy presented could be easily translated into low-cost mass-manufacturable screen-printed electrodes, with small reagent and sample volumes.
Proposing an engineering scenario to serve clinical purposes,65 our combined biosensing approach aligns with future biofilm detection strategies highlighted by leaders of the biofilm field.66 Despite an initial culture step, the application envisaged could directly assess the patient/clinical sample. Detecting biofilm growth directly from the sputum of cystic fibrotic patients could be another possibility to identify chronic infections caused by P. aeruginosa,67 enabling additional surgery procedures to be supplied where necessary and minimising the dangers of antibiotic resistance.54 With the combination of the three techniques, biofilm profiles could be added to a library to quickly identify the most common pathogens and assess treatment options. Having an “electrochemical fingerprint”68 of various strains with a hyperspectral profile would allow for quick identification and guidance for more time-consuming and accurate sequencing downstream methods.54,68,69 Adapting sample volume could help bring biofilms closer to the electrode and reduce differences in compound quantifications. Earlier and later time points, as well as more frequent monitoring would also bring a heightened understanding of biofilm dynamics and formation by providing a full understanding of sample behaviour. The combined detection by hyperspectral and electrochemical methods provides more reliability and confidence in the final result, saving time by avoiding the need to perform an array of methods to evaluate treatment options or anti-biofilm properties of pharmaceutical compounds,51 fast tracking infection treatment and drug discovery processes. Finally, the combination of EIS with HSI and SWV allows a full range of measurements capable of metabolite detection and differentiation, identification and biofilm growth assessment. While HSI characterises pigmented secondary metabolites, SWV detects redox active secondary metabolites. Having both sensing modalities along with EIS to monitor biofilms provides a greater range of sensing opportunities.
Conclusions
This study demonstrates that VNIR-HSI can be used in conjunction with electrochemical methods to detect pyocyanin within the clinically significant range for P. aeruginosa, and furthermore that regression of key spectral bands may aid in the detection and characterisation of Pseudomonas presence and growth through pyocyanin production. The wavelengths at which the greatest differentiation of pyocyanin is detected are outlined, suggesting the possibility of developing a low-cost narrowband sensor for detecting P. aeruginosa. SWV in conjunction with EIS confirmed pyocyanin presence and quantity, as well as biofilm formation over gold screen-printed electrodes in an integrated custom-built test cell. Designing and validating a method for combined use of these techniques with reference to P. aeruginosa's well-documented tendency to produce pigmented redox active compounds has the potential to translate to a wider range of biofilm-forming infectious pathogens in future. Finally, the nature of the two technologies employed (HSI and electrochemical sensing) means the system can be deployed in a range of formats, including a platform to better study biofilm development and for detection purposes at the point of care.Conflicts of interest
There are no conflicts to declare.Acknowledgements
R. D. Dunphy, P. Lasserre and L. Riordan would like to thank the Strathclyde Centre for Doctoral Training in Antimicrobial Resistance for support related to their PhD studentships.References
- National Biofilms Innovation Centre, International Biofilm Markets and Infographics, https://www.biofilms.ac.uk/international-biofilm-markets/, (accessed 17 November 2021).
- M. K. Yadav, J.-J. Song, B. P. Singh and J. E. Vidal, in New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Biofilms, ed. M. K. Yadav and B. P. Singh, Elsevier, 2020, pp. 1–13 Search PubMed.
- M. K. Yadav, J. E. Vidal and J.-J. Song, in New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Biofilms, ed. M. K. Yadav and B. P. Singh, Elsevier, 2020, pp. 15–28 Search PubMed.
- G. Gebreyohannes, A. Nyerere, C. Bii and D. B. Sbhatu, Heliyon, 2019, 5, e02192 CrossRef PubMed.
- S. Subramanian, E. I. Tolstaya, T. E. Winkler, W. E. Bentley and R. Ghodssi, ACS Appl. Mater. Interfaces, 2017, 9, 31362–31371 CrossRef CAS PubMed.
- Z. Zhang, V. E. Wagner and J. C. Victor, in Antimicrobial Coatings and Modifications on Medical Devices, ed. Z. Zhang and V. E. Wagner, Springer, Cham, 2017, pp. 1–36 Search PubMed.
- K. Lee and S. S. Yoon, J. Microbiol. Biotechnol., 2017, 27, 1053–1064 CrossRef CAS PubMed.
- K. Zhang, X. Li, C. Yu and Y. Wang, Front. Cell. Infect. Microbiol., 2020, 10, 359 CrossRef CAS PubMed.
- S. L. Percival, L. Suleman, C. Vuotto and G. Donelli, J. Med. Microbiol., 2015, 64, 323–334 CrossRef PubMed.
- D. Kour, T. Kaur, S. A. Fahliyani, A. A. Rastegari, N. Yadav and A. N. Yadav, in New and Future Developments in Microbial Biotechnology and Bioengineering, Elsevier, 2020, pp. 89–124 Search PubMed.
- E. M. Bosire and M. A. Rosenbaum, Front. Microbiol., 2017, 8, 892 CrossRef PubMed.
- S. Subramanian, R. C. Huiszoon, S. Chu, W. E. Bentley and R. Ghodssi, Biofilm, 2020, 2, 100015 CrossRef PubMed.
- K. D. Mandakhalikar, J. N. Rahmat, E. Chiong, K. G. Neoh, L. Shen and P. A. Tambyah, Sci. Rep., 2018, 8, 8069 CrossRef PubMed.
- M. M. El-Shahed, D. E. Mahmoud, N. S. Soliman, Y. A. ElMahdy and S. H. Mohamed, J. Appl. Pharm. Sci., 2020, 10, 074–080 CAS.
- K. N. Kragh, M. Alhede, L. Kvich and T. Bjarnsholt, Biofilm, 2019, 1, 100006 CrossRef PubMed.
- A. Gędas and M. A. Olszewska, in Recent Trends in Biofilm Science and Technology, ed. M. Simoes, A. Borges and L. Chaves Simoes, Academic Press, 2020, pp. 1–21 Search PubMed.
- J. Azeredo, N. F. Azevedo, R. Briandet, N. Cerca, T. Coenye, A. R. Costa, M. Desvaux, G. Di Bonaventura, M. Hébraud, Z. Jaglic, M. Kačániová, S. Knøchel, A. Lourenço, F. Mergulhão, R. L. Meyer, G. Nychas, M. Simões, O. Tresse and C. Sternberg, Crit. Rev. Microbiol., 2017, 43, 313–351 CrossRef CAS PubMed.
- K. Doll, K. L. Jongsthaphongpun, N. S. Stumpp, A. Winkel and M. Stiesch, J. Microbiol. Methods, 2016, 130, 61–68 CrossRef CAS PubMed.
- A. Signoroni, M. Savardi, A. Baronio and S. Benini, J. Imaging, 2019, 5, 52 CrossRef PubMed.
- F. McEachern, E. Harvey and G. Merle, Biotechnol. J., 2020, 15, 2000140 CrossRef CAS PubMed.
- A. C. Ward, P. Dubey, P. Basnett, G. Lika, G. Newman, D. K. Corrigan, C. Russell, J. Kim, S. Chakrabarty, P. Connolly and I. Roy, Front. Bioeng. Biotechnol., 2020, 8, 1039 CrossRef PubMed.
- J. B. J. H. van Duuren, M. Müsken, B. Karge, J. Tomasch, C. Wittmann, S. Häussler and M. Brönstrup, Sci. Rep., 2017, 7, 5223 CrossRef PubMed.
- L. D. Bharatula, E. Marsili and J. J. Kwan, Electrochim. Acta, 2020, 332, 135390 CrossRef CAS.
- H. S. Magar, R. Y. A. Hassan and A. Mulchandani, Sensors, 2021, 21, 6578 CrossRef CAS PubMed.
- D. Czerwińska-Główka and K. Krukiewicz, Bioelectrochemistry, 2020, 131, 107401 CrossRef PubMed.
- World Health Organization, Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis, World Health Organization, 2017 Search PubMed.
- I. Gandouzi, M. Tertis, A. Cernat, D. Saidane-Mosbahi, A. Ilea and C. Cristea, Materials, 2019, 12, 1180 CrossRef CAS PubMed.
- P. Karami, A. Khaledi, R. Y. Mashoof, M. H. Yaghoobi, M. Karami, D. Dastan and M. Y. Alikhani, Gene Rep., 2020, 18, 100561 CrossRef.
- A. J. Robb, S. Vinogradov, A. S. Danell, E. Anderson, M. S. Blackledge, C. Melander and E. G. Hvastkovs, Electrochim. Acta, 2018, 268, 276–282 CrossRef CAS PubMed.
- F. A. Alatraktchi, W. E. Svendsen and S. Molin, Sensors, 2020, 20, 5218 CrossRef CAS PubMed.
- F. A. Alatraktchi, J. S. Noori, G. P. Tanev, J. Mortensen, M. Dimaki, H. K. Johansen, J. Madsen, S. Molin and W. E. Svendsen, PLoS One, 2018, 13, e0194157 CrossRef PubMed.
- S. Rajamani, R. Sandy, K. Kota, L. Lundh, G. Gomba, K. Recabo, A. Duplantier and R. G. Panchal, J. Microbiol. Methods, 2019, 159, 179–185 CrossRef CAS PubMed.
- D. G. Ferris, R. A. Lawhead, E. D. Dickman, N. Holtzapple, J. A. Miller, S. Grogan, S. Bambot, A. Agrawal and M. L. Faupel, J. Low. Genit. Tract Dis., 2001, 5, 65–72 CAS.
- W. R. Windham, S.-C. Yoon, S. R. Ladely, J. A. Haley, J. W. Heitschmidt, K. C. Lawrence, B. Park, N. Narrang and W. C. Cray, J. Food Prot., 2013, 76, 1129–1136 CrossRef CAS PubMed.
- Y. Xu, Y. Dhaouadi, P. Stoodley and D. Ren, Curr. Opin. Biotechnol., 2020, 64, 79–84 CrossRef CAS PubMed.
- R. Kang, B. Park and K. Chen, Spectrochim. Acta, Part A, 2020, 224, 117386 CrossRef CAS PubMed.
- R. Kang, B. Park, M. Eady, Q. Ouyang and K. Chen, Sens. Actuators, B, 2020, 309, 127789 CrossRef CAS.
- G. Lu and B. Fei, J. Biomed. Opt., 2014, 19, 010901 CrossRef PubMed.
- W. Jun, M. S. Kim, K. Lee, P. Millner and K. Chao, Sens. Instrum. Food Qual. Saf., 2009, 3, 41–48 CrossRef.
- K. S. Kalasinsky, T. Hadfield, A. A. Shea, V. F. Kalasinsky, M. P. Nelson, J. Neiss, A. J. Drauch, G. S. Vanni and P. J. Treado, Anal. Chem., 2007, 79, 2658–2673 CrossRef CAS PubMed.
- Y. Wang, C. P. Reardon, N. Read, S. Thorpe, A. Evans, N. Todd, M. Van Der Woude and T. F. Krauss, npj Biofilms Microbiomes, 2020, 6, 1–7 CrossRef PubMed.
- J. Elliott, O. Simoska, S. Karasik, J. B. Shear and K. J. Stevenson, Anal. Chem., 2017, 89, 6285–6289 CrossRef CAS PubMed.
- J. Oziat, M. Gougis, G. G. Malliaras and P. Mailley, Electroanalysis, 2017, 29, 1332–1340 CrossRef CAS.
- N. R. Glasser, S. H. Saunders and D. K. Newman, Annu. Rev. Microbiol., 2017, 71, 731–751 CrossRef CAS PubMed.
- F. A. Alatraktchi, S. Breum Andersen, H. Krogh Johansen, S. Molin and W. E. Svendsen, Sensors, 2016, 16, 408 CrossRef PubMed.
- C. R. Santiveri, H. J. Sismaet, M. Kimani and E. D. Goluch, ChemistrySelect, 2018, 3, 2926–2930 CrossRef CAS.
- H. J. Sismaet, A. J. Pinto and E. D. Goluch, Biosens. Bioelectron., 2017, 97, 65–69 CrossRef CAS PubMed.
- T. A. Webster, H. J. Sismaet, I. J. Chan and E. D. Goluch, Analyst, 2015, 140, 7195–7201 RSC.
- P. Bouguer, Essai d'optique sur la gradation de la lumière, Claude Jombert, 1729 Search PubMed.
- A. C. Ward, P. Connolly and N. P. Tucker, PLoS One, 2014, 9, e91732 CrossRef PubMed.
- P. Dey, D. Parai, M. Banerjee, S. T. Hossain and S. K. Mukherjee, Int. J. Med. Microbiol., 2020, 310, 151410 CrossRef CAS PubMed.
- L. Flanagan, R. R. Steen, K. Saxby, M. Klatter, B. J. Aucott, C. Winstanley, I. J. S. Fairlamb, J. M. Lynam, A. Parkin and V.-P. Friman, Front. Microbiol., 2018, 9, 195 CrossRef PubMed.
- T. Das and J. Manos, in Progress in Understanding Cystic Fibrosis, ed. D. Sriramulu, InTech, 2017 Search PubMed.
- D. L. Bellin, H. Sakhtah, Y. Zhang, A. Price-Whelan, L. E. P. Dietrich and K. L. Shepard, Nat. Commun., 2016, 7, 10535 CrossRef CAS PubMed.
- D. L. Bellin, H. Sakhtah, J. K. Rosenstein, P. M. Levine, J. Thimot, K. Emmett, L. E. P. Dietrich and K. L. Shepard, Nat. Commun., 2014, 5, 3256 CrossRef PubMed.
- B. Rodriguez-Urretavizcaya, N. Pascual, C. Pastells, M. T. Martin-Gomez, L. Vilaplana and M.-P. Marco, Front. Cell. Infect. Microbiol., 2021, 11, 786929 CrossRef CAS PubMed.
- O. Simoska, M. Sans, M. D. Fitzpatrick, C. M. Crittenden, L. S. Eberlin, J. B. Shear and K. J. Stevenson, ACS Sens., 2019, 4, 170–179 CrossRef CAS PubMed.
- J. Kretzschmar and F. Harnisch, Curr. Opin. Electrochem., 2021, 29, 100757 CrossRef CAS.
- A. Price-Whelan, L. E. Dietrich and D. K. Newman, J. Bacteriol., 2007, 189, 6372–6381 CrossRef CAS PubMed.
- A. Price-Whelan, L. E. Dietrich and D. K. Newman, Nat. Chem. Biol., 2006, 2, 71–78 CrossRef CAS PubMed.
- M. Gajdács, Z. Baráth, K. Kárpáti, D. Szabó, D. Usai, S. Zanetti and M. G. Donadu, Antibiotics, 2021, 10, 1134 CrossRef PubMed.
- A. Buzid, F. Shang, F. J. Reen, E. Ó. Muimhneacháin, S. L. Clarke, L. Zhou, J. H. T. Luong, F. O'Gara, G. P. McGlacken and J. D. Glennon, Sci. Rep., 2016, 6, 30001 CrossRef CAS PubMed.
- J. W. Costerton, Z. Lewandowski, D. E. Caldwell, D. R. Korber and H. M. Lappin-Scott, Annu. Rev. Microbiol., 1995, 49, 711–745 CrossRef CAS PubMed.
- B. Behera, G. K. Anil Vishnu, S. Chatterjee, V. S. N. Sitaramgupta, V. N. Sreekumar, A. Nagabhushan, N. Rajendran, B. H. Prathik and H. J. Pandya, Biosens. Bioelectron., 2019, 142, 111552 CrossRef CAS PubMed.
- M. Magana, C. Sereti, A. Ioannidis, C. A. Mitchell, A. R. Ball, E. Magiorkinis, S. Chatzipanagiotou, M. R. Hamblin, M. Hadjifrangiskou and G. P. Tegos, Clin. Microbiol. Rev., 2018, 31, e00084 CrossRef CAS PubMed.
- T. Coenye, B. Kjellerup, P. Stoodley and T. Bjarnsholt, Biofilm, 2020, 2, 100012 CrossRef PubMed.
- N. Høiby, T. Bjarnsholt, C. Moser, P. Ø. Jensen, M. Kolpen, T. Qvist, K. Aanaes, T. Pressler, M. Skov and O. Ciofu, APMIS, 2017, 125, 339–343 CrossRef PubMed.
- J. Oziat, T. Cohu, S. Elsen, M. Gougis, G. G. Malliaras and P. Mailley, Bioelectrochemistry, 2021, 140, 107747 CrossRef CAS PubMed.
- L. Fu, Y. Zheng, P. Zhang, H. Zhang, Y. Xu, J. Zhou, H. Zhang, H. Karimi-Maleh, G. Lai, S. Zhao, W. Su, J. Yu and C.-T. Lin, Biosens. Bioelectron., 2020, 159, 112212 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Complementary HSI calibration data, electrochemical pyocyanin estimation and test cell detection. See DOI: https://doi.org/10.1039/d2sd00044j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |