Open Access Article
Open Access ArticleThrombin-linked aptamer assay for sensitive detection of immunoglobulin E in sandwich format†
Li-Min
Guo
ab and
Qiang
Zhao
 *acd
*acd
aState Key Laboratory of Environmental Chemistry and Ecotoxicology, Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing, 100085, China. E-mail: qiangzhao@rcees.ac.cn
bCollege of Traditional Chinese Medicine and Food Engineering, Shanxi University of Chinese Medicine, Jinzhong, 030619, China
cUniversity of Chinese Academy of Sciences, Beijing, 100049, China
dSchool of Environment, Hangzhou Institute for Advanced Study, UCAS, Hangzhou 310000, China
First published on 25th May 2022
Abstract
A thrombin-linked aptamer assay (TLAA) was developed with thrombin as an enzyme label for the detection of immunoglobulin E (IgE), an important biomarker for diseases. We constructed a sandwich assay for IgE using the anti-IgE antibody coated microplate and a DNA probe that contained the aptamer of IgE and the aptamer of thrombin. Taking advantage of aptamer affinity binding, the DNA probe could bind with IgE and thrombin, and the thrombin was labeled on the sandwich complex of IgE. As a protease, the thrombin catalyzed its fluorogenic peptide substrate to generate a fluorescent product. The assay showed a good linear relationship between the fluorescence signal and the IgE concentration ranging from 19 pM to 2.5 nM and a detection limit of 19 pM IgE. The detection limit further reached 0.625 pM when the TLAA was combined with rolling circle amplification (RCA) for multiple signal amplification. The TLAA exhibited high sensitivity and selectivity, and it allowed for determination of IgE in diluted human serum samples. This TLAA for IgE detection is promising for applications.
Introduction
Aptamers are short single-stranded nucleic acids selected from random sequence nucleic acid libraries by SELEX (systematic evolution of ligands by exponential enrichment) technology.1,2 They have been widely used as recognition elements for various targets with high affinity and specificity.3–5 Compared with antibodies, aptamers exhibit significant advantages such as low cost, good stability and reproducibility, facile labeling, and target versatility.6–8Thrombin is a central protease that plays an important role in the coagulation cascade. It is also related to various diseases, such as vascular diseases, metastasis, and cancers.9,10 A 15-mer DNA oligonucleotide (Kd ∼100 nM) and another 29-mer DNA oligonucleotide (Kd ∼0.5 nM)11,12 are two widely used aptamers for thrombin. Thrombin can still maintain enzymatic activity when the aptamers bind with thrombin because the binding sites of aptamers are different from the enzyme catalytic sites. Thrombin and its aptamers are often used as models in many aptamer-based assays.13–15 Most of research focused on the detection of thrombin with aptamers. Taking advantage of aptamer binding affinity and the enzymatic activity of thrombin in cleaving peptide substrates, we previously reported thrombin linked aptamer assays (TLAA) for the detection of platelet derived growth factor-BB (PDGF-BB).16,17 This strategy converted detection of PDGF-BB to the measurement of thrombin by using a DNA sequence that consists of two aptamers with the first aptamer binding to the PDGF-BB and the second aptamer binding to thrombin, the thrombin was labeled on the affinity complex, and thrombin catalyzed the peptide substrate into product, generating signals for final quantification of PDGF-BB. This TLAA contributes a new application of thrombin and its aptamer in bioanalysis, and provides a new way for signal generation.
Human immunoglobulin E (IgE) is important in human allergic responses, and it is found at low levels in human serum (∼1 nM);18,19 however, it significantly increases in patients afflicted with allergic asthma, atopic dermatitis, and other immune deficiency related diseases, such as asthma.20 IgE is biomarker for diseases. Sensitive detection of IgE is in demand in clinical diagnostics. At present, the main methods to detect IgE include antibody based immunoassays and aptamer based methods. Shin et al.21 reported electrochemical immunosensor to detect IgE in a sandwich format, and detection limit was about 4.25 pM due to signal amplification by alkaline-phosphatase (ALP). Li et al.22 presented a method based on aptamer-modified nanoparticles as detection probe and silver enhancement to improve sensitivity, allowing detection of 60 pM IgE.
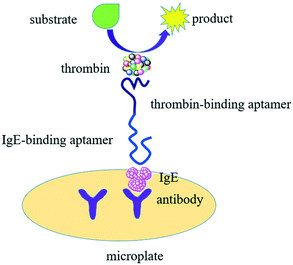
Here, we demonstrated detection of (IgE) with TLAA in a sandwich format on the microplate by using aptamers against IgE and anti-IgE antibody, taking advantage of aptamer binding, affinity labeling and thrombin catalysis. Fig. 1 shows the principle of the TLAA strategy for IgE detection. Firstly, the antibody of IgE is immobilized on the surface of microplate. The IgE is captured by antibody-coated microplate, and then it binds with a DNA probe containing the aptamer of IgE and the aptamer of thrombin, forming a sandwich complex. Then, thrombin is attached on the DNA probe through affinity interaction between aptamer and thrombin. Finally, thrombin catalyzes the cleavage of a fluorogenic peptide substrate to yield fluorescent product. With the demonstrated TLAA, we achieved sensitive and selective detection of IgE.
Experimental section
Reagents and materials
The high-binding black 96-well NUNC Maxisorp plates were used. Immunoglobulin E (IgE) was purchased from Athens Research & Technology. Anti-IgE antibody, bovine serum albumin (BSA), human immunoglobulin G (IgG), hemoglobin (Hb) and lysozyme (Lys) were obtained from Sigma. Human α-thrombin was bought from Haematologic Technologies Inc. The fluorogenic peptide substrate of thrombin, N-p-tosyl-Gly-Pro-Arg-7-amido-4-methylcoumarin hydrochloride, was purchased from Sigma. The ultrapure water was obtained through a Purelab Ultra Elga Labwater system. A microplate reader (Varioskan Flash, Thermo Fisher Scientific, Inc) was used to record the fluorescence signals. Oligonucleotides were synthesized and purified by Sangon Biotech (Shanghai, China). The sequence information of oligonucleotides was shown in Table S1 in ESI.†The following buffers were used in the experiments. Coating buffer consisted of 0.1 M Na2CO3 (pH 9.6) solution. Blocking buffer contained PBS (pH 7.5) and 10 mg mL−1 BSA. Assay buffer consisted of PBS (pH 7.5), 1 mM MgCl2 and 4 mg mL−1 BSA. SSC buffer (pH 7.2) consisted of 0.3 M NaCl and 0.03 M sodium citrate (Na3C6H5O7). Ligation buffer consisted of 30 mM Tris–HCl (pH 7.5), 4 mM MgCl2, 10 mM (NH4)2SO4, 0.1 mM nicotinamide adenine dinucleotide (NAD), 1.2 mM EDTA, and 0.05 mg mL−1 BSA. Thrombin catalysis buffer was composed of 50 mM Tris–HCl (pH 8.5) and 1 M NaCl. Three washing buffers were used, including washing buffer A (PBS and 0.1% Tween 20, pH 7.5), washing buffer B (PBS, 1 mM MgCl2 and 0.1% Tween 20, pH 7.5), and washing buffer C (SSC buffer with 0.05% Tween 20).
Assay procedure of the TLAA
Firstly, the antibody of IgE was absorbed on the cell surface of microplate. For this purpose, the antibody solution in coating buffer (2 μg mL−1, 100 μL per well) was added to the cell of microplate and incubated overnight at 4 °C. Then, the antibody coated microplate was rinsed thoroughly with washing buffer A for three times to remove any unbound antibody, followed by placing 200 μL per well blocking buffer for 1 h into the well of microplate to reduce nonspecific binding. Subsequently, various concentrations of IgE in assay buffer (100 μL per well) reacted with the antibody coated microplate for 30 min, and the microplate was then rinsed with washing buffer B for three times. Next, 100 μL of 25 nM oligonucleotide probe (IgE45-6 T-TH29) in assay buffer was added into the wells of microplate and was allowed to react for 30 min. Once the sandwich complex of antibody, IgE and oligonucleotide probe was formed, the microplate was rinsed with washing buffer B for three times. The microplate was treated with 10 nM thrombin in assay buffer (100 μL per well) for 30 min. Finally, after washing with washing buffer B for three times, 100 μL of thrombin catalysis buffer containing 0.06 mM substrate was added into the wells of microplate and incubated for 1 h. The fluorescence was measured immediately on a microplate reader with excitation and emission at 370 nm and 440 nm, respectively. The incubation was conducted at room temperature in all steps of experiments.Assay procedure of the TLAA coupled with RCA
100 μL of solution containing antibody of IgE (2 μg mL−1) in coating buffer was added in the wells of microplate and incubated overnight at 4 °C. Following by washing with washing buffer A for three times, the wells were blocked with the blocking buffer (200 μL per well) for 1 h. After decantation of blocking buffer, the wells were washed once with washing buffer A.A series of concentrations of IgE in assay buffer were added to the wells of microplate (100 μL per well) and incubated for 30 min. Unbound IgE was removed by washing with washing buffer B for three times. 100 μL of 200 nM IgE45-primer in assay buffer was added and incubated for 30 min to allow the formation of sandwich complexes in the wells of the microplate, the IgE45-primer was heated at 85 °C for 3 min and cooled to room temperature prior to introduction to the wells. After washing for three times with washing buffer A, 100 μL of 50 nM template in SSC buffer containing 0.05% Tween20 was allowed to hybridize with IgE45-primer for 1 h. The excess template was removed by rinsing with washing buffer C for three times. After that, the template was circularized via ligation by 100 μL of ligase in ligation buffer (0.02 U μL−1) for 1 h, forming the circular template for RCA. Then, 100 μL of RCA buffer containing 0.4 mM dNTPs, 0.05 U μL−1 phi29 DNA polymerase, and 0.1 mg mL−1 BSA was introduced into the wells. The RCA reaction was conducted for 1 h, followed by rinsing with washing buffer C for three times. 100 μL of 20 nM thrombin was introduced into the wells, and incubated for 30 min with the immobilized RCA products. After washing the wells with buffer B for three times, thrombin catalyzed cleavage of the fluorogenic substrate (0.059 mM) in 100 μL of thrombin catalysis buffer for 1 h, and the fluorescence was recorded by a plate reader with excitation and emission at 370 nm and 440 nm, respectively. The incubation was finished at room temperature in all steps of experiments.
Results and discussion
Feasibility of TLAA for IgE detection
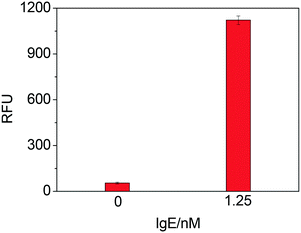
As Fig. 1 shows, a sandwich format assay was employed in our design, which involved the capture antibody immobilized on the microplate, the target IgE and an oligonucleotide probe containing the IgE aptamer and the thrombin aptamer. We first tested the feasibility of TLAA for detection of IgE. As shown in Fig. 2, in the presence of the 1.25 nM IgE, a strong fluorescence signal was observed. It indicated that the sandwich complex was formed and the thrombin was labeled on the complex. In contrast, in the absence of IgE, the sandwich complex cannot be formed, and the fluorescence signal was negligible. The result shows that the TLAA can be used to detect IgE.Optimization of experimental conditions
In order to achieve the best signal, several experimental parameters were systematically investigated. Firstly, the linker of oligonucleotide probe was a key parameter for a successful assay system for IgE. We investigated the influence of the length and type of the linker between the two aptamer sequences in the oligonucleotide probe. Based on the type of linker, eight oligonucleotide probes were classified into two distinct groups. Fig. S1† shows a comparison between four oligonucleotide probes with poly-A linker and four oligonucleotide probes with poly-T linker. The oligonucleotide probes with poly-T linker from the same group were found to have higher ΔRFU compared with that with poly-A linker. We also compared three poly-T linkers for oligonucleotide probes with 6 T's, 9 T's and 12 T's, and the oligonucleotide probes with 9 T's and 12 T's had similar ΔRFU, lower than that obtained by the oligonucleotide with 6 T's. A comparison between two oligonucleotide probes, i.e. IgE45-6 T-TH29 and TH29-6 T-IgE45, was carried out in parallel. The data showed that the ΔRFU obtained by IgE45-6 T-TH29 was higher than that from TH29-6 T-IgE45, and thus the oligonucleotide probe IgE45-6 T-TH29 consisting of 6 T linker was selected for subsequent experiments.The presence of divalent cations such as Mg2+ has been reported to be necessary for certain aptamer–protein complex formation.23 The result in the presence of MgCl2 was obviously different from that in the absence of Mg2+ (Fig. S2†). 1 mM Mg2+ was optimized to give the highest fluorescence intensity, suggesting Mg2+ at this concentration may help the aptamer to have improved affinity. Further increasing the concentration of Mg2+ decreased the intensity, possibly affecting the affinity binding, which was in agreement with the previous report.24,25
The effect of IgE antibody concentration was studied by varying the concentration of antibody to coat microplate and evaluating the ΔRFU. Fig. 3 showed that the ΔRFU increased with increase of the antibody concentration in the range of 0.5–2 μg mL−1. The ΔRFU reached a plateau when the antibody concentration exceeded 2 μg mL−1, indicating the amount of immobilized antibody has reached saturated level on the microplate. Thus, the antibody concentration of 2 μg mL−1 was used in the further experiments.
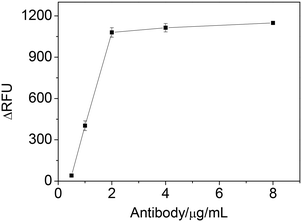 | ||
| Fig. 3 The effect of antibody concentration on ΔRFU obtained from IgE sample. Experimental conditions: 1.25 nM IgE; 25 nM IgE45-6 T-TH29; 2.5 nM thrombin; 1 mM Mg2+; 0.06 mM substrate. | ||
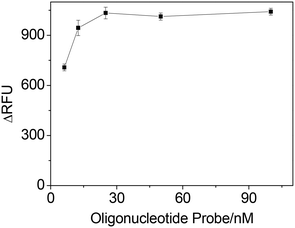
Fig. 4 shows the ΔRFU at different concentrations of oligonucleotide probe (IgE45-6 T-TH29). A significant improvement in ΔRFU was obtained when increasing the oligonucleotide probe concentration from 6.25 nM to 25 nM, which may enhanced the IgE-aptamer binding. When the concentration of oligonucleotide probe was further increased from 25 nM to 100 nM, there was no improvement in the ΔRFU. Thus, we chose 25 nM oligonucleotide probe for the next study.
In order to optimize the thrombin concentration, the ΔRFU obtained at different thrombin concentrations was examined. As shown in Fig. S3,† the ΔRFU was observed to increase at the beginning with the increase of the thrombin concentration, and then maintained an approximately steady level when thrombin concentration was greater than 2.5 nM. It suggested that the 2.5 nM thrombin was enough to catalyze the substrate.
Detection of IgE
Under the optimal experiment conditions, we examined the analytical performances of the developed method upon the addition of different concentrations of IgE and then measured the fluorescence signal. As shown in Fig. 5, the ΔRFU increased remarkably with the increasing of IgE concentrations from 0.019 to 2.5 nM, and became saturated when IgE concentration was higher than 2.5 nM. ΔRFU showed a linear relationship with the concentration of IgE in the range of 0.019–2.5 nM, represented by y = 796.16x − 17.03 (R2 = 0.993) (y is the ΔRFU, and x is the concentration of IgE with nM unit). The detection limit was calculated to be 0.019 nM (based on signal/noise [S/N] = 3).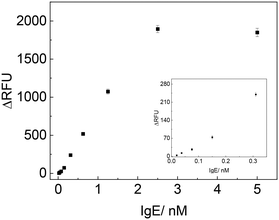 | ||
| Fig. 5 Detection of varying concentration of IgE with TLAA (the inset showed the signals corresponding to low concentrations of IgE). | ||
To further improve the assay sensitivity, we combined the TLAA with RCA, and the principle was shown in Fig. S4A.† In this TLAA coupled with RCA, IgE was captured by antibody on the microplate, and then the IgE45-primer was added to form a sandwich complex. After that, a template that was encoded with a complementary sequence of the aptamer for thrombin hybridized with the primer, and was circularized by ligase. The primer was extended around the circle template by RCA reaction to produce long nucleic acid product containing many thrombin aptamer sequence units. The RCA product captured many thrombin molecules, achieving multiple thrombin labels in the sandwich complex. Finally, thrombin catalyzed the fluorogenic peptide substrates into detectable products.
Through RCA and thrombin catalysis, this assay achieved double signal amplification, and the sensitivity was improved. As shown in Fig. S4B,† the ΔRFU increased linearly as the IgE concentration was increased from 0.625 pM to 38 pM (y = 26.75x + 10.7, R2 = 0.994; here concentration unit for x is pM). IgE concentration as low as 0.625 pM could be detected. From the slopes of the two linear fitting equations, we could estimate that the sensitivity of TLAA coupled with RCA was 34 times that of TLAA, indicating more than 34 thrombin molecules were captured by an RCA product.
The analytical performance of TLAA was comparable to or better than some previously reported methods based on aptamer for the determination of IgE,19,22,24,26–29 as the comparison results summarized in Table S2† showed.
Specificity test
To evaluate the selectivity of the TLAA, we tested some potential interfering proteins, including thrombin, platelet-derived growth factor-BB (PDGF-BB), lysozyme, hemoglobin and IgG. As shown in Fig. 6, negligible signals were obtained in the presence of thrombin, PDGF-BB, lysozyme, hemoglobin and IgG, while an obvious increase in fluorescence signal was observed in the presence of a low concentration of IgE. These results indicated that the fluorescence signal generated from the specific interaction between the aptamer and IgE. Therefore, the developed assay has high specificity toward IgE detection.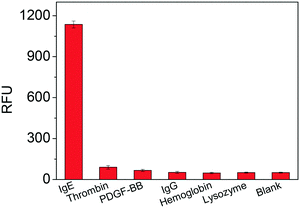 | ||
| Fig. 6 Specificity test of the proposed TLAA for IgE assay. The tested proteins included IgE (1.25 nM), thrombin (2.5 nM), PDGF-BB (2.5 nM), IgG (2 μM), hemoglobin (2 μM) and lysozyme (2 μM). | ||
Detection of IgE in dilute serum sample
In order to demonstrate the applicability of this assay to complex sample analysis, we applied TLAA to detect IgE in human serum. Serum contains many other proteins, and the components are complicated, so the serum was diluted by 100-fold with binding buffer to reduce the matrix effect. Fig. S5† showed the IgE assay performed in 1% human serum. We noticed that the fluorescence signals obtained in diluted serum samples were similar to those in buffer solutions. The ΔRFU increased upon increasing the concentration of IgE. The linear relationship between ΔRFU and the concentration of IgE was obtained in the range from 0.019 to 2.5 nM (y = 750.7x − 9.07, R2 = 0.995) and the detection limit was 0.019 nM (based on signal/noise [S/N] = 3). The recoveries of different concentrations of IgE in human serum were also calculated and shown in Table S3,† with recoveries ranging from 87.5% to 118.5%. The result indicates the proposed method can be used in the real sample analysis.Conclusions
In summary, a thrombin linked aptamer assay (TLAA) was developed for IgE detection in a sandwich format by using thrombin as label for signal amplification. The demonstrated TLAA had high sensitivity and could detect IgE at a concentration as low as 19 pM. When TLAA was coupled with RCA, a lower detection limit of 0.625 pM IgE was obtained due to multiple signal amplifications and improved sensitivity. The assay was selective, and allowed to detect IgE in diluted serum. The success of the TLAA for IgE detection in this work shows TLAA may be used for various proteins or molecules with two binding sites, and expand the application of thrombin and its aptamers.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful for the financial support from National Natural Science Foundation of China (22074156, 81803725). The serum was obtained from Zhongke Chenyu Biotechnology (Beijing, China).Notes and references
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CrossRef CAS PubMed.
- H. Nevídalová, L. Michalcová and Z. Glatz, Electrophoresis, 2020, 41, 414–433 CrossRef PubMed.
- G. Ravichandran and A. K. Rengan, ACS Appl. Nano Mater., 2020, 3, 9542–9559 CrossRef CAS.
- M. H. Ali, M. E. Elsherbiny and M. Emara, Int. J. Mol. Sci., 2019, 20, 2511 CrossRef CAS PubMed.
- Y. Saw, C. Marimuthu, G. Subash and T. T. Hock, Biosens. Bioelectron., 2015, 64, 392–403 CrossRef PubMed.
- M. Tabarzad and M. Jafar, Protein J., 2016, 35, 81–99 CrossRef CAS PubMed.
- Z. S. Pehlivan, M. Torabfam, H. Kurt, C. O. Yang, N. Hildebrandt and M. Yüce, Microchim. Acta, 2019, 186, 563 CrossRef CAS PubMed.
- B. Deng, Y. W. Lin, C. Wang, F. Li, Z. X. Wang, H. Q. Zhang, X. F. Li and X. C. Le, Anal. Chim. Acta, 2014, 837, 1–15 CrossRef CAS PubMed.
- A. S. Remiker and J. S. Palumbo, Thromb. Res., 2018, 164, S29–S33 CrossRef CAS PubMed.
- L. C. Bock, L. C. Griffin, J. A. Latham, E. H. Vermaas and J. J. Toole, Nature, 1992, 355, 564–566 CrossRef CAS PubMed.
- D. M. Tasset, M. F. Kubik and W. Steiner, J. Mol. Biol., 1997, 272, 688–698 CrossRef CAS PubMed.
- A. Gosai, B. Shin, H. Yeah, M. N. Hamilton and P. Shrotriya, Biosens. Bioelectron., 2019, 126, 88–95 CrossRef CAS PubMed.
- S. Umrao, V. Jain, Anusha, B. Chakraborty and R. Roy, Sens. Actuators, B, 2018, 267, 294–301 CrossRef CAS.
- I. Isildak, F. Navaeipour, H. Afsharan, G. S. Kanberoglu, I. Agir, T. Ozer, N. Annabi, E. E. Totu and B. Khalilzadeh, Microchim. Acta, 2020, 187, 25 CrossRef CAS PubMed.
- L. M. Guo, L. H. Hao and Q. Zhao, Anal. Bioanal. Chem., 2016, 408, 4715–4722 CrossRef CAS PubMed.
- L. M. Guo and Q. Zhao, Talanta, 2016, 158, 159–164 CrossRef CAS PubMed.
- Y. S. E. Ansari, C. Kanagaratham, O. L. Lewis and H. C. Oettgen, Adv. Immunol., 2020, 148, 93–153 Search PubMed.
- M. Y. He, N. Shang, Q. R. Zhu and J. Xu, Anal. Chim. Acta, 2021, 1143, 93–100 CrossRef CAS PubMed.
- A. C. Lessmann, E. Curto, E. F. Mateus and L. Soto, PLoS One, 2020, 15, e0228045 CrossRef PubMed.
- S. J. Oha, J. K. Ahna, H. Parka, Y. Songb, S. J. Kwonb and H. B. Shin, Anal. Methods, 2019, 11, 4410–4415 RSC.
- Y. M. Liu, J. J. Yang, J. T. Cao, J. J. Zhang, Y. H. Chen and S. W. Ren, Sens. Actuators, B, 2016, 232, 538–544 CrossRef CAS.
- E. J. Cho, J. R. Collett, A. E. Szafranska and A. D. Ellington, Anal. Chim. Acta, 2006, 564, 82–90 CrossRef CAS PubMed.
- X. X. Lin, C. Y. Yu, H. G. Lin, C. Wang, J. L. Su, J. Cheng, R. K. Kankala and S. F. Zhou, Sensors, 2019, 19, 2224 CrossRef CAS PubMed.
- X. H. Fang, Z. H. Cao, T. Beck and W. H. Tan, Anal. Chem., 2001, 73, 5752–5757 CrossRef CAS PubMed.
- A. Salimi, S. Khezrian, R. Hallaj and A. Vaziry, Anal. Biochem., 2014, 466, 89–97 CrossRef CAS PubMed.
- G. F. Shi, J. T. Cao, J. J. Zhang, Y. M. Liu, Y. H. Chen and S. W. Ren, Sens. Actuators, B, 2015, 220, 340–346 CrossRef CAS.
- Z. M. Li, Z. H. Li, Q. Q. Niu, H. Li, M. Vuki and D. K. Xu, Sens. Actuators, B, 2017, 239, 45–51 CrossRef CAS.
- B. Y. Jiang, F. Z. Li, C. Y. Yang, J. Q. Xie, Y. Xiang and R. Yuan, Anal. Chem., 2015, 87, 3094–3098 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sd00059h |
| This journal is © The Royal Society of Chemistry 2022 |