Open Access Article
Open Access ArticleMonolith-modified cellulose paper for biochemical sensing applications
Wei-Yi
Chu
a,
Chun-Hui
Yang
a,
Roman
Viter
b,
Arũnas
Ramanavičius
 c,
Shyh-Chyang
Luo
c,
Shyh-Chyang
Luo
 d and
Chien-Fu
Chen
d and
Chien-Fu
Chen
 *a
*a
aInstitute of Applied Mechanics, National Taiwan University, Taipei 10617, Taiwan. E-mail: stevechen@ntu.edu.tw
bInstitute of Atomic Physics and Spectroscopy, University of Latvia, LV-1004 Riga, Latvia
cDepartment of Physical Chemistry, Faculty of Chemistry, Vilnius University, Naugarduko 24, LT-03225 Vilnius, Lithuania
dDepartment of Materials Science and Engineering, National Taiwan University, Taipei 10617, Taiwan
First published on 11th August 2022
Abstract
In this study, we developed a microfluidic paper-based analytical device (μPAD) modified with a poly(GMA-co-EDMA) monolith for rapid tuberculosis detection. GMA-co-EDMA successfully enhanced the affinity between the paper substrate and pipetted biomolecules through covalent interactions between the opened epoxy ring and functional groups such as thiol and amino groups on the biomolecules. After synthesizing monoliths onto cellulose paper, fabrication parameters such as wax melting time duration and temperature were tested. We determined the optimized parameters for fabricating a surface-modified microfluidic-based analytical device (sPAD) after observing the modified paper surface with scanning electron microscopy (SEM) images, contact angle measurement, and colorimetric immunoassays. The optimized enzymatic reaction is 5 minutes after testing IgG from human serum (HIgG) on the sPAD. Finally, tuberculin purified protein derivative (PPD) as an analyte can be detected within 30 minutes with a detection limit of less than 0.11 ng mL−1. These results indicate that our device possesses the potential for rapid and sensitive tuberculosis detection in resource-limited settings.
Introduction
Tuberculosis (TB), a fatal infectious disease,1 led to 1.5 million deaths in 2020 according to the Global Tuberculosis Report 2021 by the World Health Organization (WHO).2 To prevent the spread of TB, detection of its etiopathogenic agent, Mycobacterium tuberculosis, is crucial for early diagnosis and treatment. Typically, a sputum sample from a suspected patient is cultured for 6 to 8 weeks to determine whether M. tuberculosis is present in the collected sample.3,4 Although the method is effective for correct diagnostics, the efficiency is considered to be low. To speed up the process, the WHO recommended nucleic acid amplification tests (NAATs) that are capable of completing the detection within a day for high-sensitivity TB diagnosis.5 However, the cost of the instrument and operational requirements, such as power sources and trained professionals, limit its applicability, especially in resource-limited regions where a large number of TB deaths occur. Thus, point-of-care (POC) devices that are sensitive, rapid in detection, and affordable have been developed for TB detection.6Microfluidic paper-based analytical devices (μPADs) are a typical type of POC devices. Because they use a cellulose-based paper substrate, μPADs have certain benefits, including being inexpensive, disposable, and biocompatible, which makes μPADs suitable for on-site detection.7–10 Typically, biomolecules such as proteins and antibodies are first added onto the cellulose paper surface and then trapped onto the surface by the porous structure of the cellulose paper. The trapped biomolecules, therefore, act as biosensors and target biomarkers in disease diagnosis.11 However, the trapping is not strong enough, and detachment of the biomolecules can occur after multiple reactions and washing steps. To fully immobilize biomolecules on the paper surface, surface modification is required since a modified layer can enhance the overall surface area for biomolecular trapping and reduce the movement of molecules in assays.12 The use of water gel,13 porous materials,14 and nanomaterials has been reported for paper surface modification.15–17 Specifically, these materials decrease biomolecule mobility by enhancing the adsorption of biomolecules onto the paper surface, wherein physical interactions such as electrostatic adsorption and hydrophobic adsorption are the most common.18,19 However, the physical adsorption interaction of biomolecules with the modified layer is only slightly stronger than that with the unmodified paper surface.20 Thus, immobilization methods based on covalent binding have been introduced to address this problem.21 In previous research, we used carboxymethyl cellulose (CMC) and 1-ethyl-3-(-3-dimethylaminopropyl) carbodiimide hydrochloride/N-hydroxysuccinimide (EDC/NHS) to modify cellulose paper. Since the interaction between proteins and the modified layer mainly involved covalent binding, the positions of the proteins did not change, leading to successful detection of the target biomarker. Despite the promising results, the degree of surface modification should be further enhanced. The presence of the same charge on cellulose paper and the modified layer leads to repulsion between them.22 Therefore, a novel surface modification that is capable of covalently binding to both cellulose paper and biomolecules is discussed.
Monoliths are a type of porous material that are commonly applied in catalytic reactions,23,24 adsorption processes,25 gas or liquid chromatography,26 electrophoresis,27 and other phase separation processes.28 For separation processes, monoliths act as stationary phases, as the well-distributed nanometer- or micrometer-sized pores enhance the surface area of the monolith and allow the trapping of molecules.29 Typically, the monolith is thermally polymerized by monomers to form a porous structure with high mechanical stability.30 Since many monomers are capable of forming monoliths with porous structures, there is high flexibility in choosing suitable monomers according to the final application.31–33 Among all types of monoliths, the monolith formed by copolymerization of glycidyl methacrylate (GMA) and ethylene glycol dimethacrylate (EDMA) is promising for use in biomolecular separation and adsorption processes since it can covalently bond with thiol, amine, and carboxyl groups in biomolecules via the epoxy groups of GMA.34,35 GMA would also form a branched structure during polymerization, preventing hydrolysis after long-term reactions or storage,36 which can increase the separation efficiency. Moreover, the poly(GMA-co-EDMA) monolith can be synthesized through only photopolymerization at room temperature, which can be completed within several minutes. In addition to the abovementioned advantages, the proposed monolith can adsorb biomolecules covalently, the interaction was strong enough to endure washing steps, and the monolith can be prepared rapidly. Thus, poly(GMA-co-EDMA) monolith has great potential in cellulose paper surface modification.
In this work, we combine a monolith and μPAD to form a novel surface-modified microfluidic-based analytical device (sPAD) with better biomolecular immobilization efficiency and stability for TB detection. The poly(GMA-co-EDMA) monolith was first synthesized in situ through photopolymerization at room temperature. To understand the changes in the properties of the paper surface, the contact angle and morphology were measured before and after photopolymerization. Then, we optimized the enzyme reaction time by testing human immunoglobulin G (HIgG) on our device to determine the optimized biomolecular immobilization time. Finally, we performed TB detection on the sPAD by spiking the test samples with different concentrations of tuberculin PPD, which was obtained from M. tuberculosis. The results showed that our device can successfully detect the presence of tuberculin PPD and had a low limit of detection (LOD). We believe that the proposed monolith surface modification method has great potential to be applied to paper surface modification in the future for the detection of different types of diseases.
Experimental section
Materials
Glycidyl methacrylate (GMA) and ethylene glycol dimethacrylate (EDMA), which are used for monolith synthesis, were purchased from Alfa Aesar (Haverhill, MA). Other chemicals used throughout the experiments, such as methanol, ethanol, 2,2-dimethoxy-2-phenylacetophenone (DMPA), bovine serum albumin (BSA), IgG from human serum (HIgG), anti-human IgG (aHIgG), anti-rabbit-IgG (aRIgG), tetramethyl-benzidine (TMB), phosphate-buffered saline (PBS), phosphate buffered saline with Tween 20 (PBST), tuberculin purified protein derivative (PPD), and Mycobacterium tuberculosis polyclonal antibody (MpAb), were all obtained from Sigma-Aldrich (St. Louis, MO). Cellulose paper produced by Whatman® (Maidstone, UK) was used as a substrate for our proposed device.Fabrication of poly(GMA-co-EDMA)-modified cellulose paper
The poly(GMA-co-EDMA) monolith was prepared in the dark by mixing reagents to form a 12 mL mixture with 24 wt% GMA, 16 wt% EMA, 30 wt% methanol, 30% ethanol, and 1 wt% DMPA.37 To thoroughly mix all the reagents, the mixture was sonicated for 10 minutes in the dark. The resulting well-mixed solution was then pipetted onto a piece of cellulose paper, which was later put under a xenon light source (LAX-C100, ASAHI Spectra, Tokyo, Japan) and exposed to 250 nm UV radiation for 6 minutes for photopolymerization. To avoid UV radiation in specific areas to prevent monolith polymerization in unwanted positions, we covered this part of cellulose paper with a glass sheet and black adhesive tape to avoid UV exposure. Subsequently, the cellulose paper was rinsed with PBST and water, dried at room temperature, and rested in a vacuum for 24 hours. The characterization was performed by X-ray photoelectron spectroscopy (XPS; PHI 5000 VersaProbe, ULVAC, Japan) after the abovementioned process to observe the chemical composition of the surface after surface modification.Fabrication of the sPAD and surface analysis
The sPAD was fabricated using a wax printer (ColorQube 8570, Xerox, Fuji, Japan) to print hole-shape solid wax onto hydrophilic circles that were 5 mm in diameter.38 After forming the poly(GMA-co-EDMA) monolith at the detection area, the paper was heated at 70 °C, 80 °C, and 90 °C for either 15 minutes or 30 minutes to determine the optimized fabrication parameters. We only modified the detection zones to prevent damage to the structure of the monolith when folding the device. To observe whether the morphology of the monolith changed, a scanning electron microscope (SEM; JSM-7600F, JEOL, Tokyo, Japan) was employed. Then, to determine that the heating process had fully melted the wax onto the back of the cellulose paper, a contact angle meter (OCA20, Dataphysics, Germany) was used to measure the testing area at the back of the cellulose paper.ELISA testing on the sPAD for enzyme reaction time determination
To optimize the parameters for enzymatic reactions, we first performed an HIgG ELISA (enzyme-linked immunosorbent assay) test on the sPAD to evaluate the performance. For this, 1 mg mL−1 HIgG solution was prepared by dissolving HIgG in a 150 mM NaCl solution. The HIgG solution was then separated into several vials, and different amounts of PBS were added to obtain solutions with HIgG concentrations ranging from 0.003 to 30 ng mL−1. Then, 3 μL of each HIgG solution was gradually pipetted to the sPAD, followed by the addition of 3 μL of 1% BSA to block the surface. Subsequently, 3 μL of 3.33 μg mL−1 anti-HIgG-HRP in PBS was added onto the sample to bind with HIgG. Washing was performed by adding 50 μL of PBST. When the sPAD had fully dried, TMB was added, and the enzyme was allowed to react for 3 to 6 minutes to obtain a result. The colorimetric results were captured by a portable microscope (UPG 650, Upmost, Taiwan). We analyzed the red and blue intensities by ImageJ (National Institutes of Health, USA) and quantified the color change by calculating the ratio between the blue and red intensities (B/R value).Comparison of tuberculosis testing on the μPAD and sPAD
To determine the difference in performance between the conventional μPAD and sPAD, tuberculosis testing was performed on both platforms. Different concentrations of tuberculin PPD, ranging between 0.004 and 40 ng mL−1, in PBS were first added to the detection area. When the substrate surface had fully dried, 3 μL of 1% BSA, 3 μL of 25 μg mL−1 MpAb in PBS, and 3 μL of 58 μg mL−1 aRIgG-HRP in PBS were pipetted onto the paper surface in sequence for the μPAD, and 3 μL of 1% BSA, 3 μL of 5 μg mL−1 Mtb in PBS, and 3 μL of 11 μg mL−1 aRIgG-HRP in PBS were pipetted onto the sPAD. Subsequently, we washed both devices with 50 μL of PBST and then added TMB to obtain the results. The colorimetric results were captured by a portable microscope, and the B/R value was calculated.Results and discussion
In this study, we developed a novel surface-modified cellulose paper-based analytical device with a poly(GMA-co-EDMA) monolith, and the device can be used for rapid tuberculosis detection, as shown in Fig. 1(a). The proposed monolith was first synthesized through photopolymerization and then covalently bonded with antigens via epoxy groups, enabling strong immobilization of antigens after multiple washing steps, as shown in Fig. 1(b). After optimizing the immune testing reaction parameters with HIgG, the detection performance of the unmodified μPAD and sPAD toward tuberculosis was determined, as shown in Fig. 1(c). The detection limit decreased due to stronger and more widespread attachment of antigens on the paper surface compared with the unmodified sPAD. Moreover, since the monolith proposed in this research is only composed of carbon, hydrogen, and oxygen, the carbonizable composition enables the surface-modified paper to be directly incinerated after use. These advantages and results indicate that the monolith-modified sPAD is a great choice for rapid on-site detection of tuberculosis.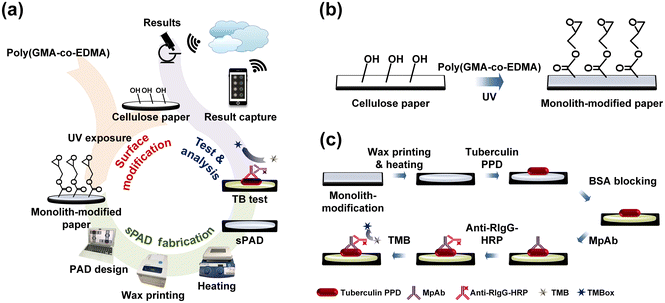 | ||
| Fig. 1 Schematic illustration of the sPAD. (a) Overall tuberculosis detection procedure. (b) Monolith surface modification mechanism. (c) Steps for detection pad fabrication and TB detection. | ||
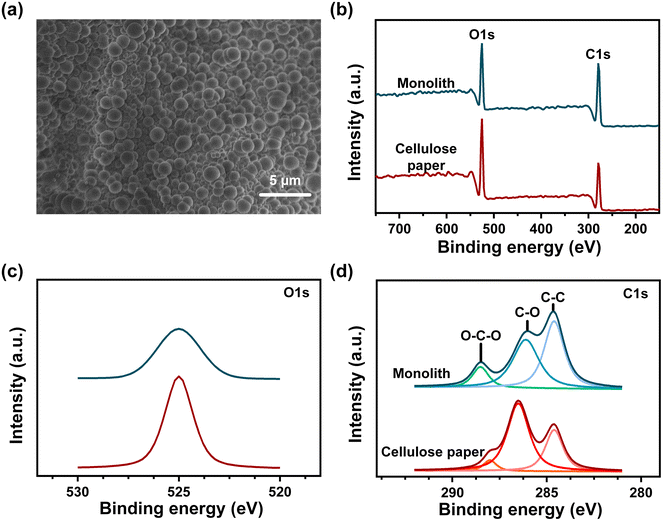
Chemical composition of monolith-modified cellulose paper
In this study, poly(GMA-co-EDMA) was utilized to enhance the detection limit of the μPAD, forming the novel sPAD. It was reported that poly(GMA-co-EDMA) is capable of covalently binding with carboxyl groups,39 therefore, this mechanism was applied to immobilize biomolecules covalently to the proposed sPAD. The synthesis of the monolith layer began with the addition of the GMA and EDMA mixture to the surface of cellulose paper. To fully cross-link the monomers into monoliths and secure the formed structure on the surface, a UV source was used for the photopolymerization reaction. The formed monolith was observed by SEM and XPS to determine the surface morphology and chemical composition before and after surface modification. Further investigation was performed at O1s (532 eV) and C 1s (286.5 eV) via XPS to obtain a detailed understanding of possible bond formation. Fig. 2(a) is an SEM image of the surface-modified cellulose paper. Circular structures were adequately distributed all over the paper surface, indicating that the monolith fully covers the surface of cellulose paper. The high coverage of monolith on the surface ensures the binding ability between the paper and biomolecule at any position of the surface-modified paper. The chemical composition of these circular structures can be better understood through the overall XPS diagram in Fig. 2(b). Under both conditions, before and after monolith modification, two peaks were present, indicative of O 1s and C 1s. The results show that the O 1s peak intensity decreased sharply as the intensity of the C 1s peak increased after surface modification. Since the monolith provides more carbon sources than oxygen sources,40 the changing ratio of oxygen to carbon leads to changes in peak intensities. Further investigation was then performed at O 1s and C 1s, as indicated in Fig. 2(c) and (d), respectively. By simply investigating the change in intensity at C 1s, the binding energy of C–O was found to decrease, while the C–C and O–C–O binding energies increased after surface modification. Since the O–C–O binding energy was approximately 288.7 eV, it is more likely that the binding represented O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–O groups,41 which were provided by GMA and EDMA during modification and therefore increased the peak intensity. Therefore, the XPS results showed that by photopolymerization, monoliths were successfully prepared to modify the cellulose paper surface.
C–O groups,41 which were provided by GMA and EDMA during modification and therefore increased the peak intensity. Therefore, the XPS results showed that by photopolymerization, monoliths were successfully prepared to modify the cellulose paper surface.
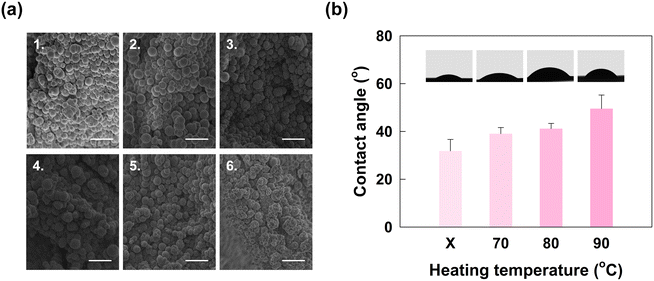
Optimization of sPAD fabrication conditions
The monolith-modified cellulose paper was then subjected to a series of typical μPAD fabrication processes so that immunoassay testing can be performed. First, wax with hollow circular patterns was printed to separate hydrophilic areas for detection. However, only the printed side was hydrophobic, which was not enough for separation between detection areas and led to crosstalk between them. Heating, therefore, was used to melt the wax so that the hydrophobicity applied to the entire paper except for the area that was left for immunoassay testing. Herein, different combinations of heating duration and heating temperature were tested. Specifically, the heating time was set at 15 minutes and 30 minutes, and the temperatures tested were 70 °C, 80 °C, and 90 °C. The optimized parameters for fabrication were later determined after observing the surface morphology and hydrophobicity on the back of the paper. The optimized parameters should not damage the surface morphology of the monolith; otherwise, the ability of the monolith to capture biomolecules during immunoassay testing would be reduced. In addition, the wax should penetrate to the back of the cellulose paper and reduce hydrophilicity at the edge of the testing area; otherwise, there would be crosstalk between testing areas. Therefore, to determine the optimized fabrication parameters, an SEM and a contact angle meter were employed to observe the surface structure and hydrophobicity, respectively. The surface morphology of the monolith after fabrication was observed by the SEM, and the results are shown in Fig. 3(a). To determine the hydrophobicity of the testing area at the back of the monolith-modified paper, the contact angle was measured, as shown in Fig. 3(b).As shown in Fig. 3(a), the original monolithic structure had a uniform and spherical shape. However, the structure of the monolith changed after heating, especially after 30 minutes of heating, as shown in Fig. 3(a4) to (a6). The originally uniform circular morphology had expanded, leading to non-uniform sizes. Specifically, heating at 90 °C for 30 minutes damaged the circularity of the monolith structure. However, according to Fig. 3(a1) to (a2), when the heating duration was shortened to 15 minutes, except for heating at 90 °C, the structure remained complete and exhibited less damage than the original structure. Since a varying and enlarged size of the monolith would lead to a lower surface area for binding with biomolecules, and there will be no other heating steps in later testing steps that will change the surface morphology, the selection of a suitable heating parameter is essential. Therefore, by carefully examining the above results, the wax melting process should be performed only at 70 °C or 80 °C with 15 minutes of heating.
We measured the contact angle of a water droplet on modified paper at different heating temperatures. As shown in Fig. 3(b), the contact angle of the paper without heating was 31.83 ± 4.82 degrees. After heating steps at 70 °C, 80 °C, and 90 °C, the contact angles were 39.05 ± 2.59, 41.20 ± 2.18, and 49.81 ± 6.43 degrees, respectively (n = 3). The results show that the monolith-modified cellulose paper was hydrophilic, similar to the findings of previous research.42 At the same time, when the heating temperature increased, the paper surface became less hydrophilic and the contact angle increased. This change possibly resulted from the slight changes in the morphology of the structure. In previous research, the structural change caused by porogens led to an enormous change in contact angle;43 therefore, it is possible that heating for 15 minutes still leads to minor changes in the structure and porosity of the monolith. Thus, the contact angle increases when a higher heating temperature is applied. However, the lower temperature could not supply enough heat for fully melting the wax to the back of cellulose paper. In such cases, 80 °C was chosen as the fabrication temperature since the wax could then penetrate the back and the surface morphology was less changed by the heat. In conclusion, heating at 80 °C for 15 minutes was the best fabrication condition for the sPAD.
HIgG test for enzyme reaction time determination
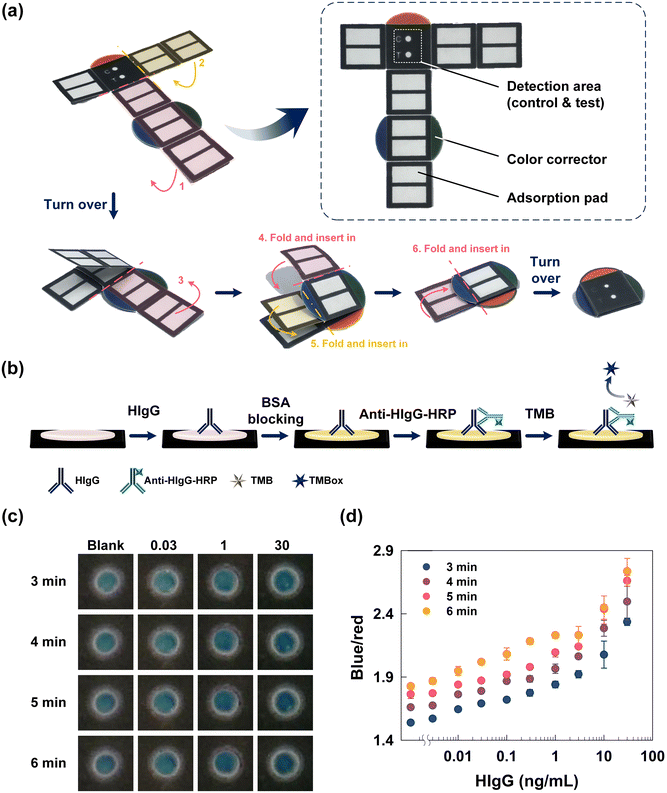
Since the proposed sPAD is a completely new device for tuberculosis immunoassays or even ELISA tests, reaction parameters such as enzymatic reaction time should be determined before performing actual detection. Generally, a long reaction time during enzymatic reactions results in darker colorimetric results and a higher B/R value, which can be detected by the naked eye. However, a long reaction time would also lead to the same shade of color even if different concentrations of analytes are used for detection. To avoid color similarity, optimization of the reaction time is crucial, allowing the user to differentiate among various concentrations of the analyte based on the darkness of the color displayed. Moreover, to better analyze the colorimetric results, we can utilize a smartphone to capture colorimetric results for further analysis and semi-quantify the results.Herein, the fabricated monolith-modified paper was first folded into a compact style to ensure that the position of the adsorbent pads was beneath the detection area, as shown in Fig. 4(a). The integrated detection and adsorption pads enabled more convenient detection and integrated flow as the reagent and washing buffer both flowed downward. The red, green, and blue color correctors can help with correction when capturing the colorimetric results and prevent interference from the light field. HIgG at concentrations between 0.003 and 30 ng mL−1 was used for optimization of the enzymatic reaction time. Then, 3.33 μg mL−1 anti-HIgG was added to detect and capture HIgG. The overall steps are shown in Fig. 4(b). Reaction times from 3 to 6 minutes were tested, followed by calculation of the B/R value for each combination of reaction time and HIgG concentration. The colorimetric results are shown in Fig. 4(c), and the calculated B/R value and standard deviation between tests are displayed in Fig. 4(d). Simple visual observation of the colorimetric results showed that the darker blue shades were obtained as the concentration of HIgG increased, which can be explained by the B/R value as well. When the enzymatic reaction time was 3 minutes, the B/R ratio increased from 1.54 to 2.34; when reaction time was 4 minutes, the B/R ratio as between 1.66 and 2.50. For 5 and 6 minute enzymatic reactions, the B/R ratio ranged from 1.76 to 2.66 and 1.83 to 2.74, respectively. Although the B/R value changed with increasing concentrations of HIgG, the difference in blueness was highest between the most diluted and the most concentrated samples. In this case, the color change along with the HIgG concentration was obvious enough clear visualization of the results. Therefore, the range of B/R values for each series of reactions was calculated to be 0.80, 0.84, 0.91, and 0.91 when the reaction time was 3, 4, 5, and 6 minutes, respectively. The results show that the range of B/R ratios increased with increasing concentrations of HIgG. Since the color change strongly relates to whether the reaction is complete, the correct color cannot be observed if the selected reaction time is not within the right range. Based on the results, both the 5 and 6 minute reactions exhibited significant changes in the B/R ratio, showing the colorimetric results can adequately indicate the concentration of HIgG. In this case, to fulfill the criteria for rapid detection, 5 minutes was chosen since it was the shortest time point at which changes in the HIgG concentration can be properly detected. Therefore, for subsequent fabrication, 5 minutes was selected as the optimized reaction parameter.
Tuberculosis PPD test
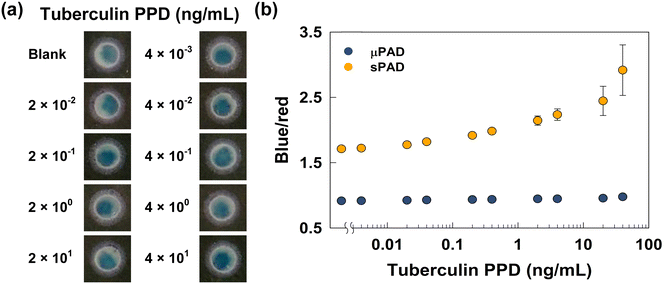
With fully optimized fabrication and reaction parameters, the proposed sPAD was ready for tuberculosis PPD tests. Different concentrations of tuberculin PPD were first spiked onto the sPAD, followed by ELISA tests. To evaluate the performance of the sPAD, a similar set of tests were also performed on the unmodified μPAD based on our previous research.22 For both series of tests, the concentrations of tuberculin PPD ranged from 0.004 ng mL−1 to 40 ng mL−1. Later, MpAb, anti-RIgG-HRP, and TMB were pipetted onto the detection pads in sequence. After 5 minutes of an enzymatic reaction, we captured the colorimetric results (Fig. 5(a)), and the calculated B/R ratios were also analyzed (Fig. 5(b)).As shown in Fig. 5(a) and results derived in our previous research, the color became bluer with increasing tuberculin PPD concentration for both sets of tests, although the color change was more obvious for the sPAD. To quantify the change in color, the B/R ratio for each colorimetric result was calculated, as shown in Fig. 5(b). We specially focused on the difference in B/R ratio between the highest concentration of tuberculin PPD and the blank. The unmodified μPAD exhibited a B/R ratio ranging from 1.07 to 1.59, which is a difference of 0.52; the sPAD modified with the monolith had a B/R ratio ranging from 1.71 to 2.92, exhibiting a difference of 1.21. Since commercial antibodies are utilized in both tests to ensure the specificity between antigen and capture antibodies, the larger B/R ratio difference for the sPAD is simply a result of the more vital interaction between antigens and the monolith-modified paper surface. The more the amount of antigen attached to the surface is, the more likely it is to oxidize TMB after completion of later steps. In contrast, the unmodified cellulose paper can not secure the antigens, which were washed away during washing steps. In such cases, the colorimetric results for the μPAD are unlikely to reflect the changing concentration of antigens, which makes the sPAD a better choice for performing tuberculosis PPD tests. Moreover, the LOD of the sPAD was calculated. According to the results shown in Fig. 5(b), the dynamic range was between 0.2 and 20 ng mL−1 with an LOD of 0.11 ng mL−1.44,45 The LOD of the sPAD was lower than that of other commercial immunoassays, with a value of 2.1 ng mL−1,46 showing that the porous structure of the monolith increased the surface area of the paper and provided higher sensitivity for detecting tuberculin PPD. However, the error was more obvious with higher tuberculin PPD concentrations. This is probably because the monolith surface was already fully occupied by the antigen, and the excess antigen can only attach to other biomolecules and can not endure the subsequent washing. Nevertheless, the dynamic range of concentrations was on the lower end; therefore, the sPAD was still stable at low concentrations of tuberculin PPD.
Conclusions
In this study, we developed a monolith-modified cellulose-based sPAD with increased stability and sensitivity for TB tests. The poly(GMA-co-EDMA) monolith was first used to modify the surface of cellulose paper, and the epoxy groups of the monolith can capture more biomolecules during ELISA and prevent the desorption of proteins during washing steps. Then, the surface-modified paper was subjected to fabrication processes, including wax printing and heating at 80 °C for 15 minutes. The sPAD thus formed was tested with HIgG to determine the optimized enzymatic reaction time, which was 5 minutes. Finally, a tuberculosis PPD test was performed with the sPAD, which exhibited a dynamic range between 0.2 and 20 ng mL−1 with an LOD of 0.11 ng mL−1. The results showed that the monolith-modified sPAD has excellent potential for TB detection in resource-limiting areas. Furthermore, the capturing antibodies used in the testing processes are changeable for other germs detection. We also expect we can use the developed device for other disease detections in the future.Author contributions
C. F. C. conceptualized the research; all authors participated in investigating and writing the paper.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This research was supported by the Ministry of Science and Technology, Taiwan (109-2223-E-002-004-MY3, 110-2923-E-002-004-MY3), the Higher Education Sprout Program at National Taiwan University (111L891504), and the Lithuania-Latvian-Taiwan joint project according to the agreement of No S-LLT-21-3 with the Research Council of Lithuania (LMTLT).References
- J. Furin, H. Cox and M. Pai, Lancet, 2019, 393, 1642–1656 CrossRef.
- W.H.O., Global Tuberculosis Report 2021, World Health Organization, Geneva, 2021 Search PubMed.
- C. Laine, S. Williams, H. C. Sox and P. Escalante, Ann. Intern. Med., 2009, 150, ITC6-1 CrossRef.
- G. E. Pfyffer and F. Wittwer, J. Clin. Microbiol., 2012, 50, 4188–4189 CrossRef.
- W.H.O., WHO operational handbook on tuberculosis, Module 3: diagnosis - rapid diagnostics for tuberculosis detention, 2021 update, World Health Organization, Geneva, 2021 Search PubMed.
- Z. Yu, Y. Tang, G. Cai, R. Ren and D. Tang, Ann. Intern. Med., 2019, 91, 1222–1226 CAS.
- C. Dincer, R. Bruch, E. Costa-Rama, M. T. Fernandez-Abedu, A. Merkoci, A. Manz, G. A. Urban and F. Guder, Adv. Mater., 2019, 31, 1806739 CrossRef PubMed.
- A. W. Martinez, S. T. Phillips, E. Carrilho, S. W. Thomas, 3rd, H. Sindi and G. M. Whitesides, Anal. Chem., 2008, 80, 3699–3707 CrossRef CAS PubMed.
- H. Yuan, J.-H. Lin, Z.-S. Dong, W.-T. Chen, Y. K. Chan, Y.-C. Yeh, H.-T. Chang and C.-F. Chen, Sens. Actuators, B, 2022, 363, 131824 CrossRef CAS.
- J.-H. Lin, S.-J. Chen, J.-E. Lee, W.-Y. Chu, C.-J. Yu, C.-C. Chang and C.-F. Chen, Chem. Eng. J., 2022, 430, 133070 CrossRef CAS.
- F. Rusmini, Z. Zhong and J. Feijen, Biomacromolecules, 2007, 8, 1775–1789 CrossRef CAS PubMed.
- H. Orelma, L. S. Johansson, I. Filpponen, O. J. Rojas and J. Laine, Biomacromolecules, 2012, 13, 2802–2810 CrossRef CAS PubMed.
- A. E. Herr, A. V. Hatch, D. J. Throckmorton, H. M. Tran, J. S. Brennan, W. V. Giannobile and A. K. Singh, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 5268–5273 CrossRef CAS.
- J. Liu, I. White and D. L. DeVoe, Anal. Chem., 2011, 83, 2119–2124 CrossRef CAS PubMed.
- L. Ren, J.-C. Wang, W. Liu, Q. Tu, R. Liu, X. Wang, J. Xu, Y. Wang, Y. Zhang, L. Li and J. Wang, Biosens. Bioelectron., 2012, 35, 147–154 CrossRef CAS PubMed.
- M. Ikami, A. Kawakami, M. Kakuta, Y. Okamoto, N. Kaji, M. Tokeshi and Y. Baba, Lab Chip, 2010, 10, 3335–3340 RSC.
- R. Burger, P. Reith, G. Kijanka, V. Akujobi, P. Abgrall and J. Ducrée, Lab Chip, 2012, 12, 1289–1295 RSC.
- S. J. Oh, B. J. Hong, K. Y. Choi and J. W. Park, OMICS, 2006, 10, 327–343 CrossRef CAS.
- D.-M. Liu, J. Chen and Y.-P. Shi, TrAC, Trends Anal. Chem., 2018, 102, 332–342 CrossRef CAS.
- F. Kong and Y. F. Hu, Anal. Bioanal. Chem., 2012, 403, 7–13 CrossRef CAS PubMed.
- M. Zhao, H. Li, W. Liu, Y. Guo and W. Chu, Biosens. Bioelectron., 2016, 79, 581–588 CrossRef CAS PubMed.
- C.-H. Yang, C.-A. Chen and C.-F. Chen, Sens. Actuators, B, 2018, 265, 506–513 CrossRef CAS.
- Q. Yu, Z. Zhang, S. Qiu, Y. Luo, Z. Liu, F. Yang, H. Liu, S. Ge, X. Zou, B. Ding, W. Ren, H.-M. Cheng, C. Sun and B. Liu, Nat. Commun., 2021, 12, 6051 CrossRef CAS PubMed.
- A. D. Salazar-Aguilar, A. Quintanilla, P. López, C. Martínez, S. M. Vega-Díaz, J. A. Casas, P. Miranzo, M. I. Osendi and M. Belmonte, ACS Appl. Mater. Interfaces, 2022, 14, 920–932 CrossRef CAS.
- S. Lawson and F. Rezaei, ACS Sustainable Chem. Eng., 2021, 9, 10902–10912 CrossRef CAS.
- J.-B. Qu, Y.-Y. Lin, Y. Liu, B.-Q. Zhu, Y.-J. Sun, W.-S. Peng and J. Li, ACS Appl. Polym. Mater., 2021, 3, 2657–2666 CrossRef CAS.
- X. Sun, W. Yang, T. Pan and A. T. Woolley, Anal. Chem., 2008, 80, 5126–5130 CrossRef CAS PubMed.
- S. Djeljadini, P. Bongartz, M. Alders, N. Hartmann, A. Oing, C. Cornelissen, F. Hesselmann, J. Arens, U. Steinseifer, J. Linkhorst and M. Wessling, Adv. Mater. Technol., 2021, 6, 2100325 CrossRef CAS.
- C. T. Tomaz and J. A. Queiroz, in Liquid Chromatography, ed. S. Fanali, P. R. Haddad, C. F. Poole, P. Schoenmakers and D. Lloyd, Elsevier, Amsterdam, 2013, pp. 121–141 Search PubMed.
- J. Liu, C.-F. Chen, C.-W. Chang and D. L. DeVoe, Biosens. Bioelectron., 2010, 26, 182–188 CrossRef CAS PubMed.
- B. Liberelle, E. J. Dil, F. Sabri, B. D. Favis, G. De Crescenzo and N. Virgilio, ACS Appl. Polym. Mater., 2021, 3, 6359–6365 CrossRef CAS.
- F. Cheng, X. Duan and K. Xie, Angew. Chem., 2021, 60, 18792–18799 CrossRef CAS.
- S. Lawson, K. Baamran, K. Newport, F. Rezaei and A. Rownaghi, ACS Appl. Mater. Interfaces, 2021, 13, 55198–55207 CrossRef CAS PubMed.
- F. Svec, J. Chromatogr. A, 2010, 1217, 902–924 CrossRef CAS PubMed.
- T. B. Tennikova, F. Svec and B. G. Belenkii, J. Liq. Chromatogr., 1990, 13, 63–70 CrossRef CAS.
- C. Tyagi, L. K. Tomar, P. Kumar, V. Pillay and H. Singh, Anal. Methods, 2014, 6, 7374–7383 RSC.
- S. H. Chuag, G. H. Chen, H. H. Chou, S. W. Shen and C. F. Chen, Sci. Technol. Adv. Mater., 2013, 14, 044403 CrossRef CAS PubMed.
- W. Dungchai, O. Chailapakul and C. S. Henry, Analyst, 2011, 136, 77–82 RSC.
- D. S. Insuasty-Cepeda, M. Maldonado, J. E. García-Castañeda and Z. J. Rivera-Monroy, RSC Adv., 2021, 11, 4247–4255 RSC.
- D. Liu, N. Song, Y.-C. Cheng, D.-X. Chen, Q. Jia and Y.-W. Yang, RSC Adv., 2014, 4, 49153–49160 RSC.
- J. Ederer, P. Janoš, P. Ecorchard, J. Tolasz, V. Štengl, H. Beneš, M. Perchacz and O. Pop-Georgievski, RSC Adv., 2017, 7, 12464–12473 RSC.
- X. Sun, J. Dong, J. Li, M. Ye, J. Ou, L. Zhang and W. Zhang, RSC Adv., 2016, 6, 113058–113065 RSC.
- Y. Lv, Y. Cao, F. Svec and T. Tan, Chem. Commun., 2014, 50, 13809–13812 RSC.
- S. F. A. Rahman, N. A. Yusof, U. Hashim, R. Hushiarian, M. Nuzaihan M. N., M. N. Hamidon, R. M. Zawawi and M. F. M. Fathil, Anal. Chim. Acta, 2016, 942, 74–85 CrossRef CAS PubMed.
- U. Z. Mohd Azmi, N. A. Yusof, N. Kusnin, J. Abdullah, S. Suraiya, P. S. Ong, N. H. Ahmad Raston, S. F. Abd Rahman and M. F. Mohamad Fathil, Sensors, 2018, 18, 3926 CrossRef.
- M. Ji, B. Cho, Y. S. Cho, S.-Y. Park, S.-N. Cho, B.-Y. Jeon and B.-S. Yoon, Yonsei Med. J., 2014, 55, 746–752 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |