Open Access Article
Open Access ArticleRapid and direct quantification of the SARS-CoV-2 virus with an ultrasensitive nanobody-based photonic nanosensor
Gisela
Ruiz-Vega
a,
Maria
Soler
 *a,
M. Carmen
Estevez
a,
Patricia
Ramirez-Priego
*a,
M. Carmen
Estevez
a,
Patricia
Ramirez-Priego
 a,
Martalu D.
Pazos
a,
María A.
Noriega
b,
Yago
Margolles
c,
Clara
Francés-Gómez
d,
Ron
Geller
d,
Giulia
Matusali
e,
Francesca
Colavita
e,
Antonino
di Caro†
e,
José M.
Casasnovas
a,
Martalu D.
Pazos
a,
María A.
Noriega
b,
Yago
Margolles
c,
Clara
Francés-Gómez
d,
Ron
Geller
d,
Giulia
Matusali
e,
Francesca
Colavita
e,
Antonino
di Caro†
e,
José M.
Casasnovas
 b,
Luis Angel
Fernández
c and
Laura M.
Lechuga
b,
Luis Angel
Fernández
c and
Laura M.
Lechuga
 a
a
aNanobiosensors and Bioanalytical Applications group (NanoB2A), Catalan Institute of Nanoscience and Nanotechnology (ICN2), CSIC, BIST, and CIBER-BBN, Bellaterra, 08193, Barcelona, Spain. E-mail: maria.soler@icn2.cat
bDepartment of Macromolecular Structure, Centro Nacional de Biotecnología, Consejo Superior de Investigaciones Científicas (CNB-CSIC), Darwin 3, Campus Cantoblanco UAM, 28049 Madrid, Spain
cDepartment of Microbial Biotechnology, Centro Nacional de Biotecnología, Consejo Superior de Investigaciones Científicas (CNB-CSIC), Darwin 3, Campus Cantoblanco UAM, 28049 Madrid, Spain
dInstitute for Integrative Systems Biology (I2SysBio), Universitat de Valencia-CSIC, 46980, Valencia, Spain
eNational Institute for Infectious Disease “L. Spallanzani” IRCCS, Via Portuense 292, 00149, Rome, Italy
First published on 14th June 2022
Abstract
The coronavirus (CoV) disease 2019 (COVID-19) is expected to become endemic in the coming years, meaning that the worldwide society shall be prepared to routinely manage the highly contagious respiratory SARS-CoV-2. A rapid and early diagnosis of the SARS-CoV-2 infection is crucial for controlling the spread of the disease, interrupting the transmission chain, and providing timely medical attention to patients. We introduce an innovative nanophotonic biosensor for the quantitative detection of viral particles in less than 20 minutes total assay time. The nanosensor, based on the bimodal waveguide (BiMW) interferometric technology, has been functionalized with novel bioengineered nanobodies (Nb) targeting the SARS-CoV-2 receptor-binding domain (RBD). Our approach relies on the direct capture of the viral particles, and the optimized methodology allows the detection of the SARS-CoV-2 virus with outstanding sensitivity, below 200 TCID50 per mL, being able to provide accurate viral load determination within a broad dynamic range (102–106 TCID50 per mL). Both the nanobodies and the sensor nanotechnology can be produced at a large scale with highly-efficient cost-effective procedures, and they are being integrated into a user-friendly point-of-care device for multiplexed and decentralized operation. The implementation of this unique biosensor in primary care assistance, hospitals, pharmacies, or private laboratories could greatly aid in the relief and descongestion of the sanitary systems and the clinical and social management of COVID-19.
Introduction
The outbreak of the Coronavirus Disease 2019 (COVID-19) and its pandemic spread have evidenced the urgent need of developing and implementing advanced diagnostic systems that enable rapid, sensitive, and accurate detection of respiratory viral infections at the point of need, being the primary and most effective way to interrupt the transmission chain and to provide timely medical attention to patients. Currently, clinical diagnosis of SARS-CoV-2 (i.e., Severe Acute Respiratory Syndrome Coronavirus 2) infection is performed through nucleic acid amplification tests based mainly on the reverse transcription-polymerase chain reaction (RT-PCR). PCR-based assays are undoubtedly one of the most robust and precise existing techniques for disease diagnosis, offering outstanding sensitivity and specificity (96–98% and 99%, respectively), standardized procedures, and a relatively facile adaptation to emerging pathogens or biomarkers.1,2 However, its out-of-the-lab deployment as a point-of-care (POC) testing system is still a major limitation, mostly restricted by the sample processing needs (e.g., RNA extraction) and the sophisticated multi-step analytical process3,4 or limitations in the field deployment of still quite complex and expensive equipment. As an alternative, immunochromatographic lateral flow assays (LFA) have been proposed and broadly employed as a rapid qualitative diagnostic test for COVID-19. LFAs targeting viral antigens have been developed for direct patient use, being affordable and not requiring any trained sanitary personnel or specialized equipment, delivering a clear yes/no response in less than 15 minutes. The convenience of such rapid diagnostic tests has been proven, especially in late pandemic stages for general population screening, and risk minimization in mass-attendance events. However, most LFAs suffer from insufficient sensitivity for early diagnosis, showing false-negative results at the initial stages of the infection (65–70% sensitivity, 97–99% specificity).5,6 Besides, the majority of COVID-19 LFAs are purely qualitative and cannot determine the viral load, which would aid in patient clinical management.Biosensors hold great promise for attaining accurate, rapid, and simple diagnosis of COVID-19. Among the different sensor modalities, photonic biosensors based on the evanescent field principle have been positioned as an ideal technology, performing ultrasensitive analysis in a label-free and real-time format.7–9 Latest research on label-free photonic biosensors has demonstrated their excellent capabilities for detection and identification of respiratory virus infections, such as influenza, respiratory syncytial virus, and coronaviruses.7 For SARS-CoV-2, few studies have been recently published fully addressing the label-free optical biosensing of viral particles,10–12 viral RNA,13 or viral antigens.14 Besides, photonic biosensors have been developed for serological assays through the detection and quantification of COVID-19 antibodies, although only a few have been clinically validated.15,16 Nonetheless, the sensitivity levels achieved so far for the direct detection of SARS-CoV-2 infection have not reached yet the clinical standards set by PCR-based assays, or they involve costly and sophisticated instrumentation and bioreagents making them unsuitable for eventual implementation for POC mass testing.
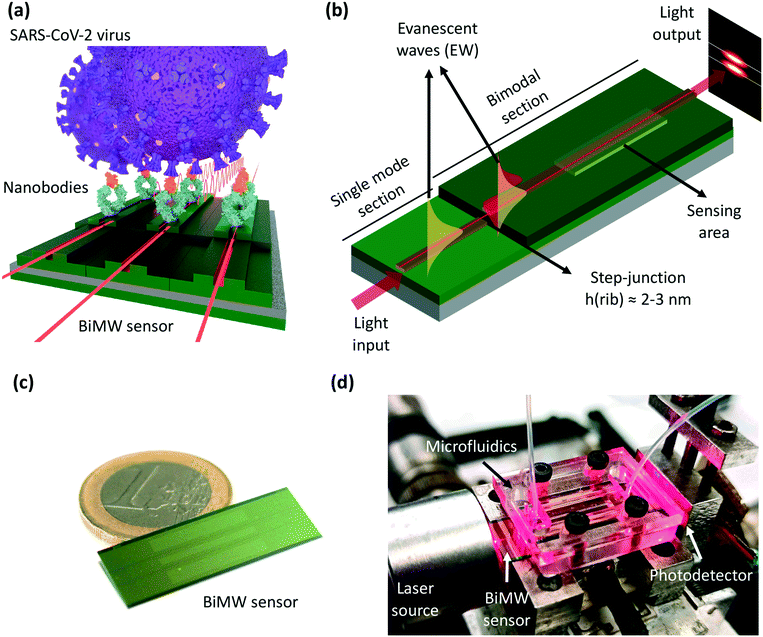
We present an innovative nanobiosensor for the direct diagnosis of COVID-19 that provides ultrasensitive detection of the SARS-CoV-2 virus over a broad range of concentrations (102–106 viral particles per mL), by direct and specific capture of viral particles, without any additional amplification step, delivering quantitative data in less than 20 minutes sample-to-result time, and importantly, with potential scalability for affordable and efficient point-of-care testing. The biosensor is based on a bimodal waveguide (BiMW) interferometer, a powerful silicon photonics sensor nanotechnology that exploits the label-free evanescent wave sensing principle (Fig. 1).17,18 The BiMW technology employs a single straight waveguide where light, at visible wavelengths, is confined and propagates, generating an evanescent field that penetrates hundreds of nanometers in the adjacent dielectric external medium. This evanescent field is highly sensitive to minute changes of the refractive index (RI), such as those caused by a biointeraction event. In particular, the bimodal waveguide allows the confinement of two light modes of the same polarization, whose respective evanescent fields behave differently to RI changes, resulting in different propagation and thus in an interferometric phase shift (Δφ) that can be readily monitored. This design involves significantly less complex fabrication processes than conventional interferometric configurations that require a split waveguide in two arms (e.g., Mach–Zehnder interferometers) and facilitates the integration in small footprint optofluidic devices, without sacrificing their remarkable sensitivity.19 In fact, the BiMW biosensor stands out among the different label-free photonic sensors due to its extraordinary refractometric resolution (i.e., 10−7–10−8 refractive index units, RIU), which is between one and two orders of magnitude better than the benchmarked surface plasmon resonance (SPR) systems.20 These nanophotonic sensor chips are currently fabricated with well-established microelectronics processes and we have implemented them in biosensor prototypes, demonstrating outstanding performance for the direct analysis of proteins and nucleic acids, with detection limits in the aM range,21–23 and also for large pathogenic bacteria, achieving direct detection of only a few cells per milliliter (i.e., 30–50 CFU mL−1).24,25
One of the most critical challenges for the direct detection of an emerging and previously unknown virus is the availability of appropriate bioreceptors.7,26 Many antibodies targeting the spike (S) protein of SARS-CoV-2, or its receptor binding domain (RBD), have been produced and commercialized in this critical task. However, the laborius and costly manufacturing process poses important limitations for their use in standard diagnostics, especially in terms of reproducibility and specificity. To demonstrate the potential of our technology to detect a small number of virus entities directly in patient samples, we have designed a detection strategy based on the direct capture of the SARS-CoV-2 virus through specific bioengineered receptors designed for this purpose. We have combined the high-performance biosensor nanotechnology with high-affinity nanobodies (Nb) recently produced against the receptor-binding domain (RBD) of the S protein.27 The Nb are single VHH domains derived from heavy chain-only antibodies (hcAb) naturally found in camelids (e.g., dromedaries, llamas, alpacas). They are the smallest antigen-binding Ab fragments known (ca. 14 kDa) and can be obtained by recombinant expression in bacteria, yeasts, and/or mammalian cell systems.28 Despite their small size, they have identical affinity and antigen specificity to conventional Ab molecules with heavy chain (i.e., immunoglobulin G, IgG). Nb have additional properties such as an enhanced stability to thermal and chemical denaturation, higher solubility, resistance to proteolysis, and the ability to bind to small protein cavities such as the active centers of enzymes and conserved inner regions on pathogen surfaces, which remain hidden to human antibodies to avoid immune neutralization.29,30 These superior characteristics provide unique advantages in the use of Nb for diagnostic applications and especially for nanobiosensor development.31,32 Moreover, they can be expressed as a monovalent molecule, or as protein fusions to other Nb and/or protein domains (e.g., Fc domain of IgG), producing bivalent and multivalent molecules with mono-, bi-, or multi-specific binding capabilities.30,33 Herein, we employed hcAb containing previously selected VHH domains with high affinity for the RBD fusioned to the Fc region of human IgG1, which have shown excellent recognition for many SARS-CoV-2 variants.27 We have compared the performance of the device incorporating the hcAb with commercial Ab targeting the RBD, and carefully optimized the conditions that allow the detection of the SARS-CoV-2 virus.
Our results represent the first report demonstrating the feasibility of employing nanophotonic BiMW sensors for direct SARS-CoV-2 virus detection and quantification in a simple, direct way, reaching limits of detection (LOD) of a few hundreds of viruses (viral load ≈ 100–200 virus per mL). The assay is completed in a few minutes, thus showing promising performance to be eventually implemented at decentralized settings in the healthcare system.
Experimental
Chemical and biological reagents
Triethoxysilane polyethylene glycol carboxylic acid (silane-PEG-COOH, 600 Da) was supplied by Nanocs (New York, US). Reagents for carboxylic acid activation, (N-(3-dimethylaminopropyl)-ethylcarbodiimide hydrochloride (EDC) and N-hydroxysulfosuccinimide (sulfo-NHS)), were provided by Sigma-Aldrich (Steinheim, Germany). Chimeric monoclonal antibodies (mAb) against RBD (anti-RBD mAb), recombinant S1 and RBD proteins were purchased from Sino Biological Europe GmbH (Germany). Human C-reactive protein (CRP) was acquired from BBI Solutions (UK). Viral transport media (VTM) were acquired at Deltalab (Spain).Production of Nb-derived hcAb (Nb–Fc)
The hcAb that contain the Nb fused to the Fc of IgG1 were expressed upon DNA transfection with the corresponding plasmid vector of suspension human embryonic kidney (HEK-293F) mammalian cell, reported previously.27 The secreted hcAb were purified from culture supernatants using protein A affinity columns (Cytiva) and further purified by size exclusion chromatography (SEC) with a Superdex75 (10/300) column (Cytiva) in HBS (20 mM HEPES, 150 mM NaCl, pH 7.4).Production of VSV-ΔG-S pseudotyped virus
A vesicular stomatitis virus pseudotyped with the Wuhan variant of the SARS-CoV-2 spike protein (VSV-ΔG-S) was produced as previously described34 at the I2SysBio facilities (University of Valencia-CSIC, Spain). Similarly, a control VSV pseudotyped with its own glycoprotein (VSV-ΔG-G) was produced by transfection with pMD2.G (Addgene plasmid 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 259). The virus was concentrated by centrifugation at 50
259). The virus was concentrated by centrifugation at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 4 hours, and resuspended in PBS at moderate/high titers (i.e., 106 focus forming units per mL (FFU mL−1)).
000 × g for 4 hours, and resuspended in PBS at moderate/high titers (i.e., 106 focus forming units per mL (FFU mL−1)).
Production of the UV-inactivated SARS-CoV-2 viral stock
SARS-CoV-2 stock preparation was performed in the biosafety level 3 (BSL-3) laboratory at the National Institute for Infectious Diseases (INMI) in Rome, Italy. Vero E6 cells (ATCC Number CRL-1586™) were used and maintained in Modified Eagle's Medium (MEM) containing 10% heat-inactivated fetal serum (FBS), 1% L-glutamine and 1% penicillin/streptomycin solutions, at 37 °C in a humidified atmosphere of 5% CO2. Vero E6 cells were infected with a SARS-CoV-2 Wuhan-D614G strain (lineage B.1 – clade G), isolated in Italy in March 2020 (GISAID accession number: EPI_ISL_568579, EVAG Ref-SKU: 008 V-04005). Specifically, cells were exposed to SARS-CoV-2 isolate at a multiplicity of infection (MOI) of 0.01 in MEM without FBS for 1 hour at 37 °C/5% CO2. At the end of the adsorption period, the inoculum was replaced with fresh MEM not containing FBS and incubated at 37 °C/5% CO2. After seventy-two hours post-infection, a cytopathic effect (CPE) was visible on about 80–90% of the cell culture and viral preparation was performed at −80 °C. Following thawing, a clarified cell supernatant was exposed to ultraviolet (UV) radiation (λ 254 nm) for 15 minutes on ice. At the end of the UV-treatment, the supernatant was aliquoted and stored at −80 °C. An aliquot of SARS-CoV-2 preparation not exposed to UV was used as a control of the inactivation process and to calculate the virus stock titer according to the method of Reed and Muench and expressed as 50% tissue culture infectious dose (TCID50 per ml).35 The virus stock titer was: 107 TCID50 per mL. The presence of residual infectivity in UV-exposed samples was assessed by infecting the Vero E6 cell culture. After five days post-infection, no CPE was observed. To further exclude residual infectivity, the related cell supernatant was back-titrated by limiting the dilution assay on Vero E6 cells; no virus growth was detected. The UV-inactivated SARS-CoV-2 stock was shipped to ICN2 (Barcelona, Spain) at controlled temperature.BiMW interferometric sensor chip and biosensor device
The BiMW sensors are fabricated at a wafer-scale through standard microelectronics technology processes in the Micro and Nanofabrication Clean Room of the National Microelectronics Center (IMB-CNM-CSIC, Barcelona, Spain). The photonic chip (1 cm width × 3 cm length) contains an array of 20 individual straight rib waveguides (rib height 1–3 nm) (Fig. 1c). Two differentiated regions are generated through the creation of a step junction that allows first the propagation of the fundamental mode (waveguide core thickness of 150 nm) and later in the second section the propagation of the fundamental and first-order modes (waveguide core thickness of 340 nm) (see Fig. 1b). In a section of the bimodal rib waveguide, the cladding is etched to generate the sensing area (15 × 0.05 mm2) where the evanescent field of the guided light can probe the surrounding dielectric environment. The chosen material of the waveguide core is silicon nitride (Si3N4) because of its high refractive index (ncore = 2.00), high density, and chemical inertness to ion species, oxygen, or moisture permeation. The core is confined by an upper and lower cladding made of silicon oxide (SiO2) with a lower refractive index (ncladd = 1.46) to ensure the guiding of light through the core with minimal loss of energy. The BiMW interferometric biosensor system employs a low-power polarized laser diode at a visible wavelength (λ = 660 nm, P = 120 mW) as a light source coupled into the rib waveguide, and a two-sectional photodiode as a detector for the interferometric signal readout (Fig. 1d). The optical system is mounted on a Peltier thermoelectric cooler, to assure thermal stability with an accuracy of 0.01 °C. The system incorporates two photodiode amplifiers to read the signal of both sections of the photodiode and an acquisition board is used for setup controlling. Data processing is performed with a customized software interface that incorporates a recently developed optical phase modulation method to transform the complex interferometric signals into a linear signal (Δφ), directly proportional to the change of the refractive index occurring at the sensor area which allows real-time monitoring.20 The sensor chip is integrated with a five-channel polydimethylsiloxane (PDMS) microfluidic system (channel dimensions = 1.25 mm wide × 500 μm height), for continuous flow and washing of the samples. The fluidic system also includes a syringe pump to guarantee a continuous flow rate of a running buffer and a 6-port injection valve connected to a loop (100 μL) that allows the sequential loading and injection of different samples and solutions.Sensor surface biofunctionalization
Before surface functionalization, the sensor chips are consecutively sonicated for 5 min in acetone, ethanol, and Milli-Q water, and 10 min in methanol/HCl 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v), to remove organic contamination. The sensor chips are then rinsed with water and dried with a stream of nitrogen. A layer of active hydroxyl groups is generated onto the sensor surface using oxygen plasma for 5 min at 45 sccm gas flow, followed by immersion in a 15% HNO3 solution at 75 °C for 25 min. After rinsing with water and drying under a N2 flow, the sensor chip is immediately immersed and incubated in a 25 mg mL−1 silane-PEG-COOH solution in ethanol/water 95
1 (v/v), to remove organic contamination. The sensor chips are then rinsed with water and dried with a stream of nitrogen. A layer of active hydroxyl groups is generated onto the sensor surface using oxygen plasma for 5 min at 45 sccm gas flow, followed by immersion in a 15% HNO3 solution at 75 °C for 25 min. After rinsing with water and drying under a N2 flow, the sensor chip is immediately immersed and incubated in a 25 mg mL−1 silane-PEG-COOH solution in ethanol/water 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (v/v) for 2 h at 4 °C. After the incubation, the sensor chip is rinsed with ethanol and water and dried with an N2 stream. A final curing step is performed by introducing the sensor chips into a glass container inside a conventional autoclave for 90 min at 120 °C at a pressure of 1.5 bar. For bioreceptor immobilization, the silanized sensor chip is placed on the experimental setup for the covalent attachment of the antibodies. The carboxyl groups are activated by flowing a solution of 0.2 M EDC/0.05 M sulfo-NHS in MES buffer (0.1 M, pH 5.5) at 20 μL min−1 over the sensor surface. The antibody solution at an optimum concentration is then injected at a flow rate of 10 μL min−1. The remaining unreacted activated groups are deactivated with an ethanolamine solution (1 M, pH 8.5) for 2 minutes at 20 μL min−1. Milli-Q water is used as the running buffer during the immobilization process.
5 (v/v) for 2 h at 4 °C. After the incubation, the sensor chip is rinsed with ethanol and water and dried with an N2 stream. A final curing step is performed by introducing the sensor chips into a glass container inside a conventional autoclave for 90 min at 120 °C at a pressure of 1.5 bar. For bioreceptor immobilization, the silanized sensor chip is placed on the experimental setup for the covalent attachment of the antibodies. The carboxyl groups are activated by flowing a solution of 0.2 M EDC/0.05 M sulfo-NHS in MES buffer (0.1 M, pH 5.5) at 20 μL min−1 over the sensor surface. The antibody solution at an optimum concentration is then injected at a flow rate of 10 μL min−1. The remaining unreacted activated groups are deactivated with an ethanolamine solution (1 M, pH 8.5) for 2 minutes at 20 μL min−1. Milli-Q water is used as the running buffer during the immobilization process.
Direct immunoassay performance and data analysis
Once the sensor chip surface is biofunctionalized, the running buffer is replaced by the selected detection buffer (PBST 0.1% for protein and pseudovirus detection, and HEPES 25 mM for SARS-CoV-2 detection). Purified samples of different concentrations of proteins, pseudovirus, or UV-inactivated SARS-CoV-2 are injected into the system at a constant flow rate of 10 μL min−1. Biosurface regeneration was performed after every detection with NaOH 10 mM. All experiments for virus detection were carried out in the ICN2 BSL2 facilities with approval of the UAB Biosafety Committee (HR-599-20). Data were analyzed using Origin 8.0 (OriginLab, MA, US) and GraphPad Prism 8 (GraphPad Software, CA, US). Calibration curves were plotted as the mean and standard deviation (mean ± SD) of the acquired biosensor response (Δφ) versus the target concentration. The phase variation (Δφ) considered was the one observed after signal stabilization, once the whole sample had completely passed through the sensor chip (i.e., t ≈ 900 s). The data were fitted to a two-site specific binding model curve. The LOD, defined as the smallest concentration distinguishable from the blank, and LOQ, defined as the minimum concentration that can be reliably detected and quantified, were determined as the concentration corresponding to three or ten times the standard deviation (SD) of the baseline of the sensor signal, respectively.Results and discussion
Evaluation of the BiMW biosensor for direct capture and detection of viral particles
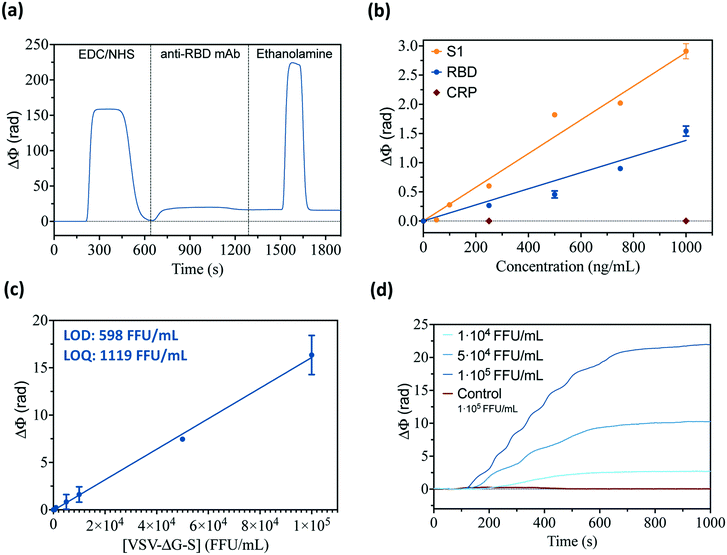
The BiMW biosensor has previously demonstrated its potential for direct, label-free detection of target biomolecules of different nature, like proteins and small molecules,20,36,37 DNA and RNA sequences,38–40 and also for the direct capture of bacteria.24,25 These results set the basis for exploring the use of such a device for the detection of viruses, much smaller entities than bacteria (i.e., around 100 nm diameter compared with considerably larger bacteria, as 1–3 μm). Commercial antibodies were initially selected to assess the preliminary conditions for sensor chip biofunctionalization, antigen detection, and efficient virus capture. We employed a commercial monoclonal antibody targeting the receptor-binding domain of the spike protein of SARS-CoV-2 (i.e., anti-RBD mAb). Monoclonal antibodies were immobilized on the BiMW sensor surface via carbodiimide crosslinking to a carboxyl-functional silanized surface (Fig. 2a). The silane employed for sensor surface functionalization includes a polyethylene glycol (PEG) chain (600 kDa) that helps prevent and minimize undesired non-specific adsorption events. Affinity toward SARS-CoV-2 was first evaluated through the direct and label-free detection of the external viral antigens (recombinant S1 and RBD proteins), employing a non-related protein (C-reactive protein) as a negative control for specificity assessment. As can be seen in Fig. 2b, the mAb showed efficient detection of both viral antigens, with limits of detection determined in the range of ng mL−1, and null cross-reactivity with the negative control. Besides, the robustness and stability of the biofunctionalized surface were confirmed by flowing a high-pH solution (NaOH 20 mM), efficiently regenerating the recognition interface by removing the captured viral antigens, without detaching or damaging the immobilized antibodies up to 12 measurement cycles (data not shown).The viability of this biofunctionalization protocol for the detection of viral particles was further assessed employing a SARS-CoV-2 pseudovirus, a non-replicative virus (i.e., vesicular stomatitis virus, VSV) specifically engineered to express the S protein of SARS-CoV-2 in its outer membrane (VSV-ΔG-S). A VSV pseudotype virus expressing its corresponding glycoprotein (G) and lacking S protein was employed as a negative control for specificity studies (i.e., VSV-ΔG-G). These pseudoviruses can mimic the SARS-CoV-2 virus in terms of size and antigen expression and can be safely used in BSL2 facilities to evaluate the BiMW biosensor because they do not encode an entry glycoprotein in their genome, and, therefore, can only perform a single round of infection. Serial dilutions of the VSV-ΔG-S pseudovirus (1 × 102–1 × 105 FFU mL−1) were directly analyzed with the BiMW sensor functionalized with specific anti-RBD mAb, demonstrating the potential of our biosensor technology for rapid detection and quantification of whole viral particles with excellent sensitivity (Fig. 2c and d). Also, the assay selectivity was confirmed by flowing a non-modified VSV pseudovirus (VSV-ΔG-G) at high concentrations, showing zero signal response (red curve in Fig. 2d). With the standard calibration curve obtained with the SARS-CoV-2 pseudovirus and the commercial monoclonal antibody, we determined the limit of detection (LOD) and limit of quantification (LOQ) of the biosensor assay at 598 FFU mL−1 and 1119 FFU mL−1, respectively. These sensitivity values already show the excellent performance and potential of our biosensor for COVID-19 diagnosis, with common levels of SARS-CoV-2 in patients found between 104 and 106 viral particles per mL.41
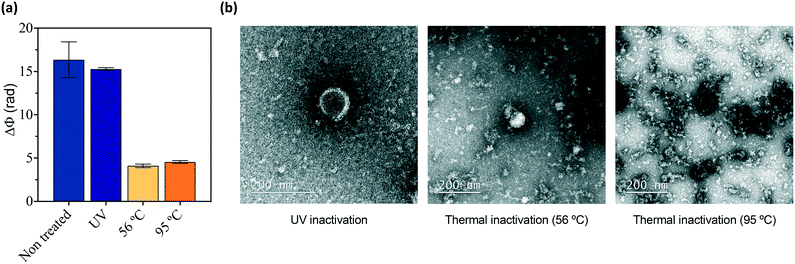
We also studied the effect of different virus inactivation methods on the integrity of the virus – and its recognition by the antibodies – taking the pseudovirus as a model. This was a necessary step before attempting the detection of SARS-CoV-2 in the BSL2 facilities, restricted to only inactivated, non-infectious specimens. Three main inactivation procedures commonly employed were tested with the non-infectious pseudovirus samples: exposure to ultraviolet (UV) radiation (15 min, λ 254 nm), mild long heat treatment (30 min, 56 °C), and harsh short heat treatment (10 min, 95 °C). The sensor response of the VSV-ΔG-S virus sample after each inactivation treatment was compared to the non-treated sample (Fig. 3a). As expected, heat treatment drastically decreased the sensor signal corresponding to the detection by the immobilized antibodies, while UV radiation did not have any significant effect on the biosensor assay. In fact, the heating of virus samples disrupts the membrane structure and denaturalizes the proteins, hampering the direct recognition of the viral antigens with antibodies. In contrast, the UV treatment is intended to only affect the genomic material, seemingly unaltering the structure of the viral particles (Fig. 3b).
Bioengineered nanobodies (Nb–Fc) for SARS-CoV-2 detection
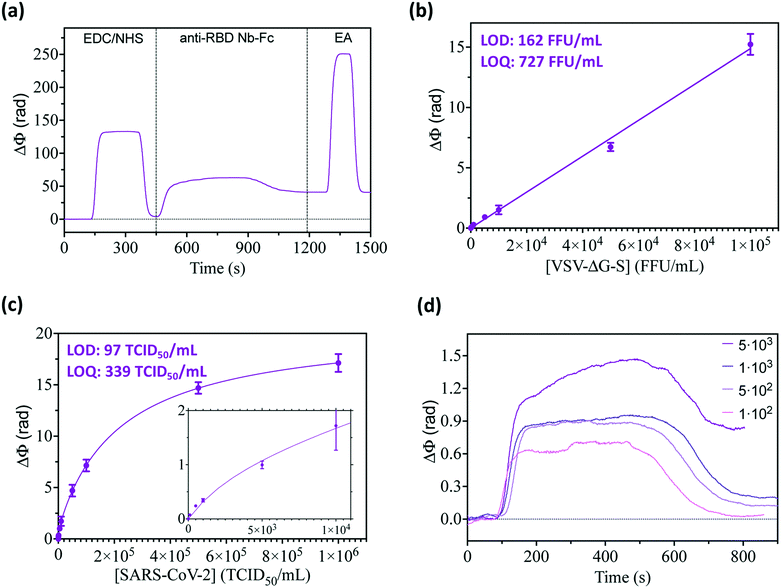
With these initial conditions for biofunctionalization and virus detection, we further evaluated the sensitivity we could achieve with high-quality, high-affinity nanobodies specifically designed and selected for binding to the RBD. The biomolecule used for virus detection was a fusion protein between the VHH domain encoding the nanobody called 1.26 and the constant fraction (Fc) of human IgG1, which has high structural similarities with conventional immunoglobulin,27 facilitating analogous biofunctionalization outcomes. The bioengineered 1.26 Nb–Fc fusions were immobilized on the silanized BiMW sensor surface following the carbodiimide chemistry procedure previously described and optimized for commercial monoclonal antibodies (Fig. 4a). We first compared the direct detection of the VSV-ΔG-S pseudovirus in analog experiments to the ones performed with commercial monoclonal antibodies. The standard calibration curve was obtained, reaching a LOD and LOQ of 162 FFU mL−1 and 727 FFU mL−1, respectively (Fig. 4b). This detection limit is more than 3 times better than the value achieved with the commercial monoclonal antibody, proving that the bioengineered 1.26 Nb–Fc, which showed excellent affinity (KD = 80 pM) for the RBD, can provide enhanced performance for diagnostics purposes.On this promising basis, we addressed the direct capture, detection, and quantification of real SARS-CoV-2 viral particles employing bioengineered Nb as recognition elements. An UV-inactivated Wuhan-D614G SARS-CoV-2 isolate was used as the standard for biosensor calibration. Fig. 4c shows the calibration curve obtained for SARS-CoV-2 virus concentrations ranging from 100 to 106 TCID50 per mL. As expected, compared to pseudoviruses, the sensor response and sensitivity for detection of real SARS-CoV-2 greatly increased, with an outstanding LOD of 97 TCID50 per mL, and a LOQ of 339 TCID50 per mL. Such remarkable values mean that we could directly visualize and quantify in real-time, and in less than 20 minutes, the presence of fewer than 100 infectious viral particles in a sample and over a wide range of concentrations (Fig. 4d). Our analytical sensitivity is comparable to standard clinical PCR-based techniques employed in hospitals (i.e., cycle threshold (Ct) values ranging between 30 and 15 approximately correspond to 102–105 TCID50) and is superior to most of the first rapid LFAs for POC diagnosis.41–43 To further confirm the excellent performance of Nb as sensor biorecognition elements, we evaluated another bioengineered Nb (2.15 Nb–Fc) showing similar RBD binding affinity (KD = 80 pM).27 From the standard calibration curve performed for direct detection of the SARS-CoV-2 virus, we determined a LOD of 87 TCID50 per mL and a LOQ of 303 TCID50 per mL (data not shown); the values very similar to the ones obtained with 1.26 Nb–Fc.
Biosensor assay performance on sample collection media
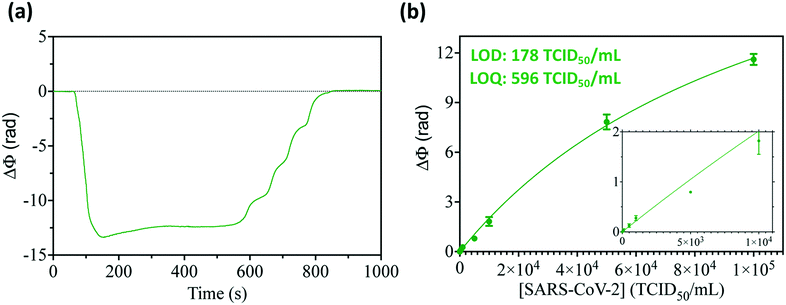
Progressing towards the clinical validation of the nanobody-based photonic biosensor technology for COVID-19 diagnostics, we have further evaluated the direct detection of the SARS-CoV-2 virus under relevant clinical conditions. In particular, we have assessed the detection of whole viruses in a common viral transport medium (VTM) employed for diluting and preserving the nasopharyngeal swab specimens from sample collection until their arrival into the laboratory. Such VTMs have different compositions including preservatives, proteins, etc. that could adsorb onto the biosensor surface, generating false-positive signals. The BiMW sensor is chemically modified with a thin layer of functional silane grafted with PEG chains, which are known to reduce nonspecific bindings and improve the antifouling resistance. The ideal effect of this biofunctionalization method was confirmed by flowing a blank sample of the VTM that resulted in a negligible signal (Fig. 5a). However, the presence of different substances in the VTM can also affect the antigen–antibody recognition interaction as well as the dispersity of the viral particles in the sample. Such an effect could be observed by an increase in the variability among the samples and virus concentrations (data not shown). To solve this, we included a specific additive to the VTM (i.e., guanidine hydrochloride 1 M), which acts as a surfactant. Thereupon, we performed the calibration for the detection of the SARS-CoV-2 virus directly in the VTM. As seen in Fig. 5b, reliable detection and quantification of SARS-CoV-2 in VTM was confirmed, achieving a LOD of 178 TCID50 per mL and a LOQ of 596 TCID50 per mL; slightly superior to the ones achieved with purified samples but still well below the clinical cut-off for COVID-19 diagnostics (103–104 TCID50 per mL).These preliminary results validate the potential of the BiMW nanobiosensor for the direct analysis of patient nasopharyngeal samples, without requiring any amplification or pretreatment process – such as the ones generally performed for nucleic acid assays (e.g., RNA extraction and purification and RT-PCR), and delivering highly accurate and precise diagnostic results in less than 20 minutes total assay time. To our knowledge, this is the first report demonstrating the application of a label-free photonic nanosensor for the direct detection of the whole SARS-CoV-2 virus in their corresponding sample collection media. Previously reported studies, addressing the quantification of SARS-CoV-2 viral particles with label-free nanoplasmonic sensors,10,11 have achieved detection limits in the range of 300 viral particles per mL, which is two times higher than our BiMW nanosensor (around 100 viruses per mL), their results were based on pseudoviruses as target samples and, in the case of Huang et al., performance is only demonstrated under standard buffer conditions. Both the biological assay approach and the analytical performance of our nanobiosensor have shown to be highly promising towards the implementation of new point-of-care technologies for improved COVID-19 diagnostics.
Conclusions
Driven by the worldwide demand for point-of-care diagnostics for COVID-19 testing, we introduce a pioneering nanophotonic biosensor that enables the quantitative analysis of the SARS-CoV-2 virus with an outstanding sensitivity – detection limits around 100 infectious viral particles per mL – in less than 20 minutes total assay time. Our nanosensor, based on robust, high-quality BiMW interferometers, is functionalized with novel bioengineered Nb against the SARS-CoV-2 RBD, and has proven superior affinity towards viral antigens and, importantly, can be produced at a large scale with affordable and highly reproducible techniques. The Nb–Fc employed as a sensor bioreceptor (1.26) has previously demonstrated good interaction affinity with different SARS-CoV-2 variants, therefore the same behavior can be expected for biosensing applications. Furthermore, SARS-CoV-2 Nb–Fc can be easily bioengineered to ensure efficient detection of different virus mutations and variants. The biosensor immunoassay has been optimized and fully demonstrated for the direct capture and detection of the whole SARS-CoV-2 virus, eliminating the need for extraction and amplification procedures, and providing a straightforward readout of not only the presence of infectious virus but also the viral load. The assay is equally effective and sensitive in viral transport media compared with laboratory standard conditions, which reinforces the potential of performing direct analysis of collected nasopharyngeal swab samples. This new nanotechnology has unique potential to be implemented in healthcare and clinical practices. Our current work is dedicated to integrating the sensor system in a compact and automated device that will be installed in a hospital laboratory for clinical validation directly with COVID-19 patient samples. Additionally, a multiplexed BiMW sensor targeting parallel detection of different respiratory viruses (e.g., influenza, human CoVs, etc.) could be easily developed to allow one-step assay identification of most common viral infections, greatly aiding in the control of these highly contagious endemic diseases.Author contributions
GRV performed the experiments for biosensor optimization and evaluation for virus detection. MS and MCE supervised the experiments, analyzed the data, and wrote the manuscript. PRP carried out the initial experiments for viral antigen detection. LAF, JMC, YM and MAN designed, produced and purified the bioengineered Nb. RG and CFG produced and purified the SARS-CoV-2 pseudovirus samples. GM, FC, and ADC produced the UV-inactivated SARS-CoV-2 samples. LML coordinates the project and supervised all experimental and discussion activities. All authors have read, revised, and approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has been mainly funded by the H2020 Research and Innovation Program of the European Commission (H2020-SC1-PHE-Coronavirus-2020, CONVAT No 101003544) and by the European Commission – NextGenerationEU (Regulation EU 2020/2094), through CSIC's Global Health Platform (PTI Salud Global – Iniciativa Estratégica de Diagnóstico). The ICN2 is funded by the CERCA Program/Generalitat de Catalunya and supported by the Severo Ochoa Centers of Excellence program, funded by the Spanish Research Agency (AEI, grant SEV-2017-0706). Work in CNB-CSIC was funded by the Spanish Ministry of Science and Innovation (MICIN) and the Spanish Research Council (CSIC) under grants PIE-RD-COVID 19 (No 202020E079) and PTI Salud Global REC_EU (No SGL 2103051, NextGenerationEU). Work at I2SysBio was funded by Ayudas de concesión directa a soluciones científico-innovadoras directamente relacionadas con la lucha contra la Covid-19 grant from the Generalitat Valenciana, Spanish National Research Council grant CSIC-COV19-082/104, Fondo Supera Covid-19 grant BlockAce, and European Commission – NextGenerationEU to Ron Geller. INMI received funds for this study from the Ministero della Salute (Ricerca Corrente - linea 1), the European Commission – Horizon 2020 (EU project 101003544 – CoNVat) and the European Virus Archive – GLOBAL (grants no. 653316 and no. 871029). We thank the Micro and Nanofabrication Clean Room of the National Microelectronics Center (IMB-CNM-CSIC, Barcelona, Spain) as a third-party partner in the CONVAT project for the fabrication of the BiMW photonic chips. We are very grateful to EPI Industries (Barcelona, Spain) for its kind donation in supporting our research on COVID-19.References
- R. Pu, S. Liu, X. Ren, D. Shi, Y. Ba, Y. Huo, W. Zhang, L. Ma, Y. Liu, Y. Yang and N. Cheng, J. Virol. Methods, 2022, 300, 114392 CrossRef CAS PubMed
.
- K. A. Walsh, K. Jordan, B. Clyne, D. Rohde, L. Drummond, P. Byrne, S. Ahern, P. G. Carty, K. K. O'Brien, E. O'Murchu, M. O'Neill, S. M. Smith, M. Ryan and P. Harrington, J. Infect. Dis., 2020, 81, 357–371 CAS
.
- N. Gupta, S. Augustine, T. Narayan, A. O'Riordan, A. Das, D. Kumar, J. H. T. Luong and B. D. Malhotra, Biosensors, 2021, 11, 141 CrossRef CAS PubMed
.
- L. Fuentes, N. Shah, S. Kelly, G. Harnett and K. A. Schulman, J. Am. Board Fam. Med., 2022, 35, 96–101 CrossRef PubMed
.
- J. Hayer, D. Kasapic and C. Zemmrich, J. Infect. Dis., 2021, 108, 592–602 CAS
.
- S. S. Khandker, N. H. H. N. Hashim, Z. Z. Deris, R. H. Shueb and M. A. Islam, J. Clin. Med., 2021, 10, 3493 CrossRef CAS PubMed
.
- M. Soler, M. C. Estevez, M. Cardenosa-Rubio, A. Astua and L. M. Lechuga, ACS Sens., 2020, 5, 2663–2678 CrossRef CAS PubMed
.
- M. G. Blevins, A. Fernandez-Galiana, M. J. Hooper and S. V. Boriskina, Photonics, 2021, 8, 342 CrossRef CAS
.
- A. Asghari, C. Wang, K. M. Yoo, A. Rostamian, X. Xu, J. D. Shin, H. Dalir and R. T. Chen, Appl. Phys. Rev., 2021, 8, 031313 CAS
.
- L. Huang, L. Ding, J. Zhou, S. Chen, F. Chen, C. Zhao, J. Xu, W. Hu, J. Ji, H. Xu and G. L. Liu, Biosens. Bioelectron., 2021, 171, 112685 CrossRef CAS PubMed
.
- Y. Zheng, S. Bian, J. Sun, L. Wen, G. Rong and M. Sawan, Biosensors, 2022, 12, 151 CrossRef CAS PubMed
.
- N. Li, X. Wang, J. Tibbs, C. Che, A. S. Peinetti, B. Zhao, L. Liu, P. Barya, L. Cooper, L. Rong, X. Wang, Y. Lu and B. T. Cunningham, J. Am. Chem. Soc., 2022, 144, 1498–1502 CrossRef CAS PubMed
.
- G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G. A. Kullak-Ublick and J. Wang, ACS Nano, 2020, 14, 5268–5277 CrossRef CAS PubMed
.
- S. L. Lee, J. Kim, S. Choi, J. Han, G. Seo and Y. W. Lee, Talanta, 2021, 235, 122801 CrossRef CAS PubMed
.
- A. Djaileb, M. Hojjat Jodaylami, J. Coutu, P. Ricard, M. Lamarre, L. Rochet, S. Cellier-Goetghebeur, D. MacAulay, B. Charron, É. Lavallée, V. Thibault, K. Stevenson, S. Forest, L. S. Live, N. Abonnenc, A. Guedon, P. Quessy, J. F. Lemay, O. Farnós, A. Kamen, M. Stuible, C. Gervais, Y. Durocher, F. Cholette, C. Mesa, J. Kim, M. P. Cayer, M. J. De Grandmont, D. Brouard, S. Trottier, D. Boudreau, J. N. Pelletier and J. F. Masson, Analyst, 2021, 146, 4905–4917 RSC
.
- O. Calvo-Lozano, M. Sierra, M. Soler, M. C. Estévez, L. Chiscano-Camón, A. Ruiz-Sanmartin, J. C. Ruiz-Rodriguez, R. Ferrer, J. J. González-López, J. Esperalba, C. Fernández-Naval, L. Bueno, R. López-Aladid, A. Torres, L. Fernández-Barat, S. Attoumani, R. Charrel, B. Coutard and L. M. Lechuga, Anal. Chem., 2022, 94, 975–984 CrossRef CAS PubMed
.
- K. E. Zinoviev, A. B. González-Guerrero, C. Domínguez and L. M. Lechuga, J. Lightwave Technol., 2011, 29, 1926–1930 Search PubMed
.
-
C. Domínguez, K. Zinoviev and L. M. Lechuga, Interferometer and sensor based on bimodal optical waveguides, and detection method, US8279445B2, 2007
.
-
D. Duval, A. B. González-Guerrero, S. Dante, J. Osmond, R. Monge, L. J. Fernández, K. E. Zinoviev, C. Domínguez and L. M. Lechuga, in Lab on a Chip, Royal Society of Chemistry, 2012, vol. 12, pp. 1987–1994 Search PubMed
.
- B. Bassols-Cornudella, P. Ramirez-Priego, M. Soler, M. C. Estévez, H. J. Díaz, M. Cardenosa-Rubio and L. M. Lechuga, J. Lightwave Technol., 2022, 40, 237–244 CAS
.
- A. Fernández Gavela, D. Grajales García, J. Ramirez, L. Lechuga, A. Fernández Gavela, D. Grajales García, J. C. Ramirez and L. M. Lechuga, Sensors, 2016, 16, 285 CrossRef
.
- C. S. Huertas, O. Calvo-Lozano, A. Mitchell and L. M. Lechuga, Front. Chem., 2019, 7, 724 CrossRef CAS PubMed
.
- C. S. Huertas and L. M. Lechuga, Methods Mol. Biol., 2022, 2393, 89–125 CrossRef PubMed
.
- J. Maldonado, A. B. González-Guerrero, C. Domínguez and L. M. Lechuga, Biosens. Bioelectron., 2016, 85, 310–316 CrossRef CAS
.
- J. Maldonado, M. C. Estévez, A. Fernández-Gavela, J. J. González-López, A. B. González-Guerrero and L. M. Lechuga, Analyst, 2020, 145, 497–506 RSC
.
- M. Soler and L. M. Lechuga, Anal. Bioanal. Chem., 2021 DOI:10.1007/s00216-021-03751-4
, in press.
- J. M. Casasnovas, Y. Margolles, M. A. Noriega, M. Guzmán, R. Arranz, R. Melero, M. Casanova, J. A. Corbera, N. Jiménez-de-Oya, P. Gastaminza, U. Garaigorta, J. C. Saiz, M. Á. Martín-Acebes and L. Á. Fernández, Front. Immunol., 2022, 1797 Search PubMed
.
- S. Muyldermans, Annu. Rev. Biochem., 2013, 82, 775–797 CrossRef CAS PubMed
.
- A. Bates and C. A. Power, Antibodies, 2019, 8, 28 CrossRef CAS PubMed
.
- S. Steeland, R. E. Vandenbroucke and C. Libert, Drug Discovery Today, 2016, 21, 1076–1113 CrossRef CAS PubMed
.
- G. Gonzalez-Sapienza, M. A. Rossotti and S. Tabares-da Rosa, Front. Immunol., 2017, 8 Search PubMed
.
- V. Salema, E. Marín, R. Martínez-Arteaga, D. Ruano-Gallego, S. Fraile, Y. Margolles, X. Teira, C. Gutierrez, G. Bodelón and L. Á. Fernández, PLoS One, 2013, 8(9), e75126 CrossRef CAS
.
- P. Bannas, J. Hambach and F. Koch-Nolte, Front. Immunol., 2017, 8, 1603 CrossRef PubMed
.
- R. Gozalbo-Rovira, E. Gimenez, V. Latorre, C. Francés-Gómez, E. Albert, J. Buesa, A. Marina, M. L. Blasco, J. Signes-Costa, J. Rodríguez-Díaz, R. Geller and D. Navarro, J. Clin. Virol., 2020, 131, 104611 CrossRef CAS PubMed
.
- L. J. Reed and H. Muench, Am. J. Epidemiol., 1938, 27, 493–497 CrossRef
.
- P. Ramirez-Priego, M. C. Estévez, H. J. Díaz-Luisravelo, J. J. Manclús, Á. Montoya and L. M. Lechuga, Anal. Chim. Acta, 2021, 1152, 338276 CrossRef CAS PubMed
.
- B. Chocarro-Ruiz, S. Herranz, A. Fernández Gavela, J. Sanchís, M. Farré, M. P. Marco and L. M. Lechuga, Biosens. Bioelectron., 2018, 117, 47–52 CrossRef CAS PubMed
.
- C. S. Huertas, S. Domínguez-Zotes and L. M. Lechuga, Sci. Rep., 2017, 7, 1–8 CrossRef PubMed
.
- C. S. Huertas, D. Fariña and L. M. Lechuga, ACS Sens., 2016, 1, 748–756 CrossRef CAS
.
- J. Maldonado, A. B. González-Guerrero, A. Fernández-Gavela, J. J. González-López and L. M. Lechuga, Diagnostics, 2020, 10, 845 CrossRef CAS PubMed
.
- N. Eshghifar, A. Busheri, R. Shrestha and S. Beqaj, Int. J. Gen. Med., 2021, 14, 435 CrossRef PubMed
.
- M. Platten, D. Hoffmann, R. Grosser, F. Wisplinghoff, H. Wisplinghoff, G. Wiesmüller, O. Schildgen and V. Schildgen, Viruses, 2021, 13, 1459 CrossRef CAS PubMed
.
- C. Buchta, I. Görzer, P. Chiba, J. V. Camp, H. Holzmann, E. Puchhammer-Stöckl, M. Mayerhofer, M. M. Müller and S. W. Aberle, Clin. Chem. Lab. Med., 2021, 59, 987–994 CrossRef PubMed
.
Footnote |
| † Current affiliation: Saint Camillus International University of Health Sciences, via di Sant'Alessandro, 8, 00131 Roma, Italy; IRCCS Sacro Cuore Don Calabria Hospital, via Don A. Sempreboni 5, 37024, Negrar di Valpolicella (Verona), Italy. |
| This journal is © The Royal Society of Chemistry 2022 |