Open Access Article
Open Access ArticleThe anticancer peptide LL-III binds with nanomolar affinity to human telomeric and cMyc G-quadruplexes†
Marco
Campanile
a,
Rosario
Oliva
a,
Pompea
Del Vecchio
a,
Roland
Winter
 b and
Luigi
Petraccone
b and
Luigi
Petraccone
 *a
*a
aDepartment of Chemical Sciences, University of Naples Federico II, Via Cintia 4, 80126, Naples, Italy. E-mail: luigi.petraccone@unina.it
bPhysical Chemistry I – Biophysical Chemistry, Department of Chemistry and Chemical Biology, TU Dortmund University, Otto-Hahn Street 4a, Dortmund 44227, Germany
First published on 21st April 2023
Abstract
LL-III is a natural anticancer peptide able to cross the membrane of cancer cells and to localize in the nucleolus, but its intracellular target is unknown. Here, we show that LL-III is able to bind with nM affinity to specific G-quadruplex structures known to be relevant anticancer targets.
Anticancer peptides (ACPs) are low-molecular-weight cationic peptides composed of 10–60 amino acids with antitumor activity.1 Most of these peptides act by inducing membrane disruption or by translocating into the cytoplasm, recognizing an intracellular target. To describe the action mechanism of ACPs, several models have been proposed.2 However, the exact mechanism strictly depends on the peptides’ physico-chemical properties (e.g., their charge and hydrophobicity), as well as the properties of the lipid bilayer (e.g., the lipids’ lateral organization, fluidity and charge density) or the intracellular targeted molecule.3,4 The peptide LL-III (VNWKKILGKIIKVVK-NH2) is a family member of Lasioglossins, three bioactive peptides extracted from the venom of the bee Lasioglossum laticeps, which exhibits a low toxicity towards healthy eukaryotic cells and is active in vitro against some cancer cell lines.5 Particularly, it has been reported that LL-III is able to translocate across the negatively charged tumour cell membranes in a non-disruptive manner and to localize in the cell nucleolus, suggesting that nuclear DNA could be a possible intracellular target.6,7 Prompted by these observations, we decided to study the interaction of this peptide with DNA sequences able to adopt a G-quadruplex structure (GQ) known to be involved in carcinogenesis. GQs are non-canonical DNA helical structures characterized by stacked G-quartets each of which consists of a planar cyclic arrangement of four guanines held together by Hoogsteen hydrogen bonds and stabilized by monovalent cations.8 These structures are thought to be implicated in important biological processes such as aging, cancer and, more generally, in the regulation of gene expression.9–11 Herein, we show that LL-III is able to bind with nanomolar affinity to mixed-type human telomeric (Tel-23) and cMyc G-quadruplex structures which are relevant anticancer targets.8,12 Interestingly, LL-III showed much lower affinity towards other parallel and anti-parallel GQs as well as towards the DNA duplex and single strands, highlighting its specificity for Tel-23 and cMyc. Overall, our results shed light on the mechanism of action of LL-III and demonstrate that its short primary structure can be used as a lead sequence for the development and application of anticancer peptides that specifically bind to G-quadruplex structures.
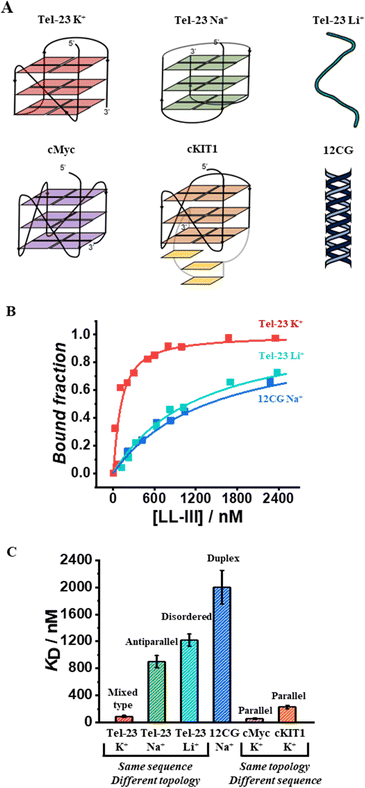
To monitor the binding of LL-III to the quadruplex forming-oligonucleotides, we performed fluorescence anisotropy measurements using labeled-oligonucleotides as previously reported (see ESI,† section 1.2).13 Briefly, we measured the variation of the fluorescence anisotropy due to the formation of the higher molecular weight peptide-DNA complex in comparison with the free DNA. First, we evaluated the affinity of LL-III for the Tel-23 (FAM-labelled) in K+ containing buffer as this DNA sequence is known to adopt a mixed-type topology under these conditions.14 Analysis of the obtained binding curve (see ESI†) reveals that LL-III binds the Tel-23 mixed-type quadruplex with a KD of 90 nM (Fig. 1). To verify whether this high binding affinity was specific for the G-quadruplex structure, we performed the same experiment in a Li+-containing buffer which prevents quadruplex formation as also confirmed by the inspection of the corresponding CD spectrum (Fig. S1, ESI†). Under these conditions, we found a 13-fold higher KD (1220 nM), indicating that LL-III specifically binds to the Tel-23 mixed-type G-quadruplex structure (Fig. 1). To further verify if the results obtained were influenced by the use of different cations in the experiments, we repeated the titration in K+ with a mutated Tel-23 (Mut_Tel-23) that does not form quadruplex even in the presence of K+ (see ESI,† Fig. S2). The LL-III affinity for Mut_Tel-23 in K+ (1110 nM) was similar to that of Tel-23 in Li+ demonstrating that the nature of the cation does not affect the binding properties of LL-III (Fig. S3, ESI†). Next, we performed the binding experiments in Na+-buffer as it is known that in this buffer Tel-23 adopts an antiparallel topology.14 In Na+-buffer, we found a 10-fold higher KD value (900 nM) with respect to the value determined in K+-buffer, revealing that LL-III is able to discriminate among the mixed-type and the antiparallel conformation of Tel-23 (Fig. 1B and Fig. S3, ESI†). Thus, among the Tel-23 topologies, the strength of the interaction follows the order: mixed-type > antiparallel > disordered.
We then checked the affinity of LL-III for the DNA duplex by using a labelled double helix with a similar number of nucleotides (and negative charges) as the Tel-23 sequence (see ESI†). Again, the affinity for the double helix was even lower (KD = 2000 nM) than the one measured for the unfolded strand and similar both in sodium and potassium buffer (Fig. 1 and Fig. S3, ESI†).
Having established the affinity of LL-III for the mixed-type and antiparallel conformation of Tel-23, we decided to evaluate the affinity of the peptide for G-quadruplex sequences known to adopt the parallel conformation in K+-buffer. Particularly, we selected cMyc and the cKIT1 GQs appropriately labelled with Tx-Red (see ESI†). We found a KD of 60 nM for cMyc, which is about 1.5-fold lower than that measured for the mixed-type Tel-23 (Fig. 1B and Fig. S3, ESI†), revealing a stronger binding for this GQ. Interestingly, the affinity for the cKIT1 was about 4-fold higher (KD = 230 nM) than for cMyc, suggesting that LL-III is able to discriminate among GQs with the same parallel topologies (Fig. 1C and Fig. S3, ESI†). To test whether the LL-III binding induces a change in the conformation of the DNA, we collected CD spectra of all the DNA-peptide complexes. We did not observe any significant spectral changes in the region 240–310 nm where only DNA contributes significantly to the CD signal (Fig. S5, ESI†), suggesting that LL-III binds to the different GQ structures without altering their conformation. To look for conformational changes of the peptide, we analysed the spectra in the 200–240 nm region after subtracting the DNA contribution. The spectrum of LL-III alone in buffer is typical of a random coil conformation, and we observed that the presence of Tel-23 in the presence of both K+ and Na+ cations does not significantly alter the spectral features, suggesting that LL-III retains a disordered conformation in the complexed state (Fig. 2A). Interestingly, the peptide spectrum changes significantly in the presence of the two parallel quadruplexes cKIT1 and cMyc, showing a minimum at 222 nm (Fig. 2A), indicating the formation of an α-helical structure. The calculated helical fraction increases from 0.15 for the peptide alone to 0.26 and 0.49 in the presence of cMyc and cKIT1, respectively (Table S1, ESI†). This result reveals that LL-III partially adopts a helical structure when binding to parallel G-quadruplexes, but not to mixed-type and antiparallel Tel-23. The formation of a partially helical conformation has been previously reported for short peptides upon binding to a parallel G-quadruplex.13,15 Interestingly, a similar mode of interaction of an α-helix with a parallel GQ was also observed in crystal structures of protein-quadruplex complexes.16,17 These observations suggest that helix formation is a general requirement for parallel G-quadruplex recognition.
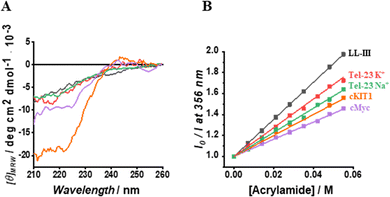 | ||
Fig. 2 (A) Normalized CD spectra of LL-III in the absence (black curve) and in the presence of Tel-23 K+ (red curve), Tel-23 Na+ (green curve), cKIT1 (orange curve) and cMyc (magenta curve) at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio. The peptide signal was obtained after subtracting the signal of the G-quadruplex from the corresponding spectrum of the complex. (B) Stern-Volmer plots for each LL-III – DNA complex obtained by following the intrinsic peptide Trp fluorescence. The above experiments were performed at 20 °C and in the proper buffer solution (see ESI†). 1 molar ratio. The peptide signal was obtained after subtracting the signal of the G-quadruplex from the corresponding spectrum of the complex. (B) Stern-Volmer plots for each LL-III – DNA complex obtained by following the intrinsic peptide Trp fluorescence. The above experiments were performed at 20 °C and in the proper buffer solution (see ESI†). | ||
On the contrary, in a previous study a loss of helical content was observed for a longer peptide upon binding to the human telomeric mixed-type GQ.18 This is consistent with our observation that binding to the mixed-type Tel-23 quadruplex does not induce helical formation in LL-III. These peptide conformational differences could be key to the future design of peptides capable of discriminating different quadruplex topologies.
Seeking further structural details of the interaction of LL-III with the various G-quadruplexes, we then investigated the solvent accessibility of the Trp residue of the peptide upon complex formation employing the steady-state fluorescence emission quenching methodology with acrylamide as quencher molecule (see ESI†).18 We found that the Trp residue in the complexes is partially hidden from the solvent compared to the free peptide (Fig. 2B). This observation clearly indicates that the Trp residue is involved in the complex formation, which is in agreement with previous studies reporting the key role of the aromatic amino acids in binding to G-quadruplexes.19,20 On the other hand, the magnitude of the KSV value differs among the quadruplexes studied, revealing subtle differences in the binding mode (Table S1, ESI†). Indeed, the highest KSV was observed for the complex of LL-III with the mixed-type Tel-23, followed by the antiparallel Tel-23, then cKIT1 and cMyc. Of note, the largest difference in KSV values was observed for the two quadruplexes cMyc and mixed-type Tel-23, with the higher and comparable affinity, revealing that the Trp residue plays a different role in the binding process with the two quadruplexes.
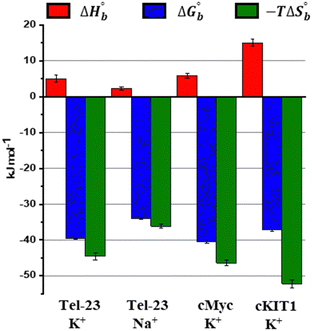
Finally, we explored the energetics of peptide-quadruplex binding by means of isothermal titration calorimetry (ITC). Particularly, we designed the ITC experiments to measure the binding enthalpy and calculated the other thermodynamic quantities using the affinity constant (see ESI†).21,22 We found for all the complexes that the binding is entropically driven (−TΔSbo < 0) with an unfavorable enthalpic contribution (ΔHbo > 0) (Fig. 3 and Table S1, ESI†). This thermodynamic signature points toward a hydrophobic driving force of the binding process (which is accompanied by a release of hydration water), suggesting that LL-III recognizes a “hydrophobic pocket” of the G-quadruplex structure, most likely represented by the G-tetrad plane most exposed to the solvent. The magnitude of the two thermodynamic contributions differs among the quadruplexes studied, revealing significant differences in the formation of the LL-III-quadruplex complexes. Interestingly, the value of the enthalpy change, ΔHbo, increases with the degree of LL-III helicity in the complexes (Table S1, ESI†), suggesting that the extent of the hydrophobic interaction increases with the helical content of the peptide. This could be due to the amphipathic nature of the LL-III helix23 which allows to maximize the contact between its aromatic and hydrophobic residues and the exposed G-tetrad plane. In conclusion, our data show that the anticancer peptide LL-III has a higher affinity for DNA G-quadruplex structures compared to single-stranded and double helical structures and demonstrate some degree of selectivity among different G-quadruplexes. Interestingly, LL-III has a higher affinity for the mixed-type Tel-23 and cMyc quadruplexes, both of which have been implicated in carcinogenesis. This information is particularly important because we have shown in a previous work that LL-III is able to selectively recognize and cross the tumor membrane.23 Putting all this information together, we can speculate on a possible mechanism of action of LL-III against cancer (Fig. 4): the peptide firstly recognizes and crosses the negatively charged tumor membrane, gaining access to the inner part of the cell. Then LL-III binds to the human telomeric and cMyc quadruplex, which are known to be excellent targets for anticancer therapy.8 In this framework, the low cytotoxicity observed for the peptide would be explained by its selectivity towards the membrane of the tumor cell rather than by its affinity to quadruplexes, which plays a central role once the peptide enters the tumor cell, however. Of note, LL-III is a natural and short peptide (15 amino acids) with a sequence that differs considerably from those of other peptides reported to bind to quadruplexes (Table S2, ESI†).13,15,24,25 Further, it belongs to a class of peptides (AMPs and ACPs) whose ability to bind to G-quadruplexes is almost unexplored. For this reason, it may represent a new lead sequence for further optimization. Overall, these findings shed new light on the mechanism of action of LL-III and open exciting new avenues for the design and application of a new class of peptide-based anticancer drugs.
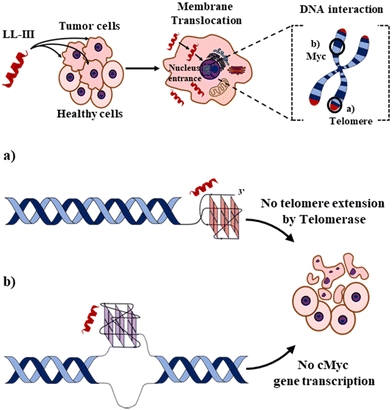 | ||
| Fig. 4 Schematic representation of the proposed action mechanism for the anticancer activity of LL-III. In a first step, the peptide selectively recognizes and interacts with the negatively charged membrane of tumor cells. Through a peptide-induced alteration of the membrane microscopic and mesoscopic properties (e.g., domain formation),23 the LL-III translocates in the cytoplasm and reaches the nucleus, where it recognizes (a) the human telomeric and (b) cMyc promoter G-quadruplexes, thereby impairing tumor cell functions and leading to its death. Figures are not to scale, and LL-III is shown as an α-helix for display purposes. | ||
The authors are grateful to the Italian MUR for granting Rosario Oliva with a research associated position (PON R&I 2014-2020, CUP: E65F21003250003). This work has in part be funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy – EXC-2033 – Projektnummer 390677874.
Conflicts of interest
There are no conflicts to declare.Notes and references
- M. Xie, D. Liu and Y. Yang, Open Biol., 2020, 10, 200004 CrossRef CAS PubMed.
- D. Gaspar, A. S. Veiga and M. A. R. B. Castanho, Front. Microbiol., 2013, 4, 1–16 CrossRef PubMed.
- V. Teixeira, M. J. Feio and M. Bastos, Prog. Lipid Res., 2012, 51, 149–177 CrossRef CAS PubMed.
- W. C. Wimley, ACS Chem. Biol., 2010, 5, 905–917 CrossRef CAS PubMed.
- V. Čeřovský, M. Buděšínský, O. Hovorka, J. Cvačka, Z. Voburka, J. Slaninová, L. Borovičková, V. Fučík, L. Bednárová, I. Votruba and J. Straka, ChemBioChem, 2009, 10, 2089–2099 CrossRef PubMed.
- F. Battista, R. Oliva, P. Del Vecchio, R. Winter and L. Petraccone, IJMS, 2021, 22, 2857 CrossRef CAS.
- J. Slaninová, V. Mlsová, H. Kroupová, L. Alán, T. Tůmová, L. Monincová, L. Borovičková, V. Fučík and V. Čeřovský, Peptides, 2012, 33, 18–26 CrossRef PubMed.
- S. Neidle, Nat. Rev. Chem., 2017, 1, 0041 CrossRef CAS.
- N. Sugimoto, Chemistry and biology of non-canonical nucleic acids, Wiley, Hoboken, 1st edn, 2020 Search PubMed.
- D. Rhodes and H. J. Lipps, Nucleic Acids Res., 2015, 43, 8627–8637 CrossRef CAS PubMed.
- D. Varshney, J. Spiegel, K. Zyner, D. Tannahill and S. Balasubramanian, Nat. Rev. Mol. Cell Biol., 2020, 21, 459–474 CrossRef CAS PubMed.
- B.-J. Chen, Y.-L. Wu, Y. Tanaka and W. Zhang, Int. J. Biol. Sci., 2014, 10, 1084–1096 CrossRef PubMed.
- A. Minard, D. Morgan, F. Raguseo, A. Di Porzio, D. Liano, A. G. Jamieson and M. Di Antonio, Chem. Commun., 2020, 56, 8940–8943 RSC.
- L. Chen, J. Dickerhoff, S. Sakai and D. Yang, Acc. Chem. Res., 2022, 55, 2628–2646 CrossRef CAS PubMed.
- K. H. Ngo, R. Yang, P. Das, G. K. T. Nguyen, K. W. Lim, J. P. Tam, B. Wu and A. T. Phan, Chem. Commun., 2020, 56, 1082–1084 RSC.
- M. C. Chen, R. Tippana, N. A. Demeshkina, P. Murat, S. Balasubramanian, S. Myong and A. R. Ferré-D’Amaré, Nature, 2018, 558, 465–469 CrossRef CAS PubMed.
- A. Traczyk, C. W. Liew, D. J. Gill and D. Rhodes, Nucleic Acids Res., 2020, 48, 4562–4571 CrossRef CAS PubMed.
- J. Jana, R. K. Kar, A. Ghosh, A. Biswas, S. Ghosh, A. Bhunia and S. Chatterjee, Mol. BioSyst., 2013, 9, 1833 RSC.
- K. Kondo, T. Mashima, T. Oyoshi, R. Yagi, R. Kurokawa, N. Kobayashi, T. Nagata and M. Katahira, Sci. Rep., 2018, 8, 2864 CrossRef PubMed.
- T. Masuzawa and T. Oyoshi, ACS Omega, 2020, 5, 5202–5208 CrossRef CAS PubMed.
- R. Oliva, S. Mukherjee, M. Manisegaran, M. Campanile, P. Del Vecchio, L. Petraccone and R. Winter, IJMS, 2022, 23, 5690 CrossRef CAS PubMed.
- R. Oliva, N. Jahmidi-Azizi, S. Mukherjee and R. Winter, J. Phys. Chem. B, 2021, 125, 539–546 CrossRef CAS PubMed.
- M. Campanile, R. Oliva, G. D’Errico, P. Del Vecchio and L. Petraccone, Phys. Chem. Chem. Phys., 2023, 25, 3639–3650 RSC.
- Z.-L. Huang, J. Dai, W.-H. Luo, X.-G. Wang, J.-H. Tan, S.-B. Chen and Z.-S. Huang, J. Am. Chem. Soc., 2018, 140, 17945–17955 CrossRef CAS.
- T. Miclot, A. Froux, L. D’Anna, E. Bignon, S. Grandemange, G. Barone, A. Monari and A. Terenzi, ChemBioChem, 2023, 24, e202200624 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc00737e |
| This journal is © The Royal Society of Chemistry 2023 |