Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Functional chromopeptide nanoarchitectonics: molecular design, self-assembly and biological applications
Rui
Chang†
a,
Luyang
Zhao†
a,
Ruirui
Xing
*ab,
Junbai
Li
 *c and
Xuehai
Yan
*c and
Xuehai
Yan
 *abd
*abd
aState Key Laboratory of Biochemical Engineering, Institute of Process Engineering, Chinese Academy of Sciences, Beijing 100190, P. R. China. E-mail: rrxing@ipe.ac.cn; yanxh@ipe.ac.cn
bSchool of Chemical Engineering, University of Chinese Academy of Sciences, Beijing 100049, P. R. China
cBeijing National Laboratory for Molecular Sciences, CAS Key Lab of Colloid, Interface and Chemical Thermodynamics Institute of Chemistry, Chinese Academy of Sciences Beijing, 100190, P. R. China. E-mail: jbli@ipe.ac.cn
dCenter for Mesoscience, Institute of Process Engineering, Chinese Academy of Sciences, Beijing 100190, P. R. China
First published on 29th March 2023
Abstract
Chromoproteins are a class of delicate natural compounds that elegantly complex photosensitive species with proteins and play a central role in important life processes, such as photosynthesis. Inspired by chromoproteins, researchers integrate simple peptides and photosensitive molecular motifs to generate chromopeptides. Compared with chromoproteins, chromopeptides exhibit a relatively simple molecular structure, flexible and adjustable photophysical properties, and a capability of programmable self-assembly. Chromopeptide self-assembly has attracted great attention as the resultant high-level architectures exhibit an ingenious combination of photofunctions and biofunctions. This review systematically summarizes recent advances in chromopeptide nanoarchitectonics with particular focus on the design strategy, assembly mechanism, and structure–function relationship. Among them, the effect of peptide sequences and the variation in photophysical performance are critically emphasized. On this basis, various applications, including biomedicine and artificial photosynthesis, are discussed together with the future prospects of chromopeptide nanoarchitectonics. This review will provide insights into chromopeptide nanoarchitectonics and corresponding materials with precise designs, flexible nanostructures and versatile functions. In addition, knowledge involving chromopeptide nanoarchitectonics may aid in the development of many other kinds of supramolecular biological materials and bioengineering techniques.
1 Introduction
Natural compounds and their organization provide inspiration for new and fascinating opportunities to develop innovative biomaterials.1–3 Chromoproteins are a class of functional proteins bearing a chromophoric group, which are used as building blocks for achieving diversified nanoarchitectonics and creating high functional systems similar to biological systems.4–6 Typical examples of natural chromoproteins include the fluorescent protein family, haemoglobin, flavoproteins, ferritin, and chloroplastin.7–9 These proteins are characterized by electronic absorption in the visible or near-infrared (NIR) spectral range and may serve as enzymes, photoreceptors, photocatalysts or electron transferring elements. Chromopeptides, which are derived from chromoproteins, integrate the programmable flexibility of peptide motifs and optical functions of chromophores, showing an ingenious combination of peptide and chromophore properties.Chromopeptides are structurally simplified analogues of chromoproteins, which perform synergistic functions similar to those of natural chromoproteins. For example, actinomycin D, also named dactinomycin, is a natural chromopeptide composed of a heterocyclic chromophore and two cyclic pentapeptide lactone rings.10 The compound functions as a transcription inhibitor and prevents RNA polymerase elongation. Chromopeptide, as its name implies, is composed of a peptide motif and a chromophore, in which the peptide motif is covalently or noncovalently coupled with a chromophore, such as porphyrins, cyanines and anthocyanins. Compared to chromoproteins, chromopeptides have relatively simple chemical structures, flexible and adjustable photophysical properties, and capabilities of programmable self-organization.
Since as early as the 1990s, very extensive attention has been directed to chromopeptides with biomimetic and phototherapeutic functions. For example, a series of synthetic analogues of heme proteins, called heme–peptide complexes, have been synthesized to elucidate the inherent structure–function relationship of natural heme proteins and their catalytic functions.11–13 As a simplified model, the heme–peptide complex provides a fundamental explanation of the molecular mechanisms of heme protein reactions and mimics the O2 binding, activation and hydrocarbon hydroxylation chemistry processes with haemoglobin, peroxidase, and cytochrome P450. During the following years, a vast number of research studies focused on the design and synthesis of covalently linked photosensitizer–peptide conjugates to achieve precise and effective phototherapy against tumours.14–18 The porphyrin and chlorin parent compounds constitute the base of many potent photosensitizers for clinical photodynamic therapy (PDT). However, many of these compounds exhibit drawbacks, such as cytotoxicity, limited permeability and low targeting ability.19–22 Peptides are most commonly used for enhancing photosensitizer targeting specificity and decreasing nonspecific phototoxicity because their synthesis process is straightforward and they are easily conjugated to photosensitizers.23,24 These chromopeptides, also called third-generation photosensitizers, have shown increased PDT efficacy compared with that of unconjugated photosensitizers.
Chromopeptide self-assembly based on the principles of nanoarchitectonics is an important strategy for designing a wide spectrum of functional biomimetic materials and devices for nanomedicine and nanobiotechnology.23,25,26 For example, in organisms, a growing number of bacterially secreted proteins self-assemble and form fibrillar amyloid-like nanostructures to fight infections. In chlorosomes, biopigments self-assemble into large supramolecular structures to capture light and transfer energy. Inspired by the self-assembly behaviour in biological systems, efforts have been devoted to exploring the self-assembly process of well-designed small biological molecules. The self-assembly process of amino acid-encoded peptides (such as ion-complementary peptides,27 amphiphilic peptides,28,29 aromatic peptides,30 and cyclic peptides31) has led to the formation of various well-ordered nanostructures that accurately mimic the function of natural proteins. However, the peptide building blocks reported previously seldom involved in the introduction of biologically derived chromophores and the fabrication of corresponding nanomaterials.28,30,32,33 As a new family of peptide building blocks, chromopeptide self-assembly provides an ingenious combination of peptide and chromophore properties as well as synergistic functions that are inaccessible to individual molecular units.34,35 In recent years, we have witnessed increasing work on the self-assembly of chromopeptides and their unique functions and potential applications, ranging from light harvesting and biomimetic catalysis to cancer diagnosis and treatment. However, despite numerous highly innovative and original studies, advances in chromopeptide self-assembly have not been summarized.
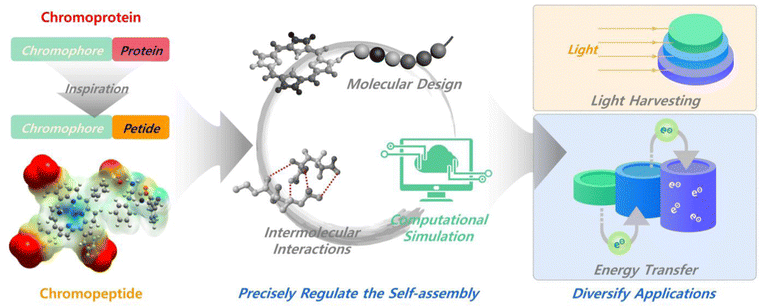
In this review, we present the self-assembly of chromopeptides, with a focus on the molecular design, assembly process control, structure regulation, function integration and biological applications of chromopeptide-based assembly materials (Fig. 1). The first section introduces the molecular strategies of designing chromopeptides, including the following typical combination types between the chromophores and peptide motifs: noncovalent combination and covalent conjugation. The second section describes principles and examples on how to regulate the self-assembly process, the resulting supramolecular structures and their functions. The following section covers the potential applications of chromopeptide assemblies, such as imaging, phototherapy, biomimetic photocatalysis and photosynthesis. With an overview of chromopeptide nanoarchitectonics, we can systematically understand this kind of new emerging peptide building blocks. More importantly, this review presents a versatile tool to develop functional biomimetic materials for a variety of applications.
 | ||
| Fig. 1 Schematic representation of the chromopeptide molecular structure, self-assembly and representative biological applications. | ||
2 Molecular design principles for chromopeptides
Chromoproteins are composed of chromophores and protein segments, mostly conjugated in a noncovalent manner.36 Chromoprotein complexes based on self-assembly or aggregation of chromoproteins are naturally occurring complexes with unique features. One typical example is the purple bacterial antenna complex system, in which 9 α/β-apoprotein pairs are assembled on a cylinder with a hollow radius of 14 Å and an outer wall radius of 34 Å. Inside each α/β-apoprotein pair are bound three NIR bacteriochlorophylls (BChl) with histidines from α-(His 31) and β-(His 30). The BChls in the complex system form an overlapped ring-like supramolecular structure, which facilitates efficient light harvesting and electron transfer.37 In many cases, the chromophores inside the chromoprotein supramolecular structure are either single molecular units or arranged in a definite but loosely-packed pattern, which depends on the functions of the chromoprotein.4,38Comparably, chromopeptides are much more flexible in molecular composition and can be obtained via both covalent and noncovalent bonds.7,39 Besides, the chromophore motifs in the chromopeptide may have stronger interaction and closer attachment with each other than those in chromoprotein. By designing chromopeptide molecules and assembling well-defined supramolecular structures, multiple unique physical properties and novel functionalities can be achieved. This section summarizes the molecular design principles together with the self-assembled structures of chromopeptides.
2.1 Noncovalent assembly of chromophores and peptides as building blocks
Similar to chromoprotein, as a biomimetic case, the chromophore and peptide building blocks are noncovalently coassembled. The noncovalent combination provides maximal freedom for each motif during assembly,40,41 and the resulting supramolecular assemblies may exhibit diverse nanostructures, morphologies and properties, which are not only dependent on the intrinsic properties of each component, but can also be regulated by the sequence of the peptide motif, the noncovalent binding site, the molar ratio between building blocks, and many other environmental conditions as well as external species.The coassembly of chromophores and polypeptides with charged repeating residues has been demonstrated to be efficient in inducing J-aggregate formation. One example is the coassembly of poly(L-Lys) and sulfonated porphyrin(tetrakis(4-sulfonatophenyl) porphine (TPPS)), yielding rod-like assemblies.42,43 In addition, the coassembly of chromophores and peptide dendrimers with well-defined and controllable end groups has been shown to produce higher ordered structures. For example, a natural zinc(ii) porphyrin derivative Zn–mesoporphyrin IX coordinates with the well-designed peptide dendrimer to generate assemblies of porphyrin–dendrimers and realizes enhanced electron transfer, which is mainly because the porphyrins stack more densely in the peptide dendrimers and the light energy delocalizes better.44,45
Importantly, the coassembly between peptides and chromophores may be controlled precisely and flexibly. Fry et al. reported that self-assembly of an amphliphilic octapeptide with an N-palmitoyl group, namely, c16-AHL3K3, in neutral aqueous media afforded micelles with an average diameter of 6 nm. Whereas by introducing a natural heme derived chromophore, (PpIX)Zn, which was chosen to mimic natural light-harvesting species, micelles still formed and their size grew from 6 nm to 30 nm along with the molar ratio between chromophores and peptides increasing from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 to 1
60 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6.46 The change in micellar size is considered to be occurring due to the competing interactions. In the absence of (PpIX)Zn, the micelles form due to the presence of a hydrophobic palmitoyl core and electrostatic repulsion between the neighboring positively charged lysine. Upon incorporating (PPIX)Zn, additional forces are introduced: (1) electrostatic attraction between lysine and the negatively charged carboxyl moieties on the macrocycle and (2) Zn–histidine coordination. Probably, the former dominantly contributes to the micelle size enlargement. Meanwhile, the coassembly morphologies were influenced by the solution pH and the peptide sequence.47 Taking the pH value as an example, the micelle at pH = 7 spontaneously converted to fibres with the pH increasing to 11. Herein, the high pH close to the pKa of lysine reduces electrostatic repulsion, shortening the distances between individual peptide molecules to a van der Waals range. Synergistically, leucines assist in the formation of long-aspect ratio nanofibers by serving as a β-sheet structural motif. These coassembly conditions have great impacts on the spin states of (PpIX)Zn. As a consequence, the photophysical properties together with its resulting catalytic activities can be finely tuned by coassembling with peptides.
6.46 The change in micellar size is considered to be occurring due to the competing interactions. In the absence of (PpIX)Zn, the micelles form due to the presence of a hydrophobic palmitoyl core and electrostatic repulsion between the neighboring positively charged lysine. Upon incorporating (PPIX)Zn, additional forces are introduced: (1) electrostatic attraction between lysine and the negatively charged carboxyl moieties on the macrocycle and (2) Zn–histidine coordination. Probably, the former dominantly contributes to the micelle size enlargement. Meanwhile, the coassembly morphologies were influenced by the solution pH and the peptide sequence.47 Taking the pH value as an example, the micelle at pH = 7 spontaneously converted to fibres with the pH increasing to 11. Herein, the high pH close to the pKa of lysine reduces electrostatic repulsion, shortening the distances between individual peptide molecules to a van der Waals range. Synergistically, leucines assist in the formation of long-aspect ratio nanofibers by serving as a β-sheet structural motif. These coassembly conditions have great impacts on the spin states of (PpIX)Zn. As a consequence, the photophysical properties together with its resulting catalytic activities can be finely tuned by coassembling with peptides.
Remarkably, the coassemly morphology is varied if the chromophore is in a relatively high concentration. Yan et al. represented a hierarchical assembly scenario of dipeptides and TPPS with a defined molar ratio.48 In an acidic solution, the dipeptide, diphenylalanine (FF), is a singly charged cation ([FF]+), and TPPS is dianionic ([H4TPPS]2−). Coassembly of TPPS and FF led to the formation of a porous microsphere with a molar ratio of 2.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, consistent with the charge-neutralized 2
1, consistent with the charge-neutralized 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dipeptide–porphyrin ionic complex. The formation of microspheres occurred in two sequential steps as follows: driven by the π–π interactions between intermolecular TPPS molecules, TPPS first assembled to form an ordered J-aggregation structure; and then, driven by the electrostatic interaction between [FF]+ and [H4TPPS]2−, a spherical structure with porosity, multiple chambers and a high degree of hydration was eventually formed (Fig. 2A–C). Interestingly, the microassembly morphology drastically varies depending on the peptide properties. Substituting amphiphilic FF with hydrophilic dilysine (KK) afforded a hierarchically organized fibre bundle.49 The coassembly of hydrophilic KK and porphyrin also preferentially formed nanorod-shaped J-aggregates via strong π–π interactions of TPPS at the beginning and then spontaneously grouped into fibre bundles with anisotropic birefringence and photoluminescence; this occurred due to the synergistic effects between long-range electrostatic repulsion and short-range van der Waals attraction (Fig. 2A, D and E). The coassembled nanoplatforms exhibited remarkable differences in their nanostructure morphologies, which was attributed to the difference in the hydrophilic and hydrophobic properties of the peptides. FF molecules show strong hydrophobicity and tend to insert into the interior of the porphyrin J-aggregate structure, while hydrophilic KK molecules tend to be fixed on the surface of porphyrin J-aggregates, thus enhancing the directivity of porphyrin nanorods and promoting the directional growth of nanorods into the fibre bundle structure (Fig. 2A). These results also indicated that organic chromophores like porphyrin at a high concentration usually form strong π–π interactions, and thus prefer aggregation to oligomers before coassembly with peptides. Compared with the porphyrin alone, the chromopeptide system exhibits much enhanced resistance to photodegradation and potential applications such as light-harvesting antennae and sustainable photocatalytic activity.
1 dipeptide–porphyrin ionic complex. The formation of microspheres occurred in two sequential steps as follows: driven by the π–π interactions between intermolecular TPPS molecules, TPPS first assembled to form an ordered J-aggregation structure; and then, driven by the electrostatic interaction between [FF]+ and [H4TPPS]2−, a spherical structure with porosity, multiple chambers and a high degree of hydration was eventually formed (Fig. 2A–C). Interestingly, the microassembly morphology drastically varies depending on the peptide properties. Substituting amphiphilic FF with hydrophilic dilysine (KK) afforded a hierarchically organized fibre bundle.49 The coassembly of hydrophilic KK and porphyrin also preferentially formed nanorod-shaped J-aggregates via strong π–π interactions of TPPS at the beginning and then spontaneously grouped into fibre bundles with anisotropic birefringence and photoluminescence; this occurred due to the synergistic effects between long-range electrostatic repulsion and short-range van der Waals attraction (Fig. 2A, D and E). The coassembled nanoplatforms exhibited remarkable differences in their nanostructure morphologies, which was attributed to the difference in the hydrophilic and hydrophobic properties of the peptides. FF molecules show strong hydrophobicity and tend to insert into the interior of the porphyrin J-aggregate structure, while hydrophilic KK molecules tend to be fixed on the surface of porphyrin J-aggregates, thus enhancing the directivity of porphyrin nanorods and promoting the directional growth of nanorods into the fibre bundle structure (Fig. 2A). These results also indicated that organic chromophores like porphyrin at a high concentration usually form strong π–π interactions, and thus prefer aggregation to oligomers before coassembly with peptides. Compared with the porphyrin alone, the chromopeptide system exhibits much enhanced resistance to photodegradation and potential applications such as light-harvesting antennae and sustainable photocatalytic activity.
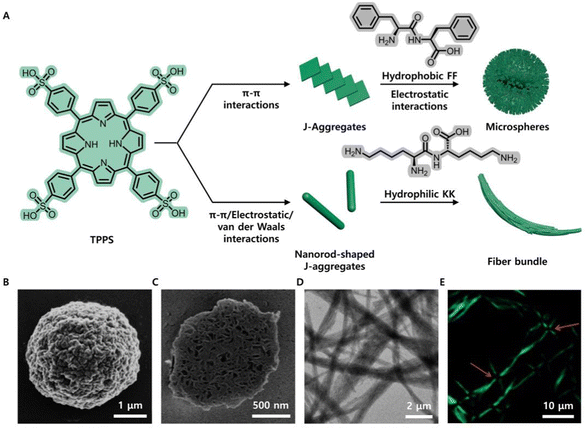 | ||
| Fig. 2 (A) Schematic illustration of the fabrication of microspheres and fibre bundles by the coassembly of TPPS with amphiphilic FF and hydrophilic KK, respectively. (B) SEM image of the porphyrin-FF microsphere, showing irregular surface texture. (C) SEM image of the microsphere cross-section, showing a porous, multichambered interior. (D) TEM image of the porphyrin-KK bundled fibres. (E) Anisotropic photoluminescence at the intersection of orthogonal fibre bundles. Reproduced from ref. 48,49 with permission from [John Wiley and Sons], copyright [2014, 2015]. | ||
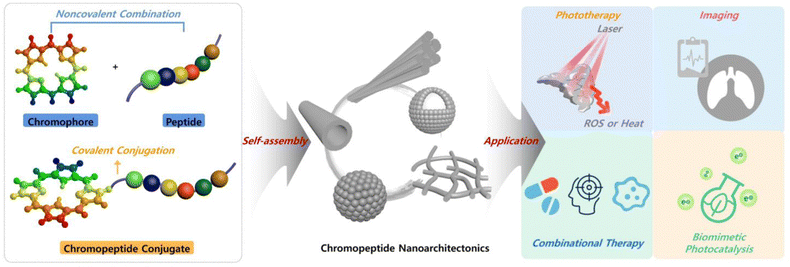
2.2 Self-assembly of molecular chromopeptide conjugates
Since chemical modification of peptides is facile, the chromophore and peptide segments could also be covalently conjugated and integrated into a new molecular building block. Herein, the chromophore is selected or designed by considering its unique functions like fluorescence, photodynamic activity, photothermal conversion, photocatalysis, and so forth. The peptide is adopted based on both its biological activity and its properties on tuning assembly. Depending on their photophysical properties and biological activities, the compounds can be directly conjugated or spaced with a linking group. Different covalent conjugation strategies lead to various assembled nanostructures, as classified in the following subsections.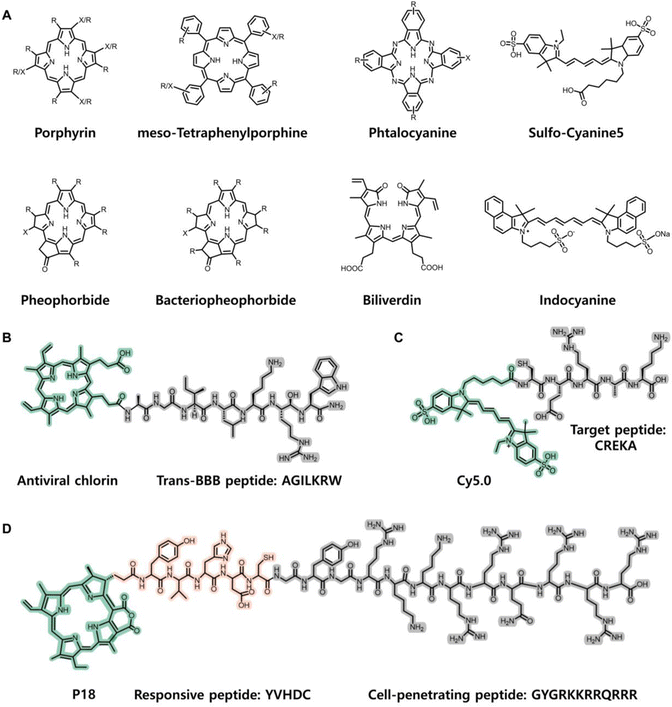 | ||
| Fig. 3 (A) Basic structures of chromophores. “R” indicates an alkyl or alkenyl substituent, and “X” indicates the functional group for covalent linkage with the peptide chain. Reproduced from ref. 39 with permission from [John Wiley and Sons], copyright [2018]. Example structures of directly conjugated chromopeptides: (B) Chromopeptide directly conjugated between antiviral porphyrin and trans-BBB peptide AGILKRW. Reproduced from ref. 50 with permission from the [American Chemical Society], copyright [2021]. (C) Chromopeptide conjugated by Cy5.0 and target peptide CREKA. Reproduced from ref. 59 with permission from the [Nature Publishing Group], copyright [2015]. (D) Chromopeptide conjugated by P18, responsive peptide YVHDC and cell penetrating peptide GYGRKKRRQRRR. Reproduced from ref. 61 with permission from the [American Chemical Society], copyright [2016]. | ||
In the field of PDT, conjugating a photosensitizer (a tetrapyrrole compound, such as porphyrin and chlorin) to natural cationic antimicrobial peptides (such as buforin II, a 21 amino acid peptide, or magainin 2, a 23 amino acid peptide) results in the formation of a porphyrin–peptide conjugate with combined antimicrobial and PDT functions.54,55 In addition to improving the solubility of hydrophobic photosensitizers, antimicrobial peptides also improve the PDT effect against different bacterial targets.56 In addition, a de novo designed six-residue oligoarginine cationic peptide has been linked to a chlorin derivative (purpurin, P18) to selectively kill Gram-positive bacteria by inserting into the cell membrane, thus improving the photodynamic activity.57 To improve the delivery of PDT for malignant tumours, cell-penetrating peptides (CPPs) have emerged as promising delivery agents for enhancing the cytosolic delivery. The short oligoarginine peptide sequence GRKKRRQRRRPPQ, which is derived from the human immunodeficient virus (HIV)-1 Tat protein residue 48–60, has been conjugated to hydrophobic porphyrins and chlorins to enhance their uptake in cancer cells.58 In addition, Toddorovski et al. synthesized a series of peptide–porphyrin conjugates by coupling a porphyrin carboxyl group to an amino group (either the N-terminal or the Lys side chain) of the peptide shuttle, exhibiting an ability to cross the blood-brain barrier (BBB) and potential antiviral activities (Fig. 3B).50
The chromopeptide can also be designed to exhibit unique properties and complicated functions. Lu et al. used a fluorescence probe CREKA-Cy5.0 to determine the binding specificity of CREKA to the fibrin–fibronectin complexes in metastatic tumours (Fig. 3C).59 Zheng et al. reported a stimulus-responsive molecular beacon composed of a pyropheophorbide (Pyro) and a singlet oxygen quencher motif which were covalently connected by a fibroblast activation protein (FAP)-responsive peptide.60 The chromopeptide compound was intrinsically nontoxic due to the quenching of ROS. By FAP activation, the peptide is cleaved, and the remaining species turn to generate 1O2 for cancer treatment. In a similar manner, Wang et al. designed a conjugate by combining P18, responsive peptide-sequenced YVHDC and CPP-sequenced GYGRKKRRQRRR (Fig. 3D).61 It can be cleaved by caspase-1 and trigger assembly from monomer to aggregates. In addition, Kim et al. reported an internalizing cyclic peptide iRGDC-conjugated pyro derivative, which was further conjugated with quencher BK01 (pyropheophorbide-iRGDC-BK01).62 The conjugate was designed as an in situ self-implantable photosensitizer. Its molecular functions include cancer cell internalization and photosensitization activated by tumour-selective proteolytic/reductive cleavage of the iRGD segment. Li et al. conjugated the cyclic pentapeptide cRGDfk and chlorin e6 (Ce6) to silk fibroin (SF) polypeptides.63 SF acts as an integrator, which not only supports the photodynamic effect of Ce6 and the tumour targeting effect of cRGDfk but also improves the biocompatibility and biosafety of the chromopeptide materials. The above works show that the variations in each functional motif and its conjugation site are limitless; thus, the chromopeptide properties and applications may be constructed on-demand.
More interestingly, some natural photosensitizers and their derivatives contain more than one carboxylic group, and a great majority of peptide molecules contain multiple binding sites, which provides more abundant possibilities for modification and is advantageous for multifunctional molecular design.64 Fernandez et al. conjugated two different chromophore molecules with dipeptide FF to obtain chromopeptide molecules via amidation. The two molecules exhibited the same one-dimensional nanofibre morphology under different assembly mechanisms.65 Varying the protective group of the chromopeptide molecules or changing the external environment, such as the solvent, can produce assemblies with different structures. Coutsolelos et al. synthesized eight peptide–chromophore conjugates and assembled them by changing the solvent ratio to obtain adjustable sizes and structures, such as spiky spheres, nanofibres, hydrogels, and nanospheres.66
The additional linker functions as a spacer that separates the peptide motif and chromophore to an appropriatedistance so that their respective functions are trivially affected by each other. 6-Aminohexanoic acid (Ahx) is an ω-amino acid with a hydrophobic and flexible structure and is a typical space linker that contains both terminal amino and carboxylic groups; in addition, a general solid phase peptide synthesis procedure similar to that of amino acids can be performed with Ahx.67 For example, by inserting an Ahx residue between a cyclic 13-mer oligopeptide (Pep42, sequenced cyclic CTVALPGGYVRVC) and a fluoroalkyl zinc phthalocyanine, both the singlet oxygen (1O2) generation of phthalocyanine and the targeting ability toward the chaperone protein GRP78 of Pep42 were maintained.68 Nevertheless, the bioconjugate is resistant to decomposition under 1O2 generation. The zinc phthalocyanine-Ahx-Pep42 bioconjugates were designed to provide access to a new class of photoactive probes that can be useful for the rational development of catalytic materials. A similar effect was observed for a designed chlorin-type photosensitizer–peptide conjugate.69 The researchers conjugated tetraphenylchlorrin (TPC) via a spacer (6-aminohexanoic acid, Ahx) to a vascular endothelial growth factor (VEGF) receptor-specific heptapeptide (ATWLPPR) (Fig. 4A). TPC was adopted as a model photosensitive chromophore that is easily available. TPC and ATWLPPR were coupled through the linker Ahx to achieve individualization. The photophysical properties of TPC remained unchanged after coupling with Ahx-ATWLPPR. In addition, the combination of ATWLPPR with Ahx does not affect the combination between the heptapeptide and neuropilin-1 (NRP-1). The presence of the Ahx spacer increased the flexibility of the molecule, and its length, and the nature of the molecule attached to it, impacted receptor affinity.
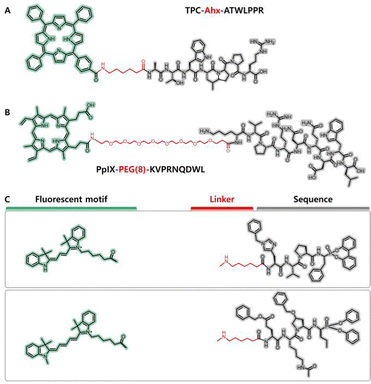 | ||
| Fig. 4 (A) Molecular structure of a chromopeptide, namely, TPC–Ahx–ATWLPPR, in which an Ahx linker (marked with blue background) was employed for conjugation between a chlorin-type photosensitizer (TPC) and peptide motif. Reproduced from ref. 69 with permission from [Elsevier], copyright [2006]. (B) Molecular structure of another chromopeptide, namely, PpIX-PEG(8)-KVPRNQDWL, in which a PEG(8) linker (marked with blue background) was employed for conjugation between the PpIX motif and peptide motif. Reproduced from ref. 75 with permission from [John Wiley and Sons], copyright [2019]. (C) Representative structures of the motifs in the toolbox, including fluorescent motifs, linkers and specific sequences. Reproduced from ref. 77 with permission from the American Chemical Society, copyright [2017]. | ||
In addition to optimizing the distance between the peptide motifs and chromophores, the linker also provides a unique avenue for tuning the amphiphilicity of the conjugate. Polyethylene glycol (PEG) has been demonstrated to be a chemically inert, biocompatible and amphiphilic substance.70,71 PEG that exhibits low degrees of polymerization and consists of a few periodic ethylene glycol units is a suitable candidate for coupling chromophores and peptide motifs for biological applications. Drag et al. designed a specific tetrapeptide sequenced Ac-Abu-DTyr-Leu-Gln-VS. for the main proteases of coronavirus disease 2019 (COVID-19) with an inhibition rate constant of 812 ± 70 M−1 s−1. To visualize the SARS-CoV main proteinase activity in patient samples, they further designed fluorescence activity-based probes (ABPs) that conjugate a dye to the tetrapeptide via a PEG(4) linker.72 Due to the adequate length and amphiphilicity of PEG(4), the conjugate almost maintained its inhibitory activity with inhibition rate constants as high as 439 ± 34 M−1 s−1, and the detection sensitivity reached as low as 5 nM with a 2.5 μM conjugate solution.
In many cases, utilizing a PEG linker can realize the controllable construction of self-assembled nanostructures. It was reported that PEG chains have an important function of improving the circulation time of assemblies in blood, and connecting the assemblies with PEG can improve their stability.73,74 Li et al. designed a self-delivery chimeric chromopepetide conjugate composed of a photosensitizer PpIX, a PEG(8) linker and a melanoma-specific antigen sequenced KVPRNQDWL (Fig. 4B).75 The PpIX intrinsically has photodynamic activity under irradiation light and facilitates apoptosis and/or necrosis of melanoma cells with an intense immune response. This chromopeptide could self-assemble into spherical nanoparticles with an average size of 102 nm and a low polydispersity index of 0.197, which facilitated tumour accumulation through an enhanced penetration and retention (EPR) effect. A similar effect was found for the PpIX-PEG(8)-RDEVDG-K(TPE)-V-CONH2 chromopeptide conjugate, in which TPE was a tetraphenylethylene derivative connected to the ε-amino group of Lys.76 The conjugate with the PEG(8) linker exhibited a switchable morphology under different pH conditions. At pH = 7.4, the conjugate self-assembled into spherical nanoparticles, and under mildly acidic conditions, the conjugate exhibited a spherocylinder morphology.
Remarkably, the design of the linker depends on the chemical properties of each molecular motif and the intended function of the conjugate. However, it is not always easy to create an excellent design. Innovatively, researchers explored a toolbox for reliable parallel imaging of active neutrophil serine proteases, which contains a set of fluorescent probes, linkers and peptides (Fig. 4C).77 The N-terminus contains fluorescent tags, and the linkers include PEG and Ahx. The researchers attempted all possible combinations of motifs for each peptide specifically identifying NE, PR3, CATG or NSP4 in complex mixtures. As a result, highly selective chromopeptides that are responsive to different protein targets were screened. The amidation coupling described in the foregoing section shows limitations, that is, the carboxylic groups are necessary on both the chromophore motifs and the linkers, which may lead to tedious chemical modifications. In addition, an analogous method was applied to conjugate carboxyl-absent phthalocyanine with bioactive peptides, which have been successfully used as receptor-targeting bifunctional agents.78
With the burgeoning of “click chemistry”, the conjugation between chromophores and peptides becomes more efficient and flexible.79 The covalent bond formation can occur under mild conditions and be accurately controlled in situ. These methods of conjugating chromophores and peptide motifs remain under development. Prospectively, the relationship between molecular structures and self-assembled nanostructures together with their functions will be valuable to investigate.
3 Effects of intermolecular interactions on the assembly of chromopeptide
Intermolecular interactions are the underlying mechanism of self-assembly.80–82 Representative intermolecular interactions include hydrogen-bonding interactions, electrostatic interactions (or Coulombic forces), hydrophobic interactions and van der Waals forces. The intensity and anisotropy of each interaction defined by a certain (combination of) molecule(s) lead to various self-assembled nanostructures. In fact, the method of peptide design has been utilized by many researchers to achieve the desired self-assembly nanostructures. There have been mature discussions on the assembly of peptides with diverse secondary structures. For example, peptides can be artificially designed as primary sequences and show the assembling tendency to certain supramolecular structures such as amyloids, hydrogels, and so forth, depending on the assembly properties of each amino acid residue and their integrated effects.83 It is also true for those broadly studied peptide derivatives like lipopeptides and peptide–drug conjugates, where the modification group of linear alkyl chains provided hydrophobic interactions to the assembled system, and the assembly of drug molecular moieties was regulated by the conjugated peptide.84,85 However, the situation becomes more complicated for the chromopeptide, as the chromophore exhibits a π-electron conjugated structure with intermolecular π–π interactions. These π–π interactions between chromopeptides are stronger and more anisotropic than those between individual peptides. Consequently, peptides bearing a chromophore show a more complicated self-assembly behaviour than that of chromophore-free peptide motifs.A series of pH-sensitive self-assembling amphiphilic dye–cyclopeptide conjugated nanoprobes were designed.86 The dye (CH1055) contained a fluorescent skeleton in the second near-infrared window (NIR-II) and was modified with a propionate group to reduce hydrophobicity. It was found that the size and structure of the assembly depend on the molecular design. When the dye and cyclopeptide were conjugated at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio, CH-c(RGDfk) solid nanospheres were formed, and the size was approximately 80 nm. When the molar ratio was 1
1 molar ratio, CH-c(RGDfk) solid nanospheres were formed, and the size was approximately 80 nm. When the molar ratio was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 (note that the dye contains four conjugatable branches), inhomogeneous CH-3c(RGDfk) nanovesicles were formed. Provided that the dye and the cyclopeptide were conjugated by a PEG(2) linker, size-reduced CH-PEG(2)-c(RGDfk) nanospheres with diameters of approximately 10 nm were formed. Clearly, the variance in these nanostructures is strictly related to the molecular structure of the chromopeptide.
3 (note that the dye contains four conjugatable branches), inhomogeneous CH-3c(RGDfk) nanovesicles were formed. Provided that the dye and the cyclopeptide were conjugated by a PEG(2) linker, size-reduced CH-PEG(2)-c(RGDfk) nanospheres with diameters of approximately 10 nm were formed. Clearly, the variance in these nanostructures is strictly related to the molecular structure of the chromopeptide.
Obtaining precise knowledge of the intermolecular interactions between chromopeptides is of paramount importance, and performing quantitative investigation on the contributions of intermolecular interactions is essential to control the self-assembled nanostructure and its functions in a more delicate way. Yan's group reported that FF-conjugated tetraphenylporphyrin (TPP-G-FF) could self-assemble into robust nanodots with a diameter of approximately 25 nm and excellent stability (Fig. 5A and B).53 Molecular dynamics simulation revealed that the colloid stability resulted from the counterbalance between hydrogen-bonding interactions, which were contributed by the carboxyl group of peptides and solvent, as well as π–π interactions contributed by the porphyrins. Notably, individual FF is a self-assembled dipeptide that may afford various assembling structures depending on the assembly process,87,88 while in contrast, the FF motif in TPP-G-FF played a role of inhibiting nanodot overassembly or overgrowth caused by the intense π–π interaction from the TPP motif, and providing aqueous stability for the nanodots (Fig. 5C). Such difference indicated that the π–π interactions between chromophore motifs were very strong and dominated the assembly. The overall effect of these interactions led to more complete fluorescence quenching of the nanodots compared to that of the TPP-FF monomeric solution; thus, the nanodots are an excellent candidate for a photothermal agent. Consequently, the photothermal conversion efficiency of the nanodots self-assembled by the peptide–porphyrin conjugate reached 54.2%, which is comparable to that of widely applied inorganic nanomaterials,89,90 such as commercial Au nanoshells (13%) and nanorods (21%) and black phosphorus quantum dots (28.4%). Furthermore, Yan et al. proposed a new mechanism coined as “supramolecular photothermal effects”, which is controlled by simple and flexible noncovalent intermolecular interactions for efficient thermal conversion.91,92
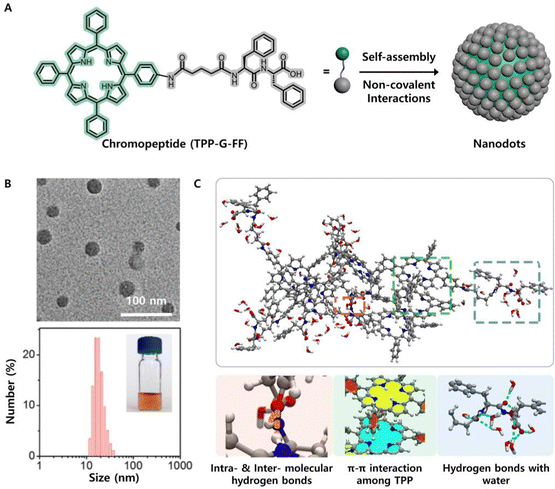 | ||
| Fig. 5 (A) Schematic illustration of the self-assembly of a peptide–porphyrin conjugate (TPP-G-FF) into photothermal nanodots. (B) TEM image and DLS size distribution of the nanodots. The inset shows a picture of the nanodots in an aqueous solution. (C) Underlying intermolecular interaction mechanism of TPP-G-FF molecules obtained by MD simulation. Reproduced from ref. 53 with permission from the American Chemical Society, copyright [2017]. | ||
Recently, Yan et al. demonstrated that the intermolecular interactions, as well as the structural, energy-conversion, and therapeutic properties of supramolecular assemblies, can be customized by using amino acid-encoded peptide sequences.93 The researchers selected pheophorbide A (PheoA), a natural chlorophyll derivative as the model chromophore because it absorbed far-red light strongly and had a carboxyl group available for coupling with peptides. Four oligopeptides sequenced as Asp (D), di-Asp (DD), FF, di-Tyr (YY) and tetraaspartic acid (DDDD), respectively, with distinct hydrophobicity were conjugated to PheoA to afford a chromopeptide building block. Remarkably, PheoA-FF self-assembled into nanoparticles, while the other conjugates self-assembled into nanofibrils. This is because FF is hydrophobic, while Y and D residues provide hydrophilic and anisotropic hydrogen-bonding interactions. All these nanoplatforms exhibited fluorescence quenching efficiencies higher than 85% and photothermal conversion efficiencies higher than 40%. The singlet oxygen quantum yields of the PheoA-D, PheoA-DD, PheoA-DDDD, PheoA-FF, and PheoA-YY nanoplatforms, in contrast, were distinguished by 0.214, 0.397, 0.494, 0.115, and 0.106, respectively. It seems that more hydrophilic nanofibres tended to exhibit higher singlet-oxygen quantum yields. In addition, these nanomaterials also exhibited different cellular internalization and disassembly behaviours in physiological environments. This study suggests that supramolecular assemblies based on chromopeptide conjugates with optimized phototherapeutic functions can be intentionally regulated by controlling noncovalent interactions through encoding the amino acid or peptide sequence.
4 Dynamically assembled nanostructures
Chromopeptide self-assembly is a strategy to produce desirable nanostructures and material properties by organizing designed molecular building blocks. Thermodynamics and kinetics of peptide self-assembly play key roles in structural modulation and function integration.33 Since the chromopeptide building blocks can be flexibly designed and their assembly process can be precisely controlled, the resulting assembled structures and functions are diverse and fascinating. This section focuses on how to modulate the nanostructures and functions of chromopeptide assemblies.The thermodynamic and kinetic conditions, including solvent, concentration, spaciotemporal variance in species, temperature, and so forth, are external environments that strongly affect the chromopeptide self-assembly process as well as the nanostructures and their functionality.
Self-assembled chromopeptide materials have a wide range of biological applications, mainly in physiological settings. Therefore, pH is among the most important thermodynamic parameters. Utilizing the pH difference between normal tissue (pH ≈ 7.4) and the tumour region (pH < 6.5), a self-transformable pH-driven chromopeptide (PpIX-AEQNPIYWARYADWLFTT-PLLLLDLALLVDADEGT, a conjugate of protoporphyrin and a membrane-anchoring peptide) was designed.94 The peptide is pH-responsive; thus, the chromopeptide remained monomeric due to hydrophilicity at pH ≈ 7.4, while the peptide motif turned into an α-helical structure and inserted into the cell membrane after the pH was reduced to 6.5. The pH-triggering dynamics of the chromopeptide facilitated the in situ generation of ROS upon light irradiation and direct damage to tumour membranes.
Notably, the thermodynamic conditions are highly related to self-assembly kinetics and lead to complexity in the self-assembly pathway. Therefore, a chromopeptide building block may self-organize into more than one kind of assembly depending on the thermodynamic and kinetic conditions. Yan and co-workers prepared a model chromopeptide molecule, FF-conjugated phthalocyanine (PF, Fig. 6A), in which phthalocyanine is an excellent self-assembling unit due to its highly symmetrical structure and unique intermolecular π–π interactions. The model building block, PF, could self-assemble into nanoparticles by rapid precipitation; specifically, a very small amount of DMSO solution of PF was simply titrated into water. However, by increasing the ratio between DMSO and water and titrating the DMSO solution of PF into water repeatedly over 72 h, a new self-assembly morphology was obtained rather than nanoparticles (Fig. 6B).95,96 The new morphology was helical nanofibrils, which exhibited a greater thermodynamic stability than that of nanoparticles. Compared with the PF monomer and nanoparticles, more interestingly, the nanofibrils exhibited a very large redshifted absorption (105 nm) and moved to the NIR region (Fig. 6C). The variance in self-assembled nanostructures occurred because rapid precipitation is an entropy-dominated process and the molecules are isotropically assembled. In contrast, the slow and fractional titration allows the molecules to find a thermodynamically favoured supramolecular structure and is an enthalpy-dominated process. Due to the closely stacked phthalocyanine motifs that contain unique π–π interactions between them, the absorption spectroscopy also drastically changes (Fig. 6D). This pathway-selective self-assembly provides more possibilities for the functionalization of chromopeptide materials.
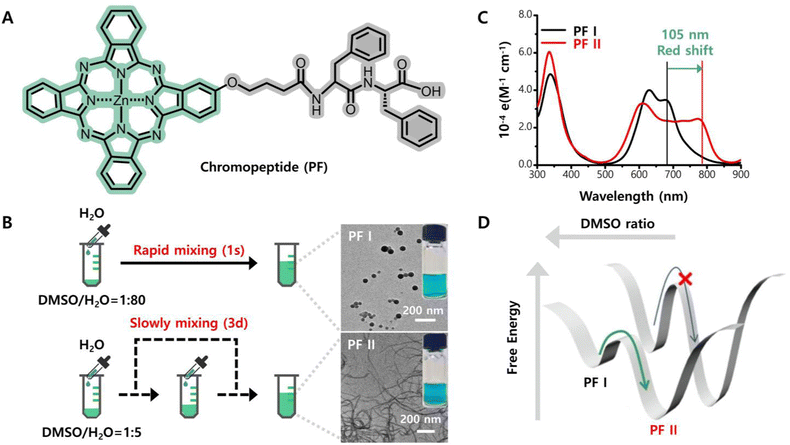 | ||
| Fig. 6 Kinetically controlled self-assembly of phthalocyanine–diphenylalanine (PF) enables a very large redshifted absorption. (A) Molecular structure of chromopeptide PF. (B) Schematic illustration of two assembly processes and the resulting assembly morphologies. (C) Electronic absorption spectra of the two assemblies. (D) Underlying thermodynamics of PF assemblies. Reproduced from ref. 95,96 with permission from the [Chinese Chemical Society], copyright [2019, 2021]. | ||
The kinetic control of chromopeptide self-assembly can be accompanied by stimulus-triggered chemical reactions. In this way, the nanostructures and functions of nanomaterials exhibit dynamic variation depending on the external environment. For example, a P18-peptide (sequenced PLGVRGRGD) was designed for cancer diagnostics and phototherapy.97 The chromopeptide diffused in the physiological environment and targeted αvβ3 integrins due to the presence of the terminal RGD ligand. Furthermore, the PLGVRG motif is enzyme responsive and can be cleaved by the overexpression of gelatinase in the tumour microenvironment, causing the chromopeptide to decompose. Consequently, the chromopeptide simultaneously self-assembled into nanofibres at tumour sites, exhibiting an assembly induced retention (AIR) effect, which resulted in an improved imaging signal and enhanced phototherapeutic efficacy.
The coupling between self-assembly and enzyme-responsive reactions usually renders a cascaded synergistic effect. A chimeric chromopeptide C16-K(PpIX)-PEG(8)-KDEVD-1MT (PpIX-1MT) was designed, in which C16 is a hydrophobic palmitoyl group, the photosensitizer PpIX is conjugated to the ε-amino group of Lys, DEVD is a caspase-3-sensitive peptide sequence, and 1MT refers to 1-methyltryptophan.98 The chromopeptide PpIX-1MT could initially form nanoparticles in phosphate buffer solution (PBS) and accumulate in tumour regions; then, reactive oxygen species (ROS) are produced and 1MT is released upon caspase-3 cleavage, which triggers an intense immune response. Similarly, a tumour extracellular acidity-responsive conjugated chromopeptide PpIX-Ahx-AEAEAKAKAEAEAKAK (PEAK) was designed, in which the ε-amino group of each Lys residue was initially protected by an acid-labile dimethylmaleic anhydride (DMA) group.99 PEAK underwent self-assembly into stable nanoparticles, which was driven by the synergistic effect of ionic complementarity (a special kind of electrostatic interaction), hydrogen-bonding interactions, van der Waals forces and hydrophobic interactions. Comparably, in a tumour environment with a low pH, the DMA groups were cleaved, baring the positively charged amino group. Due to the enhanced ionic complementarity, the nanoparticles transformed into nanorods, which facilitated the internalization of tumours, thus achieving improved phototherapeutic efficacy. Remarkably, chromopeptide design with dynamic and tunable self-assembly ability provides opportunities to realize complicated functions.
5 Applications
Self-assembled chromopeptide systems offer unique advantages for biological applications. As a key component of chromopeptides, peptide motifs exhibit good biocompatibility, biodegradability, programmability and diverse biological functionalities. Chromophores with tunable optical properties offer multiple opportunities in diagnosis, therapy, energy conversion, etc. Utilizing bioactive peptides may greatly expand the functionality of self-assembled chromopeptide systems. In addition, assembling chromopeptides offers a solution for inhibiting the enzymatic degradation and rapid metabolism of chromopeptides in the biological environment. Thus, based on the knowledge of the self-assembly mechanism detailed above, self-assembled chromopeptide systems may be promising candidates for bioimaging, phototherapy, combined therapy, biomimetic photocatalysis and photosynthesis.5.1 Imaging
Similar to chromoproteins, chromopeptides play an important role as powerful imaging tools. Some chromopeptides perform imaging functions that can be used as guidance tools for optical biopsy to specifically illuminate malignant disease areas. For example, BLZ-100 is a chromopeptide that contains a chlorotoxin peptide (36-amino acid peptide)100,101 and the NIR fluorescence imaging molecule indocyanine green (ICG) can specifically label solid tumour tissue and emit fluorescence in the NIR range for intraoperative visualization of human tumours. Importantly, chromopeptides can then self-assemble into supramolecular nanostructures with imaging capabilities (Table 1). This subsection summarizes chromopeptide nanomaterials applied for bioimaging.| Chromopeptide-based nanomaterials | Chromophore | Peptide | Morphology | Application | Ref. |
|---|---|---|---|---|---|
| “—” represents “Not provided”. | |||||
| CCNPs | Ce6 | CDP | Nanoparticles | FLI | 19 |
| TCASS | Cy | AVPIAQKDEVDKLVFFAECG | Nanofibres | FLI | 102 |
| ZB NPs | BV | His-containing | Nanoparticles | PAI | 103 |
| ZBMn NPs | BV | His-containing | Nanoparticles | PAI/MRI | |
| P18-PLG | P18 | PLGVRG | Nanofibres | PAI | 97 |
| PD-NFs | PheoA | D | Nanofibres | PAI | 93 |
| PDD-NFs | PheoA | DD | Nanofibres | — | |
| PFF-NPs | PheoA | FF | Nanoparticles | — | |
| PYY-NFs | PheoA | YY | Nanofibres | PAI | |
| PDDDD-NFs | PheoA | DDDD | Nanofibres | — | |
| PWG | Porphyrin | WG | Nanoparticles | FLI | 104 |
| TP5-ICG NFs | ICG | TP5 | Nanofibres | PAI | 105 |
| ZnPc-GGK(B)COOH NP | ZnPc | GGK(B)-COOH | Nanoparticles | FLI | 106 |
| ZnPc-GGK(B)CONH2 NP | ZnPc | GGK(B)-CONH2 | Nanoparticles | FLI | |
| PPP-NDs | Porphyrin | FF | Nanodots | PAI | 53 |
| PF NPs | ZnPc | FF | Nanoparticles | FLI/PAI | 107 |
| PEAK-DMA | PpIX | AEAEAKAKAEAEAKAK | Nanoparticles | FLI | 99 |
| ACPP-PpIX | PpIX | R9GPLGLAGE8 | — | FLI | 108 |
| PPF | Pyro | GDEVDGSGK | — | FLI | 109 |
| ELP-IR820 | IR820 | Elastin-like polypeptides (ELPs) | Nanoparticles | — | 110 |
| MRI | ICG | RADA32−melittin | Hydrogel | FLI/PAI | 111 |
Chromopeptide nanomaterials disassemble to emit fluorescence signals after reaching the target site in vivo over time, so these materials can potentially be used as bioimaging agents for cell imaging and in vivo fluorescence imaging (FLI). This is mainly because the excited pigment molecules in the chromopeptide system release energy through fluorescence emission and return to the ground state. Yan et al. utilized the hydrophobic and π–π stacking interactions between a cationic diphenylalanine (CDP) derived from FF and hydrophobic photosensitive Ce6 to self-assemble into chromopeptide spherical nanoparticles (CCNPs).19 The self-assembled chromopeptide nanospheres were taken up by MCF-7 cells and decomposed over time to release fluorescent signals, thereby realizing FLI of cells in vitro and dynamic fluorescence distribution in mice in vivo. Compared with the molecular state of photosensitive Ce6, chromopeptide nanoassemblies show enhanced structural stability, thus achieving longer imaging sessions in vivo. When the peptide motif of the chromopeptide is endowed with specific biological activity, controllable intelligent imaging becomes a reality. For example, a chromopeptide conjugate was designed, which consisted of a tumour-specific recognition motif (sequenced AVPIAQK), an enzymatic cleavage linker (sequenced DEVD), a self-assembly motif (sequenced KLVFFAECG) and a cyanine dye Cy.102 Under the action of the tumour recognition motif, chromopeptide molecules entered tumour cells and were cleaved by caspase-3/7 to trigger self-assembly in vivo, while the remaining residues self-assembled into β-sheet nanofibres under the action of hydrogen bonds. Due to the significant permeability of chromopeptide molecules, these self-assembled nanofibres in vivo exhibited high cumulative efficiency, and the fluorescent signals used for ex vivo imaging of intact patient bladders exhibited high specificity and sensitivity, which had an important clinical value in image-guided surgery for bladder cancer.
In addition, chromopeptide nanomaterials are also applied in photoacoustic imaging (PAI), a nonionizing imaging modality based on the photoacoustic (PA) effect,112–114 which depends on the physiochemical properties of the chromophore segment. Yan et al. constructed a series of supramolecular assemblies based on well-designed chromopeptides. For example, the researchers designed nanoparticles with broad NIR absorption using the endogenic biopigment biliverdin (BV) and a His-containing peptide (Z-His-Obzl, ZHO), which can be used as PAI contrast agents for diagnosing tumours with high spatial resolution and penetration depth.103 More importantly, the metal ion Mn2+ was integrated into the chromopeptide nanoparticles, which enhanced the photostability of the nanoparticles and endowed the nanoparticles with magnetic resonance imaging (MRI) capabilities. Dual-modal imaging, which integrates PAI and MRI, offers precise imaging guidance for photothermal therapy (PTT) of tumours. Chromopeptide assemblies are widely used in FLI and PAI because they contain natural chromophores. In addition, chomopeptide assemblies are also considered to have great potential in other imaging fields. On the one hand, the assemblies can be used as a carrier of other imaging agents; on the other hand, the assemblies with other imaging capabilities such as ultrasound imaging can be obtained by designing the molecular structure of chromopeptides. However, it still requires researchers to explore the application of chromopeptide assemblies much more in the imaging field.
5.2 Phototherapy
Phototherapy is a clinically approved technique that involves treating diseases with a laser, which is used as a scalpel.115–118 Compared with conventional therapeutic methods, phototherapy exhibits several advantages, including minimal invasiveness, convenient operation, fast recovery and strong specificity. For phototherapeutic purposes, the chromophore plays a key role in converting light energy to reactive oxidative species (ROS, for PDT, Section 5.2.1) or to local tumour hyperthermia (for photothermal therapy, Section 5.2.2), which leads to disease suppression under different mechanisms. The photoconversion mechanism depends on both the intrinsic physiochemical properties of the chromophore and the supramolecular structure of the chromopeptide assemblies. Coincidentally, the peptide motifs in the chromopeptide system interact with chromophores to form assembled materials with different structures and flexible functions, providing the possibility to tune the physical and chemical properties of phototherapeutic agents (Table 2); thus, the system exhibits high potential for phototherapy applications.| Chromopeptide-based nanomaterials | Chromophore | Peptide | Morphology | Application | Ref. |
|---|---|---|---|---|---|
| “r” represents d-Arg and “Fx” represents l-cyclohexylalanine; “—” represents “Not provided”. | |||||
| CCNPs | Ce6 | CDP | Nanoparticles | PDT | 19 |
| ZB NPs | BV | His-containing | Nanoparticles | PTT | 103 |
| ZBMn NPs | BV | His-containing | Nanoparticles | PTT | |
| P18-PLGVRGRGD | P18 | PLGVRGRGD | Nanofibres | Targeted therapy combined with PTT | 97 |
| PD-NFs | PheoA | D | Nanofibres | PTT | 93 |
| PDD-NFs | PheoA | DD | Nanofibres | — | |
| PFF-NPs | PheoA | FF | Nanoparticles | — | |
| PYY-NFs | PheoA | YY | Nanofibres | PTT | |
| PDDDD-NFs | PheoA | DDDD | Nanofibres | — | |
| PWG | Porphyrin | WG | Nanoparticles | PDT | 104 |
| TP5-ICG NFs | ICG | TP5 | Nanofibres | Immunotherapy combined with PTT | 105 |
| ZnPc-GGK(B)COOH NP | ZnPc | GGK(B)-COOH | Nanoparticles | Chemotherapy combined with PDT | 106 |
| ZnPc-GGK(B)CONH2 NP | ZnPc | GGK(B)-CONH2 | Nanoparticles | Chemotherapy combined with PDT | |
| PPP-NDs | Porphyrin | FF | Nanodots | PTT | 53 |
| PF NPs | ZnPc | FF | Nanoparticles | PDT and PTT | 107 |
| PEAK-DMA | PpIX | AEAEAKAKAEAEAKAK | Nanoparticles | PDT | 99 |
| ACPP-PpIX | PpIX | R9GPLGLAGE8 | — | PDT | 108 |
| PPF | Pyro | GDEVDGSGK | — | PDT | 109 |
| ELP-IR820 | IR820 | Elastin-like polypeptides (ELPs) | Nanoparticles | PTT | 110 |
| MRI | ICG | RADA32−melittin | Hydrogel | PTT | 111 |
| Pep@Ce6 | Ce6 | Cationic polypeptide | Spherical micelles | PDT | 119 |
| APP | Ce6 | GKRWWKWWRRPLGVRGC | — | PDT | 120 |
| ELTST | Ppa | GPLGLAG | Nanovesicles | Chemotherapy combined with PDT | 121 |
| Ce6-DEVD-MMAE | Ce6 | DEVD | Nanoparticles | Chemotherapy combined with PDT | 122 |
| MEL/Ce6@HA | Ce6 | Melittin | Nanoparticles | Chemotherapy combined with PDT | 123 |
| PAPP-DMA | PpIX | PKKKRKV | Nanoparticles | Targeted therapy combined with PDT | 124 |
| UCNPs@TiO2-Ce6-TAT | Ce6 | TAT | Core/shell | Targeted therapy combined with PDT | 125 |
| pnPNP | PpIX | PKKKRKV | Nanorods | Targeted therapy combined with PDT | 126 |
| M-ChiP | PpIX | rFxrFxrFxr | Spherical micelles | Targeted therapy combined with PDT | 127 |
| HB-PA | HB | CRGDKGPDC | Nanovesicles | Targeted therapy combined with PDT | 128 |
| Ppa-iRGDC-BK01 | Ppa | iRGDC | — | Targeted therapy combined with PDT | 62 |
| ChiP-Exo | PpIX | PKKKRKV | Exosomes | Targeted therapy combined with PDT | 129 |
| TPC-Ahx-ATWLPPR | TPC | ATWLPPR | — | Targeted therapy combined with PDT | 69 |
| ZnPc-BBN | ZnPc | YQRLGNWAVGHLM | — | Targeted therapy combined with PDT | 78 |
| RGD-L-Pyro | Pyro | cRGDfK | — | Targeted therapy combined with PDT | 130 |
| RGD-L-Glu-Pyro | Pyro | cRGDfK | — | Targeted therapy combined with PDT | |
| PPMA | PpIX | KVPRNQDWL | Nanoparticles | Immunotherapy combined with PDT | 75 |
| PpIX-1MT | PpIX | KDEVD | Nanoparticles | Immunotherapy combined with PDT | 98 |
| mPEG-Pep-IDOi/ICG | ICG | PVGLIG | Core–shell | Immunotherapy combined with PTT | 131 |
| IR780-M-APP | IR780 | NYSKPTDRQYHF-NH2 | Nanoparticles | Immunotherapy combined with PDT | 132 |
5.2.1.1 Photodynamic antitumour therapy. Chromopeptide assemblies can effectively treat tumours in terms of the PDT effect. Inspired by the multicomponent self-organization of natural proteins, pigments and metal ions in metalloproteins,133–136 Yan et al. chose metal-binding amino acids or peptides, photosensitizer Ce6 and metal ions Zn2+ to construct multicomponent coordination chromopeptide-based assemblies (Fig. 7A).137 Metal coordination is a directional and reversible chemical bond, and its bonding strength is stronger than that of noncovalent interactions.138,139 Therefore, the chromopeptide-based assemblies formed by metal coordination show strong colloidal stability, which is not exhibited by those formed through nonmetal coordination. More importantly, the assemblies showed that the photosensitizers exhibited a capacity for ultrasensitive pH-responsive release. This is closely related to metal coordination. The protonation of the imidazole group at pH values below 5.0 effectively weakens the coordination between His and Zn2+. The sensitive response to the pH increase can be attributed to the improved solubility of His-containing amino acids or peptides at pH values above 8.5, which diminished the self-assembly hydrophobic interactions. In addition, since the coordination inhibited intramolecular motion, the emission path of Ce6 was effectively enhanced, and the nonradiative heat release path was suppressed, showing high ROS yield in vitro (Fig. 7B) and highly efficient PDT in vivo (Fig. 7C and D). By purposefully designing chromopeptides, mimicking the composition of metalloproteins, and utilizing a coordinated self-assembly approach, it is possible to successfully combine robust blood circulation and targeted burst release into a single chromopeptide system for efficient photodynamic cancer treatment.
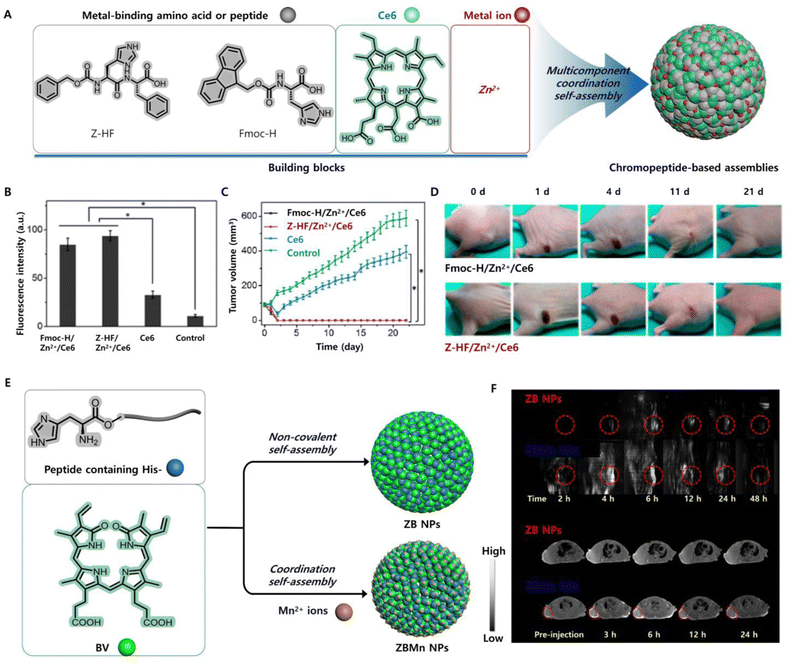 | ||
| Fig. 7 (A) Schematic illustration of a chromopeptide system formed by multicomponent self-assembly. (B) Fluorescence intensity of cells, showing the ROS produced by different groups of drugs after irradiation. (C) Tumour growth profiles during the observation in vivo. (D) Photographs of all mice or tumours in different groups after treatment in vivo. Reproduced from ref. 137 with permission from the [American Chemical Society], copyright [2018]. (E) Schematic illustration of the formation of chromopeptide-based nanoparticles. (F) PAI (upper) and (below) T1-weighted MRI of chromopeptide-based nanoparticles in mice. Reproduced from ref. 103 with permission from [John Wiley and Sons], copyright [2019]. | ||
In many cases, the photodynamic therapeutic efficacy is remarkably influenced by the nanoparticle morphology. Inspired by the dynamic changes in the morphology of molecular assemblies in organisms to complete different life processes, van Hest et al. prepared pH-responsive transformable chromopeptide-based nanomaterials using Try–Gly–porphyrin conjugates (PWG) as the assembly motif.104 These nanomaterials appeared like a nanoparticle under neutral conditions. Comparably, when exposed to acidic conditions, such as the tumour microenvironment, the nanoparticles converted into nanofibres, which was mainly because the protonation of PWG promoted the formation of intermolecular hydrogen bonds. These chromopeptide nanofibres enhanced intersystem crossing through fibrillar transformation to promote the generation of singlet oxygen, realized the effective accumulation and long-term retention of photosensitive drugs at the tumour site, and thus exhibited excellent antitumour therapeutic effects in vivo. Surprisingly, researchers used PpIX and low pH insertion peptides (pHLIP) to design a chromopeptide self-assembled material with an ability to anchor membranes through pH-driven secondary structure transformation, which was not affected by tumour heterogeneity.94 Specifically, the material showed a structural transition from random coils at pH 7.4 to α-helices at pH 6.5 or 5.5 through the action of pHLIP; thus, the material can spontaneously insert into the lipid bilayer without cell internalization to achieve tumour-specific accumulation and realize in situ PDT on the cell membrane. In addition, Yu et al. developed tumour microenvironment-adaptable chromopeptide-based superhelices and nanoparticles by coassembling pentapeptide FF-Amp-FF (AmpF) with Ce6.140 The adaptation occurred because the 4-amino-proline amide bond in AmpF was trans-isomerized at a low pH (6.5) and cis-isomerized at a high pH (7.2). More importantly, compared with nanoparticles, nanofibres formed by reversible transformation of nanoparticles in the cytoplasm produced higher levels of ROS, so the therapeutic effect could be maintained even with a reduction in dosage. Although the chromopeptide assemblies used for anti-tumour have increased intelligence, most of them are based on the passive transport of nanomaterials, which requires nanomaterials to have strong structural stability. In addition, the active targeting of the lesion needs to be strengthened, especially the chromopeptide assemblies applied in photodynamic therapy of cancer.
5.2.1.2 Photodynamic antibacterial therapy. Chromopeptide assemblies are not only used in photodynamic anti-tumour therapy, but also used in antibacterial therapy. PDT for bacterial infection has attracted great attention because it has the advantage of rare drug resistance, but it is also limited by the short life of ROS and the poor sensitivity to Gram-negative bacteria. The researchers used chlorin e6 (Ce6)-conjugated α-cyclodextrin (α-CD) and a polyethylene glycol (PEG)ylated polypeptide (Pep) as assembly units to obtain chromopeptide-based micelles (Pep@Ce6), which showed good therapeutic effects in bacterial infections caused by Gram-negative bacteria.119 This was because cationic chromopeptide-based micelles had enhanced bacterial membrane capture ability, and could effectively play a photodynamic role in eliminating biofilms and inactivate persistent cells.
In addition, Staphylococcus aureus infection is also the most common type of bacterial infection, and the increase in drug resistance is also a major threat to the treatment of these infections. Xia and other workers designed a chromopeptide based on the covalent coupling of Ce6 with the polycationic antimicrobial peptide (GKRWWKWRR) modified by the gelatinase cutting peptide sequence (PLGVRG).141 The chromopeptide could eliminate bacterial infection and inhibit the formation of bacterial biofilms in vitro under laser irradiation. When Ce6 was coupled with dimer peptides, the wound healing speed and quality of mice infected with Staphylococcus aureus were also improved. More importantly, they found that after assembling the chromopeptide with gold nanoclusters (AuNc), a synergistic effect between gelatinase responsive release of chromopeptides and photodynamic therapy was achieved.120 Because the gelatinase secreted by Staphylococcus aureus effectively promoted the release of chromopeptide molecules containing antibacterial peptides from AuNc, the therapeutic effect of Staphylococcus aureus was superior to that of Escherichia coli. The chromopeptide assemblies are expected to provide guidance for the development of materials for the treatment of bacterial infection and the promotion of wound healing. At present, many chromopeptides have been designed, but whether they can be assembled and how they can be released at specific lesion sites after assembly need further study. In addition, some research studies has found that the peptide itself has antibacterial activity,142,143 which greatly promotes the application of chromopeptides in antibacterial therapy.
Inspired by light-absorbing haemoglobin photothermal biological macromolecules, Yan et al. proposed a method for the self-assembly of chromopeptide conjugates to prepare nanodots for photothermal antitumour therapy.53 Chromopeptide conjugates (TPP-G-FF) were formed by connecting FF and porphyrin with glutaric anhydride as a linker. These conjugates self-assembled in water to produce nanodots with a uniform distribution. The formation mechanism of chromopeptide nanodots was studied by molecular dynamics simulation, and the results showed that TPP tended to form J-aggregates due to π–π interactions, and the strong hydrophilic interaction between FF and solvent was beneficial for increasing the nanodot stability. Therefore, the fluorescence emission path and singlet oxygen generation path of the chromopeptide-based nanodots are nearly suppressed, so the photoenergy tends to be converted to the local high temperature. Hence, chromopeptide-based nanodots can achieve more efficient photothermal transformation, generate higher local temperatures and enhance antitumour effects and can also be used as ideal PAI agents. However, although the chromopeptide-based nanodots greatly improved the stability, there were still problems, i.e., an insufficient absorption wavelength and insufficient laser penetration. Specifically, Yan and co-workers assembled chromophore BV and peptide motif ZHO into chromopeptide-based nanoparticles (ZB NPs) with NIR broadband wavelength absorption through noncovalent interactions.103 ZBMn NPs were obtained by integrating Mn2+ into ZB NPs to further improve the stability and MRI capabilities (Fig. 7E). As expected, the obtained ZBMn NPs broadened the absorption spectrum to a longer NIR region and exhibited efficient photothermal conversion. Moreover, compared with nanoparticles without metal ions, ZBMn NPs exhibited excellent stability after illumination, mainly due to the coordination of metal ions and peptide chain imidazoles. PAI and MRI showed that the maximum accumulation of nanoparticles at the tumour site occurred 6 h after tail vein administration (Fig. 7F). These chromopeptide-based nanoparticles utilize a clear metabolic mechanism, which provides the possibility for clinically transforming chromopeptide-based nanoparticles to photothermal agents under the guidance of multimodal imaging. The PTT effect of ZBMn NPs was verified in mice. The results showed that the tumours were ablated without recurrence in the PTT group during the whole observation period, while the tumour volume of the control group increased rapidly over time. The tumour photographs recorded over time clearly showed that after photothermal treatment with ZBMn NPs, the tumour first scabbed and then slowly fell off without recurrence.
Chromopeptide photothermal agents have shown advantages for solving the poor penetration and low accumulation efficiency of nanomaterials at tumour sites. For example, Wang et al. developed a molecular-level deep tissue penetration and NIR laser-guided in situ self-assembly strategy, which greatly improved the penetration depth and accumulation of nanomaterials at tumour sites.147 This was achieved by integrating a thermally responsive scaffold (poly(β-thioester)) into a chromopeptide system that contained the photothermal molecule ICG and functional peptides (cytotoxic and cell penetrating peptides). Specifically, the obtained molecular-level substances reached the interior of the tumour at normal body temperature and generated heat under laser irradiation, resulting in thermoresponsive in situ self-assembly into spherical nanoparticles, which was beneficial for intratumoral aggregation and cell killing. Given the in situ self-assembly effect of chromopeptides under NIR laser irradiation, many other drugs with high anticancer activity can be integrated into the chromopeptides to achieve enhanced antitumour effects. In addition, Yan et al. designed five chromopeptide conjugates by encoding peptide sequences with amino acids, which can self-assemble into assemblies with different optical properties.93 By regulating the self-assembly process, assemblies with a stable structure and excellent photothermal performance were finally optimized, and the tumours in mice were successfully eliminated. PTT can ablate tumour lesions, and in antibacterial therapy, there are also some reports of using inorganic photothermal materials to resist bacteria.148 However, few chromopeptide assemblies are used for photothermal antibacterial therapy. It is believed that with the attention paid to chromopeptides, chromopeptide assemblies will also have rapid development in photothermal antibacterial and other biomedical fields.
5.3 Combinational phototherapy
A combination of phototherapy and other theranostic methods may lead to synergistic effects, which will be a future trend in cancer therapeutics. The combined methods mainly include chemotherapy, targeted therapy, immunotherapy and other therapies. For combinational phototherapeutic purposes, it is necessary to design chromopeptides more precisely and ingeniously and endow them with multiple functions.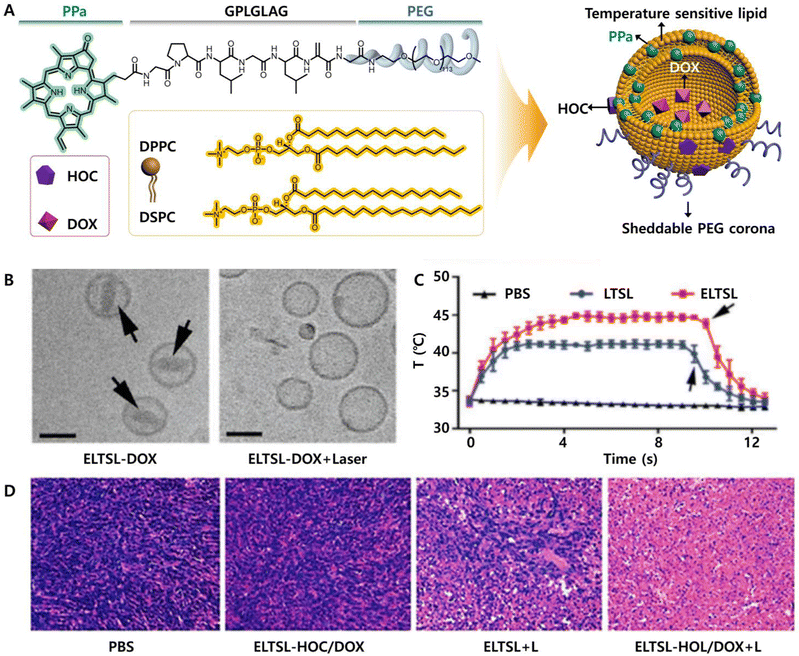 | ||
| Fig. 8 (A) Chemical structure of the chromopeptide-based chemotherapy combined with phototherapy material with a programmed stimulus-responsive liposome vesicle structure. (B) Cryo-TEM images of the chromopeptide-based material before and after laser irradiation. (C) Photothermal profile of the chromopeptide-based vesicles in vivo. (D) H&E staining of the tumour sections in different groups of mice. Reproduced from ref. 121 with permission from [John Wiley and Sons], copyright [2017]. | ||
In addition, the combination of chemotherapy with phototherapy has overcome the limitations of PDT, i.e., the poor tissue penetration and rapid consumption of oxygen. Kim et al. designed a chromopeptide conjugate using the chromophore Ce6, a caspase 3 cleavable peptide (DEVD) and the anticancer drug monomethyl auristatin E (MMAE), which self-assembled into nanoparticles with an average size of 90.8 ± 18.9 nm.122 Compared with PDT alone, this combined treatment method induced cancer cell apoptosis with a lower irradiation intensity and activated caspase 3 to cleave the anticancer drug MMAE, completing the additional process of killing cancer cells in the absence of a laser.
In the chromopeptide system, some peptides exhibit antitumour effects similar to those of chemotherapy drugs, so these chromopeptide assemblies can also realize the combined effects of chemotherapy and phototherapy without being additionally loaded with chemotherapeutic drugs. Melittin is among the most effective therapeutic peptides and is used as a chemical medicine for treating many diseases, such as cancer.152,153 However, melittin involves serious haemolysis problems and exhibits nonspecific cytotoxicity. Based on this, Wu et al. constructed a chromopeptide supramolecular assembly using melittin, photosensitizer Ce6 and hyaluronic acid (HA).123 The assembly effectively reduced the side effects of melittin, and Ce6 synergistically promoted the membrane lysis efficiency of melittin with laser irradiation, thus realizing chemotherapy combined with photodynamic antitumour therapy. Although chromopeptide-based nanomaterials can play an important role in the combined treatment of chemotherapy and phototherapy, these materials are useless against highly malignant tumours with metastatic and recurrent tendencies, mainly because these chromopeptide-based nanomaterials cannot effectively accumulate in tumour tissues. Therefore, performing in-depth exploration and development of the active targeting functionality of chromopeptide-based materials is the next goal of this research.
As the most important cell structure, the nucleus is often the target during material design.155 Han and co-workers designed a nucleus-targeting amphiphilic chimeric chromopeptide conjugate, including PpIX, a PEG linker and a DMA-modified nuclear localization peptide sequence PKKKRKV, which self-assembled into spherical nanoparticles.124 The chromopeptide assemblies exhibited a negative charge on the surface and remained stable under physiological conditions. In a tumour microenvironment, because the DMA group responded to acidity, the charge on the assemblies reversed and the surface exhibited a positive charge for nuclear targeting, which accelerated tumour cell interaction. It was observed that many PAPP-DMA nanoparticles entered the nucleus after the cells were incubated for 24 h, while nanoparticles that changed the sequence of targeted peptides remained in the cytoplasm. These chromopeptide assemblies in which the nucleus was the target exhibited a prolonged blood circulation time, achieving maximum drug accumulation in the tumour cell nucleus and effectively reducing the toxic side effects after PDT. Similarly, another group used the nuclear targeting peptide TAT in chromopeptide assemblies to prevent the efflux of light-sensitive chromophore molecules, which occurred due to the presence of P-glycoprotein in multidrug-resistant cancers.125
To further improve the efficiency of targeted drug delivery, carrier-free self-delivery chromopeptide-based nanorods were designed that target the plasma membrane and cell nucleus.126 The nanorods were self-assembled by chromopeptide conjugates containing a hydrophobic alkyl chain that can target the plasma membrane, PpIX, a peptide sequence PKKKRKV for nuclear localization, and a hydrophilic PEG chain. Plasma membrane-based targeting of PDT directly induced cell necrosis, enhanced membrane permeability, and increased the cellular uptake of nanorods. Subsequently, with the participation of the nuclear targeting peptide, the in situ generation of ROS caused oxidative damage to the DNA chain of cells, achieving an enhanced PDT effect. To improve the biocompatibility of targeted drug delivery systems, researchers also used exosome-modified chromopeptide assemblies (ChiP-Exo) to reduce immunogenicity and systemic toxicity.129 In addition, the researchers designed a self-delivery chromopeptide-based nanoparticle (M-ChiP) to dually target mitochondria and plasma membranes towards photodynamic tumour therapy.127
Some proteins that are overexpressed on the surface of tumour cells are also commonly used as targets for drug delivery. Xu et al. chose an amphiphilic iRGD peptide (internalizing RGD, CRGDKGPDC) and chemically modified with hydrophilic Arg, a hydrophobic alkyl chain and a Pro sequence to obtain a new molecule, which self-assembled into spherical nanovesicles.128 The target receptor of iRGD is the integrin receptor (αvβ3, αvβ5 and αvβ1), which is overexpressed in tumour endothelial cells and tumour cells. After binding to the receptor, iRGD was cleaved by tumour-related proteases, thereby exposing its C-terminus motif (CRGDK/R). Then, the sequence bound to tumour endothelial cells and the tumour cell transmembrane receptor glycoprotein neuropilin-1 receptor (NRP-1) to mediate the penetration of vesicles into the tumour. Therefore, the photosensitizer hypocrellin B (HB) was loaded into iRGD-based vesicle structures to produce chromopeptide assemblies, achieving enhanced photosensitizer accumulation, penetration and PDT. In addition, due to repeated administration, PDT also exhibits side effects that cannot be ignored. Based on iRGD targeted therapy, a chromopeptide conjugate (Ppa-iRGDC-BK01) was designed to solve the potential issue through an iRGD peptide.62 Chromopeptide conjugates formed a reservoir by self-aggregation at the injection site, while a supermolecular depot was formed after the targeting, penetration, reactivation of photosensitive molecules and the heat-promoted assembly of body temperature were complete; as a result, the effect of the long-released photosensitizer was achieved and repeated administrations were not necessary.
As a channel for drug delivery systems, tumour blood vessels have also been used as another target by researchers to administer drugs in recent years.156 As a part of tumour blood vessels, tumour vascular endothelial cells are widely used as targets; the cells exhibit more stable gene expression compared with that of tumour cells and are not prone to drug resistance. In tumour vascular endothelial cells, the vascular endothelial growth factor and its receptors are closely related to tumour angiogenesis and subsequent development.157,158 Aided by the killing effect of phototherapy, some researchers combined the targeted peptide of vascular endothelial cells with a photosensitive chromophore to obtain chromopeptide conjugates for cancer treatment.69,159 However, little research has been performed on this vascular-targeted chromopeptide assembly. Although many materials have been designed to target tumour cells, the efficiency of drugs reaching cells after passing through layers of barriers in the body is less than 1%.160,161 Due to this low drug availability, repeated dosing is necessary, which increases the side effects. Therefore, the material design of chromopeptide assemblies based on tumour vascular targeting can still be further developed.
As a class of immunomodulators, peptides with multiple immunoregulatory functions have the ability to enhance or suppress the body's immune system.163,164 Yan et al. selected two clinically approved water-soluble small molecules, thymopentin TP5 (RKDVY) and ICG, which coassembled into chromopeptide-based supramolecular assemblies (TP5-ICG NFs) for localized combined photothermal immunotherapy against pancreatic cancer (Fig. 9A).105 A TEM image showed that the assemblies were nanofibre structures (Fig. 9B), which was also consistent with an atomic force microscopy (AFM) image (Fig. 9C). In addition, Fourier transform infrared (FTIR) spectroscopy showed the movement of characteristic wavebands, especially the sulfonic acid group of ICG and the amide group of TP5, which verified that this fibre structure was dominated by hydrogen bonding (Fig. 9D and E). Due to the presence of hydrogen bonds, most of the ICG in the nanostructure prevented unstable alkenes from oxidizing, thereby improving the light stability (Fig. 9F) and photo-to-heat conversion efficiency. Two pancreatic tumour cell lines, Pan02 and BxPC-3, were used to evaluate the in vitro photothermal treatment effects of nanofibres. Compared with the laser group alone, the two cell lines showed obvious cell necrosis and apoptosis in the nanofibre treatment group, which demonstrated that the nanofibres are strongly phototoxic (Fig. 9G). The immune analysis results showed that after TP5-ICG NFs treatment, the frequency CD3+ T cells, CD3+CD4+ T cells and CD3+CD8+ T cells in the mouse spleen was significantly enhanced compared with those of other controls, indicating that the immunity of the mice was enhanced. The changes in cytokine levels were also measured by flow cytometry. Compared with the control group, the frequency of CD4+ IL-4+ T cells and CD8+ INF-γ+ T cells produced by mice in the TP5-ICG NFs group was the highest, which was caused by TP5, demonstrating that TP5 can be effectively delivered through nanofibres to enhance the immune effect. The results of haematoxylin and eosin (H&E) staining showed that organs in the TP5-ICG NFs group did not exhibit metastatic or inflammatory lesions under the effect of immune regulation, while organs in the control group contained metastatic tumour lesions in the liver and spleen. Therefore, the combination of immunotherapy and photothermal therapy can effectively inhibit tumour growth and eliminate metastasis (Fig. 9H). The use of clinically approved drugs in this work has further promoted the practical transformation and application of chromopeptide assemblies, but in addition to its application in the treatment of pancreatic cancer, whether it can be extended to experimental models closer to clinical ones needs further research.
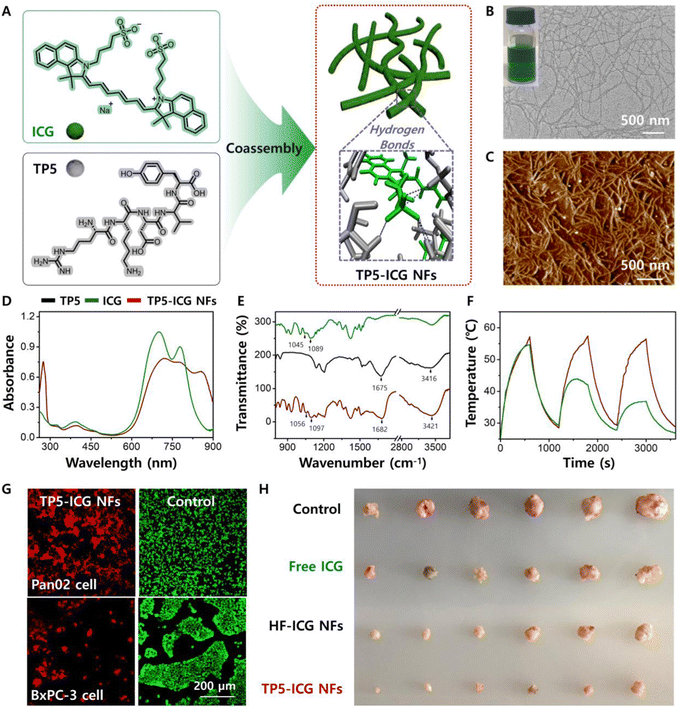 | ||
| Fig. 9 (A) Schematic illustration of chromopeptide-based supramolecular nanofibres (TP5-ICG NFs) towards localized photothermal immunotherapy. (B) TEM image and (C) AFM image of TP5-ICG NFs. (D) UV-vis absorption spectra and (E) FTIR spectra of free TP5, free ICG and TP5-ICG NFs. (F) Photostability results of free ICG and TP5-ICG NFs. (G) CLSM images of the phototoxicity of TP5-ICG NFs. (H) Photograph of tumours harvested from mice in different groups at 21 days. Reproduced from ref. 105 with permission from [John Wiley and Sons], copyright [2021]. | ||
In addition to targeting and immunomodulatory functions, chromopeptide systems can act as antigens. A chromopeptide conjugate (PpIX-PEG(8)-KVPRNQDWL) was prepared by connecting PpIX with an antigen peptide using a PEG(8) linker.75 The conjugates self-assembled into spherical nanoparticles (PPMA) that exhibited good dispersity and stability and effectively generated ROS. After laser irradiation, nanoparticles in tumour cells produced ROS, which induced necrosis or apoptosis of tumour cells and then further generated an immune response under the stimulation of PDT. In addition, introducing a melanoma-specific antigen peptide (KVPRNQDWL) activated specific cytotoxic T cells, which significantly enhanced the antitumour immune response and effectively inhibited the growth of melanoma in mice. This work shows a good tumour treatment effect, but whether the presence and length of the linker will affect the treatment effect of the chromopeptide has not yet been thoroughly studied. Although progress has been made with chromopeptide-based nanomaterials in phototherapy combined with immunotherapy, in-depth exploration of peptide functions remains necessary. For example, more specific tumour antigen peptides could be found to design more suitable and powerful materials to treat malignant tumours that are difficult to resect or highly metastatic.
In addition, immunotherapy drugs can be integrated with the chromopeptide system into a single platform. Recently, a new molecule was formed by linking an immune checkpoint inhibitor to a chromopeptide conjugate containing the enzyme-responsive peptide sequence DEVD and chromophore PpIX.98 These molecules self-assembled into nanoparticles with a particle size of approximately 50 nm and achieved a cascade of synergistic immunotherapy and phototherapy antitumour effects. The nanoparticles exhibited good stability in different solutions that simulated physiological environments. In a mouse model of lung tumour metastasis established by colon cancer cells, the nanoparticles were passively transported to the tumour site under the EPR effect and generated ROS under laser irradiation to induce cell apoptosis. Subsequently, under the action of caspase-3 in tumour cells, 1MT was released to inhibit the indoleamine-2,3-dioxygenase (IDO) pathway and activate CD8+ T cells. A chromopeptide-based nanoprodrug platform with a core–shell nanostructure was constructed by coassembling the chromophore ICG with a conjugate composed of PEG, a peptide sequence (PVGLIG) and an indoleamine 2,3-dioxygenase (IDO) inhibitor (trade name Epacadostat).131 In a tumour microenvironment (TME), the PEG layer with a core–shell structure was stripped by matrix metalloproteinase-2 (MMP-2), and its size changed from 140 nm to under 40 nm, which enhanced cellular uptake. Under near-infrared laser irradiation, small-scale nanodrugs convert light energy into heat energy to directly kill tumour cells and generate tumour-related antigens to trigger the immune response. In addition, IDO drugs were released to regulate the T-cell response in the TME, realizing the combination of phototherapy and immune checkpoint blocking therapy. To solve the problems with anti-PD-L1 (the low stability and inability to precisely control the drug loading of the antibody), Luan et al. designed a chromopeptide molecule (IR780-M-APP), which was connected by the anti-PD-L1 peptide (APP) and photosensitizer IR780 through peptide segments, and replaced the antibody with APP.132 These molecules self-assembled into nanoparticles with accurate and controllable APP loading. Under laser irradiation, the nanoparticles exert a photodynamic effect, leading to immunogenic cell death, which activates the immune response and blocks the PD-1/PD-L1 pathway to promote immunotherapy.
5.4 Biomimetic photocatalysis and photosynthesis
Photocatalysis technology is called “artificial photosynthesis”, which involves the conversion of light energy into chemical energy in the presence of a catalyst.7,165–167 The reaction process involves several basic processes, excitation activation, energy transfer, charge separation, electron transfer, and catalytic reaction. Chromopeptides exhibit several advantages, including programmable sequences, tunable self-assembly architecture and ability to easily mimic natural light-harvesting complexes. The light-harvesting, energy transfer, and electron transfer properties generated by chromophore fragments on chromopeptides can be flexibly tuned by peptide-modulated self-assembly (Table 3).| Chromopeptide-based nanomaterials | Chromophore | Peptide | Morphology | Application | Ref. |
|---|---|---|---|---|---|
| FF/porphyrin/Pt | THPP | FF | Nanotubes | Light-harvesting and biomimetic photosynthesis | 168 |
| TPPS/FF | TPPS | FF | Hierarchical porous Microsphere | Light-harvesting and photocatalysis | 48 |
| TPPS/KK/Pt | TPPS | KK | Fibre | Photocatalysis | 49 |
| TPPS/KK/TiO2/Pt | TPPS | KK | Fibre | Biomimetic photobacteria and H2 evolution | 169 |
| TPPS/Cystine/ADH | TPPS | Cystine | Microsphere | Biomimetic photoenzyme and fuel synthesis | 170 |
| TPPS/Fmoc-Leu | TPPS | Fmoc-Leu | Urchin-like microsphere | Light-harvesting | 171 |
| MCpP-FF | TPP | FF | Multiporous microsphere | Photosynthesis | 172 |
To imitate natural photosynthesis, Park et al. selected tetra(hydroxyphenyl) porphyrin (THPP), which exhibits similar optical and electrochemical properties to chlorophyll a, as the model light-collecting molecule and adsorbed it onto nanotubes with FF through hydrophobic interactions to simulate natural photosynthetic system I.168 Compared with a single FF nanotube, the J-aggregate formed by the THPP layer on the surface of the FF nanotube realized stronger exciton coupling between THPP monomers, and its structure is similar to the natural light capture complex containing chlorophyll molecules. Furthermore, Pt nanoparticles (nPt) were deposited on light-harvesting peptide nanotubes through a self-mineralization method to mimic the charge separation function of quinone in natural photosystems. The results also showed that the nanotubes with added Pt successfully promoted the regeneration of NADH and the synthesis of oxidoreductase under visible light. In addition, Yan and Mann et al. designed multifunctional biomimetic chromopeptide microspheres by coassembling FF and TPPS to simulate the natural light-harvesting system.48 To demonstrate the photocatalytic reduction ability of the microspheres, the microsphere solution was irradiated under light for 150 min, and the results showed that 4-nitrophenol (4-NP) could be reduced to 4-aminophenol (4-AP). More importantly, the microsphere structure introduced a higher stability, effectively prevented photodegradation, and could be used as a reasonable photosynthesis model to simulate the energy capture and transformation of primitive abiotic cells.
Inspired by the self-assembly of chromophores that is regulated by natural proteins, Yan et al. proposed a strategy to construct a bionic functional system through a peptide-regulated process of porphyrin self-assembly. The researchers constructed a series of chromopeptide systems, manufactured artificial photosystems via a molecular self-assembly strategy, and mimicked the architectural principles and functional mechanisms of the photosynthetic system.48,49,169–171,173,174 For example, they constructed a hierarchical fibre bundle chromopeptide structure with an organization similar to that of pigments in chlorosomes through coassembling peptide KK and TPPS.49 The surface of the chromopeptide fibre bundle was positively charged, which contributed to the mineralization of TiO2 nanoparticles. In addition, after the photoreduction of platinum potassium(ii) tetrachloride, the chromopeptide assemblies also promoted the in situ mineralization of Pt nanoparticles on the fibre (Fig. 10A). More importantly, when TiO2/Pt was introduced into the chromopeptide fibre bundle, the whole system generated hydrogen, which was similar to the original reaction centre model, indicating that the chromopeptide assemblies realized the biomimetic structure of the green sulfur bacteria (Fig. 10B).169 In another example, inspired by pathological processes of cystine stones, the researchers introduced Zn2+ metal ions into the chromopeptide system and obtained hierarchically ordered microspheres via coordination and hydrogen bonds. These microspheres are formed from the splitting growth of nanorods and can be effectively encapsulated with TPPS and alcohol dehydrogenase (ADH) through electrostatic effects (Fig. 10C). These chromopeptide-based microspheres simultaneously mimicked the structure of grana and the light-dark coupled reactions in chloroplasts, resulting in enhanced sustainability in photocatalytic fuel production (Fig. 10D).170
 | ||
| Fig. 10 (A) Schematic diagram of molecular evolution towards mimicking primitive hydrogen-producing photobacteria by self-organization of TPPS and peptides. (B) TEM image of the hybrid fibres showing mineralized nanoparticles. Reproduced from ref. 169 with permission from [John Wiley and Sons], copyright [2015]. (C) Schematic representation of the hierarchical chloroplast mimics fabricated by the Zn2+-directed assembly of cystine with encapsulation of porphyrin and enzyme molecules for photoenzymatic reactions. (D) Time dependence of H2 production on the microspheres in the presence of K2PtCl4 and NADH (red line) or K2PtCl4, NADH and ethanol (black line). Reproduced from ref. 170 with permission from [John Wiley and Sons], copyright [2016]. | ||
Compared with other biomimetic light-trapping nanostructures, chromopeptide-based materials exhibit good biocompatibility and biodegradability. The chromophore is responsible for light absorption, and the peptide regulates the structure and solubility. Self-assembled nanomaterials of chromophores and peptide conjugates with fixed stoichiometric ratios are also used to enrich biomimetic photosynthetic materials. Collini and co-workers designed a nanoarchitecture by the self-assembly of pyropheophorbide–peptide conjugates, which acted as artificial light harvesters.175 Among them, the self-assembly of peptide fragments promoted the formation of an exciton network structure that exhibited broad spectral absorption of pigment molecules. The excitation energy of the system could be gathered into a variety of long-life dark states at an ultrafast speed, which showed that the system exhibited great potential for capturing light efficiently. In addition, Gazit et al. constructed nanofibre-based multiporous microspheres that exhibited broad-spectrum photosensitivity and remarkable electron transfer characteristics, and these microspheres were obtained by the self-assembly of a conjugate composed of porphyrin and phenylalanine.172 Therefore, the microsphere could also be used as an antenna for artificial photosynthesis. Although current biomimetic materials, including chromopeptide-based materials, show high efficiency in light capture, the overall energy efficiency for photosynthesis still remains low. However, their roles in pushing this field forward cannot be ignored. Chromopeptide-based materials provide a reference for further designing simple functional materials to imitate the complex process of natural photocatalysis and photosynthesis. In addition to the chromopeptide assemblies based on porphyrin and its derivatives, the biomimetic photocatalysis and photosynthesis potential of other chromophores such as cyanines still need to be explored.
6. Conclusions
Research studies on chromopeptides and chromopeptide self-assembly have emerged as an attractive and on-developing field (Fig. 11). Chromopeptides are a biologically-derived building block inspired by natural chromoprotein, which integrate the functions of both chromophores and peptides. Because of their elegant molecular structure and flexible design, chromopeptides have been used to construct a variety of self-assembled nanostructures with fascinating functions. The molecular structure of the chromopeptides can be formed by both covalent bonds and noncovalent bonds. Cooperative intermolecular interactions between chromopeptide assemblies lead to particular structures and functions. By precisely and ingeniously designing the molecular structure of the chromopeptides and controlling the key factors in the self-assembly process (thermodynamics and kinetics), the expected nanostructures and material properties can be achieved. The strategy of chromopeptide self-assembly has promoted the development of peptide-based biomedical nanomaterials and biomimetic nanomaterials with integration of optical and biological functions. Their applications in imaging, phototherapy and combination therapy as well as in biomimetic photocatalysis and photosynthesis have been well demonstrated.Despite the abundant achievements in the study of chromopeptides, challenges still remain in several aspects. One is how to precisely regulate the self-assembly process and chromopeptide structures. Due to the presence of both chromophores and peptide motifs in the chromopeptide system, their assembly behaviors are drastically different due to the variance in the types and strengths of intermolecular interactions. Therefore, regulating the assembly of chromopeptides requires integration of the interaction of the two types of motifs, chromophores and peptides. This undoubtedly greatly increases the technical difficulty of regulating the assembly. In order to achieve the purpose of precise regulation, we believe that, on one hand, it is necessary to pay attention to the design of the chromophore motif, that is, to concentrate on the functionality of the choromophores and their assembly properties, for example, to match their intermolecular interaction strengths with the peptide. Molecular design and synthesis may be required in certain cases. On the other hand, we recommended to make sufficient use of computational simulation to assist molecular design. Both classical molecular dynamics and artificial-intelligence-based prediction have been widely used in peptide sequence design and assembly,176,177 but rarely in chromopeptides. Establishing a model suitable for the self-assembly of chromopeptides through computational simulation for refining a set of more quantitative structure–activity relationships will greatly benefit. The second challenge is to diversify the application of chromopeptides. Theoretically, chromopeptides can be applied to a wider range of scenarios such as light harvesting, biological charge transport, and energy transfer, in addition to the fields mentioned in this review. However, these applications depend on the properties and functions of chromopeptide assemblies, which in turn are closely related to their supramolecular structures. Limited by the current assembly regulation technique, it is still difficult to realize the applications that require rigorous supramolecular structures of chromopeptides. Thankfully, however, there have been a handful of reports of this kind, albeit abiotic ones.178–180 Therefore, along with improving precise assembly, it is suggested to fully utilize the biological advantages of natural pigment derivatives and peptides to expand their application scenarios, thereby promoting the diversified development of chromopeptide self-assembly from biomedical and artificial photosynthesis materials to bioengineering technology and industrial products. We believe that chromopeptide assemblies will be designed more flexibly and intelligently as researchers focus more on this burgeoning field.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Science Fund for Distinguished Young Scholars of China (no. 22025207), the National Natural Science Foundation of China (project no. 22232006, 22072154 and 22077122), the Youth Innovation Promotion Association of Chinese Academy of Sciences (no. 2021048) and the National Natural Science Foundation of Hebei Province (no. B2020103025).Notes and references
- R. R. Naik and S. Singamaneni, Chem. Rev., 2017, 117, 12581–12583 CrossRef CAS PubMed
.
- M. D. Ward and P. R. Raithby, Chem. Soc. Rev., 2013, 42, 1619–1636 RSC
.
- P. Yin, H. M. Choi, C. R. Calvert and N. A. Pierce, Nature, 2008, 451, 318–322 CrossRef CAS PubMed
.
- K. Ariga, X. Jia, J. Song, J. P. Hill, D. T. Leong, Y. Jia and J. Li, Angew. Chem., Int. Ed., 2020, 59, 15424–15446 CrossRef CAS PubMed
.
- K. Ariga, Int. J. Mol. Sci., 2022, 23, 3577 CrossRef CAS PubMed
.
- K. Ariga, Nanoscale Horiz., 2021, 6, 364–378 RSC
.
- Q. Zou, K. Liu, M. Abbas and X. Yan, Adv. Mater., 2016, 28, 1031–1043 CrossRef CAS PubMed
.
- N. G. Gurskaya, A. F. Fradkov, A. Terskikh, M. V. Matz, Y. A. Labas, V. I. Martynov, Y. G. Yanushevich, K. A. Lukyanov and S. A. Lukyanov, FEBS Lett., 2001, 507, 16–20 CrossRef CAS PubMed
.
- V. V. Verkhusha and K. A. Lukyanov, Nat. Biotechnol., 2004, 22, 289–296 CrossRef CAS PubMed
.
- E. Reich, R. M. Franklin, A. J. Shatkin and E. L. Tatum, Science, 1961, 134, 556–557 CrossRef CAS PubMed
.
- M. Sono, M. P. Roach, E. D. Coulter and J. H. Dawson, Chem. Rev., 1996, 96, 2841–2887 CrossRef CAS PubMed
.
- D. L. Huffman, M. M. Rosenblatt and K. S. Suslick, J. Am. Chem. Soc., 1998, 120, 6183–6184 CrossRef CAS
.
- M. M. Rosenblatt, J. Y. Wang and K. S. Suslick, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 13140–13145 CrossRef CAS PubMed
.
- R. Chang, Q. Zou, R. Xing and X. Yan, Adv. Therap., 2019, 2, 1900048 CrossRef
.
- H. You, H. E. Yoon, P. H. Jeong, H. Ko, J. H. Yoon and Y. C. Kim, Bioorg. Med. Chem., 2015, 23, 1453–1462 CrossRef CAS PubMed
.
- L. Y. Zhao, Q. L. Zou and X. H. Yan, Bull. Chem. Soc. Jpn., 2019, 92, 70–79 CrossRef CAS
.
- L. Luan, W. Fang, W. Liu, M. Tian, Y. Ni, X. Chen and X. Yu, Org. Biomol. Chem., 2016, 14, 2985–2992 RSC
.
- G. R. Geier and T. Sasaki, Tetrahedron Lett., 1997, 38, 3821–3824 CrossRef CAS
.
- K. Liu, R. Xing, Q. Zou, G. Ma, H. Mohwald and X. Yan, Angew. Chem., Int. Ed., 2016, 55, 3036–3039 CrossRef CAS PubMed
.
- H. Abrahamse and M. R. Hamblin, Biochem. J., 2016, 473, 347–364 CrossRef CAS PubMed
.
- R. Bonnett, Chem. Soc. Rev., 1995, 24, 19–33 RSC
.
- A. P. Castano, T. N. Demidova and M. R. Hamblin, Photodiagn. Photodyn. Ther., 2004, 1, 279–293 CrossRef CAS PubMed
.
- M. Abbas, Q. Zou, S. Li and X. Yan, Adv. Mater., 2017, 29, 1605021 CrossRef PubMed
.
- S. Li, R. Xing, R. Chang, Q. Zou and X. Yan, Curr. Opin. Colloid Interface Sci., 2018, 35, 17–25 CrossRef CAS
.
- R. V. Ulijn and D. N. Woolfson, Chem. Soc. Rev., 2010, 39, 3349–3350 RSC
.
- M. J. Webber, E. A. Appel, E. W. Meijer and R. Langer, Nat. Mater., 2016, 15, 13–26 CrossRef CAS PubMed
.
- S. Zhang, Nat. Biotechnol., 2003, 21, 1171–1178 CrossRef CAS PubMed
.
- S. Fleming and R. V. Ulijn, Chem. Soc. Rev., 2014, 43, 8150–8177 RSC
.
- Y. C. Yu, P. Berndt, M. Tirrell and G. B. Fields, J. Am. Chem. Soc., 1996, 118, 12515–12520 CrossRef CAS
.
- L. Adler-Abramovich and E. Gazit, Chem. Soc. Rev., 2014, 43, 6881–6893 RSC
.
- Q. Song, Z. Cheng, M. Kariuki, S. C. L. Hall, S. K. Hill, J. Y. Rho and S. Perrier, Chem. Rev., 2021, 121, 13936–13995 CrossRef CAS PubMed
.
- R. P. Cheng, S. H. Gellman and W. F. DeGrado, Chem. Rev., 2001, 101, 3219–3232 CrossRef CAS PubMed
.
- J. Wang, K. Liu, R. Xing and X. Yan, Chem. Soc. Rev., 2016, 45, 5589–5604 RSC
.
- M. Aoudia and M. A. J. Rodgers, J. Am. Chem. Soc., 1997, 119, 12859–12868 CrossRef CAS
.
- M. Wehner and F. Würthner, Nat. Rev. Chem., 2020, 4, 38–53 CrossRef CAS
.
- G. D. Scholes, G. R. Fleming, A. Olaya-Castro and R. van Grondelle, Nat. Chem., 2011, 3, 763–774 CrossRef CAS PubMed
.
- R. J. Cogdell, N. W. Isaacs, A. A. Freer, J. Arrelano, T. D. Howard, M. Z. Papiz, A. M. Hawthornthwaite-Lawless and S. Prince, Prog. Biophys. Mol. Biol., 1997, 68, 1–27 CrossRef CAS PubMed
.
- X. Qin, M. Suga, T. Kuang and J. R. Shen, Science, 2015, 348, 989–995 CrossRef CAS PubMed
.
- F. Biscaglia and M. Gobbo, Pept. Sci., 2018, 110, e24038 CrossRef
.
- K. Muller-Dethlefs and P. Hobza, Chem. Rev., 2000, 100, 143–167 CrossRef CAS PubMed
.
- E. R. Johnson, S. Keinan, P. Mori-Sanchez, J. Contreras-Garcia, A. J. Cohen and W. Yang, J. Am. Chem. Soc., 2010, 132, 6498–6506 CrossRef CAS PubMed
.
- A. S. R. Koti and N. Periasamy, Chem. Mater., 2003, 15, 369–371 CrossRef CAS
.
- L. Zhang and M. Liu, J. Phys. Chem. B, 2009, 113, 14015–14020 CrossRef CAS PubMed
.
- M. Sakamoto, A. Ueno and H. Mihara, Chem. Commun., 2000, 1741–1742 RSC
.
- M. Sakamoto, A. Ueno and H. Mihara, Chem. – Eur. J., 2001, 7, 2449–2458 CrossRef CAS PubMed
.
- H. C. Fry, L. A. Solomon, B. T. Diroll, Y. Liu, D. J. Gosztola and H. M. Cohn, J. Am. Chem. Soc., 2020, 142, 233–241 CrossRef CAS PubMed
.
- L. K. Solomon, J. B. Kronenberg and H. C. Fry, J. Am. Chem. Soc., 2017, 139, 8497–8507 CrossRef CAS PubMed
.
- Q. Zou, L. Zhang, X. Yan, A. Wang, G. Ma, J. Li, H. Mohwald and S. Mann, Angew. Chem., Int. Ed., 2014, 53, 2366–2370 CrossRef CAS PubMed
.
- K. Liu, R. Xing, C. Chen, G. Shen, L. Yan, Q. Zou, G. Ma, H. Mohwald and X. Yan, Angew. Chem., Int. Ed., 2015, 54, 500–505 CAS
.
- D. A. Mendonca, M. Bakker, C. Cruz-Oliveira, V. Neves, M. A. Jimenez, S. Defaus, M. Cavaco, A. S. Veiga, I. Cadima-Couto, M. Castanho, D. Andreu and T. Todorovski, Bioconjugate Chem., 2021, 32, 1067–1077 CrossRef CAS PubMed
.
- M. Ethirajan, Y. Chen, P. Joshi and R. K. Pandey, Chem. Soc. Rev., 2011, 40, 340–362 RSC
.
- B. C. Kovaric, B. Kokona, A. D. Schwab, M. A. Twomey, J. C. de Paula and R. Fairman, J. Am. Chem. Soc., 2006, 128, 4166–4167 CrossRef CAS PubMed
.
- Q. Zou, M. Abbas, L. Zhao, S. Li, G. Shen and X. Yan, J. Am. Chem. Soc., 2017, 139, 1921–1927 CrossRef CAS PubMed
.
- R. Dosselli, R. Ruiz-Gonzalez, F. Moret, V. Agnolon, C. Compagnin, M. Mognato, V. Sella, M. Agut, S. Nonell, M. Gobbo and E. Reddi, J. Med. Chem., 2014, 57, 1403–1415 CrossRef CAS PubMed
.
- F. Moret, M. Gobbo and E. Reddi, Photochem. Photobiol. Sci., 2015, 14, 1238–1250 CrossRef CAS PubMed
.
- R. Dosselli, C. Tampieri, R. Ruiz-Gonzalez, S. De Munari, X. Ragas, D. Sanchez-Garcia, M. Agut, S. Nonell, E. Reddi and M. Gobbo, J. Med. Chem., 2013, 56, 1052–1063 CrossRef CAS PubMed
.
- J. Zhou, G. B. Qi and H. Wang, J. Mater. Chem. B, 2016, 4, 4855–4861 RSC
.
- M. Sibrian-Vazquez, T. J. Jensen, R. P. Hammer and M. G. H. Vicente, J. Med. Chem., 2006, 49, 1364–1372 CrossRef CAS PubMed
.
- Z. Zhou, M. Qutaish, Z. Han, R. M. Schur, Y. Liu, D. L. Wilson and Z. R. Lu, Nat. Commun., 2015, 6, 7984 CrossRef CAS PubMed
.
- P.-C. Lo, J. Chen, K. Stefflova, M. S. Warren, R. Navab, B. Bandarchi, S. Mullins, M. Tsao, J. D. Cheng and G. Zheng, J. Med. Chem., 2009, 52, 358–368 CrossRef CAS PubMed
.
- L. L. Li, Q. Zeng, W. J. Liu, X. F. Hu, Y. Li, J. Pan, D. Wan and H. Wang, ACS Appl. Mater. Interfaces, 2016, 8, 17936–17943 CrossRef CAS PubMed
.
- H. J. Cho, S. J. Park, W. H. Jung, Y. Cho, D. J. Ahn, Y. S. Lee and S. Kim, ACS Nano, 2020, 14, 15793–15805 CrossRef CAS PubMed
.
- B. Mao, C. Liu, W. Zheng, X. Li, R. Ge, H. Shen, X. Guo, Q. Lian, X. Shen and C. Li, Biomaterials, 2018, 161, 306–320 CrossRef CAS PubMed
.
- A. Orosz, G. Mezo, L. Herenyi, J. Habdas, Z. Majer, B. Mysliwa-Kurdziel, K. Toth and G. Csik, Biophys. Chem., 2013, 177–178, 14–23 CrossRef CAS PubMed
.
- G. Ghosh, K. K. Kartha and G. Fernandez, Chem. Commun., 2021, 57, 1603–1606 RSC
.
- E. Nikoloudakis, K. Mitropoulou, G. Landrou, G. Charalambidis, V. Nikolaou, A. Mitraki and A. G. Coutsolelos, Chem. Commun., 2019, 55, 14103–14106 RSC
.
- A. Markowska, A. R. Markowski and I. Jarocka-Karpowicz, Int. J. Mol. Sci., 2021, 22, 12122 CrossRef CAS PubMed
.
- E. N. Carrion, J. Santiago, D. Sabatino and S. M. Gorun, Inorg. Chem., 2017, 56, 7210–7216 CrossRef CAS PubMed
.
- L. Tirand, C. Frochot, R. Vanderesse, N. Thomas, E. Trinquet, S. Pinel, M. L. Viriot, F. Guillemin and M. Barberi-Heyob, J. Controlled Release, 2006, 111, 153–164 CrossRef CAS PubMed
.
- S. Zalipsky, Adv. Drug Delivery Rev., 1995, 16, 157–182 CrossRef CAS
.
- K. Knop, R. Hoogenboom, D. Fischer and U. S. Schubert, Angew. Chem., Int. Ed., 2010, 49, 6288–6308 CrossRef CAS PubMed
.
- W. Rut, K. Groborz, L. Zhang, X. Sun, M. Zmudzinski, B. Pawlik, X. Wang, D. Jochmans, J. Neyts, W. Mlynarski, R. Hilgenfeld and M. Drag, Nat. Chem. Biol., 2021, 17, 222–228 CrossRef CAS PubMed
.
- A. D'Souza A and R. Shegokar, Expert Opin. Drug Delivery, 2016, 13, 1257–1275 CrossRef PubMed
.
- A. L. Klibanov, K. Maruyama, V. P. Torchilin and L. Huang, FEBS Lett., 1990, 268, 235–237 CrossRef CAS PubMed
.
- H. Cheng, G. L. Fan, J. H. Fan, R. R. Zheng, L. P. Zhao, P. Yuan, X. Y. Zhao, X. Y. Yu and S. Y. Li, Macromol. Biosci., 2019, 19, 1800410 CrossRef PubMed
.
- K. Han, W. Y. Zhang, Z. Y. Ma, S. B. Wang, L. M. Xu, J. Liu, X. Z. Zhang and H. Y. Han, ACS Appl. Mater. Interfaces, 2017, 9, 16043–16053 CrossRef CAS PubMed
.
- P. Kasperkiewicz, Y. Altman, M. D'Angelo, G. S. Salvesen and M. Drag, J. Am. Chem. Soc., 2017, 139, 10115–10125 CrossRef CAS PubMed
.
- E. Ranyuk, N. Cauchon, K. Klarskov, B. Guerin and J. E. van Lier, J. Med. Chem., 2013, 56, 1520–1534 CrossRef CAS PubMed
.
- C. L. Schreiber and B. D. Smith, Nat. Rev. Chem., 2019, 3, 393–400 CrossRef CAS PubMed
.
- C. A. Hunter, Angew. Chem., Int. Ed., 2004, 43, 5310–5324 CrossRef CAS PubMed
.
- A. E. Reed, L. A. Curtiss and F. Weinhold, Chem. Rev., 1988, 88, 899–926 CrossRef CAS
.
- C. Yuan, W. Ji, R. Xing, J. Li, E. Gazit and X. Yan, Nat. Rev. Chem., 2019, 3, 567–588 CrossRef CAS
.
- J. Li, J. Wang, Y. Zhao, P. Zhou, J. Carter, Z. Li, T. A. Waigh, J. R. Lu and H. Xu, Coord. Chem. Rev., 2020, 421, 213418 CrossRef CAS
.
- R. W. Chakroun, F. Wang, R. Lin, Y. Wang, H. Su, D. Pompa and H. Cui, ACS Nano, 2019, 13, 7780–7790 CrossRef CAS PubMed
.
- X. Zhao, F. Pan, H. Xu, M. Yaseen, H. Shan, C. A. E. Hauser, S. Zhang and J. R. Lu, Chem. Soc. Rev., 2010, 39, 3480–3498 RSC
.
- H. Chen, K. Shou, S. Chen, C. Qu, Z. Wang, L. Jiang, M. Zhu, B. Ding, K. Qian, A. Ji, H. Lou, L. Tong, A. Hsu, Y. Wang, D. W. Felsher, Z. Hu, J. Tian and Z. Cheng, Adv. Mater., 2021, 33, 2006902 CrossRef CAS PubMed
.
- C. Chen, K. Liu, J. Li and X. Yan, Adv. Colloid Interface Sci., 2015, 225, 177–193 CrossRef CAS PubMed
.
- J. Zhao, R. Huang, W. Qi, Y. Wang, R. Su and Z. He, Prog. Chem., 2014, 26, 1445–1459 CAS
.
- Z. Sun, H. Xie, S. Tang, X. F. Yu, Z. Guo, J. Shao, H. Zhang, H. Huang, H. Wang and P. K. Chu, Angew. Chem., Int. Ed., 2015, 54, 11526–11530 CrossRef CAS PubMed
.
- C. M. Hessel, V. P. Pattani, M. Rasch, M. G. Panthani, B. Koo, J. W. Tunnell and B. A. Korgel, Nano Lett., 2011, 11, 2560–2566 CrossRef CAS PubMed
.
- L. Zhao, Y. Liu, R. Chang, R. Xing and X. Yan, Adv. Funct. Mater., 2019, 29, 1806877 CrossRef
.
- L. Zhao, Y. Liu, R. Xing and X. Yan, Angew. Chem., Int. Ed., 2020, 59, 3793–3801 CrossRef CAS PubMed
.
- R. Chang, Q. Zou, L. Zhao, Y. Liu, R. Xing and X. Yan, Adv. Mater., 2022, 34, 2200139 CrossRef CAS PubMed
.
- G. F. Luo, W. H. Chen, S. Hong, Q. Cheng, W. X. Qiu and X. Z. Zhang, Adv. Funct. Mater., 2017, 27, 1702122 CrossRef
.
- L. Zhao, S. Li, Y. Liu, R. Xing and X. Yan, CCS Chem., 2019, 1, 173–180 CAS
.
- L. Zhao, X. Ren and X. Yan, CCS Chem., 2021, 3, 678–693 CrossRef
.
- D. Zhang, G. B. Qi, Y. X. Zhao, S. L. Qiao, C. Yang and H. Wang, Adv. Mater., 2015, 27, 6125–6130 CrossRef CAS PubMed
.
- W. Song, J. Kuang, C. X. Li, M. Zhang, D. Zheng, X. Zeng, C. Liu and X. Z. Zhang, ACS Nano, 2018, 12, 1978–1989 CrossRef CAS PubMed
.
- K. Han, J. Zhang, W. Zhang, S. Wang, L. Xu, C. Zhang, X. Zhang and H. Han, ACS Nano, 2017, 11, 3178–3188 CrossRef CAS PubMed
.
- J. Fidel, K. C. Kennedy, W. S. Dernell, S. Hansen, V. Wiss, M. R. Stroud, J. I. Molho, S. E. Knoblaugh, J. Meganck, J. M. Olson, B. Rice and J. Parrish-Novak, Cancer Res., 2015, 75, 4283–4291 CrossRef CAS PubMed
.
- J. Parrish-Novak, K. Byrnes-Blake, N. Lalayeva, S. Burleson, J. Fidel, R. Gilmore, P. Gayheart-Walsten, G. A. Bricker, W. J. Crumb, Jr., K. S. Tarlo, S. Hansen, V. Wiss, E. Malta, W. S. Dernell, J. M. Olson and D. M. Miller, Int. J. Toxicol., 2017, 36, 104–112 CrossRef CAS PubMed
.
- H. W. An, L. L. Li, Y. Wang, Z. Wang, D. Hou, Y. X. Lin, S. L. Qiao, M. D. Wang, C. Yang, Y. Cong, Y. Ma, X. X. Zhao, Q. Cai, W. T. Chen, C. Q. Lu, W. Xu, H. Wang and Y. Zhao, Nat. Commun., 2019, 10, 4861 CrossRef PubMed
.
- R. Xing, Q. Zou, C. Yuan, L. Zhao, R. Chang and X. Yan, Adv. Mater., 2019, 31, 1900822 CrossRef PubMed
.
- B. Sun, R. Chang, S. Cao, C. Yuan, L. Zhao, H. Yang, J. Li, X. Yan and J. C. M. van Hest, Angew. Chem., Int. Ed., 2020, 59, 20582–20588 CrossRef CAS PubMed
.
- S. Li, W. Zhang, R. Xing, C. Yuan, H. Xue and X. Yan, Adv. Mater., 2021, 33, 2100595 CrossRef CAS PubMed
.
- Y. Li, R. C. H. Wong, X. Yan, D. K. P. Ng and P. C. Lo, ACS Appl. Bio Mater., 2020, 3, 5463–5473 CrossRef CAS PubMed
.
- S. Li, L. Zhao, R. Chang, R. Xing and X. Yan, Chem. – Eur. J., 2019, 25, 13429–13435 CrossRef CAS PubMed
.
- S. Y. Li, H. Cheng, W. X. Qiu, L. H. Liu, S. Chen, Y. Hu, B. R. Xie, B. Li and X. Z. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 28319–28329 CrossRef CAS PubMed
.
- K. Stefflova, H. Li, J. Chen and G. Zheng, Bioconjugate Chem., 2007, 18, 379–388 CrossRef CAS PubMed
.
- K. Huang, M. Gao, L. Fan, Y. Lai, H. Fan and Z. Hua, Biomater. Sci., 2018, 6, 2925–2931 RSC
.
- H. Jin, G. Zhao, J. Hu, Q. Ren, K. Yang, C. Wan, A. Huang, P. Li, J. P. Feng, J. Chen and Z. Zou, ACS Appl. Mater. Interfaces, 2017, 9, 25755–25766 CrossRef CAS PubMed
.
- H. F. Zhang, K. Maslov, G. Stoica and L. V. Wang, Nat. Biotechnol., 2006, 24, 848–851 CrossRef CAS PubMed
.
- M. H. Xu and L. H. V. Wang, Rev. Sci. Instrum., 2006, 77, 041101 CrossRef
.
- L. V. Wang and S. Hu, Science, 2012, 335, 1458–1462 CrossRef CAS PubMed
.
- Y. Liu, L. Zhang, R. Chang and X. Yan, Chem. Commun., 2022, 58, 2247–2258 RSC
.
- S. Li, W. Zhang, H. Xue, R. Xing and X. Yan, Chem. Sci., 2020, 11, 8644–8656 RSC
.
- J. Li and K. Pu, Chem. Soc. Rev., 2019, 48, 38–71 RSC
.
- C. Liang, L. Xu, G. Song and Z. Liu, Chem. Soc. Rev., 2016, 45, 6250–6269 RSC
.
- Q. Gao, D. Huang, Y. Deng, W. Yu, Q. Jin, J. Ji and G. Fu, Chem. Eng. J., 2021, 417, 129334 CrossRef CAS
.
- L. Qiu, C. Wang, X. Lei, X. Du, Q. Guo, S. Zhou, P. Cui, T. Hong, P. Jiang, J. Wang, Y. Q. Li and J. Xia, Biomater. Sci., 2021, 9, 3433–3444 RSC
.
- F. Zhou, B. Feng, T. Wang, D. Wang, Q. Meng, J. Zeng, Z. Zhang, S. Wang, H. Yu and Y. Li, Adv. Funct. Mater., 2017, 27, 1606530 CrossRef
.
- W. Um, J. Park, H. Ko, S. Lim, H. Y. Yoon, M. K. Shim, S. Lee, Y. J. Ko, M. J. Kim, J. H. Park, D. K. Lim, Y. Byun, I. C. Kwon and K. Kim, Biomaterials, 2019, 224, 119494 CrossRef CAS PubMed
.
- H. R. Jia, Y. X. Zhu, K. F. Xu and F. G. Wu, Adv. Healthcare Mater., 2018, 7, 1800380 CrossRef PubMed
.
- K. Han, W. Zhang, J. Zhang, Q. Lei, S. Wang, J. Liu, X. Zhang and H. Han, Adv. Funct. Mater., 2016, 26, 4351–4361 CrossRef CAS
.
- Z. Yu, W. Pan, N. Li and B. Tang, Chem. Sci., 2016, 7, 4237–4244 RSC
.
- H. Cheng, P. Yuan, G. Fan, L. Zhao, R. Zheng, B. Yang, X. Qiu, X. Yu, S. Li and X. Zhang, Appl. Mater. Today, 2019, 16, 120–131 CrossRef
.
- H. Cheng, R. R. Zheng, G. L. Fan, J. H. Fan, L. P. Zhao, X. Y. Jiang, B. Yang, X. Y. Yu, S. Y. Li and X. Z. Zhang, Biomaterials, 2019, 188, 1–11 CrossRef CAS PubMed
.
- Y. Jiang, X. Pang, R. Liu, Q. Xiao, P. Wang, A. W. Leung, Y. Luan and C. Xu, ACS Appl. Mater. Interfaces, 2018, 10, 31674–31685 CrossRef CAS PubMed
.
- H. Cheng, J. H. Fan, L. P. Zhao, G. L. Fan, R. R. Zheng, X. Z. Qiu, X. Y. Yu, S. Y. Li and X. Z. Zhang, Biomaterials, 2019, 211, 14–24 CrossRef CAS PubMed
.
- W. Li, S. Tan, Y. Xing, Q. Liu, S. Li, Q. Chen, M. Yu, F. Wang and Z. Hong, Mol. Pharmaceutics, 2018, 15, 1505–1514 CrossRef CAS PubMed
.
- Y. Liu, Y. Lu, X. Zhu, C. Li, M. Yan, J. Pan and G. Ma, Biomaterials, 2020, 242, 119933 CrossRef CAS PubMed
.
- N. Wang, Y. Zhou, Y. Xu, X. Ren, S. Zhou, Q. Shang, Y. Jiang and Y. Luan, Chem. Eng. J., 2020, 400, 125995 CrossRef CAS
.
- J. Liu, S. Chakraborty, P. Hosseinzadeh, Y. Yu, S. Tian, I. Petrik, A. Bhagi and Y. Lu, Chem. Rev., 2014, 114, 4366–4469 CrossRef CAS PubMed
.
- E. Y. Tshuva and S. J. Lippard, Chem. Rev., 2004, 104, 987–1012 CrossRef CAS PubMed
.
- F. A. Armstrong, H. A. Heering and J. Hirst, Chem. Soc. Rev., 1997, 26, 169–179 RSC
.
- Y. Lu, N. Yeung, N. Sieracki and N. M. Marshall, Nature, 2009, 460, 855–862 CrossRef CAS PubMed
.
- S. Li, Q. Zou, Y. Li, C. Yuan, R. Xing and X. Yan, J. Am. Chem. Soc., 2018, 140, 10794–10802 CrossRef CAS PubMed
.
- G. J. Kubas, Acc. Chem. Res., 1988, 21, 120–128 CrossRef
.
- T. J. Meyer, Pure Appl. Chem., 1986, 58, 1193–1206 CrossRef CAS
.
- M. Li, Y. Ning, J. Chen, X. Duan, N. Song, D. Ding, X. Su and Z. Yu, Nano Lett., 2019, 19, 7965–7976 CrossRef CAS PubMed
.
- X. Lei, L. Qiu, M. Lan, X. Du, S. Zhou, P. Cui, R. Zheng, P. Jiang, J. Wang and J. Xia, Biomater. Sci., 2020, 8, 6695–6702 RSC
.
- H. G. Boman, J. Intern. Med., 2003, 254, 197–215 CrossRef CAS PubMed
.
- Y. Liu, S. Ding, J. Shen and K. Zhu, Nat. Prod. Rep., 2019, 36, 573–592 RSC
.
- S. Lal, S. E. Clare and N. J. Halas, Acc. Chem. Res., 2008, 41, 1842–1851 CrossRef CAS PubMed
.
- Y. Liu, P. Bhattarai, Z. Dai and X. Chen, Chem. Soc. Rev., 2019, 48, 2053–2108 RSC
.
- X. H. Huang, I. H. El-Sayed, W. Qian and M. A. El-Sayed, J. Am. Chem. Soc., 2006, 128, 2115–2120 CrossRef CAS PubMed
.
- F. H. Liu, Y. Cong, G. B. Qi, L. Ji, Z. Y. Qiao and H. Wang, Nano Lett., 2018, 18, 6577–6584 CrossRef CAS PubMed
.
- J. Chen, T. Dai, J. Yu, X. Dai, R. Chen, J. Wu, N. Li, L. Fan, Z. Mao, G. Sheng and L. Li, Biomater. Sci., 2020, 8, 4447–4457 RSC
.
- G. Early Breast Cancer Trialists’ Collaborative, Lancet, 2005, 365, 1687–1717 CrossRef PubMed
.
- Y. Matsumura and H. Maeda, Cancer Res., 1986, 46, 6387–6392 CAS
.
- L. Kelland, Nat. Rev. Cancer, 2007, 7, 573–584 CrossRef CAS PubMed
.
- H. Raghuraman and A. Chattopadhyay, Biosci. Rep., 2007, 27, 189–223 CrossRef CAS PubMed
.
- B. Bechinger, J. Membr. Biol., 1997, 156, 197–211 CrossRef CAS PubMed
.
- Z. Xie, T. Fan, J. An, W. Choi, Y. Duo, Y. Ge, B. Zhang, G. Nie, N. Xie, T. Zheng, Y. Chen, H. Zhang and J. S. Kim, Chem. Soc. Rev., 2020, 49, 8065–8087 RSC
.
- L. Pan, Q. He, J. Liu, Y. Chen, M. Ma, L. Zhang and J. Shi, J. Am. Chem. Soc., 2012, 134, 5722–5725 CrossRef CAS PubMed
.
- D. Hallahan, L. Geng, S. M. Qu, C. Scarfone, T. Giorgio, E. Donnelly, X. Gao and J. Clanton, Cancer Cell, 2003, 3, 63–74 CrossRef CAS PubMed
.
- S. Soker, S. Takashima, H. Q. Miao, G. Neufeld and M. Klagsbrun, Cell, 1998, 92, 735–745 CrossRef CAS PubMed
.
- D. T. Connolly, D. M. Heuvelman, R. Nelson, J. V. Olander, B. L. Eppley, J. J. Delfino, N. R. Siegel, R. M. Leimgruber and J. Feder, J. Clin. Invest., 1989, 84, 1470–1478 CrossRef CAS PubMed
.
- L. Tirand, N. Thomas, M. Dodeller, D. Dumas, C. Frochot, B. Maunit, F. Guillemin and M. Barberi-Heyob, Drug Metab. Dispos., 2007, 35, 806–813 CrossRef CAS PubMed
.
- L. M. Ellis and D. J. Hicklin, Nat. Rev. Cancer, 2008, 8, 579–591 CrossRef CAS PubMed
.
- C. M. Overall and O. Kleifeld, Nat. Rev. Cancer, 2006, 6, 227–239 CrossRef CAS PubMed
.
- C. W. Ng, J. Li and K. Pu, Adv. Funct. Mater., 2018, 28, 1804688 CrossRef
.
- K. Zhao, R. Xing and X. Yan, Pept. Sci., 2020, 113, e24202 Search PubMed
.
- R. Chang and X. Yan, Small Struct., 2020, 1, 2000068 CrossRef
.
- A. J. Bard and M. A. Fox, Acc. Chem. Res., 1995, 28, 141–145 CrossRef CAS
.
- M. R. Wasielewski, Chem. Rev., 1992, 92, 435–461 CrossRef CAS
.
- K. Liu, Y. Kang, G. Ma, H. Möhwald and X. Yan, Phys. Chem. Chem. Phys., 2016, 18, 16738–16747 RSC
.
- J. H. Kim, M. Lee, J. S. Lee and C. B. Park, Angew. Chem., Int. Ed., 2012, 51, 517–520 CrossRef CAS PubMed
.
- K. Liu, R. Xing, Y. Li, Q. Zou, H. Mohwald and X. Yan, Angew. Chem., Int. Ed., 2016, 55, 12503–12507 CrossRef CAS PubMed
.
- K. Liu, C. Yuan, Q. Zou, Z. Xie and X. Yan, Angew. Chem., Int. Ed., 2017, 56, 7876–7880 CrossRef CAS PubMed
.
- K. Liu, M. Abass, Q. Zou and X. Yan, Green Energy Environ., 2017, 2, 58–63 CrossRef
.
- K. Tao, B. Xue, S. Frere, I. Slutsky, Y. Cao, W. Wang and E. Gazit, Chem. Mater., 2017, 29, 4454–4460 CrossRef CAS PubMed
.
- K. Liu, H. Zhang, R. Xing, Q. Zou and X. Yan, ACS Nano, 2017, 11, 12840–12848 CrossRef CAS PubMed
.
- J. Sun, R. Yendluri, K. Liu, Y. Guo, Y. Lvov and X. Yan, Phys. Chem. Chem. Phys., 2017, 19, 562–567 RSC
.
- E. Meneghin, F. Biscaglia, A. Volpato, L. Bolzonello, D. Pedron, E. Frezza, A. Ferrarini, M. Gobbo and E. Collini, J. Phys. Chem. Lett., 2020, 11, 7972–7980 CrossRef CAS PubMed
.
- R. Batra, T. D. Loeffler, H. Chan, S. Srinivasan, H. Cui, L. V. Korendovych, V. Nanda, L. C. Palmer, L. A. Solomon, H. C. Fry and S. K. R. S. Sankaranarayanan, Nat. Chem., 2022, 14, 1427–1435 CrossRef CAS PubMed
.
- P. W. J. M. Frederix, G. G. Scott, Y. M. Abul-Haija, D. Kalafatovic, C. G. Pappas, N. Javid, N. T. Hunt, R. V. Ulijn and T. Tuttle, Nat. Chem., 2015, 7, 30–37 CrossRef CAS PubMed
.
- G. Bullard, F. Tassinari, C. H. Ko, A. K. Mondal, R. Wang, S. Mishra, R. Naaman and M. J. Therien, J. Am. Chem. Soc., 2019, 141, 14707–14711 CrossRef CAS PubMed
.
- M. Di Valentin, M. Albertini, E. Zurlo, M. Gobbo and D. Carbonera, J. Am. Chem. Soc., 2014, 136, 6582–6585 CrossRef CAS PubMed
.
- Q. Song, S. Goia, J. Yang, S. C. L. Hall, M. Staniforth, V. G. Stavros and S. Perrier, J. Am. Chem. Soc., 2021, 143, 382–389 CrossRef CAS PubMed
.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |