Open Access Article
Open Access ArticleGitHub as an open electronic laboratory notebook for real-time sharing of knowledge and collaboration
Kymberley R.
Scroggie
 ab,
Klementine J.
Burrell-Sander
ab,
Klementine J.
Burrell-Sander
 ab,
Peter J.
Rutledge
ab,
Peter J.
Rutledge
 ab and
Alice
Motion
ab and
Alice
Motion
 *ab
*ab
aSchool of Chemistry, The University of Sydney, NSW, Australia. E-mail: alice.motion@sydney.edu.au
bDrug Discovery Initiative, The University of Sydney, NSW, Australia
First published on 18th July 2023
Abstract
Electronic laboratory notebooks have expanded the utility of the paper laboratory notebook beyond that of a simple record keeping tool. Open electronic laboratory notebooks offer additional benefits to the scientific community including increased transparency, reproducibility, and integrity. A key element underpinning these benefits is facile and expedient knowledge sharing which aids communication and collaboration. In previous projects, we have used LabTrove and LabArchives as open electronic laboratory notebooks, in partnership with GitHub (an open-source web-based platform originally developed for collaborative coding) for communication and discussion. Here we present our personal experiences using GitHub as the central platform for many aspects of the scientific process, including version-controlled recording of experiments, results and interpretation, data storage, project management, workflows, communication, and collaboration. We report on the utility of GitHub as an open electronic laboratory notebook for chemistry research, and discuss our experiences employing it with the Open Source Mycetoma and Open Source Tuberculosis consortia. By outlining its features and shortcomings through their implementation in our work, we demonstrate how using GitHub as a central platform can aid the real-time sharing of knowledge and collaboration, and further democratise scientific research within both open and traditional research models.
Introduction
Technological advances have allowed scientists to move beyond the primitive utility of the paper laboratory notebook as a record-keeping tool. In 1994, Borman noted that electronic laboratory notebooks (ELNs) “could revolutionise how scientists record their research, manage their data and share their information with others”.1 ELNs have indeed been integrated into laboratory information management systems (LIMS) and electronic laboratory environments (ELEs), but they have also revolutionised the way in which scientists disseminate knowledge, particularly through the internet.ELNs enable knowledge sharing, facilitating faster transfer of knowledge and collaboration, which in turn expedites future knowledge generation and improves research efficiency.2,3 The digital storage of information further increases efficiency with greater longevity, readability and searchability. Despite these benefits, the shift away from paper to electronic has been an evolutionary process rather than revolutionary and scientists, particularly those in academia, have been slow to accept and adopt ELNs.4
The ability of scientists to move to electronic documentation of their work with minimal disruption has been identified as the key factor for broader acceptance of ELNs in an academic setting.5 However, the highly diverse nature of different disciplines within academia leads to a broad range of specific needs that require highly specialised or custom ELNs to affect a seamless transition. While some commercial ELNs can support many specialised requirements, their licensing and maintenance costs often put them out of reach for individual academic research groups.6,7 Instead, many have made use of generic, freely available platforms such as OneNote,8 EverNote9,10 or Google Docs,11 with others developing their own ELNs to reap the specific benefits they require.12–14
We have successfully used several different ELNs for our own work as part of different open source drug discovery consortia, including Open Source Malaria15 (http://opensourcemalaria.org/), Open Source Mycetoma16 (https://github.com/OpenSourceMycetoma) and Open Source Tuberculosis (http://opensourcetb.org/). Open source drug discovery is a new approach to drug discovery in which all aspects of research are shared publicly and in real-time (i.e. immediately as it is produced) to facilitate collaboration and knowledge sharing.17 These consortia follow the principles of open science, in which scientific knowledge is developed collaboratively and made freely accessible to any interested parties,18 and more specifically Todd's Six Laws of Open Science.19
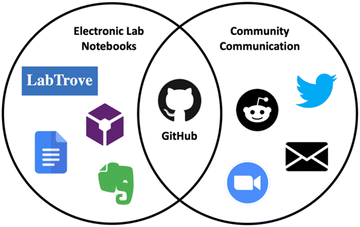
In line with openly sharing our research, we have hosted ELNs on the open source platform LabTrove20 and the commercial ELN LabArchives (https://www.labarchives.com/) while simultaneously using GitHub (http://www.github.com/) to support discussion and collaboration. To bring together the sharing of knowledge and collaboration into a single open and central location, we have now explored the use of GitHub itself as the ELN (Fig. 1). Using GitHub as both an ELN and a hub for instant communication elevates it to the status of a “collaboratory” as envisioned by Wulf – a “centre without walls, in which the nation's researchers can perform their research without regard to geographical location, interacting with colleagues, accessing instrumentation, sharing data and computational resource, and accessing information in digital libraries”.21 This article draws on the experiences of two of the authors using GitHub as an ELN for various synthetic chemistry projects and provides preliminary findings into its usability. We report on the utility of GitHub as an open ELN, detail its features in this dimension, and discuss its implementation for open source drug discovery. We also share an ELN template GitHub repository for those considering alternative ELNs. While we have used GitHub as an open ELN and repositories are open by default we note that for projects that require confidentiality or follow a traditional research methodology, information and data can be held within closed repositories with access limited to only invited users.
GitHub
GitHub is a web-based graphical interface for Git, an open source version control system. It was originally designed for software developers to work collaboratively on open source code, however, in recent years the GitHub community has expanded. After software development, education, and data, science now represents the fourth largest category of users.22 Examples range from machine-learning programs like the tuberculosis and lung cancer screening initiative AiAi.care Project, to organic chemistry applications including reaction visualisers, spectroscopic databases, and chemistry learning tools. Open Source Malaria, Open Source Mycetoma, Open Source Tuberculosis and Open Source Antibiotics (https://github.com/opensourceantibiotics) are four open source drug discovery examples hosted on the platform which make use of its forum-like structure to facilitate open, real-time collaboration and discussion among teams of scientists all around the world.The version control enabled by Git is directly transferable to ELNs. Importantly for the validity and verifiability of scientific research, using Git enables users to keep track of the who, what, when and even why: when saving changes, GitHub offer the option to provide a short description of what was changed and why the change was made. This record-keeping enables greater transparency, making it easy to see if an edit was made to fix typos, add information, or alter data, and is crucial in maintaining integrity and preventing misunderstanding or misuse of data.23 Furthermore, all activities are attributed to the user via their display name, bestowing a level of accountability and responsibility, while also ensuring that contributors receive attribution for their work.
A number of user interfaces (UIs) for Git exist, including GitHub, GitLab and Gitea, each offering slightly different user experiences. Each can be used as an ELN as described in this article however, Github is more openly accessibly and offers additional UI features (e.g.Discussions) enabling public discussion making it more suitable for hosting open source and collaborative projects.
GitHub's accessibility is also important to the open science ethos. No account or subscription is required to view work within a public GitHub repository, allowing people to access data without concerns of cost or association with institutions. Through a standard internet browser, anyone can view content as soon as it is published without the researcher needing to “share” their work, or the reader having to access any proprietary products. In contrast, to view content on GitLab it is required to have an account and be signed in, while Gitea is a self-hosted UI.
Not only is the content on GitHub openly accessible, but users can connect to content on GitHub in different ways: from the web-based site, desktop app, or mobile app. The mobile app is available for Android and iOS and is easy to use on a standard smartphone or tablet. Many popular ELNs are primarily laptop-based,23 and while no research has yet specifically examined the use of mobile apps for ELNs, we envision that this mode of access will improve record-keeping in laboratory settings due to the ease of access, portability, and ubiquity of mobile devices. Most, if not all, researchers are able to access the GitHub app on their device to swiftly read through past methods, add details and observations in the moment, or snap a photo for the ELN. A similar sentiment has been expressed by others who suggest that many researchers are likely to prefer mobile-based ELNs for their portability and extra features, like the built-in camera and option to annotate images using a stylus.24,25
We have used GitHub repositories as an ELN for both laboratory-based synthetic projects and computer-based social science projects and describe our experiences using it in the synthetic chemistry laboratory as a case study below.
ELN structure and utility
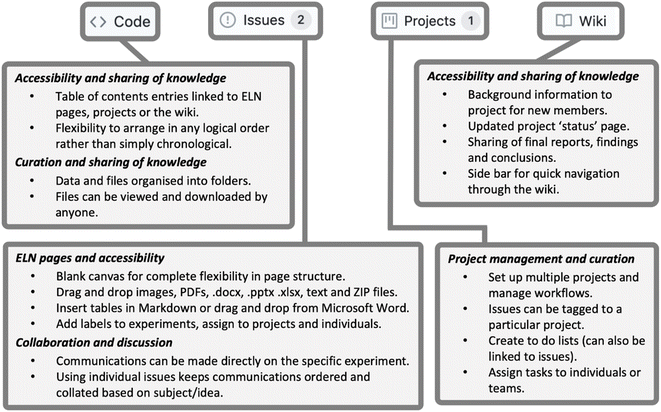
At the top layer, GitHub uses repositories to organise and store data and information. Each repository has Code, Issues, Discussions, Projects and Wiki tabs, all of which contribute to the ELN workflow (Fig. 2). Repositories also contain Pull Requests, Actions and Insight tabs, which are currently not used in our ELN workflow, along with Security and Settings tabs which are not discussed here.Repositories can be set up either by an individual or an organisation (e.g. research group) and assigned to individuals. Within Open Source Mycetoma and Open Source Tuberculosis there are topic-specific repositories which support discussion and collaboration, while ELN repositories are created by individuals and linked to the relevant organisation's repositories. This gives researchers the freedom to organise ELN repositories in a way that suits their individual needs. For example, while multiple projects can be contained in a single repository, a researcher may choose to have multiple repositories, one for each project they are involved in. Alternatively, a research group could set up repositories for each project with all researchers working on the project contributing to the single repository. Either way, an overview of all repositories can be viewed on both the individual's and organisation's profile. This interconnectivity of related work and segregation of distinct topics makes GitHub a useful tool, not only as an ELN, but also as a platform for the presentation of research and collaboration.
Notebook pages
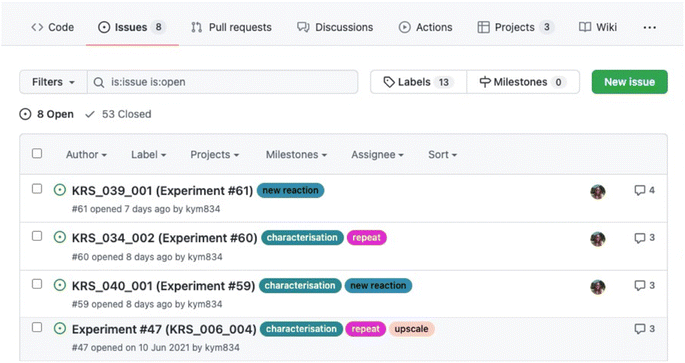
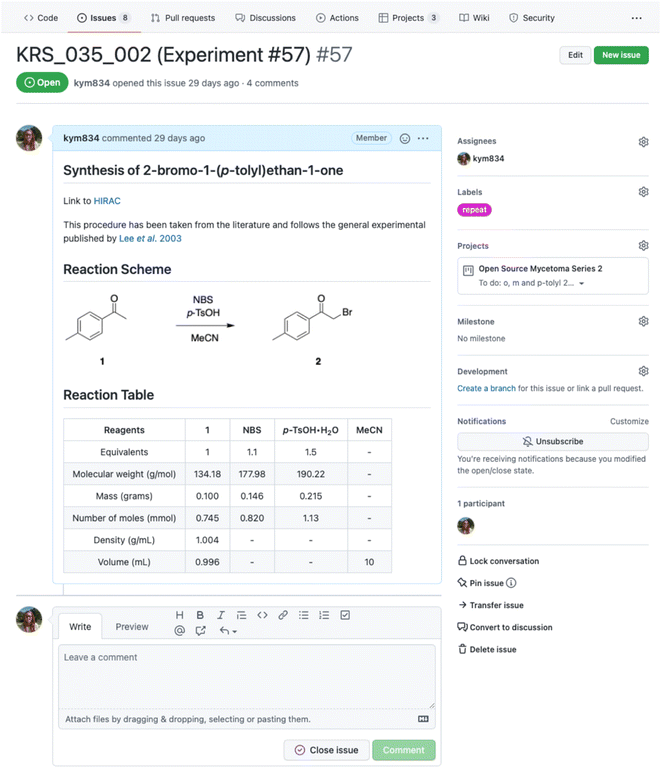
Issues are used as notebook pages: each represents an individual experiment and contains all the essential information, including the title, aim, quantity of reagents, methods, results, discussion and conclusions, along with linked references. In creating a new issue, the title, hyperlink to the risk assessment, reaction scheme (uploaded as an image) and table of reagents are posted. Plain text is formatted using Markdown, creating headings, tables and hyperlinks to aid clarity and readability. Making use of the forum-like structure, each subsequent addition to the notebook page is posted as a comment and conveniently time stamped. All experimental, observational and analytical data are also uploaded to the relevant issue. Once an experiment is completed, the issue is ‘closed’. This keeps the Issues landing page free of clutter and ‘open’ issues (active experiments) easily accessible, as open and closed issues are segregated. Examples of a typical Issues landing page and notebook page are shown in Fig. 3 and 4 respectively.Data management
GitHub supports numerous common file types including Microsoft Office files, PDFs, image and video files, and ZIP files. The ability to upload ZIP files to an issue is particularly advantageous as it allows both processed and raw data to be included, promoting best practice in data storage and facilitating reuse.26–28 Data files are easily added via the web browser version using a simple drag-and-drop method that should be intuitive to most computer users. Files can be uploaded to a relevant issue, or under the Code tab for centralised data storage. Files stored centrally on this tab can then be hyperlinked to relevant issues, so that users can rapidly access data related to the topic at hand. Furthermore, GitHub offers a desktop application, which can be used to add and curate files within the Code tab in a system analogous to typical file management operating systems.Metadata and curation
The use of metadata and curation aids the organisation and accessibility of the ELN to those beyond of the individual ELN user.29 GitHub contains numerous forms of metadata, including timestamps, indication of contributors, specific, customisable descriptors in the form of Labels and categorisation according to projects and status. Labels, analogous to the hash tags used ubiquitously on social media sites, are short descriptive statements such as “new experiment”, “upscale” or “help needed”. GitHub automatically suggests labels, drawing either from a default list (in the case of new users) or from previously used ones (added by a user). These labels are added to each issue and used to filter experiments by their various attributes. As issues are created in the order experiments are planned or performed and appear on the landing page in this order, labels make it easier to find relevant issues.Along with labels, issues are sorted into projects, consolidating all experimental work relevant to a given branch of investigation in one central location. Each project has its own landing page, accessible from the Projects tab, which contains links to all issues assigned to it as cards. Cards can be further sorted into columns and categorised. The authors favour the division into To do, In Progress, and Done categories. Using this system to organise their work, it is easy to keep track of planned, ongoing, and completed experiments and assess progress. The process can also be automated, so that performing specific actions automatically shifts cards into a new column within its assigned project. For example, one author has a workflow whereby assigning a newly created issue to a project adds the respective card to do and closing an issue moves it to Done. As with issues, projects are ‘closed’ and archived once the line of investigation is completed. This capacity for curation is an important workflow tool, as it prevents landing pages from being cluttered with obsolete links or information.
This method of automated workflow integrated into the capture of metadata at the source (the initial creation of a new issue) helps reduce the burden of curation.30 Previous work has noted the “blank canvas effect”, whereby researchers fail to add metadata due to unfamiliarity, rather than unwillingness.31 GitHub actively encourages the assignment of metadata through labels and projects categorisation, and the capture of metadata at the source. Upon creation of a new issue GitHub prompts users to add labels. This comparatively strong metadata support and active encouragement may be more effective than expecting users to create and curate their own labels without prompting.32 We suggest that the project, status and label features offered by GitHub facilitate individual project management, thus making researchers more likely to incorporate them into their ELNs for strategic reasons rather than because the system requires it.
Another important aspect of curation which is especially useful for making open-source work accessible to those not already involved in a project is the Wiki tool. This provides a place for a formal presentation of the work contained within the ELN, with pages organised according to topic. In this synthetic chemistry case study, a page in the Wiki has been dedicated to every different reaction, with this reaction landing page housing links to the notebook page for each attempt at the reaction, both successful and unsuccessful, alongside optimised methods, exemplar characterisation data and other relevant notes. These pages provide meta context and information that is often not present within the notebook pages of a typical ELN and are easily cross-linked, so that each page refers to multiple other relevant pages within the Wiki. This makes it easier for other researchers to find useful methods and data, while maintaining a high level of transparency, which improves both research integrity and reproducibility of results.
It is important to note that this method of organising data means that all attempts at a reaction are made publicly available, not just the successful experiments. This not only allows other researchers to come to their own conclusions about the results obtained, it also prevents duplication of unsuccessful efforts, as researchers can easily see what has and hasn't worked. While such complete transparency about the scientific process can appear intimidating to some researchers, it is a valuable asset of open ELNs and a powerful tool to support research integrity.29,33,34
Real-time sharing of knowledge and collaboration
Arguably GitHub's biggest advantage over other currently available ELNs is its capacity for rapid, real-time sharing of knowledge and collaboration. In the traditional scientific model, new work is typically shared only after significant positive results are found and through avenues that entail considerable delays, including conference presentations that may occur infrequently or the famously lengthy peer review process. In this context, real-time sharing means making experimental data available as soon as it is produced. Openly sharing knowledge in this way increases transparency, reproducibility, integrity,35,36 collaboration37,38 and impact,39 and groups that collaborate on their research recognise the benefits of sharing and discussing their work.40 In our use of GitHub, the Issues and Wiki tabs are used for the real-time sharing of knowledge while collaboration is facilitated through comments on specific issues. Publications in preparation are maintained through third party platforms that allow real-time collaborative editing, such as Google Drive, with links posted to GitHub. This simplifies the process of keeping track of successive versions of manuscripts. These functions allow researchers to communicate in real-time, discussing their work in a format that is rapid, direct and accessible.The immediate publication of additions to notebook or wiki pages, along with the ease of sharing these updates via email or social media sites allows new ideas and experiments to be made available to the broader public almost immediately, and avoids the delays typically seen when research is shared through formal channels like research papers and conference presentations. Indeed, the real-time sharing of research through wiki pages and specific issues was particularly useful for work conducted as part of the Open Source Tuberculosis project, which involved using Twitter to seek advice and suggestions on synthetic procedures. Both specific attempts at a reaction and a proposed synthetic scheme could be easily shared online, making it straightforward for interested parties to read more and make informed recommendations based on the experimental work already completed. Sharing this work resulted in a number of useful suggestions on alternative reagents and reaction conditions for experiments, further evidence that openly sharing knowledge in real-time is a powerful collaboration tool.
Unlike other UIs for Git, GitHub also facilitates public collaboration in a forum-like structure through the Discussions tab. Discussions is a relatively new feature, and before its introduction, the authors conduced discussions through dedicated issues. However, as it is typical of any online forum, we propose that its major application in collaboration lies in its ease of access for GitHub users. Having separate, dedicated spaces for the ELN and discussion within a single central system will conceivably facilitate conversations which might not be related to a specific experiment, such as conversations about a project's overall direction, organisation of meetings, or general brainstorming and sharing of ideas. Each Discussion can be organised into appropriate categories, so that users can quickly find the conversations they are interested in without having to sift through irrelevant topics, with links to relevant issues and wiki pages as appropriate.
Additionally, the customisability of GitHub allows repositories to be set up in a way that makes it easy for unfamiliar readers to quickly acquaint themselves with both the overall project and any recent updates, aiding the collaborative process. When accessing a repository, visitors are initially directed to the Code tab, making this the ideal place for introducing the ELN's owner(s) or curator(s), and the project(s) it relates to, through a README.md file. Hyperlinks guide visitors to other relevant sites, such as researchers' websites and social media profiles, and other GitHub repositories related to the project.
Shortcomings
In using GitHub as an ELN, the authors experienced several benefits as we've discussed above, but we also faced challenges. In the sections below we address these challenges and provide recommendations and workarounds we have found useful.Markdown learning curve
Unlike many ELNs currently available to researchers, GitHub is not specifically designed for scientists. While this is part of what makes it such a versatile ELN and useful for diverse types of research, it also creates some hurdles for researchers wishing to move an established workflow to GitHub. As individuals not familiar with computer science or programming languages, the biggest hurdle we have experienced is the need to use Markdown to format text. Markdown is a software used to format text using a plain-text editor, whereby text and images can be formatted by inserting extra characters or commands (e.g. to bold text, insert two asterisks before and two asterisks after that text). Unless a user is already familiar with this syntax, it can be a challenging learning curve, as the new syntax must be learned and applied for users to create clear, easy-to-read entries in their ELN and fully benefit from GitHub's potential.Fortunately, several factors and workarounds make Markdown a less imposing challenge than it first seems. First, GitHub provides users with a truncated menu of “clickable” formatting options which insert the characters or commands required to render common format styles. In addition, there is a Preview tab available for all text entries, which shows the user how the rendered and formatted text will appear, and so speeds the learning process. Users can start by clicking a desired formatting option, and gradually learn the necessary text entry, much the same as learning a hot-key for a formatting option in Microsoft Office. Secondly, because Markdown is a commonly used markup language, many guides to using it are available online (e.g.https://www.markdownguide.org/, https://docs.github.com/en/get-started/writing-on-github/getting-started-with-writing-and-formatting-on-github). There are also apps that enable users to write and format as they normally would in programs like Microsoft Word, with the text automatically converted to a Markdown format which can then be copied across to GitHub.
Another way to circumvent the challenges inherent in learning Markdown is to build and share templates. GitHub offers the option of creating issue templates which facilitates quicker creation of new issues. This works particularly well for synthetic experiments, with the same basic template being followed when writing up most experiments. The chosen template contains the desired formatting with space for researchers to add their own methods, results and data in the appropriate sections. A template may be created by researchers themselves, sourced from other group members, or from online resources – we have created and shared a number of templates to be used in different settings. Templates are already used in other ELNs to expedite the creation of lab entries with similar or near-identical information, as occurs with repeated or parallel reactions and procedures.13,27,41 Although previous work suggests templates are not always widely adopted,20 GitHub's requirement that users work with Markdown to create posts makes them more attractive.
Data storage limitation
GitHub supports file sizes up to 25 MB when attaching files via a browser, and files up to 100 MB when uploaded using the desktop application. Files greater than 100 MB in size can be uploaded if they are first converted to multiple smaller files, which can be re-assembled after download, but this requires both uploader and downloader to have the relevant programs and technical knowledge to perform these conversions and re-assemblies. This is a significant issue for certain disciplines in which data files may often be larger than this threshold, such as X-ray spectroscopy.20 However, this problem of data storage is not unique to GitHub. Many ELNs have limits on file size,7 for instance, LabArchives has an accumulative limit of 100 GB per user. It is also not static: as computing progresses and large files become more prevalent, it is likely that GitHub and other cloud based ELN providers will update their capacity and increase data size limits. A workaround for research involving large data files is to upload data to data repositories like Zenodo or Open Source Framework, and share the appropriate links on GitHub.Integration of discipline-specific applications
Many ELNs are tailored to particular disciplines, and include integration of applications commonly used within those disciplines. For instance, Chemotion is a free, open source ELN designed for organic chemists, and features chemical drawing tools, mole calculators and integration with SciFinder and PubChem. GitHub does allow integration with many applications, but as yet does not offer chemistry-specific tools. Instead, reaction schemes can be uploaded as images, and Excel files take the place of reaction tables. Raw and processed data from chemical drawing programs and chemical analysis software can also be uploaded, allowing users to access original files. Finally, while these discipline-specific inclusions make Chemotion and similar ELNS attractive options for organic chemists, they are not well suited to any other field of research. This shortcoming is further compounded by the lack of a dedicated space for discussion, making such ELNs less appropriate for multidisciplinary knowledge sharing and collaboration.Outlook
Perhaps the most immediately applicable future use of GitHub is to connect with new collaborators. GitHub's keyword tagging system extends beyond intra-repository linking, as each repository itself can be tagged with relevant terms which are discoverable to the broader userbase. This means that anybody interested in a given topic can quickly find others working on the same subject by following these links. Furthermore, GitHub-based ELNs are very easily shared online, encouraging participation from an audience beyond fellow GitHub users. Our work often involves sharing experimental work on Twitter and other social networks to solicit advice and publicise the project. Our GitHub ELN enables expedient sharing of both individual experiments (issues) and overviews of general synthetic approaches (wikis) with a simple URL. More broadly, GitHub's extensive array of options for communication and discussion, as well as the minimal barrier to using the site, make it straight-forward for new collaborators to get involved at whatever level they wish. We also believe that GitHub has potential to extend beyond the functionality of an ELN that ‘seamlessly integrates the process of data collection, data processing and data publication with minimal overheads for the researcher’ as recently envisioned and outlined by Jablonka et al.42Many discussions of ELNs also envisage an extension of online programs to encompass the broader experimental environment, in so-called ELEs.43 These may include integration of certain workflows into an ELE, so that certain experimental parameters, conditions and results can be automatically updated in and between connected ELNs.41 Future applications in this space could expand capabilities to include functionalities of specific use for researchers, for example integration of commonly used chemistry programs like molecular structure drawing tools would make it easier to add relevant data to GitHub. More ambitious proposals include incorporating a LIMS or existing browser-based sites like Reaxys and SciFinder, allowing researchers to quickly scan the web for information on specific substances or reactions from within an interlinked ELE. While these converging functionalities are currently beyond our capacity and in some part contingent on GitHub becoming more established as a site for hosting ELNs and other aspects of scientific research, GitHub does currently offer integration with many applications and actions to automate workflows.† Furthermore, GitHub is home to software developers and thus an ideal location to recruit collaborators with the requisite knowledge to develop code-based solutions.
Conclusions
Many ELNs have been developed by first studying how a given discipline uses paper notebooks or other record-keeping tools, then using this information to design a fit-for-purpose ELN.40 Conversely, GitHub was designed for a different purpose, and we have appropriated it for use as an ELN. Our experience to date demonstrates the versatility of GitHub for use as an ELN, and showcases the practical implications of its features in a synthetic chemistry context, along with the flexibility it offers researchers seeking to share their work and collaborate with others in real-time.Importantly, it offers version control, encourages and enables the inclusion of metadata and curation, and expedites the sharing of knowledge with real-time updates and very low barriers to discussion and collaboration between interested parties. The fact that GitHub has not been designed with a single field of research in mind also makes it ideal for cross-discipline collaboration, as each discipline can adapt different elements of GitHub's functionality for their own use while maintaining the same core GitHub infrastructure. Instead of having to familiarise themselves with new ELN software and layouts, or swap between multiple ELN providers, researchers working on multidisciplinary projects can use a single, centralised service with consistent controls and familiar structure.
While there are some features which are undeniably more oriented towards coders, such as the Actions tab in which users can set up workflows using code, these features do not detract from GitHub's usefulness as an ELN, which lies mainly in its adaptability and capacity for knowledge-sharing and collaboration. To overcome the potential impediment of using Markdown, we offer guides for those looking to trial GitHub for scientific research, as well as a template repository containing an issue template with appropriate labels, and a wiki template with suggested headings and formatting. Additionally, although this work features GitHub's application in the context of open science, we note that repositories can also be made private. Such repositories are accessible only by invitation, and thus appropriate for settings in which confidentiality is required.
GitHub's practical features and free, open source nature make it an attractive alternative not just to paper-based laboratory notebooks, but also to other ELNs, which can be expensive, inflexible, exclusive, and unsuitable for openly accessible research. We therefore encourage researchers in all disciplines to trial GitHub as an ELN, and to share their experiences in using it for their own projects (https://github.com/TheBreakingGoodProject/ELN-Templates/discussions/2).
Data availability
This paper describes open source notebooks using GitHub as an ELN. All data relates to those shared on GitHub and can be found: https://github.com/TheBreakingGoodProject and at links within: https://github.com/TheBreakingGoodProject/ELN-Templates, https://github.com/KlementineJBS/USYD_PhD_ELN, https://github.com/TheBreakingGoodProject/ELN-Kymberley-Scroggie, https://github.com/alintheopen/SCOPE/issues/23, https://github.com/TheBreakingGoodProject/GitHub-How-To-Guide.Author contributions
K. R. S. and K. J. B.-S. contributed equally. All authors; conceptualisation. A. M.; funding acquisition. K. R. S. and K. J. B.-S.; investigation. K. R. S. and A. M.; project administration. K. R. S.; visualisation. K. R. S. and K. J. B.-S.; writing – original draft. All authors; writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge and pay respect to the Gadigal people of the Eora Nation, the traditional owners of the land on which we research, teach, and collaborate at the University of Sydney. This work was supported by funding from the Westpac Research Fellowship (Motion), Google Impact Challenge and the University of Sydney Drug Discovery Initiative. KJB-S gratefully acknowledges the support of the John A Lamberton Research Scholarship. The authors acknowledge the facilities and the scientific and technical assistance of staff within the Sydney Analytical Core Research Facility and the School of Chemistry at the University of Sydney. Additionally, we acknowledge the broader members of the Open Source Malaria, Open Source Mycetoma and Open Source Tuberculosis communities of which we are part and with whom we collaborate using GitHub.Notes and references
- S. Borman, Chem. Eng. News, 1994, 72, 10–20 CrossRef.
- J. M. Jeschke, S. Lokatis, I. Bartram and K. Tockner, FACETS, 2019, 4, 423–441 CrossRef.
- M. Woelfle, P. Olliaro and M. H. Todd, Nat. Chem., 2011, 3, 745–748 CrossRef CAS PubMed.
- F. Kloeckner, R. Farkas, T. Franken and T. Schmitz-Rode, Biomed. Eng. Biomed. Tech., 2014, 59, 95–102 Search PubMed.
- S. Kanza, C. Willoughby, N. Gibbins, R. Whitby, J. G. Frey, J. Erjavec, K. Zupančič, M. Hren and K. Kovač, J. Cheminformatics, 2017, 9, 31 CrossRef PubMed.
- F. Rudolphi and L. J. Goossen, J. Chem. Inf. Model., 2012, 52, 293–301 CrossRef CAS PubMed.
- S. G. Higgins, A. A. Nogiwa-Valdez and M. M. Stevens, Nat. Protoc., 2022, 17, 179–189 CrossRef CAS PubMed.
- S. Guerrero, A. López-Cortés, J. M. García-Cárdenas, P. Saa, A. Indacochea, I. Armendáriz-Castillo, A. K. Zambrano, V. Yumiceba, A. Pérez-Villa, P. Guevara-Ramírez, O. Moscoso-Zea, J. Paredes, P. E. Leone and C. Paz-y-Miño, PLOS Comput. Biol., 2019, 15, e1006918 CrossRef CAS PubMed.
- A. R. Van Dyke and J. Smith-Carpenter, J. Chem. Educ., 2017, 94, 656–661 CrossRef CAS.
- E. Walsh and I. Cho, SLAS Technol, 2013, 18, 229–234 CrossRef PubMed.
- D. Bromfield Lee, J. Chem. Educ., 2018, 95, 1102–1111 CrossRef CAS.
- P. Tremouilhac, A. Nguyen, Y.-C. Huang, S. Kotov, D. S. Lütjohann, F. Hübsch, N. Jung and S. Bräse, J. Cheminformatics, 2017, 9, 54 CrossRef PubMed.
- A. J. Milsted, J. R. Hale, J. G. Frey and C. Neylon, PLOS ONE, 2013, 8, e67460 CrossRef CAS PubMed.
- L. Patiny, M. Zasso, D. Kostro, A. Bernal, A. M. Castillo, A. Bolaños, M. A. Asencio, N. Pellet, M. Todd, N. Schloerer, S. Kuhn, E. Holmes, S. Javor and J. Wist, Magn. Reson. Chem., 2017, 56, 520–528 CrossRef PubMed.
- A. E. Williamson, P. M. Ylioja, M. N. Robertson, Y. Antonova-Koch, V. Avery, J. B. Baell, H. Batchu, S. Batra, J. N. Burrows, S. Bhattacharyya, F. Calderon, S. A. Charman, J. Clark, B. Crespo, M. Dean, S. L. Debbert, M. Delves, A. S. M. Dennis, F. Deroose, S. Duffy, S. Fletcher, G. Giaever, I. Hallyburton, F.-J. Gamo, M. Gebbia, R. K. Guy, Z. Hungerford, K. Kirk, M. J. Lafuente-Monasterio, A. Lee, S. Meister, C. Nislow, J. P. Overington, G. Papadatos, L. Patiny, J. Pham, S. A. Ralph, A. Ruecker, E. Ryan, C. Southan, K. Srivastava, C. Swain, M. J. Tarnowski, P. Thomson, P. Turner, I. M. Wallace, T. N. C. Wells, K. White, L. White, P. Willis, E. A. Winzeler, S. Wittlin and M. H. Todd, ACS Cent. Sci., 2016, 2, 687–701 CrossRef CAS PubMed.
- W. Lim, Y. Melse, M. Konings, H. Phat Duong, K. Eadie, B. Laleu, B. Perry, M. H. Todd, J.-R. Ioset and W. W. J. van de Sande, PLoS Negl. Trop. Dis., 2018, 12, e0006437 CrossRef PubMed.
- M. N. Robertson, P. M. Ylioja, A. E. Williamson, M. Woelfle, M. Robins, K. A. Badiola, P. Willis, P. Olliaro, T. N. C. Wells and M. H. Todd, Parasitology, 2014, 141, 148–157 CrossRef PubMed.
- R. Vicente-Saez and C. Martinez-Fuentes, J. Bus. Res., 2018, 88, 428–436 CrossRef.
- M. H. Todd, ChemMedChem, 2019, 14, 1804–1809 CrossRef CAS PubMed.
- K. A. Badiola, C. Bird, W. S. Brocklesby, J. Casson, R. T. Chapman, S. J. Coles, J. R. Cronshaw, A. Fisher, J. G. Frey, D. Gloria, M. C. Grossel, D. B. Hibbert, N. Knight, L. K. Mapp, L. Marazzi, B. Matthews, A. Milsted, R. S. Minns, K. T. Mueller, K. Murphy, T. Parkinson, R. Quinnell, J. S. Robinson, M. N. Robertson, M. Robins, E. Springate, G. Tizzard, M. H. Todd, A. E. Williamson, C. Willoughby, E. Yang and P. M. Ylioja, Chem. Sci., 2015, 6, 1614–1629 RSC.
- R. T. Kouzes, J. D. Myers and W. A. Wulf, Computer, 1996, 29, 40–46 CrossRef.
- GitHub, The 2020 State of the Octoverse, 2020 Search PubMed.
- C. L. Bird and J. G. Frey, Chem. Soc. Rev., 2013, 42, 6754 RSC.
- R. Kwok, Nature, 2018, 560, 269–270 CrossRef CAS PubMed.
- K. Colabroy and J. K. Bell, in Biochemistry Education: From Theory to Practice, American Chemical Society, 2019, vol. 1337, pp. 173–195 Search PubMed.
- E. M. Hart, P. Barmby, D. LeBauer, F. Michonneau, S. Mount, P. Mulrooney, T. Poisot, K. H. Woo, N. B. Zimmerman and J. W. Hollister, PLOS Comput. Biol., 2016, 12, e1005097 CrossRef PubMed.
- M. D. Wilkinson, M. Dumontier, Ij. J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J.-W. Boiten, L. B. da Silva Santos, P. E. Bourne, J. Bouwman, A. J. Brookes, T. Clark, M. Crosas, I. Dillo, O. Dumon, S. Edmunds, C. T. Evelo, R. Finkers, A. Gonzalez-Beltran, A. J. G. Gray, P. Groth, C. Goble, J. S. Grethe, J. Heringa, P. A. C. ’t Hoen, R. Hooft, T. Kuhn, R. Kok, J. Kok, S. J. Lusher, M. E. Martone, A. Mons, A. L. Packer, B. Persson, P. Rocca-Serra, M. Roos, R. van Schaik, S.-A. Sansone, E. Schultes, T. Sengstag, T. Slater, G. Strawn, M. A. Swertz, M. Thompson, J. van der Lei, E. van Mulligen, J. Velterop, A. Waagmeester, P. Wittenburg, K. Wolstencroft, J. Zhao and B. Mons, Sci. Data, 2016, 3, 160018 CrossRef PubMed.
- S. Bechhofer, I. Buchan, D. De Roure, P. Missier, J. Ainsworth, J. Bhagat, P. Couch, D. Cruickshank, M. Delderfield, I. Dunlop, M. Gamble, D. Michaelides, S. Owen, D. Newman, S. Sufi and C. Goble, Future Gener. Comput. Syst., 2013, 29, 599–611 CrossRef.
- D. Solle, Anal. Bioanal. Chem., 2020, 412, 3961–3965 CrossRef CAS PubMed.
- J. Frey, Int. J. Digit. Curation, 2008, 3, 44–62 CrossRef.
- C. Willoughby, C. L. Bird, S. J. Coles and J. G. Frey, J. Chem. Inf. Model., 2014, 54, 3268–3283 CrossRef CAS PubMed.
- C. Willoughby, T. A. Logothetis and J. G. Frey, J. Cheminformatics, 2016, 8, 9 CrossRef PubMed.
- S. Buck, Science, 2015, 348, 1403 CrossRef CAS PubMed.
- D. B. Resnik and A. E. Shamoo, Account. Res., 2017, 24, 116–123 CrossRef PubMed.
- M. Schapira, The Open Lab Notebook Consortium and R. J. Harding, F1000Research, 2019, 8, 87 Search PubMed.
- M. R. Munafò, B. A. Nosek, D. V. M. Bishop, K. S. Button, C. D. Chambers, N. Percie du Sert, U. Simonsohn, E.-J. Wagenmakers, J. J. Ware and J. P. A. Ioannidis, Nat. Hum. Behav., 2017, 1, 0021 CrossRef PubMed.
- D. Speicher and A. B. Cremers, IPSI Transactions on Internet Research, 2020, vol. 16, pp. 38–44 Search PubMed.
- A. Y. Wang, A. Mittal, C. Brooks and S. Oney, Proc. ACM Hum.-Comput. Interact., 2019, 3, 1–30 CAS.
- R. J. Harding, PLOS Biol., 2019, 17, e3000120 CrossRef PubMed.
- C. L. Bird, C. Willoughby and J. G. Frey, Chem. Soc. Rev., 2013, 42, 8157 RSC.
- P. M. Piccione, Educ. Chem. Eng., 2020, 31, 42–53 CrossRef.
- K. M. Jablonka, L. Patiny and B. Smit, Nat. Chem., 2022, 14, 365–376 CrossRef CAS PubMed.
- K. T. Taylor, in Collaborative Computational Technologies for Biomedical Research, John Wiley & Sons, Ltd, 2011, pp. 301–320 Search PubMed.
Footnote |
| † As of April 1, 2022. Information obtained from https://github.com/marketplace |
| This journal is © The Royal Society of Chemistry 2023 |