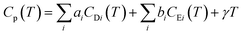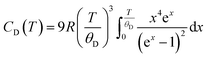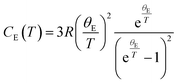Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The Liverpool materials discovery server: a suite of computational tools for the collaborative discovery of materials
Samantha
Durdy†
 ab,
Cameron J.
Hargreaves†
c,
Mark
Dennison
d,
Benjamin
Wagg
d,
Michael
Moran
bc,
Jon A.
Newnham
ab,
Cameron J.
Hargreaves†
c,
Mark
Dennison
d,
Benjamin
Wagg
d,
Michael
Moran
bc,
Jon A.
Newnham
 c,
Michael W.
Gaultois
c,
Michael W.
Gaultois
 bc,
Matthew J.
Rosseinsky
bc,
Matthew J.
Rosseinsky
 bc and
Matthew S.
Dyer
bc and
Matthew S.
Dyer
 *bc
*bc
aDepartment of Computer Science, University of Liverpool, Ashton Street, Liverpool, L69 3BX, UK. E-mail: msd30@liverpool.ac.uk
bLeverhulme Research Centre for Functional Materials Design, University of Liverpool, 51 Oxford Street, Liverpool, L7 3NY, UK
cDepartment of Chemistry, University of Liverpool, Crown St, Liverpool, L69 7ZD, UK
dServers and Storage Team, University of Liverpool, University of Liverpool Computing Services, Crown Street, Liverpool, L3 5UE, UK
First published on 15th September 2023
Abstract
The discovery of new materials often requires collaboration between experimental and computational chemists. Web based platforms allow more flexibility in this collaboration by giving access to computational tools without the need for access to computational researchers. We present Liverpool materials discovery server (https://lmds.liverpool.ac.uk/), one such platform which currently hosts six state of the art computational tools in an easy to use format. We describe the development of this platform, highlighting the advantages and disadvantages the methods used. In addition, we provide source code, a tutorial example, and setup scripts, and an application programming interface (API) to enable other research groups to create similar platforms, to promote collaboration both within and between research groups.
1 Introduction
The development of computational tools to accelerate experimental workflows in materials discovery has received significant investment in recent years. However, how best to incorporate these tools into the materials discovery workflow is still an active research area. While sharing these tools is possible through code repositories such as GitHub, or through sharing compiled code,1 this requires technical expertise, which can act as a barrier in the uptake of these tools by experimental researchers who may not have this expertise. Therefore, minimising the technical expertise required for synthetic researchers to use computational tools is crucial for these tools to be successfully adopted.Direct collaboration with computational experts offsets this need for computational expertise. Computational chemists are able to identify state of the art tools and can develop bespoke applications where needed. This type of collaboration may require organisational restructuring to most effectively accommodate computational developers and trained technical users identifying separate priorities.
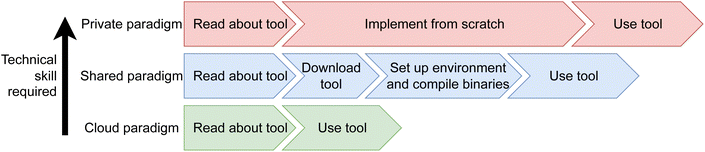
Three distinct paradigms for sharing computational tools may be observed; the private, shared, and cloud paradigms (Fig. 1). Private tools are those described in literature, but not made publicly available. Shared tools are accessible when run on local hardware, and cloud tools are accessible for remote usage. Notably, both shared and cloud paradigms may or may not be open source and/or free. This paper focuses on the cloud and shared paradigms, with the aims of promoting use of cloud paradigms to ease collaboration between experimental and computational chemists.
While often nebulously defined, “the cloud” typically refers to the global network of computer servers on which computation is executed non-locally. Cloud based tools (sometimes referred to as applications, apps, services, or micro-services) described here are broadly algorithms or functions which take user input communicated via web protocols (such as HTTP2), process this data on demand, and return the output to the user in a presentable fashion (i.e., in a graphical web browser). Examples of such tools for materials scientists range from state of the art ML models to predict material properties,3 and vast libraries of DFT calculations,4 to simple utilities that assert the charge neutrality of a chemical formula.5
Web applications are a prominent example of software which promotes collaboration. Use of web browsers is ubiquitous, and graphical user interfaces (GUIs) are the typical method of interacting with software. As such, accessing computational tools through a GUI in a web browser minimises the technical expertise required to use such tools.
While it is possible for web applications to be run locally (and thus fall under the private or shared paradigm), this often demands programming and networking expertise, which may be outside the past experience of researchers with familiarity in other technical domains. Further, designing bespoke applications for internal use clearly limits the audience which can interact with a tool. Locally hosting python notebooks6 is a common solution to sharing tools within research groups, however this still requires some technical knowledge to setup, and does not provide an accessible interface for non-technical users. Publicly accessible web applications,3,5,7,8 which fall under the cloud paradigm, can be designed towards a specific use case to improve usability, and allow many researchers to interact with an application.
A collection of these tools together forms a cloud platform. Cloud platforms offer computational researchers the opportunity to share their tools with a wider audience. These platforms can also include related resources, for example, both AFLOW8 and the Materials Cloud3 platforms host datasets, and provide a front-end to access to these datasets.
While cloud platforms offer many benefits, they may not offer suitable extensibility for computational researchers who want to share newly developed tools. Some platforms, like Materials Atlas,5 allow developers to upload new tools; however, developers may be reticent to launch a tool with these platforms for a number of reasons. Vendor lock-in can arise when relying on third parties to host tools, direct access to live code may be limited, use of certain code libraries or programming languages may be restricted, there may be limited control over end of life provisioning, and a culture of collaboration may not have been established between teams. An alternative solution is to host applications in house. Using modern frameworks, this can be done easily and securely while minimally increasing maintenance duties for the research team.
We present a new cloud platform, the “Liverpool Materials Discovery Server” (LMDS). We detail the applications currently accessible on this platform and our approach to enable researchers flexibility when deploying new applications. The aim of this platform is not only to share tools created by local researchers, but also to provide frameworks for other research groups to launch bespoke platforms while minimising the technical debt associated with such a task.
LMDS is designed to be simple and easily replicated, with an emphasis on reducing technical overhead rather than computational overhead. Source code and architectural information is provided, allowing easy adoption by other research groups to share their ML models either on their local intranet or on a public facing website. Giving each team personal ownership of their work promotes diversity in the field, and allows each group to discover their own optimal workflow as well as share their findings to the wider community. The approaches outlined in this paper should allow the launch of new cloud platforms with minimal time and financial investment. In the following discussion we address considerations that must be taken into account when sharing computational tools, and the role of such tools in the materials discovery workflow.
2 Available tools
This section will discuss the tools currently publicly available on the LMDS server (https://lmds.liverpool.ac.uk/). We start with tools to explore chemical space using a mathematically driven chemical distance metric, the element movers distance.9 A tool predicting the porosity of metal organic frameworks (MOFs) is then discussed before three tools to investigate the transport and thermal properties of inorganic crystals. These are tools to explore Li ion conductivity, thermal conductivity, and heat capacity. Finally we briefly detail the application programming interfaces (APIs) which are available for these applications.2.1 ElMTree
The Element Movers Distance (ElMD)9 has been applied to assess the compositional similarity of both hypothetical and previously reported compounds.10–12 The comparatively high computational complexity of the metric does not impede usage for small numbers of comparisons, but it does make it less suitable for use in high throughput similarity searches. Nevertheless, due to its clear utility, a brute force search interface to return the most similar composition reported in materials databases is included as a part of the Materials Atlas suite.5 Here the intensive nature of the search is given as the justification for limiting the search to restricted datasets (http://materialsatlas.org/tool_similarcomposition), but even with this constraint, brute force searches take up to a minute to execute on modern hardware, which is not fast enough for general usage.The ElMTree improves on this by using a metric indexing data structure,13,14 the List of Clusters (LC),15 where randomly selected objects of the search space are designated as routing objects. The remaining objects are assigned to their closest routing object, with the distance from each routing object to its furthest child stored as the routing objects' covering radius. This ensures that objects which are in close proximity to one another in the metric space are found in similar clusters of the indexed LC. When similarity searches are performed, large regions of the LC may be completely disregarded via the covering radius property. As the metric must obey the triangle inequality, we can use the distance to the routing object and its covering radius to ignore any children of routing objects which must be further away than the desired cut-off value of proximity. This allows significantly fewer comparisons to be made in total, enabling search times which increase logarithmically in proportion to the total number of items in the search space. The ElMTree has been indexed with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 367
367![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 526 unique compositions across 5
526 unique compositions across 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 410
410![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 119 records from 57 materials datasets16–61via the public APIs for Optimade62 and Matminer,63 and indexed with permission using the licensed APIs for the ICSD,58 CSD,59 Pearsons,60 and the MPDS.61 Each of these datasets give the composition as well as a reported property or structure of a material, with both experimental and computationally characterised materials indexed.
119 records from 57 materials datasets16–61via the public APIs for Optimade62 and Matminer,63 and indexed with permission using the licensed APIs for the ICSD,58 CSD,59 Pearsons,60 and the MPDS.61 Each of these datasets give the composition as well as a reported property or structure of a material, with both experimental and computationally characterised materials indexed.
Given the range of datasets, searches on the ElMTree may be further refined by the user, where results may be filtered to only include materials which have had crystal structures reported, to exclude materials which have only been characterised computationally, or to force each of the search results to include or exclude certain elements. As this is based on the ElMD, which uses the 103 elements from the modified Pettifor scale64 as its underlying elemental metric, unexpected behaviour may occur for the unstable elements which are not in this index. Despite the large search space, similarity queries typically find the 100 closest matches in under a second. These user friendly response times enable researchers to make comparisons against a wider range of reported compositions in high throughput computational workflows.
2.2 ElM2D
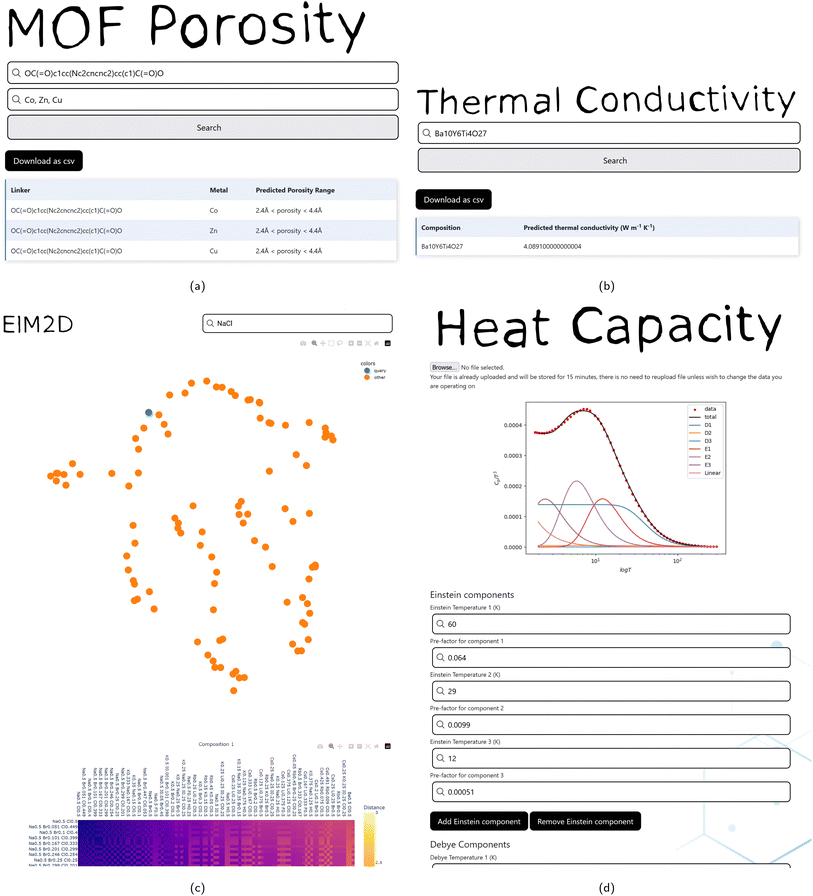
A method of generating ElM2D scatter plots of compositional similarity65 is provided, which allows the results of querying the ElMTree application to be visualised in 2 dimensions. A matrix of ElMD distances may be constructed between a query and the 100 most similar compositions returned by the ElMTree (Fig. 2c). Alternatively, a list of compositions separated by commas may be input to the search box, and the distance matrix will be formed between these specific compositions only. This distance matrix is embedded to the plane using UMAP,66 with an interactive scatter plot and distance matrix generated using the plotly library.67 This allows ElMTree queries and datasets of compositions to be placed within chemical context, providing an intuitive representation of the chemical distribution.2.3 Metal organic framework porosity prediction
The pore size of a MOF determines its guest accessibility, however it is not immediately apparent which metal-linker combinations will result in desired pore size. As such the ability to predict MOF porosity from just metal and linker is useful in aiding experimental chemists in choosing a metal linker combination to synthesise.This application is a front end for a previously reported ML model which predicts the porosity of a MOF from a metal and a linker SMILES string.68 These inputs are sequentially passed through three random forests (RFs). Each RF is trained to perform a binary classification of the porosity of MOFs as above or below an increasing threshold of pore limiting diameter (PLD). The first RF classifies the porosity of a MOF as being porous or non-porous (PLD of greater or less than 2.4 Å). The second classifies porosity as having large or small pores (PLD greater or less than 4.4 Å given porosity > 2.4 Å). The last model classifies porosity pores as being large or very large (PLD greater or less than 5.9 Å given porosity > 4.4 Å). In combination these random forests assign a MOF porosity as one of four categories (porosity < 2.4 Å, porosity < 4.4 Å, porosity < 5.9 Å, and porosity ≥ 5.9 Å), this classification was be 80.5% accurate on a random test set. 80% (5912 MOFs) of the data in the dataset was used for training and 20% (1479 MOFs) for the test set.
The user has the option to download the results or view them in the web page (Fig. 2a). In order to suit a variety of potential use cases, this tool can take a variety of inputs, being; two equal length comma separated lists (one of metals, one of linkers); one linker and a list of metal symbols (or vice versa); or a single metal and linker combination can be input. Due to limitations in the training set of this model, only certain metals are supported, a list of these can be found in the associated code repository,69 and is linked to on the application itself.
2.4 Composition based ML predictions of Li-electrolyte conductivity
The commercial interest of solid state electrolyte materials has driven a range of studies which construct statistical models to predict the ionic conductivity of materials. These are typically used in screening purposes for assessing the potential conductivity of novel formulations before the lengthy process of synthesis.70–72 Here we present one such model12 which aims to predict the ionic conductivity of candidate electrolyte materials based solely on composition. As synthesising a compound and then characterising its structure is both time consuming and error prone, it is advantageous to perform virtual screens of much broader compositional spaces before focusing on formulations of interest for further investigation. Regression and classification CrabNet models73 are trained on the 403 compositions in the Liverpool ionics dataset (https://pcwww.liv.ac.uk/%7Emsd30/lmds/LiIonDatabase.html) to predict specific values of conductivity in log10 (S cm−1) or whether a compound is likely to possess a conductivity greater than 10−4 S cm−1 at room temperature. In 5-fold cross validation on the Liverpool ionics dataset, regression models were found to possess a mean absolute error of 0.85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 (S cm−1), with classification models achieving a mean accuracy of 0.81 across the 5 folds. The interface takes in a chemical composition, or a comma separated list of compositions, processes these to be machine readable, generates predictions, and returns these predictions to the user as a downloadable table. As this uses the ElMD package for parsing compositions, valid predictions are limited to compositions which only contain elements reported in the modified Pettifor scale. This architecture and user interface can be extended into other domains by replacing the trained conductivity model with a bespoke model predicting other materials properties.
log10 (S cm−1), with classification models achieving a mean accuracy of 0.81 across the 5 folds. The interface takes in a chemical composition, or a comma separated list of compositions, processes these to be machine readable, generates predictions, and returns these predictions to the user as a downloadable table. As this uses the ElMD package for parsing compositions, valid predictions are limited to compositions which only contain elements reported in the modified Pettifor scale. This architecture and user interface can be extended into other domains by replacing the trained conductivity model with a bespoke model predicting other materials properties.
2.5 Thermal conductivity prediction for thermoelectric materials
Thermoelectric materials generate an electrical potential in response to a difference in temperature across the material and vice versa. Thermoelectric materials have many applications, with one of the most notable being a solid state replacement for chemical refrigerants. The suitability of materials as thermoelectrics is a combination of a low thermal conductivity, high electrical conductivity, and a high Seebeck coefficient. To screen materials for use as thermoelectrics, all three properties are of importance, we present a front end to a RF model of the thermal conductivity of thermoelectrics.The model presented here is an updated version of a previously reported thermal conductivity prediction model74 for probing the Ba–Y–Ti–O phase space of thermoelectric materials. The RF was trained on a mixture of experimentally measured56 and DFT computed75 thermal conductivities for a range of 1958 different thermoelectric materials. The RF is trained using a composition based featurization, and as such predictions can be obtained from a chemical formula only.
Users can input chemical compositions in plain text and receive a prediction for the thermal conductivity at room temperature and atmospheric pressure. This model was found to have an R2 of 0.71 and a root mean squared error of 0.55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10(W m−1 K−1) in predicting the logarithm of the thermal conductivity using 5-fold cross validation on the available data (note that training and validation was done on the previously reported thermoelectric materials dataset,74 so outside of the thermoelectric material domain performance may differ). This can be useful for mapping out chemical phase spaces to find candidate regions with useful thermal properties.
log10(W m−1 K−1) in predicting the logarithm of the thermal conductivity using 5-fold cross validation on the available data (note that training and validation was done on the previously reported thermoelectric materials dataset,74 so outside of the thermoelectric material domain performance may differ). This can be useful for mapping out chemical phase spaces to find candidate regions with useful thermal properties.
2.6 Heat capacity modelling
Modelling the heat capacity (Cp) of materials is important as it allows for the extraction of parameters such as the Debye and Einstein temperatures. To extract these parameters, Cp can be modelled by a linear combination of the Debye and Einstein functions:where CD and CE represent the Debye and Einstein contributions to the heat capacity, respectively. γ is a linear term, and ai and bi are prefactors that are normalised to sum to 1. The Debye contributions to the heat capacity are modelled by:
in which R is the ideal gas constant, and θD is the Debye temperature.76 Whereas, the Einstein contributions are modelled by:
where θE is the Einstein temperature.77
One linear component, any number of Debye terms, and any number of Einstein terms (along with their respective prefactors) can be manually adjusted in order to identify optimal parameters for modelling heat capacity data. The outputted heat capacity models can be viewed as Cp(T) or Cp/Tn(T) plots in linear, log, and log–log scales in order to aid fitting in the high and low temperature regions.
2.7 APIs
Each tool also has an associated (API) to enable programmatic access to underlying models. This allows developers to integrate these models into automated workflows, or other web applications. The exception to this is the ElM2D app, as it would be more appropriate to make a query to the ElMTree API, and visualise those results locally. Details of each API may be found on the web page for each tool.3 Discussion
3.1 Architectural considerations
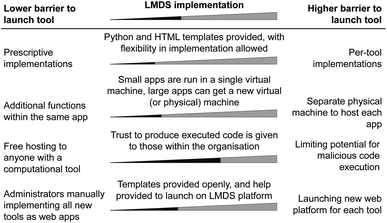
A key consideration in the LMDS was the ease of extending the platform through publication of new tools. While ML experts may have limited knowledge in computer networking, frameworks and examples (such as those found in the associated code repositories78) can reduce the technical complexity of deploying new tools. New web frameworks are frequently released, and balancing the technical debt of learning how to interface and maintain each new library with networking and computational resource issues becomes non-trivial (Fig. 3). We outline some of the technical design choices which were taken when designing the LMDS architecture.One such consideration was the level of restriction on the range of technologies that technical users may employ in their development cycle. Constraining developers to certain libraries enforces a greater degree of homogeneity in a codebase, allowing for a larger quantity of each application's code to be reused and reducing the work required to deploy new tools. However, restricting to specific libraries may impose limits on newer approaches which could be deployed, or may simply not align with developers' personal preferences. Each of the provided tools is written in Python (when applicable) owing to its popularity, but otherwise collaborators are not constrained by which external libraries they may use. Were researchers to propose tools based in other programming languages or frameworks, existing HTML and styling are provided for reuse.
The degree of separation between applications in a cloud platform is a notable design decision. Malfunction or security compromise of one tool should not effect other tools on the platform. In the past, applications were isolated their own physical server, in a process referred to as server segmentation. Current best practice is to isolate applications virtually for efficiency purposes.
One such method to separate applications from each other is virtualisation, which allows multiple virtual “machines” (VMs) to operate on a single physical machine through the use of hardware emulation. A host operating system runs a virtualisation program (hypervisor) which manages the computational resources of each VM. Each VM runs its own operating system, which may be selected depending on the task at hand, with Linux distributions often chosen for web applications. Virtualisation allows for dynamic scheduling of resources, while ensuring that a single application's malfunction does not affect other tools on the platform. VMs are easy to deploy, and updating VMs remains similar to updating to physical machines, although VMs are accessed through a hypervisor. Each VM does carry some overhead, as each operating system needs to store its own data in memory for each application, but the capacity of modern systems means this is generally not a concern.
Where the capacity of a system is a concern, containerisation provides a similar method of isolating applications from one another, with Docker79 and Kubernetes80 being two popular tools for this. Containerisation comes with a lower computational overhead than VMs (in particular with regards to memory consumption).81,82 However this becomes yet another technology for developers to learn when deploying new tools. As a low barrier to entry for new application deployment is a key goal for the LMDS, we have opted not to use containerisation technology.
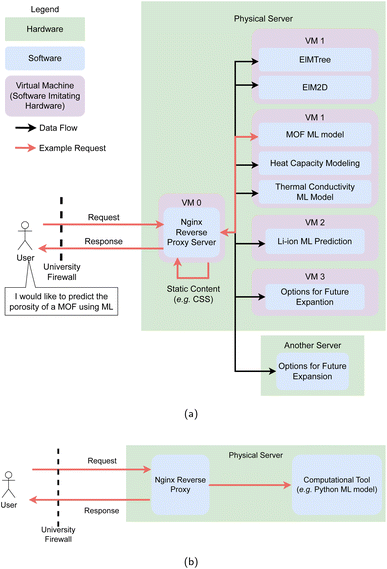
To balance the increased memory cost of VMs over containers such as docker and the need for the isolation of apps, the LMDS hosts smaller applications (for example the MOF prediction tool and heat capacity modelling tool) on a single VM. Meanwhile, individual VMs are allocated to larger tools (such as ElMTree) to provide a level of isolation.
Managing these different VMs requires a reverse proxy server to direct requests with different web addresses the correct tool without requiring a separate domain name or subdomain for each tool. To create this reverse proxy HTML requests to each application are routed through a VM running Nginx,83 which enables each of the separate applications to be accessed through a single domain name. Internally this server resolves each request to the internal IP address the specific application is hosted on. The reverse proxy provides some protection from direct denial of service (DDOS) attacks by enabling rate limiting functionality. This Nginx server also encrypts HTTP traffic into HTTPS traffic84 (Fig. 4), which provides a security assertion to users that their data has not been seen or interfered with by any third parties. Apache HTTP server is an application historically popular for this task,85,86 however Nginx was selected due to its wide market adoption, strong performance,87 and simple configuration. Nginx can redirect requests to additional physical machines external to the hypervisor that the LMDS is currently hosted on, providing flexibility in future expansion (Fig. 4a). Setup scripts have been provided to configure this proxy for new tools.78
3.2 Development and maintenance of cloud platforms
Creation of web applications as outlined here may require some knowledge outside the main domain of a computational researcher, such as HTML and cascading style sheets (CSS). While ample learning materials for such skills exist online, this adds a barrier to the launch of new web applications. While styling presented here65,69,88 can be used as a template, it is likely research groups will wish to develop their own aesthetic with which to brand their website. While heavily adjusted, the front end seen here was outsourced commercially. Other research groups may find outsourcing front end web development to be a cheap and fast way to overcome the limitations in front end design skills.Where work is outsourced, basic HTML skills would still be helpful to make ongoing changes to web platforms. Maintenance of cloud platforms is a major consideration which should be made, not just to ensure the longevity of a platform, but also to ensure its security. Using scheduled commands (such as Linux's Crontab) generated files (plots, output data etc.) can be deleted and operating system updates can be installed. Updates such as these do run the risk of breaking existing code, but for simple apps this is not a major risk. Going without updates creates a large security risk, as such best practice is to apply updates regularly.
With scheduled updates maintenance of cloud platforms can be minimal, but manual intervention will eventually be needed. Fixing broken applications, launching new applications, or taking applications offline all require some technical skill, and knowledge of the cloud platforms architecture. As such considerations should be made as to how systems will be maintained including following changes in staffing. While no website can stay online 100% of the time, good documentation, and forward planning can help to minimise issues when they arise.
3.3 Research context and significance
As the user complexity and model complexity of ML projects grow, it is important that access to, and understanding of, ML tools do not become a barrier for their use. Cloud platforms such as the LMDS provide easier access to these tools, however it is on a per-practitioner basis to portray the understanding of best use and interpretation of such tools.For example many ML models (such as that used in the thermal conductivity prediction model) take composition as input, but have no mechanisms to check the chemical viability of such compositions. Without adequately communicating this limitation with collaborators, the trust in such a model may be hindered, and the interpretation of its results will be incomplete. While explainability and uncertainty estimates in ML are active research areas,89,90 a good understanding of the limits and correct usage of ML models by those who use them is also important. Drawing up interesting counter examples when presented with predictive models to demonstrate their limits is a valid method of testing the capabilities of models. This may not be a particularly useful test in many cases, as we know that statistical models will under perform on chemical domains they have never been exposed to, and this may not be how the model should be used in practice. For example, the thermal conductivity model (Section 2.5) is trained only on thermoelectric materials, while it may be used to predict thermal conductivity for other materials, it may under-perform outside the thermoelectric domain.
Similarly, it is a per-application question as to whether a tool will actually be useful in the materials discovery workflow. While the MOF porosity prediction tool (Section 2.3) may offer a variety of input options for flexible usage by experimental researchers, if no such researcher exists, the usefulness of such a tool is limited. As such, while cloud based tools do provide ease of access, it remains vital that collaborations between experimental and computational researchers involve open communication channels.
Cloud platforms are an excellent supplement to existing communications methods, and need not be a prohibitively expensive to deploy. While ML models may be costly to train and require significant compute resources, after training is performed the models can often be deployed using lower end hardware and still make inferences in a timely manner. Consequently, the LMDS platform with the architecture outlined above may be deployed on relatively cheap hardware. We provide a minimum framework for launch of platforms (Fig. 4b), as well as how we expand this for isolation of applications (Fig. 4). The frameworks and implementation details provided here should provide a reasonable starting point for other researchers to share their tools with the wider community.
While the LMDS is hosted in house, the frameworks provided could be used for platforms deployed to commercial cloud providers, such as Amazon Web Services or Microsoft Azure. Third party cloud providers alleviate concerns over server maintenance and hardware failure. However, each commercial cloud platform requires bespoke training to use, which may be a niche skill for computational materials science researchers. Concerns may be raised over vendor lock in, as such services may become more expensive or less reliable in the future. Further, the monthly billing cycle commercial cloud providers often demand is not compatible long term with the fixed consumable budgets that are typically provided as part of a research grant. Depending on funding and available hardware, a commercial cloud provider may be the best solution for rapid delivery. If the mission critical up time that is guaranteed by server hardware is not a driving design choice, then many computational research groups may find they already have the necessary resources to hand, as this architecture may be run on an underused workstation.
In house solutions, such as those presented here, may be deployed onto new or existing hardware, and tailored to suit the team's existing technical specialities, but this approach is not without disadvantages. Local hardware needs ongoing maintenance in the case of equipment failure, such as hard drives, which will have an associated cost. Networked applications operating under the framework of an institution will have to comply with the organisation's pre-existing networking and security protocols, especially if accessible from the public internet, which may introduce further tasks which must be satisfied to launch a new cloud tool. By working with the University of Liverpool's servers and storage team to test the architecture throughout the development process, we were able to ensure that the final product is robust and secure.
Releasing the LMDS as a simple technology stack with limited functionality means other research groups can extend this framework to rapidly prototype bespoke applications to suit their specific requirements. Cloud platforms enhance partnerships between computational and experimental research teams, and provide an additional interactive medium for accessing research.
However, monolithic platforms are by definition less integrated with experimental researchers than bespoke platforms. Enhancing the interface between technical developers and their expert users allows new tools to be integrated into materials discovery workflows. The process of constructing a new cloud platform requires technical expertise, but the barrier to entry is low enough that many computational researchers do in fact possess these skills. We hope the tools provided here reduce this technical gap further to make creation of cloud platforms simpler for others. Future undertakings could investigate methods of unifying multiple cloud platforms into singular portals, or developing frameworks that require even less technical expertise to create new cloud applications. Centralised or monolithic systems risk excluding researchers who wish to share computational tools with the wider community if the technical or organisational processes to host such tools remain unclear.
Creating new methods to access computational tools through cloud platforms is one way of exploring how computational methods may be adopted by experimental researchers. As computational methods continue to develop, so too will their place in the discovery of new materials. Future research may lead to more cloud platforms, new frameworks to ease the creation of such platforms, or focus on entirely novel collaborative techniques. While advancing the accuracy of the predictions made by ML models remains a dominant research area in this field, the concurrent development of tools which interface with these models is a crucial piece of supporting work to ensure wide and effective adoption.
4 Methods
All bespoke programs presented here are implemented in Python. The MOF porosity prediction and thermal conductivity tools use random forests implemented in sci-kit learn.91 MOF porosity prediction tool was featurised using mordred and RDKit.92,93 The Li-ion conductivity tool uses CrabNet73 to predict the conductivities of compositions. ElMTree uses the ElMD9 library and a simple implementation of the list of clusters. ElM2D uses the aforementioned ElMTree application with the UMAP66 and plotly67 libraries. The modeling of heat capacity was carried forward using the SciPy library.94 Each of the web applications are implemented using Flask, with the gunicorn process manager used to spawn Flask processes, examples of these implementations have been provided,69,88 including a basic Flask app, which can be used as templates for other researchers.95 VMs are run through VMWare vSphere ESXi 7,96 with Nginx used to serve HTTP responses to users and route URLs to each VM.83 Example setup scripts for gunicorn and Nginx can are available.78 The linux utility crontab is used to ensure regular updates are executed and to remove temporary files in relevant LMDS applications.5 Conclusions
We have presented the LMDS, a cloud platform for experimental researchers to use in discovering new materials, available at https://lmds.liverpool.ac.uk/. The LMDS platform allows for easy access to previously published computational models,68 as well as novel tools to help experimental researchers.Making computational tools easily accessible is crucial to maximise their benefit. Thus, we developed the LMDS platform with the objective of simplifying the sharing of computational tools, ensuring they are readily available to researchers with minimal computational expertise. We provide considerations that lead to the production of this platform, justifying key design considerations.
Difficulties in applying computational methods in experimental research are discussed, as are the barriers for deployment of such methods to cloud platforms. Examples of the tools discussed in this manuscript have been provided,69,88 as well as scripts setting these up with an Nginx reverse proxy server, and Python process manager,78 enabling other researchers to reproduce this tool chain and share their own methods, either internally or on the open internet.
We believe minimising organisational overhead in collaborations between computational and experimental researchers promotes the incorporation of computational methods in the synthesis of new materials. Access to state of the art computational methods, such as in ways presented here, accelerates research and improves the prediction, analysis, and realisation of new materials.
Data availability
Setup scripts for Nginx and gunicorn are available: https://github.com/lrcfmd/LMDS_helper_scriptsInformation about tools listed in Section 2 are as follows:
• Section 2.1: the code for the ElMTree tool is available at https://github.com/lrcfmd/ElMTree
• Section 2.2: the code for the ElM2D tool is available at https://github.com/lrcfmd/ElM2D
• Section 2.3: the code for the metal organic framework porosity prediction tool application is available at https://github.com/lrcfmd/LMDS_MOF_Porosity_Tool. The data associated with the previous publication of these models is available at https://datacat.liverpool.ac.uk/1494/
• Section 2.4: code to reproduce the ML models associated with composition based ML predictions of Li-Electrolyte Conductivity is available at https://github.com/lrcfmd/LiIonML
• Section 2.5: thermal conductivity prediction for thermoelectric materials. Publication of model available at https://onlinelibrary.wiley.com/doi/10.1002/anie.202102073. Code and data for reproduction of model for web app available at https://github.com/lrcfmd/thermal_conductivity_RF
• Section 2.6: the code associated with the heat capacity modelling can be found at https://github.com/lrcfmd/LMDS_heat_capacity_modelling
Author contributions
Conceptualisation – C. J. H.; M. W. G.; M. J. R.; M. S. D.; data curation – S. D.; C. J. H.; formal analysis – S. D.; C. J. H.; M. M.; J. N.; funding acquisition – M. J. R.; M. S. D.; project administration – S. D.; C. J. H.; M. S. D.; software – S. D.; C. J. H.; M. D.; B. W.; supervision – D. A.; M. W. G.; M. J. R.; M. S. D.; visualisation – C. J. H.; CloudPloys; writing, original draft – S. D.; C. J. H.; writing, review & editing; S. D.; C. J. H.; M. W. G.; M. J. R.; M. S. D.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the University of Liverpool (studentship to C. J. H.), by the Faraday Institution (SOLBAT, grant number FIRG007), and by EPSRC under EP/V026887 and EP/R018472/1. The authors thank the Leverhulme Trust for funding this research via the Leverhulme Research Centre for Functional Materials Design (RC-2015-036). MWG thanks the Ramsay Memorial Fellowships Trust for funding through a Ramsay Trust Memorial Fellowship. This work was undertaken on Barkla, part of the High Performance Computing facilities at the University of Liverpool, UK. We would like to thank the company CloudPloys for their initial CSS design.References
- L. Ward, A. Dunn, A. Faghaninia, N. Zimmermann, S. Bajaj, Q. Wang, J. Montoya, J. Chen, K. Bystrom, M. Dylla, K. Chard, M. Asta, K. Persson, G. Snyder, I. Foster and A. Jain, Comput. Mater. Sci., 2018, 152, 60–69 CrossRef.
- R. Fielding, J. Gettys, J. Mogul, H. Frystyk, L. Masinter, P. Leach and T. Berners-Lee, RFC 2616, Hypertext Transfer Protocol – HTTP/1.1, 1999, http://www.rfc.net/rfc2616.html Search PubMed.
- L. Talirz, S. Kumbhar, E. Passaro, A. V. Yakutovich, V. Granata, F. Gargiulo, M. Borelli, M. Uhrin, S. P. Huber, S. Zoupanos, C. S. Adorf, C. W. Andersen, O. Schütt, C. A. Pignedoli, D. Passerone, J. VandeVondele, T. C. Schulthess, B. Smit, G. Pizzi and N. Marzari, Sci. Data, 2020, 7, 1–12 CrossRef PubMed.
- A. Jain, J. Montoya, S. Dwaraknath, N. E. R. Zimmermann, J. Dagdelen, M. Horton, P. Huck, D. Winston, S. Cholia, S. P. Ong and K. Persson, in The Materials Project: Accelerating Materials Design Through Theory-Driven Data and Tools, Springer International Publishing, 2018, pp. 1–34 Search PubMed.
- J. Hu, S. Stefanov, Y. Song, S. S. Omee, S. Y. Louis, E. M. Siriwardane, Y. Zhao and L. Wei, npj Comput. Mater., 2022, 8, 1–12 CrossRef.
- Jupyter, Jupyter Lab and Jupyter Notebook, 2015, https://jupyter.org/ Search PubMed.
- J. O'Mara, B. Meredig and K. Michel, JOM, 2016, 68, 2031–2034 CrossRef.
- S. Curtarolo, W. Setyawan, G. L. Hart, M. Jahnatek, R. V. Chepulskii, R. H. Taylor, S. Wang, J. Xue, K. Yang, O. Levy, M. J. Mehl, H. T. Stokes, D. O. Demchenko and D. Morgan, Comput. Mater. Sci., 2012, 58, 218–226 CrossRef CAS.
- C. J. Hargreaves, M. S. Dyer, M. W. Gaultois, V. A. Kurlin and M. J. Rosseinsky, Chem. Mater., 2020, 32, 10610–10620 CrossRef CAS.
- L. Wei, N. Fu, E. M. Siriwardane, W. Yang, S. S. Omee, R. Dong, R. Xin and J. Hu, Inorg. Chem., 2022, 61, 8431–8439 CrossRef CAS PubMed.
- S. G. Baird, T. Q. Diep and T. D. Sparks, Digital Discovery, 2022, 1, 226–240 RSC.
- C. J. Hargreaves, M. W. Gaultois, L. M. Daniels, E. J. Watts, V. A. Kurlin, M. Moran, Y. Dang, R. Morris, A. Morscher and K. Thompson, et al. , npj Comput. Mater., 2023, 9, 9 CrossRef CAS.
- H. Samet, Foundations of Multidimensional and Metric Data Structures (The Morgan Kaufmann Series in Computer Graphics and Geometric Modeling), Morgan Kaufmann Publishers Inc., San Francisco, CA, USA, 2005 Search PubMed.
- L. Chen, Y. Gao, X. Song, Z. Li, Y. Zhu, X. Miao and C. S. Jensen, ACM Comput. Surv., 2022, 55, 1–39 Search PubMed.
- E. Sadit Tellez and E. Chávez, in The list of clusters revisited, 2012, pp. 187–196 Search PubMed.
- J. Zhou, L. Shen, M. D. Costa, K. A. Persson, S. P. Ong, P. Huck, Y. Lu, X. Ma, Y. Chen, H. Tang and Y. P. Feng, Sci. Data, 2019, 6, 86 CrossRef PubMed.
- A. Marrazzo, M. Gibertini, D. Campi, N. Mounet and N. Marzari, Nano Lett., 2019, 19, 8431–8440 CrossRef CAS PubMed.
- J. Schmidt, H.-C. Wang, T. F. T. Cerqueira, S. Botti and M. A. L. Marques, Sci. Data, 2022, 9, 64 CrossRef CAS PubMed.
- V. Vitale, G. Pizzi, A. Marrazzo, J. R. Yates, N. Marzari and A. A. Mostofi, npj Comput. Mater., 2020, 6, 66 CrossRef.
- A. Mansouri Tehrani, A. O. Oliynyk, M. Parry, Z. Rizvi, S. Couper, F. Lin, L. Miyagi, T. D. Sparks and J. Brgoch, J. Am. Chem. Soc., 2018, 140, 9844–9853 CrossRef CAS PubMed.
- Computational Materials Repository, https://cmr.fysik.dtu.dk/ Search PubMed.
- I. Castelli, D. Landis, K. S. Thygesen, S. Dahl, I. Chorkendorff, T. Jaramilo and K. W. Jacobsen, Energy Environ. Sci., 2012, 9034–9043 RSC.
- D. Ongari, A. V. Yakutovich, L. Talirz and B. Smit, Materials Cloud Archive, 2021 Search PubMed.
- I. Petousis, D. Mrdjenovich, E. Ballouz, M. Liu, D. Winston, W. Chen, T. Graf, T. D. Schladt, K. A. Persson and F. B. Prinz, Sci. Data, 2017, 4, 160134 CrossRef CAS PubMed.
- G. Pilania, A. Mannodi-Kanakkithodi, B. P. Uberuaga, R. Ramprasad, J. E. Gubernatis and T. Lookman, Sci. Rep., 2016, 6, 19375 CrossRef CAS PubMed.
- M. de Jong, W. Chen, T. Angsten, A. Jain, R. Notestine, A. Gamst, M. Sluiter, C. Krishna Ande, S. van der Zwaag, J. J. Plata, C. Toher, S. Curtarolo, G. Ceder, K. A. Persson and M. Asta, Sci. Data, 2015, 2, 150009 CrossRef CAS PubMed.
- G. Kim, S. V. Meschel, P. Nash and W. Chen, Sci. Data, 2017, 4, 170162 CrossRef PubMed.
- O. Kubaschewski, C. Alcock and P. Spencer, Materials Thermochemistry, Pergamon Press, 6th edn, 1993 Search PubMed.
- M. W. Chase, NIST-JANAF Thermochemical Tables, 1998, http://kinetics.nist.gov/janaf/ Search PubMed.
- K. Rzyman, Z. Moser, A. Miodownik, L. Kaufman, R. Watson and M. Weinert, Calphad, 2000, vol. 24, pp. 309–318 Search PubMed.
- CRC Handbook of Chemistry and Physics, ed. W. M. Haynes, CRC Press, 88th edn, 2007 Search PubMed.
- S. Grindy, B. Meredig, S. Kirklin, J. E. Saal and C. Wolverton, Phys. Rev. B: Condens. Matter Mater. Phys., 2013, 87, 1–8 CrossRef.
- Y. Zhuo, A. Mansouri Tehrani and J. Brgoch, J. Phys. Chem. Lett., 2018, 9, 1668–1673 CrossRef CAS PubMed.
- F. Faber, A. Lindmaa, O. A. von Lilienfeld and R. Armiento, Int. J. Quantum Chem., 2015, 115, 1094–1101 CrossRef CAS.
- Y. T. Sun, H. Y. Bai, M. Z. Li and W. H. Wang, J. Phys. Chem. Lett., 2017, 8, 3434–3439 CrossRef CAS PubMed.
- F. Ren, L. Ward, T. Williams, K. J. Laws, C. Wolverton, J. Hattrick-Simpers and A. Mehta, Sci. Adv., 2018, 4, 4 Search PubMed.
- Y. Kawazoe, T. Masumoto, A.-P. Tsai, J.-Z. Yu and T. Aihara Jr, Datasheet from Landolt-Börnstein – Group III Condensed Matter, https://materials.springer.com/lb/docs/sm_lbs_978-3-540-47679-5_2 Search PubMed.
- University of Alabama Heusler database, https://citrination.com/datasets/150561/ Search PubMed.
- S. Gražulis, D. Chateigner, R. T. Downs, A. F. T. Yokochi, M. Quiros, L. Lutterotti, E. Manakova, J. Butkus, P. Moeck and A. LeBail, J. Appl. Crystallogr., 2009, 42, 726–729 CrossRef PubMed.
- K. Choudhary, I. Kalish, R. Beams and F. Tavazza, Sci. Rep., 2017, 7, 5179 CrossRef PubMed.
- K. Choudhary, B. DeCost and F. Tavazza, Phys. Rev. Mater., 2018, 2, 083801 CrossRef CAS PubMed.
- M. F. Cover, O. Warschkow, M. M. M. Bilek and D. R. McKenzie, J. Phys.: Condens. Matter, 2009, 21, 305403 CrossRef CAS PubMed.
- L. Ward, A. Agrawal, A. Choudhary and C. Wolverton, npj Comput. Mater., 2016, 2, 16028 CrossRef.
- G. Petretto, S. Dwaraknath, H. P. C. Miranda, D. Winston, M. Giantomassi, M. J. van Setten, X. Gonze, K. A. Persson, G. Hautier and G.-M. Rignanese, Sci. Data, 2018, 5, 180065 CrossRef CAS PubMed.
- Mechanical properties of some steels, https://citrination.com/datasets/153092/ Search PubMed.
- D. Campi, N. Mounet, M. Gibertini, G. Pizzi and N. Marzari, The Materials Cloud 2D database (MC2D), 2022, https://archive.materialscloud.org/record/2022.84 Search PubMed.
- S. Huber, M. Bercx, N. Hörnmann, M. Uhrin, G. Pizzi and N. Marzari, The Materials Cloud three-dimensional crystals database (MC3D), 2022, https://archive.materialscloud.org/record/2022.38 Search PubMed.
- P. Villars, K. Cenzual, R. Gladyshevskii and S. Iwata, in Pauling File: Toward a Holistic View, John Wiley & Sons, Ltd, 2019, ch. 3, pp. 55–106 Search PubMed.
- A. Jain, S. P. Ong, G. Hautier, W. Chen, W. D. Richards, S. Dacek, S. Cholia, D. Gunter, D. Skinner, G. Ceder and K. A. Persson, APL Mater., 2013, 1, 011002 CrossRef.
- A. Morris, ODBX is a public database of crystal structures from the group of Dr Andrew Morris at the University of Birmingham, 2023, https://odbx.science/ Search PubMed.
- F. P. Kinik, A. Ortega-Guerrero, D. Ongari, C. P. Ireland and B. Smit, Pyrene-based metal organic frameworks, 2020, https://archive.materialscloud.org/record/2020.156 Search PubMed.
- V. Stanev, C. Oses, A. G. Kusne, E. Rodriguez, J. Paglione, S. Curtarolo and I. Takeuchi, npj Comput. Mater., 2018, 4, 1 CrossRef.
- M. I. S. National Institute of Materials Science, SuperCon, http://supercon.nims.go.jp/index_en.html Search PubMed.
- A. Merkys, N. Mounet, A. Cepellotti, N. Marzari, S. Gražulis and G. Pizzi, J. Cheminf., 2017, 9, 56 Search PubMed.
- C. Tholander, C. Andersson, R. Armiento, F. Tasnadi and B. Alling, J. Appl. Phys., 2016, 120, 225102 CrossRef.
- M. W. Gaultois, T. D. Sparks, C. K. H. Borg, R. Seshadri, W. D. Bonificio and D. R. Clarke, Chem. Mater., 2013, 25, 2911–2920 CrossRef CAS.
- A. A. Emery and C. Wolverton, Sci. Data, 2017, 4, 170153 CrossRef CAS PubMed.
- I. Levin, NIST Inorganic Crystal Structure Database (ICSD), Type: dataset, 2018, DOI:10.18434/M32147, https://data.nist.gov/od/id/mds2-2147.
- C. R. Groom, I. J. Bruno, M. P. Lightfoot and S. C. Ward, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2016, 72, 171–179 CrossRef CAS PubMed.
- P. Villars, K. Cenzual, et al., Pearson's Crystal Database, 2007, https://www.crystalimpact.com/pcd/, accessed September 2023 Search PubMed.
- E. Blokhin and P. Villars, Handbook of Materials Modeling: Methods: Theory and Modeling, 2020, pp. 1837–1861 Search PubMed.
- C. W. Andersen, R. Armiento, E. Blokhin, G. J. Conduit, S. Dwaraknath, M. L. Evans, Á Fekete, A. Gopakumar, S. Gražulis, A. Merkys, F. Mohamed, C. Oses, G. Pizzi, G.-M. Rignanese, M. Scheidgen, L. Talirz, C. Toher, D. Winston, R. Aversa, K. Choudhary, P. Colinet, S. Curtarolo, D. Di Stefano, C. Draxl, S. Er, M. Esters, M. Fornari, M. Giantomassi, M. Govoni, G. Hautier, V. Hegde, M. K. Horton, P. Huck, G. Huhs, J. Hummelshø, A. Kariryaa, B. Kozinsky, S. Kumbhar, M. Liu, N. Marzari, A. J. Morris, A. A. Mostofi, K. A. Persson, G. Petretto, T. Purcell, F. Ricci, F. Rose, M. Scheffler, D. Speckhard, M. Uhrin, A. Vaitkus, P. Villars, D. Waroquiers, C. Wolverton, M. Wu and X. Yang, Sci. Data, 2021, 8, 217 CrossRef.
- L. Ward, A. Dunn, A. Faghaninia, N. E. Zimmermann, S. Bajaj, Q. Wang, J. Montoya, J. Chen, K. Bystrom and M. Dylla, et al. , Comput. Mater. Sci., 2018, 152, 60–69 CrossRef.
- H. Glawe, A. Sanna, E. Gross and M. A. Marques, New J. Phys., 2016, 18, 093011 CrossRef.
- C. J. Hargreaves, ElM2D compositional plotting library, https://github.com/lrcfmd/ElM2D, 2022 Search PubMed.
- L. McInnes, J. Healy, N. Saul and L. Großberger, J. Open Source Softw., 2018, 3, 861 CrossRef.
- Plotly, The interactive graphing library for Python (includes Plotly Express), https://github.com/plotly/plotly.py, 2023 Search PubMed.
- R. Pétuya, S. Durdy, D. Antypov, M. W. Gaultois, N. G. Berry, G. R. Darling, A. P. Katsoulidis, M. S. Dyer and M. J. Rosseinsky, Angew. Chem., Int. Ed., 2022, 61, e202114573 CrossRef.
- S. Durdy and C. J. Hargreaves, LMDS MOF porosity prediction tool, 2022, https://github.com/lrcfmd/LMDS_MOF_Porosity_Tool Search PubMed.
- F. A. L. Laskowski, D. B. McHaffie and K. A. See, Identification of Potential Solid-State Li-Ion Conductors with Semi-Supervised Learning, 2022, https://resolver.caltech.edu/CaltechAUTHORS:20220711-653076000 Search PubMed.
- A. D. Sendek, G. Cheon, M. Pasta and E. J. Reed, J. Phys. Chem. C, 2020, 124, 8067–8079 CrossRef CAS.
- E. D. Cubuk, A. D. Sendek and E. J. Reed, J. Chem. Phys., 2019, 150, 214701 CrossRef PubMed.
- A. Y. T. Wang, S. K. Kauwe, R. J. Murdock and T. D. Sparks, npj Comput. Mater., 2021, 7, 1–10 CrossRef.
- C. M. Collins, L. M. Daniels, Q. Gibson, M. W. Gaultois, M. Moran, R. Feetham, M. J. Pitcher, M. S. Dyer, C. Delacotte, M. Zanella, C. A. Murray, G. Glodan, O. Pérez, D. Pelloquin, T. D. Manning, J. Alaria, G. R. Darling, J. B. Claridge and M. J. Rosseinsky, Angew. Chem., Int. Ed., 2021, 60, 16457–16465 CrossRef CAS PubMed.
- P. Gorai, D. Gao, B. Ortiz, S. Miller, S. A. Barnett, T. Mason, Q. Lv, V. Stevanović and E. S. Toberer, Comput. Mater. Sci., 2016, 112, 368–376 CrossRef.
- P. Debye, Ann. Phys., 1912, 344, 789–839 CrossRef.
- A. Einstein, Ann. Phys., 1907, 327, 180–190 CrossRef.
- S. Durdy, LMDS Server setup tools, https://github.com/lrcfmd/LMDS_helper_scripts, 2022 Search PubMed.
- D. Merkel, Linux J., 2014, 2014, 2 Search PubMed.
- Kubernetes Manual, 2017, https://kubernetes.io/docs/concepts/overview/what-is-kubernetes/, accessed 19-Aug-2022 Search PubMed.
- J. Watada, A. Roy, R. Kadikar, H. Pham and B. Xu, Emerging Trends, Techniques and Open Issues of Containerization: A Review, 2019 Search PubMed.
- Z. Li, M. Kihl, Q. Lu and J. A. Andersson, in Performance overhead comparison between hypervisor and container based virtualization, Institute of Electrical and Electronics Engineers Inc., 2017, pp. 955–962 Search PubMed.
- W. Reese, Linux J., 2008, 2008, 1 Search PubMed.
- R. T. Fielding, M. Nottingham and J. Reschke, HTTP Semantics, RFC 9110, 2022, https://www.rfc-editor.org/info/rfc9110 Search PubMed.
- Web Server Market Share, https://w3techs.com/technologies/history_overview/web_server/ms/y, accessed: 2022-12-22 Search PubMed.
- Netcraft, Web Server Survey, 2022, https://news.netcraft.com/archives/category/web-server-survey/, accessed: 2022-12-22 Search PubMed.
- D. Kunda, S. Chihana and M. Sinyinda, Computer Engineering and Intelligent Systems, 2017, 8, 43–52 Search PubMed.
- S. Durdy, C. J. Hargreaves and J. A. Newnham, LMDS heat capacity modeling tool, 2022, https://github.com/lrcfmd/LMDS_heat_capacity_modelling Search PubMed.
- P. P. Angelov, E. A. Soares, R. Jiang, N. I. Arnold and P. M. Atkinson, Wiley Interdiscip. Rev. Data Min. Knowl. Discov., 2021, 11, 5 Search PubMed.
- P. Linardatos, V. Papastefanopoulos and S. Kotsiantis, Entropy, 2021, 23, 18 CrossRef PubMed.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot and E. Duchesnay, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- H. Moriwaki, Y. S. Tian, N. Kawashita and T. Takagi, J. Cheminf., 2018, 10, 4 Search PubMed.
- RDKit Development Team, RDKit: Open-source cheminformatics, https://www.rdkit.org, accessed February 20, 2023 Search PubMed.
- P. Virtanen, R. Gommers, T. E. Oliphant, M. Haberland, T. Reddy, D. Cournapeau, E. Burovski, P. Peterson, W. Weckesser, J. Bright, S. J. van der Walt, M. Brett, J. Wilson, K. J. Millman, N. Mayorov, A. R. J. Nelson, E. Jones, R. Kern, E. Larson, C. J. Carey, İ. Polat, Y. Feng, E. W. Moore, J. VanderPlas, D. Laxalde, J. Perktold, R. Cimrman, I. Henriksen, E. A. Quintero, C. R. Harris, A. M. Archibald, A. H. Ribeiro, F. Pedregosa, P. van Mulbregt and SciPy 1.0 Contributors, Nat. Methods, 2020, 17, 261–272 CrossRef CAS.
- S. Durdy, Example Flask App Tutorial, https://github.com/lrcfmd/LMDS_example, 2023 Search PubMed.
- B. Walters, Linux J., 1999, 1999, 6 Search PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |