Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Unveiling the synthesis patterns of nanomaterials: a text mining and meta-analysis approach with ZIF-8 as a case study†
Joseph R. H.
Manning
 * and
Lev
Sarkisov
* and
Lev
Sarkisov
 *
*
Department of Chemical Engineering, University of Manchester, M13 9PL, UK. E-mail: Joseph.Manning@manchester.ac.uk; Lev.Sarkisov@manchester.ac.uk
First published on 27th September 2023
Abstract
With the continuously growing number of scientific articles on the synthesis of nanomaterials, it becomes impossible for researchers to grasp and comprehend the landscape of synthetic protocols available for a particular material. The aim of this study is to explore the feasibility of extracting the collective knowledge on the synthesis of a particular material accumulated over the years from the published corpus of articles and organizing it in a systematic manner. Accordingly, we developed methods to perform detailed text mining on a single nanomaterial target for the purposes of methodology optimisation. Taking the common material ZIF-8 as a case study, we analysed 1600 synthesis protocols to identify trends in parameters, such as reagents, concentrations, and reaction time/temperature. We used this information to find the distribution of synthesis parameters and their relationships to one another, identifying the limits of common reaction parameters and revealing subtle details, such as insolubility of metal acetate reagents in alcoholic solvents, or the occurrence of amorphous oxides at low stoichiometric ratios. We then clustered similar synthesis protocols together, using their relative popularity to identify promising regions of the synthesis phase space for optimisation, reducing the need for brute force synthesis optimisation. The techniques developed here are a general tool accelerating the synthesis development of a wide range of nanomaterials by aggregating existing research trends, averting the need for laborious manual comparison of existing synthesis protocols or repetition of previously-developed techniques.
Introduction
The number of chemical syntheses reported is large and growing exponentially.1 While naturally indicative of greater scientific progress, this leads to two significant challenges. Firstly, researchers are confronted with the growing difficulty of maintaining a comprehensive overview and understanding of the diverse landscape of synthesis routes and conditions accessible for a particular group of compounds. Secondly, although the repository of published synthesis data contains an immense wealth of information, its potential for systematic development of new synthesis methods remains largely untapped and underutilized. In response to this, various informatics approaches have been adopted to standardise the data produced during chemical research. For example, the creation of chemical synthesis ontologies2–4 and automated reactionware5,6 has enabled new procedures to be directly compared against previously-published data or shared openly through chemical “programming languages”.7,8 However, the nature of reporting synthesis methods – as unformatted prose in a written report – has remained largely unchanged.As a result, most new publications and the entire body of prior chemical synthesis reports remains unlabelled, with the potential for far broader data mining and informatics research if these reports could be standardised. Accordingly, with the advent of text mining methods and natural language processing (NLP),9 software has been developed to interpret chemical details from the plain text within chemistry publications10,11 including compound structure,12 reaction stoichiometry,13 and performance.14 Using these tools, large databases of organic14,15 and inorganic16–19 chemicals and reactions have been developed and used for novel materials discovery. For example, Cole and co-workers created a database of organic dyes to identify ideal mixtures for broad-spectrum light absorption in dye-sensitized solar cells, regardless of the intension of the original studies.15 Similar strategies have been used by Olivetti and co-workers to analyse how synthesis gel composition and organic structure directing agent can dictate crystal polymorphs for a range of zeolite syntheses.16
One weakness of these text mining approaches is their reliance on unambiguous identification of the chemical entities in question, using named-entity recognition (NER)9,20 and the programmatic naming conventions defined by IUPAC21 to succeed. In the absence of such well-accepted naming schemes – as is the case for a variety of emerging nanomaterial families like porous silicas, polymers of intrinsic microporosity, and covalent organic framework materials – large scale data mining becomes far less practical. An excellent example of this is metal–organic framework (MOF) materials – infinite condensation polymers of various organic ligands and metal ions or clusters. There are millions of possible MOFs,22–25 and hundreds of thousands of frameworks already synthesized,26–29 necessitating data-driven approaches to accelerate progress in the field. However, unambiguous naming conventions for MOFs have yet to be fully adopted,30 frustrating text-mining of the primary publications themselves. Instead, informatics methods have largely been driven by the creation of a subset of the Cambridge Structural Database (CSD)31 focused on MOF materials,28 as these resources allow researchers to analyse the full range of experimentally known MOF structures, identifying the best experimentally-realised materials for future research and development.
To accelerate development of experimental procedures to make MOFs, however, data-mining approaches must look beyond structure into the synthesis protocols – unique sets of synthesis parameters varying from one another in any way – used to make them. By understanding the relationships between protocol and eventual material, new synthesis methods can be digitally generated, obviating the need for arduous trial-and-error or intuition-based approaches.6 To this end, large-scale post hoc analyses of experimental MOF synthesis protocols have recently been developed.32,33 These studies apply NLP to the underlying publications in the CSD MOF subset to interpret their synthesis protocols, identifying such details as solvents used, specific reagents, solvents, and reaction parameters. As a result, broad descriptive statistics about the synthesis strategies to produce MOFs have been developed,33 and even predictive models to suggest synthesis parameters for novel MOF materials when given a hypothetical structure.32
While these approaches give an excellent overview of the field of MOFs in general, they are vulnerable to bias in the papers submitting to the CSD. As the database focuses of chemical structure rather than synthesis protocols, only 1–2 synthesis examples of each framework are included. Further, the synthesis protocols are generally submitted from initial studies reporting the discovery of a material, rather than exploring the full range of potential approaches to a single target, meaning that only a very vague understanding of any individual MOF can be generated with this approach. For example, while candidate solvents and reaction parameters can be suggested, other salient parameters such as reagent ratios, product isolation methods, and alternative synthesis strategies (e.g. hydrothermal or mechanochemical versus solvent crystallisation) cannot. Deeper insight into individual MOFs and the peculiarities of their synthesis protocols can be gained through targeted meta-analysis of studies focusing on that particular material,34 enabling regression of product properties like defect density against synthesis details. However, challenges of manually comparing synthesis protocols against one another severely limit the scale of such meta-analyses, preventing their widespread use.
To address these issues, in this article we pose the following questions: can we leverage previously-developed chemistry text mining tools to analyse the synthesis protocols for a single target nanomaterial? If so, can we develop methods to process the extracted information on a uniform basis, enabling like-for-like comparison regardless of original format? Finally, can we harness this information to accelerate synthesis refinement of the material e.g. by generating proposed synthesis conditions correlated to high material quality and yield?
As a case study, we consider ZIF-8, a commonly synthesized MOF material which has been extensively studied within the literature. ZIF-8 is constructed from a combination of zinc ions and 2-methylimidazole in the sodalite topology, held together with metal–amine bonds rather than the more common metal–carboxylate bonds, thus rendering the material both hydrophobic and water-stable.35,36 Accordingly, ZIF-8 has garnered significant interest in the literature for applications including gas storage and separation, adsorptive refrigeration,37 biomolecule encapsulation,38 catalysis,39 and sensing.40 Further, ZIF-8 can be synthesized from a number of strategies – for example using protic or aprotic solvents,41 a range of temperatures,42 reagent concentrations,43 modulators and crystal growth modifiers,44 and acid/base conditions.45 In sum, over 7500 papers have been published regarding ZIF-8 to date. Given the breadth of synthesis protocols established for ZIF-8, it practically impossible to manually compare all possible synthesis methodologies to one another. Applying text mining methods to automatically and quantitively analyse ZIF-8 synthesis protocols would enable larger-scale analysis and the identification of promising synthesis strategies.
In this study we developed methods to extract and aggregate synthesis protocols in a uniform format. We studied 1600 synthesis protocols of ZIF-8 and related materials from 3197 original articles, performing an automated meta-analysis of the synthesis methods contained. We analysed the chemical identities used alongside quantities and reaction conditions to provide a systematic design space for ZIF-8, identifying key trends in the approaches used. Finally, we group similar synthesis protocols together with unsupervised clustering techniques, identifying hidden patterns in the data.
Methodology
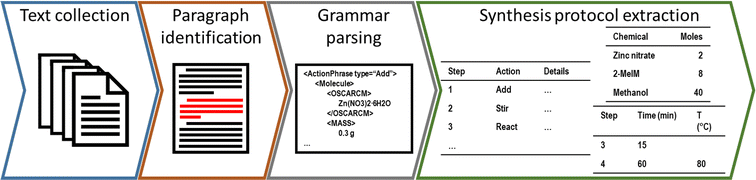
The workflow of extracting and analysing synthesis protocols was split into four overarching steps: text collection, where a corpus of research papers is identified and downloaded; paragraph identification, where raw synthesis protocols are identified within the prose; grammar parsing, where the natural language is converted into hierarchical data for later interpretation; and synthesis protocol extraction, where the extracted data is standardised to produce a structured “recipe” for each synthesis protocol. Key steps in the workflow are depicted in Fig. 1. The first three steps have been widely described elsewhere, and only a brief description is provided in this section (with associated code provided by the authors on GitHub at https://github.com/SarkisovTeam/SynOracle-preprocessing). The final stage of the workflow was developed in this study using python 3.9,46 and is made freely available by the authors on GitHub at https://github.com/SarkisovTeam/SyntheticOracle.Text collection, paragraph identification, and grammar parsing
To produce a corpus of ZIF-8 synthesis protocols, we initially followed established methods to download collections of papers and identify synthesis protocols within them.32,33 Synthesis papers were identified by searching the SCOPUS database using Elsevier's elsapy software (https://github.com/ElsevierDev/elsapy). Papers were identified using the search term “ZIF OR zeolitic imidazole* AND synthesis”, returning 4198 results published between June 2010 and April 2022. These were then categorised by publisher, from which the three largest groups were targeted for downloading (ACS, RSC, and Elsevier), reducing the total corpus to 3179 papers. XML or HTML versions of each paper were then downloaded according to their publisher's specifications – using elsapy in the case of Elsevier, web scraping in the case of the RSC, and through the text and data mining service at the ACS.Once downloaded, synthesis paragraphs were identified using ChemDataExtractor2.1 (ref. 10) according to previously developed protocols for identifying MOF synthesis methods.32,33 In this procedure, chemical named entity recognition was performed using BERT47 to identify potential reagents, and part-of-speech (POS) tagging was carried out on the remaining tokens to interpret sentence grammar. Chemical quantities were identified from the POS tags as CD-NN bigrams (phrases consisting of a cardinal number followed by a noun), and regex matching of the noun against a library of SI units. Synthesis paragraphs were identified as containing three or more chemical named entities and three or more chemical quantities, after which each paragraph was extracted as plain text for manual confirmation and later analysis.
Once confirmed that each extracted paragraph contained a synthesis procedure, hierarchical grammar parsing was performed in the ChemicalTagger software11 to associate chemical named entities with quantities and specific synthesis actions (termed ActionPhrases). These were stored as nested tags within an XML document. No further analysis was used to compensate for incorrect or missing values in the original text (e.g. unreported drying temperatures).
Synthesis protocol extraction
To interpret and compare synthesis protocols against one another, data about synthesis steps, conditions, and chemicals involved had to be converted from nested XML data into useful information using the software developed in this study. To perform this, XML data extracted from ChemicalTagger was recursively parsed into strings within a pandas48,49 DataFrame object such that each row consisted of a single ActionPhrase, its associated time and temperature, and details of any chemical entity involved.Chemical identities were first confirmed by cross-referencing identified chemical names against the PubChem database50 using the pubchempy python library (https://pubchempy.readthedocs.io/en/latest/index.html). From this, a unique identifier for each individual chemical was generated, enabling extraction of key information about each chemical and summation of identical chemicals together. To prevent semantically identical reagents from being considered separately (e.g. zinc nitrate and their hydrates), PubChem identifiers were supplemented with structural information gathered from the cheminformatics tool RDkit.51 Specifically, chemicals whose formulae contained the elements zinc or cobalt, as well as the nitrate, acetate, sulfate, and imidazole substructures were separately identified.
Then, numerical quantities associated with each chemical were calculated. To do this, chemical quantities were categorised by type from the structured XML output of ChemicalTagger (e.g. by volume, moles, mass etc.), and parsed into physically meaningful units with the pint python library (https://pint.readthedocs.io/en/0.20.1/index.html). To prevent double-counting in situations where two units were mentioned, e.g. by the common phrase “5 g of [reagent] (0.8 mmol)”, only a single unit type was considered for each chemical entity according to the priority list (moles > mass > volume). These units were then converted into moles using the molecular mass identified from the PubChem identity. In the case of converting volume to moles, densities were estimated from the ChEDL database of critical point properties52 using the COSTALD method.53 Once chemical identities and quantities had been fully converted, these were aggregated into a single bill of materials for each synthesis (visualised in Table 1). Conditions (i.e. time and temperature values) were similarly parsed from strings into meaningful units using the pint python library, and stored as minutes and degrees kelvin, respectively.
| PubChem identifier | Chemical name | Original quantities | Amount (millimol) |
|---|---|---|---|
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 749 749 |
2-Methylimidazole | 0.24 g, 3.4 mmol | 3.4 |
15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 865 865![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 313 313 |
Zn(NO3)2·6H2O | 0.956 g, 3.2 mmol | 3.2 |
| 6212 | Chloroform | 40 mL | 500 |
| 6228 | DMF | 70 mL | 1210 |
Finally, to reduce the effect of original authors' writing styles on the interpretation of synthesis sequences, synthesis actions were condensed into a smaller vocabulary than originally defined by ChemicalTagger using a similar technique to the recently developed ULSA for inorganic nanomaterials syntheses.55 Synthesis labels from ChemicalTagger were categorised as either being related to the set up stage of the synthesis (labelled “addition”), the synthesis itself (labelled “reaction”), or reaction workup (labelled “extraction”), as described in Table 2. Some synthesis actions could reasonably occur during any of those reaction stages, e.g. changing the temperature, therefore a fourth “other” category for these ambiguous actions was also defined. A fifth category, “start”, was used to signify opening statements of synthesis protocols (e.g. “ZIF-8 was produced by our previously published method”), which would otherwise be miscategorised as an “extraction” or “other” action. “Start” actions were then excluded from further analysis.
| Action type | ActionPhrase |
|---|---|
| “Addition” | Add, dissolve, stir |
| “Reaction” | ApparatusAction, synthesize, wait |
| “Extraction” | Degass, dry, extract, filter, partition, precipitate, purify, quench, recover, remove, yield |
| “Other” | Concentrate, cool, heat |
Grouping similar synthesis protocols together
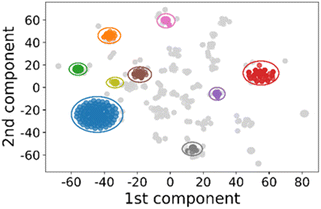
To group synthesis protocols together, we related individual syntheses to one another by the identity of the reagents used only. To calculate the mathematical relationship between different synthesis protocols the list of chemicals was first vectorised, creating a numerical representation of the chemical combination used in each synthesis. Briefly, an M × N matrix was created, where M is the number of synthesis protocols, and N is the number of unique chemicals present across all of synthesis protocols studied. To reduce noise in the data, only synthesis protocols containing 2-methylimidazole were considered, and metal sources were grouped by chemical substructures as described previously. In total, 139 unique chemicals were identified across 1134 synthesis protocols.For each synthesis protocol, a vector was generated using the term frequency–inverse document frequency algorithm (TF-IDF), a commonly used text mining method to estimate the importance of words in a group of documents.56 The TF-IDF algorithm weights the frequency of a word used in each document against its frequency across the group of documents – words present in many documents are given a low weight, while words occurring in only rarely are given a high weight. This is shown in eqn (1), which calculates the weight of word t in the individual document d as part of the group of documents D (containing n total documents), where f is the frequency the word occurs. As in this study the “words” are chemical names, common chemicals like methanol are afforded a low weight, while rarer chemicals like CTAB are afforded a relatively higher weight.
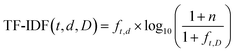 | (1) |
Once the chemical identities had been vectorised, similarity was calculated by the DBSCAN clustering method.57 DBSCAN calculates the local density of data points in Euclidean space (synthesis protocols in the case of this study), defined as the number of neighbours closer than a threshold distance from each data point. Clusters are identified as disconnected regions containing a high density of data points, while isolated data points with no connection to a larger cluster as identified as noise.
To visualise the results of the clustering analysis, the high dimensional data were projected into two dimensions using the t-distributed stochastic neighbour embedding (t-SNE) method.58 To do this the algorithm calculates the distances between each datapoint in high dimensional space, and estimates low-dimensional coordinates for each datapoints which preserves the distance between each point and its neighbours.
Results and discussion
Validation against manually-extracted information
To perform a quantitative meta-analysis of ZIF-8 synthesis, we first demonstrate the validity of the information extracted by comparing the performance of our text mining approach against a manually identified “ground truth” from a small number of papers sourced from the NIST database of emerging adsorbent materials. Using this database served two purposes: it was sufficiently small to provide a tractable number of articles for high-fidelity analysis, and each synthesis report was confirmed to contain ZIF-8 by the isotherm data provided. Overall, 44 publications describing ZIF-8 synthesis were identified, of which full information could be extracted for 43. The manuscripts were downloaded from their publisher, synthesis paragraphs manually identified, and synthesis information extracted both manually and using our software. In all cases, data reported within the paper and manually collated were considered as the ground truth. Full information on the specific publications and paragraphs identified for validation are provided in the ESI.†From these paragraphs, three key parameters were extracted: a sequence of synthesis actions taken, a table of constituent chemicals, and the reaction conditions (i.e. temperatures and quoted times). For each parameter, the F1-score was calculated providing a numeric score for each text mining task compared against the manually-extracted ground truth. Extracted chemical identities were cross-referenced against the PubChem database of compounds to act as both a unique identifier and source of key information about each species. Finally, physical quantities – the values of time, temperature, and chemical quantity – were converted from plain text to numerical units using the pint python library and compared against their manually extracted counterparts. These data are summarised in Table 3, with further details provided in the ESI.†
| Metric | Precision | Recall | F 1-score | Matching quantities |
|---|---|---|---|---|
| Synthesis actions | 59 | 77 | 66 | — |
| Aggregated actions | 83 | 89 | 84 | — |
| Reagent identification | 82 | 96 | 87 | 82 |
| Temperature parsing | 76 | 83 | 77 | 74 |
| Time parsing | 72 | 74 | 72 | 69 |
Identification of individual synthesis actions performed similarly to the original ChemicalTagger benchmarking, with an F1-score of 66% cf. 55–63% agreement in the original study.11 We believe this is due to the relatively large vocabulary of synthesis actions which led to sensitivity during human labelling due to the resultant ambiguity; for example, introduction of reagents at the start of the reaction could reasonably be assigned the “add” or “dissolve” action labels due to their semantic similarities. This conclusion was supported when ChemicalTagger's performance was compared against ChatGPT (Table S4 and Fig. S2†), which had an almost ideal F1-score of 99%. When synthesis actions were converted to their conceptual types and aggregated, the F1-score between manual identification and ChemicalTagger increased significantly to over 80% indicating that all synthesis stages were identified even if the specific ActionPhrases themselves were not. Therefore, we conclude that the text mining captures the essence of the synthesis protocol, but is unable to fully summarise the semantics of synthesis due to “linguistic noise” i.e. variability between different authors writing styles.
In terms of synthesis parameters, F1-scores and quantity matching were between 60 and 80% in all cases. These range of scores are slightly lower than previous text-mining efforts, which generally score between 60 and 98%.1,60 We ascribe this relatively low score to more stringent criteria used in this study: as we define true positive to be the successful identification of a PubChem database entry, precision is lowered when cross-referencing fails. This is further exacerbated by the presence of typographical errors and colloquial chemical names which are not recognised by an automated PubChem database search (e.g. 2-methylinidazole or 2-MeIM, rather than 2-methylimidazole). Failure to successfully convert numerical quantities similarly reduced the F1-score during time and temperature parsing.
In sum, while individual synthesis features could be reliably extracted using the methods developed here, it is currently impossible to reliably reproduce the entirety of any specific synthesis protocol. To achieve such high-fidelity reproduction, methods would have to be developed to estimate the completeness of a synthesis protocol, requiring a much larger set of manually-labelled synthesis sequences, similar to that developed by Wang et al. for individual synthesis actions.55 Efforts to create such a dataset are ongoing in our research group. Instead, further analysis in this study is performed by compiling a group of similar synthesis protocols to extract a representative aggregate of synthesis details, hence enabling quantitative meta-analysis.
Interpreting ZIF-8 synthesis strategies
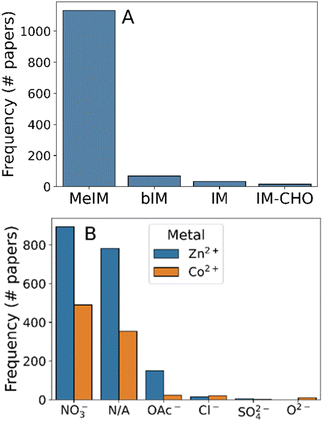
Given the effectiveness of our text mining methods to extract synthesis information from text, we progressed to a larger dataset of 3179 experimental synthesis reports of ZIF-8. From this dataset we processed 1600 synthesis protocols, enabling strong statistical analysis of the synthesis options which have been explored.We first analysed the reagent compounds used during synthesis, which should consist of 2-methylimidazole and Zn salts only. As can be seen in Fig. 2, this is not the case: while methylimidazole was by far the most common linker molecule mentioned (Fig. 2A), 34% of the synthesis protocols mentioned cobalt salts. In fact, 32% of the synthesis protocols omitted zinc entirely, indicating that these were synthesis protocols of ZIF-67 instead – the cobalt equivalent of ZIF-8. The remaining cobalt-mentioning synthesis protocols also contained zinc, indicating that they may be mixed-metal systems. This ambiguity highlights some of the key nomenclature issues with MOF materials – ZIF-8 and -67 are practically the same material in terms of synthesis protocol but this proximity is not reflected in the common name. The use of unambiguous naming algorithms such as MOFid30 can avoid this linguistic ambiguity, even accurately describing the continuous transition between the two frameworks.
To further analyse the reagents used we grouped the metal salts used by anion type (Fig. 2B), assuming that there was no consequence of using anhydrous versus hydrated salts. Nitrate was the most commonly used counterion, being present in 75% of syntheses. Ambiguous mentions of zinc and cobalt compounds were present in 17.2% of the 1600 protocols, encompassing minor zinc salts (e.g. Zn(OH)2 in the case of ref. 61), indirect reference to zinc precursors in synthesis (e.g. “the sample obtained with Zn”62), or mis-identified zinc compounds due to word tokenisation errors (e.g. “firstly, 645 mg (2.469 mmol) of Zn (NO3)2·4H2O was dissolved”,63 where the space character between “Zn” and its counterions causes incorrect chemical parsing). Aside from nitrates and ambiguous mentions, the only other commonly-mentioned metal salt was zinc acetate (present in 11.5% of synthesis protocols). The presence of chloride, acetate, and oxide precursors indicate that the synthesis is compatible to a range of electrolyte environments, agreeing with experimental reports which have shown that counterion choice significantly alters crystal nucleation and growth rates.64,65 Despite the utility of these other salts, the overwhelming popularity of nitrate counterions found during our analysis indicates that other factors e.g. cost may have been prohibitive to their widespread adoption.
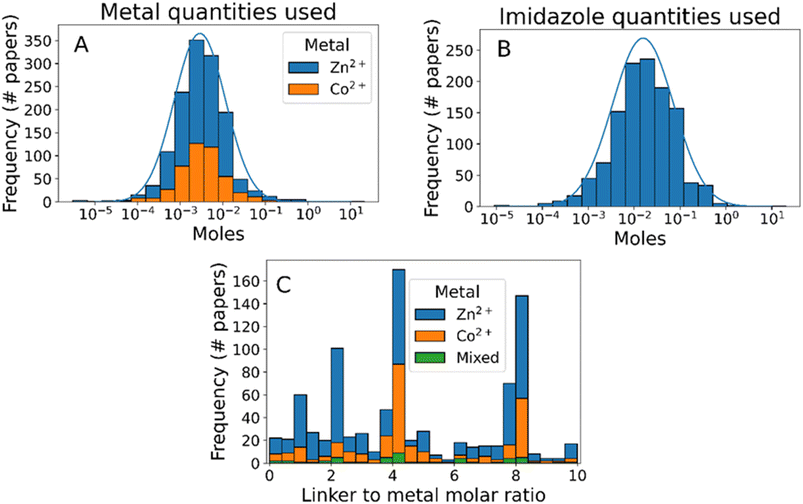
In addition to reagent identity, our text mining method provides information about the quantity of each reagent used, enabling analysis of synthesis protocol scale and reaction stoichiometry (Fig. 3). The scale of ZIF-8 synthesis follows approximately a log-normal distribution, with 95% of synthesis using 0.18–46 millimol of metal ions and 0.73–330 millimol of 2-methylimidazole (Fig. 3A and B, respectively), demonstrating the flexibility of ZIF-8 synthesis with respect to scale. In terms of reaction stoichiometry, most synthesis protocols use an excess of linkers compared to the stoichiometric ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 3C). This excess has been shown to control particle sizes by slowing the rate of crystal growth,38,66–68 although few synthesis protocols use a higher ratio than 8
1 (Fig. 3C). This excess has been shown to control particle sizes by slowing the rate of crystal growth,38,66–68 although few synthesis protocols use a higher ratio than 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Interestingly, despite clear evidence that excess concentration of metal ions forms undesired by-products such as Zn(OH)(NO3)(H2O),43,68–70 6% of the synthesis protocols analysed used a molar ratio of 1
1. Interestingly, despite clear evidence that excess concentration of metal ions forms undesired by-products such as Zn(OH)(NO3)(H2O),43,68–70 6% of the synthesis protocols analysed used a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 or lower.
1 or lower.
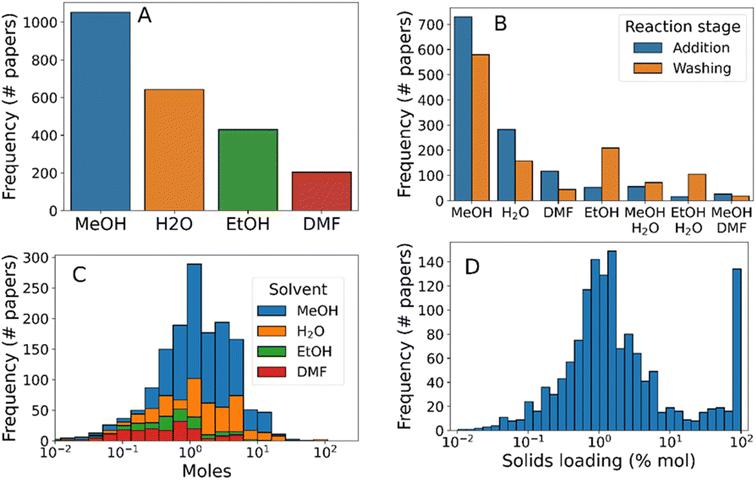
After considering reagents, the next most import aspect of a synthesis protocol lies in the choice of solvent environment for the reaction. Solvent choice has ramifications on the reaction mixture dielectric constant, in turn dictating factors such as reagent solubility and reaction kinetics. Further, the choice between protic and aprotic solvents, can accelerate reaction mechanisms relying on proton transfer, such as the linker deprotonation present during ZIF-8 synthesis.66 Finally, overall reaction concentration is critical for determining whether the reaction mixture will act as an ideal solution, and in terms of the relative mass efficiency of the synthesis, both of which have consequences in terms of synthesis protocol viability in terms of scaleup to process-level manufacture.
The vast majority of synthesis protocols studied here contain one of methanol, ethanol, water, and DMF. Methanol was by far the most frequently mentioned solvent, present in 66% of synthesis protocols (Fig. 4A), followed by water (40% of synthesis protocols), ethanol (27%), and finally DMF (12%). Less frequently used solvents included chloroform (1.4%), toluene (1.0%), and ethylene glycol (0.88%). To analyse the usage of each solvent present, we separated them by “synthesis” and “workup” procedure steps, as well as incorporating binary solvent mixtures (Fig. 4B). This analysis revealed that, while ethanol was the third most prevalent solvent overall, it was the second most common solvent used for washing and purification (and the fifth most common reaction solvent). Mixed solvent systems, primarily methanol–water, were present in 8% of syntheses presumably to tune the reaction dielectric and proton transfer catalysis rate.71
The distribution of solvent quantities used within the syntheses studied (Fig. 4C) showed that each solvent followed approximately lognormal distributions. Both DMF and ethanol were used in smaller quantities than methanol or water (means of 0.4, 0.6, 1.4, and 1.6 mol per synthesis, respectively), indicating that the latter two solvents were more appropriate for scaling up the synthesis. Finally, we analysed the total solids concentration of synthesis protocols by dividing total reagent amounts by the solvent amounts used (Fig. 4D). As with individual reagent concentrations, the total solids concentration followed an approximately log-normal distribution between 0.1 and 10% mol. Separately, 7.7% of synthesis protocols had a solids loading of approximately 100% mol – signifying mechanochemical synthesis protocols. Although mechanochemistry is a promising synthesis route due to its high yields72 and low environmental impact73 compared to conventional solvent synthesis methods, the relatively low popularity may be explained due to practical difficulties of mechanochemical synthesis e.g. prevention of hot-spot formation in the reaction vessel.74
In addition to reagents and solvents, ancillary chemicals such as surfactants, pH modifiers, and modulators are often key to ensure the success of MOF syntheses as well as dictating secondary particle characteristics such as size and crystal form. Three chemical types were prevalent within the synthesis protocols studied: acids, bases, and surfactant compounds. Unlike solvents and reagents, no individual ancillary chemical was identified in more than 3.5% of synthesis protocols (Fig. 5). However, bases were present in 18% of all the synthesis protocols analysed, carrying out the important role of deprotonating the linker molecule in the reaction mixture. From the variety of distinct molecules used for this role, it appears that no molecular recognition occurs, simply pH control. Despite the requirement for methylimidazole deprotonation for the reaction to progress, acids were detected in 6.3% of syntheses, however from inspection of the individual synthesis protocols acids only appeared during post-synthetic modification of the ZIF-8 materials e.g. after carbonisation75 or impregnation into silicas.76 Finally, surfactants like cetyltrimethylammonium bromide (CTAB) or sodium dodecylsulfate (SDS) were present in 4.6% of synthesis protocols, being used to slow the growth of individual ZIF-8 crystals and therefore control the particle shape.59,77
 | ||
| Fig. 5 Histograms of ancillary chemical prevalence in ZIF-8 synthesis. (A) Acids, (B) bases, and (C) surfactants. | ||
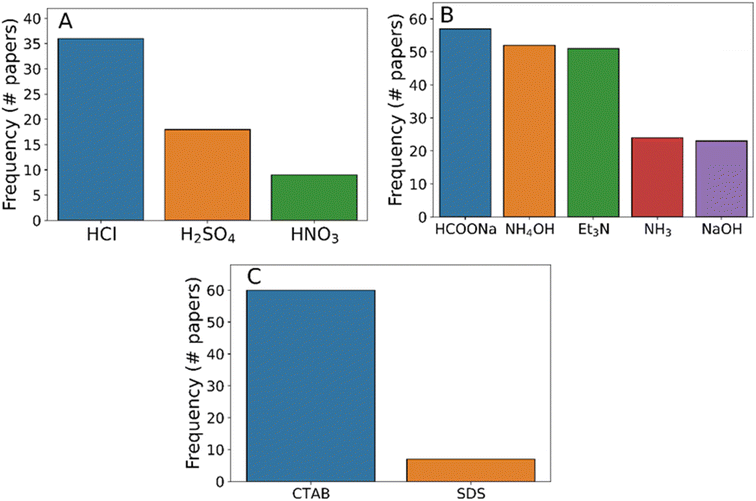
While it is possible to identify broad differences in synthesis strategy from feedstock compounds alone, it is impossible to understand why one chemical is chosen over another without further detail about the synthesis protocol being described. For example, the modulator sodium formate has been shown to perform different roles in room-temperature syntheses compared to hydrothermal alternatives.44,78 In the first instance, we also consider the conditions (i.e. time and temperature) during the process. These are shown in Fig. 6, demonstrating that the majority of protocols have synthesis times under six hours. Even after disregarding protocols with a reported synthesis time of 0 minutes as being spurious, it is clear that synthesis can be completed very quickly. In terms of synthesis temperature, the majority of the extracted temperatures were found to be room temperature indicating that thermal driving forces were not necessary for the formation of ZIF-8. This is further corroborated by the relative lack of procedures mentioning heated reaction conditions compared to heated drying conditions (Fig. 6B).
 | ||
| Fig. 6 Histograms of conditions during ZIF-8 synthesis processes. (A) Total time elapsed and (B) temperatures used during synthesis. Annotational on (B) indicate the boiling points of the four most common solvents identified. Data are broken down by reaction step type as defined in Table 2. Where multiple variables are plotted, data bars are stacked on top of one another. | ||
Overall, the tools developed in this study provide wide-ranging descriptive statistics of various ZIF-8 synthesis routes. The data generated are an excellent addition to existing literature review methods, facilitating the interpretation of different synthesis aspects e.g. reagent choices, stoichiometric ratios and reaction conditions. From these data we are able to identify gaps in the existing literature or synthesis conditions most likely to succeed, as well as providing useful input data for later technoeconomic analysis.
Harnessing synthesis information for accelerated methodology development
While the analysis performed is useful as a means of understanding the ZIF-8 reaction system, a key aim of this study was to systematize the collective synthesis knowledge for the material, thereby connecting synthesis protocols to some key performance indicators either of the synthesis (e.g. yield) or material (e.g. crystal form, surface area). One crucial barrier to this goal was the correlation of material performance data with synthesis protocol information: research papers are inconsistent in reporting of material properties (primarily as different quality metrics are used depending on the motivation of the original research), and the sample naming conventions used within research articles prevent unambiguous linking between the described protocols and materials produced. For example, while a synthesis paragraph might detail the synthesis of “nano-sized ZIF-8”, later mentions in the text may be labelled differently e.g. “ZIF-8nano”,79 confounding attempts for automated identification of reaction products using regular expressions.33 While this issue will undoubtably be resolved by the adoption of transformer-based language models such as BERT80 and GPT-4,81 such models became available only recently and the scientific community,82 including our group, is in the process of probing their extension to scientific data mining. In fact, the current study highlighted a number of issues with the current structure and completeness of reported synthetic protocols, understanding of which will be very helpful in engineering and fine-tuning GPT-based models.As a result, the analysis performed in this study can only provide insight into how the MOF material is made rather than linking different synthesis features to specific outcomes like yield or quality. In the absence of such synthesis outcome information, we instead focus on how best to prepare the information gathered in this study for the generation of predictive models for ZIF-8 materials quality. A key challenge when attempting to optimise synthesis protocols either through systematic experimentation5 or by training machine learning models6 is the high dimensionality of the information contained in each synthesis. For example, 8 unique reagent chemicals were discussed in the previous section – 3 metal sources, 1 linker, and 4 solvents. Although intuitively only 3 chemicals are typically required for synthesis – a metal salt, linker, and solvent – recent automated optimisation of HKUST-1 synthesis included protocols containing anywhere from 1 to 5 different solvents.6 Therefore, to fully explore the 8-dimensional chemical space and find the globally optimum set of synthesis parameters, N8 experiments would be required (where N is the number of quantity values tested for each variable). While theoretically this dimensionality would scale with the number of synthesis steps used, we were unable to identify meaningfully distinct groups of synthesis actions (data not shown here, for brevity) and hence did not consider the sequence as impacting the synthesis outcome.
Exhaustive searching an 8-dimensional synthesis space is highly impractical, however, requiring many hundreds of experiments even for N = 2. Therefore, although we cannot use the database of synthesis protocols to identify the quality of the ZIF-8 produced, it can be used to identify patterns in the synthesis protocols published hence reducing the synthesis space for optimisation. First, we reduced the chemical space by identifying the most complex synthesis protocols published, revealing that few synthesis protocols in our database contained more than 2 different solvents or metal sources. If the protocols considered in the synthesis space were limited to this level of complexity, the dimensionality would be reduced to 18(N5) i.e. a 5-dimensional combinatorial space with 18 different combinations of reagents.
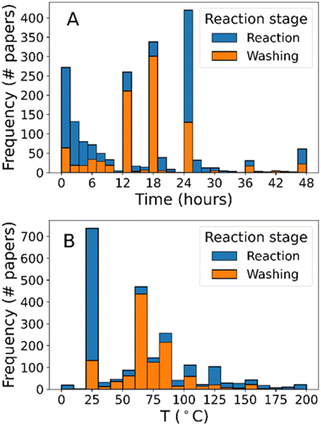
Second, we also considered the frequency of different synthesis routes, reasoning that only successful synthesis protocols are generally published therefore simple synthesis routes that are never published are likely to be unsuccessful. To this end, we used clustering to identify lower-dimensional sub-regions of the synthesis phase space which have been widely researched in experimental papers – essentially using a chemical combination's popularity as a proxy for its importance. The chemical identities used were encoded using TF-IDF vectorisation, then similar synthesis protocols were grouped by their density in the encoded space. The outcome of this clustering analysis is visualised using a 2-d projection in Fig. 7 and summarised in Table 4, where the distance between points is indicative of each protocol's similarity to its neighbours. Eight clusters of reagent combinations were identified each containing 2–4 chemicals of a total of 6 reagents. We posit that these clusters represent well defined strategies to synthesize ZIF-8, which can be explored separately, therefore reducing the total amount of information required to explore these regions of the synthesis space.
 | ||
| Fig. 7 2-Dimensional representation of the chemical combination space for ZIF-8 synthesis, generated using the t-SNE algorithm. Major synthesis pathways are identified using the DBSCAN clustering method and colour coded, while noise data is shown in light grey. Clusters are circled and described in Table 4. | ||
| Cluster number (colour) | Common chemicals | Number of protocols in cluster (number with unique features) |
|---|---|---|
| 1 (blue) | Zinc, nitrate, methanol | 225 (177) |
| 2 (red) | Cobalt, nitrate, methanol | 147 (119) |
| 3 (brown) | Zinc, nitrate, water | 50 (42) |
| 4 (orange) | Zinc, nitrate | 39 (38) |
| 5 (green) | Zinc, cobalt, nitrate, methanol | 31 (31) |
| 6 (pink) | Cobalt, nitrate, water | 28 (27) |
| 7 (grey) | Zinc, nitrate, DMF | 25 (25) |
| 8 (purple) | Zinc, acetate, water | 24 (23) |
| 9 (olive) | Zinc, nitrate, methanol, water | 21 (20) |
The well-defined synthesis strategies clustered in Fig. 7 are notably different from the analysis performed in the previous section. In the first instance, ethanol was fully absent signifying its insignificance as a reaction solvent and matching the earlier analyses. Separately, acetate salts are only identified in one cluster and only associated with water. This association is due to the lack of solubility of zinc acetate in methanol (ca. 15 g L−1cf. 430 g L−1 in water), information which can only otherwise be gained by specific knowledge of the chemistry of zinc acetate. While obvious to those who already are aware of the system, this information may otherwise be overlooked by chemists naive to the intricacies of ZIF-8 synthesis – an example of chemical intuition.6 Therefore, clustering of similar synthesis protocols together can help users to avoid some common pitfalls when planning experiments for the first time.
Finally, to demonstrate the benefit of this approach towards synthesis optimisation, we consider the reduction in the combinatorial space that would be required to fully optimise the identified popular sub-regions of the synthesis space. From the clustering analysis, we identified 6 sub-regions with only 3 chemicals of interest – clusters 1, 2, 3, 6, 7, and 8 in Table 4, containing only a single metal salt, 2-methylimidazole, and a single solvent – and a further 2 sub-regions with 4 chemicals of interest: clusters 5 and 8 containing either mixed salts or solvents. Accordingly, instead of exhaustively exploring all combinatorial options, or all combinatorial options up to a certain number of synthesis reagents, full optimisation of the commonly-reported ZIF-8 synthesis routes would only require 6(N3) + 2(N4) ≈ N4.4 experiments. To illustrate the extent of dimensionality reduction in real terms, the number of experiments required to explore the synthesis space are shown in Table 5 for various values of N. In combination with the quantity distributions shown in Fig. 3 and 4, text mining and data reduction tools demonstrated in this paper will provide excellent initial values for efficient searching of chemical synthesis space, thereby accelerating methodology refinement for a range of nanomaterials.
| Exhaustive exploration (N8) | Limited experimental complexity (18(N5)) | Identified clusters only (6(N3) + 2(N4)) | |
|---|---|---|---|
| N = 3 | 6500 | 4400 | 320 |
| N = 5 | 390![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 300 |
2000 |
| N = 10 | 1 × 108 | 1.8 × 106 | 2.6 × 104 |
Conclusions
In this study, we used text mining to study the synthesis methodology landscape of a single MOF material, exploring to what extent the previously accumulated collective knowledge of a particular nanomaterial can accelerate the development of reliable and scalable synthesis protocols. As the first step toward this objective, in this study, we posed three research questions: first, is it possible to use text mining tools to provide deep insight into a single synthetic target, rather than a comprehensive overview of a family of materials? Second, is it possible to standardise the synthesis details extracted as a means of performing like-for-like comparison between different studies? Finally, is it possible to use this analysis to suggest optimal synthesis conditions, thereby accelerating methodology development?To this end, we developed software to systematically analyse nanomaterials synthesis methods based on established text mining protocols. We extracted structured data to describe the details of each synthesis protocol, enabling large-scale statistical analysis of the synthesis parameter space and clustering of similar methods together to identify well-explored regions of the synthesis space. We believe that this progress represents the first step in creating a closed feedback loop for the automated optimisation of experimental nanomaterials synthesis, visualised in Fig. 8. In this feedback loop text mined information can identify common limits to parameters as well as low-dimensional sub-regions of interest in the synthesis space. By using this information as initial conditions for iterative high-throughput experimentation, the search for synthesis protocols optimised against any target material quality metric can be greatly accelerated.
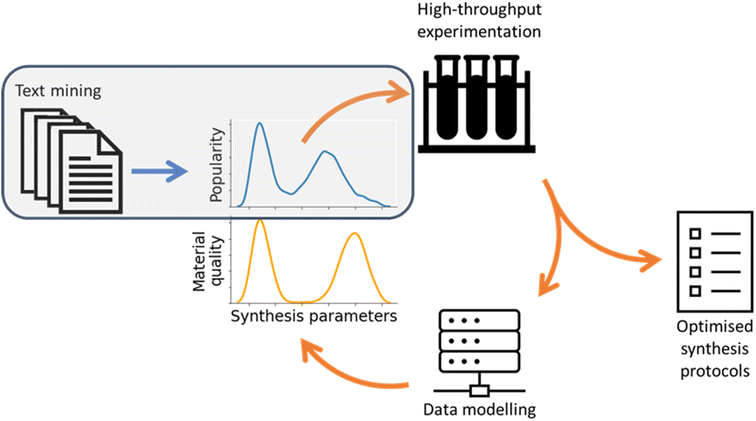 | ||
| Fig. 8 Scheme of a synthesis protocol optimisation feedback loop. Work carried out in this study is shaded in grey. | ||
As a case study to demonstrate the utility of this approach, we performed a quantitative meta-analysis of 1600 synthesis methods for the common MOF ZIF-8. Using this framework, we identified key aspects of the synthesis including the range of chemicals used as reagents, solvents, and ancillary modulators/pH modifiers. We extracted information about the quantity of each reagent used during the synthesis, enabling us to identify the distribution of synthesis scales, reagent ratios, and reaction mixture solids concentration, as well as reaction times and temperatures. Further insight was gathered by cross-referencing chemicals mentioned against the stage they were introduced into the synthesis protocol – for example identifying that ethanol is primarily used as a washing solvent rather than in the reaction medium. We demonstrated how the quantitative meta-analysis performed here can assist in systematic searches of the synthesis phase space by identifying both low-dimensional regions of interest and the distribution of synthesis parameters. As a result, we were able to reduce the number of hypothetical experiments required to optimise ZIF-8 significantly. Notably, while we considered MOF materials as a case study in this work, the methods developed here do not depend on any specific structural identifiers e.g. CSD reference numbers, indicating that they are general to any synthesis type. Particularly, we envisage they will be useful the systematising understanding of other emerging nanomaterial systems such as mesoporous (organo)silicas, covalent organic frameworks, and polymers of intrinsic microporosity.
Despite the deep insight we were able to gain into the synthesis system of ZIF-8, the current study also identified significant challenges associated with developing a true “synthetic oracle” for predicting the ideal synthesis parameters for any given material. While we were able to identify and extract information about the synthesis, we were unable to reliably connect the quality of the material produced to the methods themselves (e.g. by identifying specific yield or surface area). A crucial next step is therefore to adopt state of the art transformer-based methods e.g. BERT or GPT-4 to better interpret the entire research article as a single unit and therefore identify implicitly described synthesis protocols (e.g. tabulated changes to individual synthesis parameters). A second challenge lies in the estimating the viability of synthesis parameters extracted during text mining or proposed by generative models, preventing automated reproduction of a synthesis protocol without human oversight and validation. Finally, as has been discussed elsewhere, the synthesis protocol extraction methods developed here can only build from published information, which is biased towards the most successful synthesis methods only. More comprehensive reporting of synthesis information using structured formats akin to the crystallographic information file format would enable far more wide-reaching analysis to be performed.
In summary, the methods developed in this study acts as a preliminary approach for the large-scale standardisation and analysis of experimental synthesis data, representing the first step in creating a closed feedback loop for the automated optimisation of experimental nanomaterials synthesis. By interfacing with automated and high throughput reactionware e.g. through integration of the XDL chemical programming language, methodology development will be significantly accelerated thereby easing the adoption of nanomaterials at larger scales and in new settings.
Data availability
All software and papers used in this study are made available under an MIT license at https://github.com/SarkisovTeam/SynOracle-preprocessing (commit b9c3627) and https://github.com/SarkisovTeam/SyntheticOracle (commit 01898b7). Data used for generating figures in this study are provided in .json format in the publication figures directory, alongside Jupyter notebook files detailing the generation of all figures in the publication. Scripts to generate the .json synthesis information data are contained within the “worked example” directories of each repository.Conflicts of interest
There are no conflicts to declare.Notes and references
- O. Kononova, T. He, H. Huo, A. Trewartha, E. A. Olivetti and G. Ceder, iScience, 2021, 24, 102155 CrossRef PubMed.
- E. Kim, K. Huang, O. Kononova, G. Ceder and E. Olivetti, Matter, 2019, 1, 8–12 CrossRef.
- D. R. Baer and I. S. Gilmore, J. Vac. Sci. Technol., A, 2018, 36, 068502 CrossRef.
- D. R. Baer, P. Munusamy and B. D. Thrall, Biointerphases, 2016, 11, 04B401 CrossRef PubMed.
- B. Burger, P. M. Maffettone, V. V. Gusev, C. M. Aitchison, Y. Bai, X. Wang, X. Li, B. M. Alston, B. Li, R. Clowes, N. Rankin, B. Harris, R. S. Sprick and A. I. Cooper, Nature, 2020, 583, 237–241 CrossRef CAS PubMed.
- S. M. Moosavi, A. Chidambaram, L. Talirz, M. Haranczyk, K. C. Stylianou and B. Smit, Nat. Commun., 2019, 10, 539 CrossRef CAS PubMed.
- L. Wilbraham, S. H. M. Mehr and L. Cronin, Acc. Chem. Res., 2021, 54, 253–262 CrossRef CAS PubMed.
- A. J. S. Hammer, A. I. Leonov, N. L. Bell and L. Cronin, JACS Au, 2021, 1, 1572–1587 CrossRef CAS PubMed.
- D. M. Jessop, S. E. Adams, E. L. Willighagen, L. Hawizy and P. Murray-Rust, J. Cheminf., 2011, 3, 1–12 Search PubMed.
- J. Mavračić, C. J. Court, T. Isazawa, S. R. Elliott and J. M. Cole, J. Chem. Inf. Model., 2021, 61, 4280–4289 CrossRef PubMed.
- L. Hawizy, D. M. Jessop, N. Adams and P. Murray-Rust, J. Cheminf., 2011, 3, 1–13 Search PubMed.
- M. Vazquez, M. Krallinger, F. Leitner and A. Valencia, Mol. Inf., 2011, 30, 506–519 CrossRef CAS PubMed.
- J. Guo, A. S. Ibanez-Lopez, H. Gao, V. Quach, C. W. Coley, K. F. Jensen and R. Barzilay, J. Chem. Inf. Model., 2022, 62, 2035–2045 CrossRef CAS PubMed.
- E. J. Beard, G. Sivaraman, Á. Vázquez-Mayagoitia, V. Vishwanath and J. M. Cole, Sci. Data, 2019, 6, 1–11 CrossRef PubMed.
- C. B. Cooper, E. J. Beard, Á. Vázquez-Mayagoitia, L. Stan, G. B. G. Stenning, D. W. Nye, J. A. Vigil, T. Tomar, J. Jia, G. B. Bodedla, S. Chen, L. Gallego, S. Franco, A. Carella, K. R. J. Thomas, S. Xue, X. Zhu and J. M. Cole, Adv. Energy Mater., 2019, 9, 1–10 Search PubMed.
- Z. Jensen, S. Kwon, D. Schwalbe-Koda, C. Paris, R. Gómez-Bombarelli, Y. Román-Leshkov, A. Corma, M. Moliner and E. A. Olivetti, ACS Cent. Sci., 2021, 7, 858–867 CrossRef CAS PubMed.
- E. J. Beard and J. M. Cole, Sci. Data, 2022, 9, 1–19 CrossRef PubMed.
- J. Zhao and J. M. Cole, Sci. Data, 2022, 9, 1–11 CrossRef PubMed.
- O. Kononova, H. Huo, T. He, Z. Rong, T. Botari, W. Sun, V. Tshitoyan and G. Ceder, Sci. Data, 2019, 6, 1–11 CrossRef PubMed.
- T. Isazawa and J. M. Cole, J. Chem. Inf. Model., 2022, 62, 1207–1213 CrossRef CAS PubMed.
- R. Klinger, C. Kolářik, J. Fluck, M. Hofmann-Apitius and C. M. Friedrich, Bioinformatics, 2008, 24, 268–276 CrossRef PubMed.
- S. Majumdar, S. M. Moosavi, K. M. Jablonka, D. Ongari and B. Smit, ACS Appl. Mater. Interfaces, 2021, 13, 61004–61014 CrossRef CAS PubMed.
- P. G. Boyd and T. K. Woo, CrystEngComm, 2016, 18, 3777–3792 RSC.
- C. E. Wilmer, M. Leaf, C. Y. Lee, O. K. Farha, B. G. Hauser, J. T. Hupp and R. Q. Snurr, Nat. Chem., 2012, 4, 83–89 CrossRef CAS PubMed.
- S. Y. S. Lee, B. Kim, H. Cho, H. Lee, S. Y. S. Lee, E. S. Cho and J. Kim, ACS Appl. Mater. Interfaces, 2021, 13, 23647–23654 CrossRef CAS PubMed.
- S. M. Moosavi, A. Nandy, K. M. Jablonka, D. Ongari, J. P. Janet, P. G. Boyd, Y. Lee, B. Smit and H. J. Kulik, Nat. Commun., 2020, 11, 4068 CrossRef CAS PubMed.
- P. Z. Moghadam, A. Li, X. W. Liu, R. Bueno-Perez, S. D. Wang, S. B. Wiggin, P. A. Wood and D. Fairen-Jimenez, Chem. Sci., 2020, 11, 8373–8387 RSC.
- P. Z. Moghadam, A. Li, S. B. Wiggin, A. Tao, A. G. P. Maloney, P. A. Wood, S. C. Ward and D. Fairen-Jimenez, Chem. Mater., 2017, 29, 2618–2625 CrossRef CAS.
- D. Ongari, L. Talirz and B. Smit, ACS Cent. Sci., 2020, 6, 1890–1900 CrossRef CAS PubMed.
- B. J. Bucior, A. S. Rosen, M. Haranczyk, Z. Yao, M. E. Ziebel, O. K. Farha, J. T. Hupp, J. I. Siepmann, A. Aspuru-Guzik and R. Q. Snurr, Cryst. Growth Des., 2019, 19, 6682–6697 CrossRef CAS.
- C. R. Groom and F. H. Allen, Angew. Chem., Int. Ed., 2014, 53, 662–671 CrossRef CAS PubMed.
- Y. Luo, S. Bag, O. Zaremba, A. Cierpka, J. Andreo, S. Wuttke, P. Friederich and M. Tsotsalas, Angew. Chem., Int. Ed., 2022, 61, e202200242 CrossRef CAS PubMed.
- K. Gubsch, R. Bence, L. T. Glasby and P. Z. Moghadam, Chem. Mater., 2023, 35, 4510–4524 CrossRef PubMed.
- C. S. Cox, E. Slavich, L. K. Macreadie, L. K. McKemmish and M. Lessio, Chem. Mater., 2023, 35, 3057–3072 CrossRef CAS.
- H. Zhang and R. Q. Snurr, J. Phys. Chem. C, 2017, 121, 24000–24010 CrossRef CAS.
- S. Bhattacharyya, R. Han, W. G. Kim, Y. Chiang, K. C. Jayachandrababu, J. T. Hungerford, M. R. Dutzer, C. Ma, K. S. Walton, D. S. Sholl and S. Nair, Chem. Mater., 2018, 30, 4089–4101 CrossRef CAS.
- M. F. De Lange, B. L. Van Velzen, C. P. Ottevanger, K. J. F. M. Verouden, L. C. Lin, T. J. H. Vlugt, J. Gascon and F. Kapteijn, Langmuir, 2015, 31, 12783–12796 CrossRef CAS PubMed.
- M. Kinoshita, S. Yanagida, T. Gessei and A. Monkawa, J. Cryst. Growth, 2022, 600, 126877 CrossRef CAS.
- Y. R. Lee, X. H. Do, S. S. Hwang and K. Y. Baek, Catal. Today, 2021, 359, 124–132 CrossRef CAS.
- A. Paul, I. K. Banga, S. Muthukumar and S. Prasad, ACS Omega, 2022, 7, 26993–27003 CrossRef CAS PubMed.
- A. Lewis, F. S. Butt, X. Wei, N. A. Mazlan, Z. Chen, Y. Yang, S. Yang, N. Radacsi, X. Chen and Y. Huang, Results Eng., 2023, 17, 100751 CrossRef CAS.
- C.-W. Tsai and E. H. G. Langner, Microporous Mesoporous Mater., 2016, 221, 8–13 CrossRef CAS.
- K. Kida, M. Okita, K. Fujita, S. Tanaka and Y. Miyake, CrystEngComm, 2013, 15, 1794–1801 RSC.
- J. Cravillon, C. A. Schröder, H. Bux, A. Rothkirch, J. Caro and M. Wiebcke, CrystEngComm, 2012, 14, 492–498 RSC.
- N. A. H. M. Nordin, A. F. Ismail, A. Mustafa, P. S. Goh, D. Rana and T. Matsuura, RSC Adv., 2014, 4, 33292–33300 RSC.
- G. Van Rossum and F. L. Drake, Python 3 Reference Manual, CreateSpace, Scotts Valley, CA, 2009 Search PubMed.
- J. Devlin, M.-W. Chang, K. Lee and K. Toutanova, arXiv, 2019, preprint, arXiv:1810.04805, DOI:10.48550/arXiv.1810.04805.
- W. McKinney, in Proceedings of the 9th Python in Science Conference, ed. S. van der Walt and J. Millman, 2010, vol. 1, pp. 56–61 Search PubMed.
- The pandas development team, pandas-dev/pandas: Pandas, 2023, DOI:10.5281/zenodo.8364959.
- S. Kim, J. Chen, T. Cheng, A. Gindulyte, J. He, S. He, Q. Li, B. A. Shoemaker, P. A. Thiessen, B. Yu, L. Zaslavsky, J. Zhang and E. E. Bolton, Nucleic Acids Res., 2023, 51, D1373–D1380 CrossRef PubMed.
- G. Landrum, RDKit Software, 2010, https://www.rdkit.org Search PubMed.
- C. Bell and Y. R. Cortes-Pena, CalebBell/chemicals: 1.1.3, 2016, DOI:10.5281/zenodo.7857398.
- G. H. Thomson, R. W. Hankinson and G. H. Thomson, AIChE J., 1979, 25, 653–663 CrossRef.
- Z. Zhang, S. Xian, Q. Xia, H. Wang, Z. Li and J. Li, AIChE J., 2013, 59, 2195–2206 CrossRef CAS.
- Z. Wang, K. Cruse, Y. Fei, A. Chia, Y. Zeng, H. Huo, T. He, B. Deng, O. Kononova and G. Ceder, Digital Discovery, 2022, 1, 313–324 RSC.
- K. S. Jones, J. Doc., 1972, 28, 11–21 CrossRef.
- M. Esther, H.-P. Kriegel, J. Sander and X. Xu, in Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, 1996, pp. 226–231 Search PubMed.
- L. van der Maaten and G. Hinton, J. Mach. Learn. Res., 2008, 9, 2579–2605 Search PubMed.
- Y. Pan, D. Heryadi, F. Zhou, L. Zhao, G. Lestari, H. Su and Z. Lai, CrystEngComm, 2011, 13, 6937 RSC.
- M. C. Swain and J. M. Cole, J. Chem. Inf. Model., 2016, 56, 1894–1904 CrossRef CAS PubMed.
- J. Liu, R. Li, Y. Wang, Y. Wang, X. Zhang and C. Fan, J. Alloys Compd., 2017, 693, 543–549 CrossRef CAS.
- V. V. Butova, V. A. Polyakov, E. A. Erofeeva, I. S. Yahia, H. Y. Zahran, A. F. Abd El-Rehim, A. M. Aboraia and A. V. Soldatov, Inorganica Chim. Acta, 2020, 509, 119678 CrossRef CAS.
- A. Samadi-Maybodi, S. Ghasemi and H. Ghaffari-Rad, Electrochim. Acta, 2015, 163, 280–287 CrossRef CAS.
- A. Schejn, L. Balan, V. Falk, L. Aranda, G. Medjahdi and R. Schneider, CrystEngComm, 2014, 16, 4493–4500 RSC.
- M. Jian, B. Liu, R. Liu, J. Qu, H. Wang and X. Zhang, RSC Adv., 2015, 5, 48433–48441 RSC.
- Z. Öztürk, M. Filez and B. M. Weckhuysen, Chem.–Eur. J., 2017, 23, 10915–10924 CrossRef PubMed.
- N. F. Hamidon, M. I. M. Tahir, M. A. M. Latif and M. B. A. Rahman, J. Coord. Chem., 2022, 75, 1180–1192 CrossRef CAS.
- D. Yamamoto, T. Maki, S. Watanabe, H. Tanaka, M. T. Miyahara and K. Mae, Chem. Eng. J., 2013, 227, 145–150 CrossRef CAS.
- B. Chen, F. Bai, Y. Zhu and Y. Xia, Microporous Mesoporous Mater., 2014, 193, 7–14 CrossRef CAS.
- Y. Zhang, Y. Jia, M. Li and L. Hou, Sci. Rep., 2018, 8, 1–7 Search PubMed.
- P. S. Albright and L. J. Gosting, J. Am. Chem. Soc., 1946, 68, 1061–1063 CrossRef CAS PubMed.
- Y. R. Lee, M. S. Jang, H. Y. Cho, H. J. Kwon, S. Kim and W. S. Ahn, Chem. Eng. J., 2015, 271, 276–280 CrossRef CAS.
- W. Xia, S. K. Lau and W. F. Yong, J. Clean. Prod., 2022, 370, 133354 CrossRef CAS.
- A. W. Tricker, G. Samaras, K. L. Hebisch, M. J. Realff and C. Sievers, Chem. Eng. J., 2020, 382, 122954 CrossRef CAS.
- N. L. Torad, J. Kim, M. Kim, H. Lim, J. Na, S. M. Alshehri, T. Ahamad, Y. Yamauchi, M. Eguchi, B. Ding and X. Zhang, J. Hazard. Mater., 2021, 405, 124248 CrossRef CAS PubMed.
- C. Zhou, Z. Li, J. Li, T. Yuan, B. Chen, X. Ma, D. Jiang, X. Luo, D. Chen and Y. Liu, Chem. Eng. J., 2020, 385, 123835 CrossRef CAS.
- G. Zheng, Z. Chen, K. Sentosun, I. Pérez-Juste, S. Bals, L. M. Liz-Marzán, I. Pastoriza-Santos, J. Pérez-Juste and M. Hong, Nanoscale, 2017, 9, 16645–16651 RSC.
- J. Cravillon, R. Nayuk, S. Springer, A. Feldhoff, K. Huber and M. Wiebcke, Chem. Mater., 2011, 23, 2130–2141 CrossRef CAS.
- J. M. Tuffnell, J. K. Morzy, N. D. Kelly, R. Tan, Q. Song, C. Ducati, T. D. Bennett and S. E. Dutton, Dalton Trans., 2020, 49, 15914–15924 RSC.
- S. Huang and J. M. Cole, Chem. Sci., 2022, 13, 11487–11495 RSC.
- K. M. Jablonka, P. Schwaller, A. Ortega-guerrero and B. Smit, ChemRxiv, 2023, preprint, DOI:10.26434/chemrxiv-2023-fw8n4-v2.
- Z. Zheng, O. Zhang, C. Borgs, J. T. Chayes and O. M. Yaghi, J. Am. Chem. Soc., 2023, 145, 18048–18062 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3dd00099k |
| This journal is © The Royal Society of Chemistry 2023 |