Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Orchestrating nimble experiments across interconnected labs
Dan
Guevarra
 *ab,
Kevin
Kan
ab,
Yungchieh
Lai
ab,
Ryan J. R.
Jones
*ab,
Kevin
Kan
ab,
Yungchieh
Lai
ab,
Ryan J. R.
Jones
 ab,
Lan
Zhou
ab,
Phillip
Donnelly†
a,
Matthias
Richter‡
ab,
Lan
Zhou
ab,
Phillip
Donnelly†
a,
Matthias
Richter‡
 ab,
Helge S.
Stein
c and
John M.
Gregoire
ab,
Helge S.
Stein
c and
John M.
Gregoire
 *ab
*ab
aDivision of Engineering and Applied Science, California Institute of Technology, Pasadena, CA 91125, USA. E-mail: guevarra@caltech.edu; gregoire@caltech.edu
bLiquid Sunlight Alliance, California Institute of Technology, Pasadena, CA, USA
cTUM School of Natural Sciences, Department of Chemistry, Munich Data Science Institute, Technical University of Munich, Munich, Germany
First published on 3rd October 2023
Abstract
Advancements in artificial intelligence (AI) for science are continually expanding the value proposition for automation in materials and chemistry experiments. The advent of hierarchical decision-making also motivates automation of not only the individual measurements but also the coordination among multiple research workflows. In a typical lab or network of labs, workflows need to independently start and stop operation while also sharing resources such as centralized or multi-functional equipment. A new paradigm in instrument control is needed to realize the combination of independence with respect to periods of operation and interdependence with respect to shared resources. We present Hierarchical Experimental Laboratory Automation and Orchestration with asynchronous programming (HELAO-async), which is implemented via the Python asyncio package by abstracting each resource manager and experiment orchestrator as a FastAPI server. This framework enables coordinated workflows of adaptive experiments, which will elevate Materials Acceleration Platforms (MAPs) from islands of accelerated discovery to the AI emulation of team science.
Introduction
Materials Acceleration Platforms (MAPs)1 aim to leverage modern artificial intelligence (AI) algorithms to accelerate the discovery of molecular and solid state materials. For solid state materials, automation and integration with computation2 has been historically achieved via combinatorial methods3,4 and recently implemented using autonomous or self-driving labs.5,6 The pace of advancement in experiment automation is staggering and motivates rethinking of how instruments are made and controlled. Initial efforts in AI-guided automated workflows have naturally focused on achieving super-human efficiency of data acquisition. Here, automated experiment selection alleviates reliance on human researchers but does not supplant human governance of the scientific line of inquiry and the strategy for its exploration.7 Recent developments in physics and chemistry-aware models8,9 as well as hypothesis learning algorithms10 exemplify the ever-increasing sophistication of automated experiment design. While such algorithms do not yet rival the decision making of human experts, the trajectory of AI algorithms clearly indicates that the frontier of experiment automation is not the automation of individual experiments but rather entire workflows and their ensembles.Expanding upon the vision of interconnected workflows for AI emulation of team science, Bai et al.11 have envisioned world-wide coordination of self-driving labs driven by the rapidly evolving fields of knowledge graphs, semantic web technologies, and multi-agent systems. Ren et al.12 emphasize the critical need for interconnected laboratories to leverage resources and learn epistemic uncertainties. To realize this collective vision, experiment automation software that builds upon the state of the art13–24 must be developed to interconnect laboratories and their research workflows. A hallmark of human scientific research is on-the-fly adaption of experimental workflows based on recent observations. Human scientists also interleave workflows spanning materials discovery to device prototyping.25 The interleaved execution of multiple workflows typically involves shared resources, which is often a practical necessity for minimizing the capital expense of establishing any experimental workflow. These considerations require “nimble” experiment automation, and in the present work we describe our approach to automating nimble, interconnected workflows via asynchronous programming.
Results and discussion
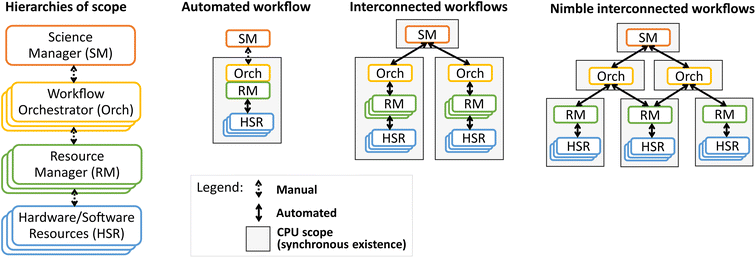
Fig. 1 illustrates the hierarchical components of a scientific workflow, with the highest hierarchy being a Science Manager that designs research projects and strategies for their implementation. The Workflow Orchestrator implements these strategies as (dynamic) series of experiments. Execution of the workflows requires management of laboratory resources by Resource Managers, which interface directly with the Hardware/Software Resources of the lab. During execution of an “automated workflow,” a human scientist typically serves as Science Manager, overseeing the execution of automation software that encompasses a tightly-integrated Workflow Orchestrator and Resource Manager, which collectively control the workflow's suite of lab resources.Parallel execution of automated workflows can be realized via “interconnected workflows” wherein a human or machine Science Manager oversees multiple automated workflows that each run on their own central processing unit (CPU). This scheme is appropriate when each workflow can operate independently. If two workflows share an experiment resource, one strategy for their automation is to integrate them into a hybrid workflow that is executed from a single CPU. The shared runtime among all processes in a single CPU inherently limits operational flexibility, a runtime interdependence that is impractical for interconnected labs. In addition to addressing the needs of shared resources, asynchronous programming provides the requisite flexibility for experiment automation tasks such as passing messages, writing data to files, and polling devices. These needs can be partially fulfilled with multi-thread programming, although we find asynchronous programming to be a more natural solution. For a more technical discussion of the difference between asynchronous programming and threading, we refer to the reader to ref. 26.
As an example of a shared resource, consider a lab in which a central piece of equipment such as an X-ray diffractometer or reactive annealing chamber is used in several distinct workflows. Traditional methods of experiment automation would involve each workflow taking ownership of that equipment during the workflow's runtime. Combining all workflows in a single instance of automation software limits the ability of different workflows to start and stop as dictated by science management and/or equipment maintenance needs. Human researchers address this challenge by creating a system to schedule the requested usage of shared equipment, and the automation analogue is to have the shared equipment operated by a broker whose runtime is independent of all other workflow automation software. This is the central tenet of “Nimble interconnected workflows” as depicted in Fig. 1, wherein each resource family is controlled by an asynchronous Resource Manager. The runtime independence of Resource Managers and Workflow Orchestrators enables each Orchestrator to maintain focus on a single research workflow while empowering a Science Manager to coordinate efforts across many workflows in any number of physical laboratories.
The series of workflow automation capabilities illustrated by Fig. 1 has been largely mirrored by the evolution of reported automation software for materials chemistry. Seminal demonstrations of software for automating an experiment workflow include ARES,13 ChemOS,14 and Bluesky.15 Continual development of these platforms have resulted in new capabilities such as the generalized ARES-OS16 and remote operation with Bluesky.17 ChemOS14 and ESCALATE27 have increasingly incorporated ancillary aspects of automation such as encoding design of experiments and interfacing with databases. These efforts have built toward multi-workflow integration in HELAO18,19 and NIMS-OS20 as well as object-oriented, modular frameworks for co-development of multiple MAPs21,23 and multi-agent automation frameworks.22 Enabling independent operation of workflow components ultimately requires asynchronous programming, as envisioned by the present work and ChemOS2.0.24 The abstraction of lab equipment as asynchronous web servers is implemented in HELAO-async using FastAPI,28 a performant, standards-based web framework for writing Application Programming Interfaces (APIs) in Python. SiLA2 (ref. 29) is an alternative framework that may be used to realize the HELAO-style communication within and among multiple workflows. To best of our knowledge, HELAO-async is the first instrument control software platform that realizes the “Nimble interconnected workflows” paradigm illustrated in Fig. 1, where workflows can be independently started and stopped while sharing resources such as centralized or multi-functional equipment. These capabilities are agnostic to whether workflow operation decisions are determined via human or artificial intelligence, while the independent execution of multiple workflows is critical for fully realizing the value of hierarchical active learning and multi-modal AI algorithms.
The implementation of “Nimble interconnected workflows” in HELAO-async is outlined in Fig. 2. A Science Manager (see Fig. 1) is implemented as an Operator for active science management and Observer for passive science management. The Orchestrator manages workflow-level automation, which generally involves launching a series of actions on the workflow's suite of Action Servers. In the parlance of Fig. 1, Action Servers are the Resource Managers, which execute actions via Device Drivers that comprise the Hardware/Software Resources. The design principle of this framework is to enable asynchronous launching of workflows via Operator–Orchestrator communication, as well as asynchronous execution of workflows via Orchestrator–Action Server communication. When multiple Orchestrators share a resource, queuing and prioritization are managed by the respective Action Server.
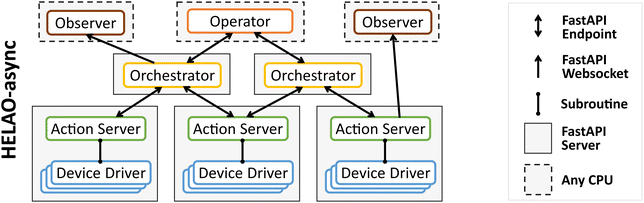 | ||
| Fig. 2 The HELAO-async framework is outlined as a specific implementation of the “Nimble interconnected workflow” concept from Fig. 1. Orchestrators manage workflows by controlling Action Servers that manage resources via Device Drivers. By establishing an independent FastAPI server for each Orchestrator and Action Server, workflows have independent runtimes while sharing resources as needed. A human or AI Operator manages the collection of Orchestrators, and an Observer can consume data streams from any server. The use of FastAPI endpoints and websockets for these interactions creates flexibility for the implementation of Operators and Observers. | ||
The HELAO-async implementation described herein is intended to be agnostic with respect to the type of Operator and Observer, which may involve any combination of a human researcher, an autonomous operator selecting experiments via an AI-based acquisition function, or a more general broker19 for coordinating experiments across many workflows. The scope of an Orchestrator is that of a single workflow, and by implementing each Orchestrator as a FastAPI server, the parameterized workflow is exposed to the Operator via custom FastAPI endpoints. For example, “global_status” is an endpoint of an Orchestrator FastAPI server that is programmed as follows to enable any program to request and receive the Orchestrator's status:
The Orchestrator executes a workflow via Action Server FastAPI endpoints, for example this abridged version of the “acquire_data” endpoint, which includes parameters for the duration and rate of the acquisition:
Fig. 2 also depicts Observers, which can subscribe to the FastAPI websockets established by an Orchestrator and/or Action Server. Each Orchestrator and Action Server must be programmed to publish data of interest to websockets, which enables any number of Observers to listen-in as needed. Our common implementation to-date is a web browser-based Observer (a.k.a. Visualizer) that researchers can launch to monitor quasi-real-time data streams, a critical capability for experiment quality control. For example, the Orchestrator web socket “ws_status” enables any program to subscribe to the Orchestrator's status messages:
The asynchronous operation at the Orchestrator and Action Server levels was particularly motivated by hierarchical active learning schemes, for example human-in-the-loop30,31 or fully autonomous hierarchical active learning.32 When workflow-level decisions are made by a human with autonomous workflow execution, a community of asynchronous Orchestrators enables humans to execute on any available workflow. We envision that this mode of operation will be critical for integrating physically-separated laboratories and realizing cloud laboratories.11,12,33
HELAO-async is being actively developed in two public repositories: https://github.com/High-Throughput-Experimentation/helao-core encompasses the API data structures and https://github.com/High-Throughput-Experimentation/helao-async contains instrument drivers, API server configurations, and experiment sequences. These repositories contain drivers for our suite of experimental resources spanning motion control, liquid handling, electrochemistry, analytical chemistry, and optical spectroscopy. Our Orchestrator-level implementations include scanning droplet electrochemistry,34 scanning optical spectroscopy,35 electrochemical cells with scheduled electrolyte aliquots for monitoring corrosion,36 and several methods of coupling electrochemical transformations with analytical detection of the chemical products.37 These latter two examples share the need for liquid and/or gas aliquoting from operational electrochemical cells, for which we typically use a Tri Plus robotic sample handling system (CTC Analytics), which is a shared resource across multiple workflows.
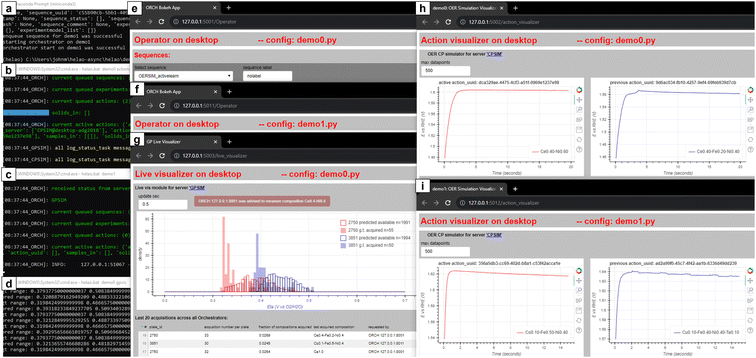
Given our safety and security protocols, readers of this manuscript may not execute HELAO-async code with our laboratory equipment. While we encourage the duplication and adaption of our hardware and/or software for operation in other labs, we have built a virtual demo for the present purposes of introducing HELAO-async. To create a minimal implementation of Fig. 2, the demo contains two independent Orchestrators, each with a dedicated Action Server that simulates the acquisition of electrochemistry data to characterize the overpotential for the oxygen evolution reaction (OER). We have packaged previously-acquired electrochemistry data with the demo.38 The two Orchestrators share a common resource, which in practice may be the robotic sample handling system. In the demo, the shared Action Server is an active learning agent that manages requests for new acquisition instructions from the two Orchestrators. This shared-resource Action Server runs independently and is unaffected by the runtime of each Orchestrator, which is demonstrated in the demo by independently starting and stopping the Orchestrators, representing the asynchronous operation of research workflows within one or across multiple laboratories. Running the demo batch script will open five user interface browsers, two Operators that control the respective Orchestrators, two Visualizers (Observers) that show the data streams from the respective electrochemistry Action Server, and a Visualizer (Observer) for the shared active learning Action Server. This Visualizer shows the progress of the active learning campaign, including the contributions from each of the independent Orchestrators. A snapshot of these five web browser interfaces is shown in Fig. 3. Due to the use of static random seeding of the active learning, the demo runs deterministically, where the contents of Fig. 3 show the status approximately 17 minutes after launching the demo batch script.
While the HELAO-async schematic of Fig. 2 indicates the intended role and scope of each FastAPI server with respect to the universal research roles summarized in Fig. 1, there remains flexibility in how to implement HELAO-async for a given workflow or ensemble of workflows. Regarding the scope of a single Action Server, a set of resources may be bundled in a single FastAPI server based on (i) their intended use as a grouping of shared resources, (ii) the need for synchronization among the resources, or (iii) safety-related interdependencies. As examples, consider (i) an autosampler for a piece of equipment and the piece of equipment itself, which may have distinct drivers but will always be used together so it is best to code their joint actions in an Action Server and abstract the joint action of sampling and measuring as a single FastAPI endpoint; (ii) an isolation valve and a pump where the isolation valve needs to be opened before the pump starts and the pump needs to stop before the isolation valve is closed, which are couplings of driver steps that are best hard coded within a single Action Server; (iii) a set of motors where the limits of the first motor depend on the position of the second motor, for which evaluating the safety of a given motor movement is best done within a single Action Server. While these examples illustrate why multiple resources should be bundled in a single Action Server, the primary counter examples involve resource sharing and hardware/software modularity. Programming the action queuing and prioritization for an Action Server that is shared among multiple Orchestrators is best done by minimizing the set of resources in the Action Server. To best leverage Action Server code for multiple physical instantiations of resources, the scope of an Action Server should be limited to the set of resources that are always implemented collectively into a workflow. This practice also facilitates error handling, where code or instrument failure within a given Action Server will result in the Orchestrator losing access to these capabilities. However, this type of single-point failure does not inherently crash the Orchestrator, so other aspects of the workflow may continue operating. Once the failure is resolved, restarting the Action Server will make its endpoints available to the Orchestrator to restore full workflow operation. We note that programming Orchestrators for automated error recovery is nontrivial, and the present work focuses on providing the capabilities to implement such strategies via automaton of workflows as networks of FastAPI servers.
The multi-Orchestrator demo described above additionally illustrates optionality with respect to the implementation of AI-guided design of experiments. If the AI agent is intended to be a Science Manager across multiple workflows, it should be implemented as an Operator in HELAO-async. However, in the demo, the active learning engine is implemented as an Action Server that is a shared resource for the two Orchestrators. In this case the Science Manager is a human who configured each Orchestrator to receive guidance from the active learning Action Server, which is a prudent mode of operation when the human will routinely switch an Orchestrator from executing experiments according to AI vs. human guidance. As such, this demo may be prescient in the context of instrument control using large language models that integrate human and AI design of experiments.39 While self-driving labs have traditionally been constructed with the AI acquisition function as the Operator, the future of MAPs will likely relegate active learning to be a resource that facilitates but not governs operation of automated experimental workflows. The asynchronous programming and implementation of workflows as networked servers in HELAO-async are designed to enable development of individual automated workflows followed by their seamless interconnection with additional workflows that can be managed by any combination of human and artificial intelligence.
Conclusions
The advent of self driving labs as a fully autonomous implementation of a materials acceleration platform has broadened the purview of experimentation automation beyond traditional high throughput experimentation. Concomitantly, the materials automation community has envisioned international networks of laboratories that may be controlled by multi-agent AI systems and/or human-in-the-loop active learning. As new automated workflows are developed in isolation and then fed into broader networks of capabilities, instrument control software must be prepared to make the transition from single-workflow orchestration to participation in many-workflow automation schemes. Traditional lab automation software cannot effectively manage sharing of resources among workflows while maintaining an independent runtime environment for each workflow. We introduce HELAO-async as a framework for facilitating the automation of individual resources, their incorporation into workflows, and the interconnection of workflows. Combining object-oriented and asynchronous programming, HELAO-async abstracts Resource Managers and Workflow Orchestrators as FastAPI servers. Communication, both between servers and with additional programming instances such as web user interfaces and AI-driven experiment brokers, is implemented using FastAPI websockets and endpoints. In addition to a demo virtual instrument to facilitate learning about HELAO-async, the present work introduces the open source code repositories that house the automation software for a suite of materials acceleration platforms in the high throughput experimentation group at the California Institute of Technology.Data availability
The data for the virtual workflow automation demo is available in the code repository and was previously released along with publication of ref. 38.Code availability
The source code for HELAO-async is available at https://github.com/High-Throughput-Experimentation/helao-core and https://github.com/High-Throughput-Experimentation/helao-async. The instructions for establishing the HELAO Python environment are provided in https://github.com/High-Throughput-Experimentation/helao-async/blob/main/readme.md. The demo is embedded in the repository, and the instructions for launching the demo are available at https://github.com/High-Throughput-Experimentation/helao-async/blob/main/helao/demos/multi_orch_demo.md.• Project name: HELAO-async.
• Project home page: https://github.com/High-Throughput-Experimentation/helao-async.
• Operating system(s): Windows 7, Windows 10, Linux (limited driver functionality).
• Programming language: Python 3.8+.
• License: MIT.
• DOI: https://doi.org/10.22002/q2984-04886.
Author contributions
D. G. is the primary architect and programmer for HELAO-async, implementing the vision established by J. M. G., H. S. S., and D. G.; P. D. prototyped asyncio routines; M. R., Y. L., R. J. J., and K. K. helped implement HELAO-async for specific workflows under guidance from D. G. and J. M. G.; L. Z. contributed to Observer visualization code.Conflicts of interest
J. M. G. consults for companies that aim to automate experiments.Acknowledgements
This material is primarily based on work performed by the Liquid Sunlight Alliance, which is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, Fuels from Sunlight Hub under Award DE-SC0021266. Software development was also supported by Toyota Research Institute and by the Air Force Office of Scientific Research under award FA9550-18-1-0136.References
- M. M. Flores-Leonar, L. M. Mejía-Mendoza, A. Aguilar-Granda, B. Sanchez-Lengeling, H. Tribukait, C. Amador-Bedolla and A. Aspuru-Guzik, Curr. Opin. Green Sustainable Chem., 2020, 25, 100370 CrossRef.
- R. Vescovi, R. Chard, N. D. Saint, B. Blaiszik, J. Pruyne, T. Bicer, A. Lavens, Z. Liu, M. E. Papka, S. Narayanan, N. Schwarz, K. Chard and I. T. Foster, Patterns, 2022, 3, 100606 CrossRef PubMed.
- M. L. Green, C. L. Choi, J. R. Hattrick-Simpers, A. M. Joshi, I. Takeuchi, S. C. Barron, E. Campo, T. Chiang, S. Empedocles, J. M. Gregoire, A. G. Kusne, J. Martin, A. Mehta, K. Persson, Z. Trautt, J. V. Duren and A. Zakutayev, Applied Physics Reviews, 2017, 4, 011105 CrossRef.
- J. M. Gregoire, L. Zhou and J. A. Haber, Nat. Synth., 2023, 2, 493–504 CrossRef.
- E. Stach, B. DeCost, A. G. Kusne, J. Hattrick-Simpers, K. A. Brown, K. G. Reyes, J. Schrier, S. Billinge, T. Buonassisi, I. Foster, C. P. Gomes, J. M. Gregoire, A. Mehta, J. Montoya, E. Olivetti, C. Park, E. Rotenberg, S. K. Saikin, S. Smullin, V. Stanev and B. Maruyama, Matter, 2022, 4, 2702–2726 CrossRef.
- M. Seifrid, R. Pollice, A. Aguilar-Granda, Z. Morgan Chan, K. Hotta, C. T. Ser, J. Vestfrid, T. C. Wu and A. Aspuru-Guzik, Acc. Chem. Res., 2022, 55, 2454–2466 CrossRef CAS PubMed.
- H. S. Stein and J. M. Gregoire, Chem. Sci., 2019, 10, 9640–9649 RSC.
- G. E. Karniadakis, I. G. Kevrekidis, L. Lu, P. Perdikaris, S. Wang and L. Yang, Nat. Rev. Phys., 2021, 3, 422–440 CrossRef.
- D. Chen, Y. Bai, S. Ament, W. Zhao, D. Guevarra, L. Zhou, B. Selman, R. B. van Dover, J. M. Gregoire and C. P. Gomes, Nature Machine Intelligence, 2021, 3, 1–11 CrossRef.
- M. A. Ziatdinov, Y. Liu, A. N. Morozovska, E. A. Eliseev, X. Zhang, I. Takeuchi and S. V. Kalinin, Adv. Mater., 2022, 34, 2201345 CrossRef CAS PubMed.
- J. Bai, L. Cao, S. Mosbach, J. Akroyd, A. A. Lapkin and M. Kraft, JACS Au, 2022, 2, 292–309 CrossRef CAS PubMed.
- Z. Ren, Z. Ren, Z. Zhang, T. Buonassisi and J. Li, Nat. Rev. Mater., 2023, 1–2 Search PubMed.
- P. Nikolaev, D. Hooper, F. Webber, R. Rao, K. Decker, M. Krein, J. Poleski, R. Barto and B. Maruyama, npj Comput. Mater., 2016, 2, 1–6 CrossRef.
- L. M. Roch, F. Häse, C. Kreisbeck, T. Tamayo-Mendoza, L. P. E. Yunker, J. E. Hein and A. Aspuru-Guzik, Sci. Robot., 2018, 3, eaat5559 CrossRef PubMed.
- D. Allan, T. Caswell, S. Campbell and M. Rakitin, Synchrotron Radiation News, 2019, 32, 19–22 CrossRef.
- J. R. Deneault, J. Chang, J. Myung, D. Hooper, A. Armstrong, M. Pitt and B. Maruyama, MRS Bull., 2021, 46, 566–575 CrossRef.
- T. Konstantinova, P. M. Maffettone, B. Ravel, S. I. Campbell, A. M. Barbour and D. Olds, Digital Discovery, 2022, 1, 413–426 RSC.
- F. Rahmanian, J. Flowers, D. Guevarra, M. Richter, M. Fichtner, P. Donnely, J. M. Gregoire and H. S. Stein, Adv. Mater. Interfaces, 2022, 9, 2101987 CrossRef.
- M. Vogler, J. Busk, H. Hajiyani, P. B. Jørgensen, N. Safaei, I. E. Castelli, F. F. Ramirez, J. Carlsson, G. Pizzi, S. Clark, F. Hanke, A. Bhowmik and H. S. Stein, Matter, 2023, 6(7), 2095–2245 CrossRef.
- R. Tamura, K. Tsuda and S. Matsuda, Sci. Technol. Adv. Mater.: Methods, 2023, 3, 2232297 Search PubMed.
- C. J. Leong, K. Y. A. Low, J. Recatala-Gomez, P. Q. Velasco, E. Vissol-Gaudin, J. D. Tan, B. Ramalingam, R. I. Made, S. D. Pethe, S. Sebastian, Y.-F. Lim, Z. H. J. Khoo, Y. Bai, J. J. W. Cheng and K. Hippalgaonkar, Matter, 2022, 5, 3124–3134 CrossRef.
- A. G. Kusne and A. McDannald, Matter, 2023, 6, 1880–1893 CrossRef.
- R. Vescovi, T. Ginsburg, K. Hippe, D. Ozgulbas, C. Stone, A. Stroka, R. Butler, B. Blaiszik, T. Brettin, K. Chard, M. Hereld, A. Ramanathan, R. Stevens, A. Vriza, J. Xu, Q. Zhang and I. Foster, arXiv, 2023 DOI:10.48550/arXiv.2308.09793.
- M. Sim, M. Ghazi Vakili, F. Strieth-Kalthoff, H. Hao, R. Hickman, S. Miret, S. Pablo-García and A. Aspuru-Guzik, ChemRxiv, 2023 DOI:10.26434/chemrxiv-2023-v2khf.
- H. S. Stein, A. Sanin, F. Rahmanian, B. Zhang, M. Vogler, J. K. Flowers, L. Fischer, S. Fuchs, N. Choudhary and L. Schroeder, Curr. Opin. Electrochem., 2022, 35, 101053 CrossRef CAS.
- Asyncio vs. Threading in Python, 2023, https://superfastpython.com/asyncio-vs-threading/ Search PubMed.
- I. M. Pendleton, G. Cattabriga, Z. Li, M. A. Najeeb, S. A. Friedler, A. J. Norquist, E. M. Chan and J. Schrier, MRS Commun., 2019, 9, 846–859 CrossRef CAS.
- M. Lathkar, High-Performance Web Apps with FastAPI: The Asynchronous Web Framework Based on Modern Python, Apress, 2023 Search PubMed.
- L. Bromig, D. Leiter, A.-V. Mardale, N. v. d. Eichen, E. Bieringer and D. Weuster-Botz, SoftwareX, 2022, 17, 100991, DOI:10.1016/j.softx.2022.100991.
- A. Biswas, Y. Liu, N. Creange, Y.-C. Liu, S. Jesse, J.-C. Yang, S. V. Kalinin, M. A. Ziatdinov and R. K. Vasudevan, A dynamic Bayesian optimized active recommender system for curiosity-driven human-in-the-loop automated experiments, 2023, http://arxiv.org/abs/2304.02484 Search PubMed.
- J. H. Montoya, M. Aykol, A. Anapolsky, C. B. Gopal, P. K. Herring, J. S. Hummelshøj, L. Hung, H.-K. Kwon, D. Schweigert, S. Sun, S. K. Suram, S. B. Torrisi, A. Trewartha and B. D. Storey, Applied Physics Reviews, 2022, 9, 011405 CrossRef CAS.
- S. Ament, M. Amsler, D. R. Sutherland, M.-C. Chang, D. Guevarra, A. B. Connolly, J. M. Gregoire, M. O. Thompson, C. P. Gomes and R. B. v. Dover, Sci. Adv., 2021, 7, eabg4930 CrossRef CAS PubMed.
- J. Li, J. Li, R. Liu, Y. Tu, Y. Li, J. Cheng, T. He and X. Zhu, Nat. Commun., 2020, 11, 2046 CrossRef CAS PubMed.
- J. M. Gregoire, C. Xiang, X. Liu, M. Marcin and J. Jin, Rev. Sci. Instrum., 2013, 84, 024102 CrossRef PubMed.
- S. Mitrovic, E. W. Cornell, M. R. Marcin, R. J. Jones, P. F. Newhouse, S. K. Suram, J. Jin and J. M. Gregoire, Rev. Sci. Instrum., 2015, 86, 013904 CrossRef PubMed.
- L. Zhou, A. Shinde, J. H. Montoya, A. Singh, S. Gul, J. Yano, Y. Ye, E. J. Crumlin, M. H. Richter, J. K. Cooper, H. S. Stein, J. A. Haber, K. A. Persson and J. M. Gregoire, ACS Catal., 2018, 8, 10938–10948 CrossRef CAS.
- R. J. R. Jones, Y. Wang, Y. Lai, A. Shinde and J. M. Gregoire, Rev. Sci. Instrum., 2018, 89, 124102 CrossRef PubMed.
- H. S. Stein, D. Guevarra, A. Shinde, R. J. R. Jones, J. M. Gregoire and J. A. Haber, Mater. Horiz., 2019, 1251–1258 RSC.
- Z. Ren, Z. Zhang, Y. Tian and J. Li, ChemRxiv, 2023 DOI:10.26434/chemrxiv-2023-tnz1x.
Footnotes |
| † Present address: Good Terms LLC, CO, USA. |
| ‡ Present address: deepXscan GmbH, Dresden, Germany. |
| This journal is © The Royal Society of Chemistry 2023 |