Open Access Article
Open Access ArticleDistinct breast milk microbiota, cytokine, and adipokine profiles are associated with infant growth at 12 months: an in vitro host–microbe interaction mechanistic approach†
Erika
Cortés-Macías
 a,
Marta
Selma-Royo
a,
Karla
Rio-Aige
a,
Marta
Selma-Royo
a,
Karla
Rio-Aige
 bc,
Christine
Bäuerl
bc,
Christine
Bäuerl
 a,
María José
Rodríguez-Lagunas
a,
María José
Rodríguez-Lagunas
 bc,
Cecilia
Martínez-Costa
bc,
Cecilia
Martínez-Costa
 de,
Francisco J.
Pérez-Cano
de,
Francisco J.
Pérez-Cano
 bc and
Maria Carmen
Collado
bc and
Maria Carmen
Collado
 *a
*a
aDepartment of Biotechnology, Institute of Agrochemistry and Food Technology-, National Research Council (IATA-CSIC), Valencia, Spain. E-mail: mcolam@iata.csic.es
bPhysiology Section, Department of Biochemistry and Physiology, Faculty of Pharmacy and Food Science, University of Barcelona (UB), 08028 Barcelona, Spain
cNutrition and Food Safety Research Institute (INSA-UB), 08921 Santa Coloma de Gramenet, Spain
dDepartment of Pediatrics, School of Medicine, University of Valencia, Valencia, Spain
ePediatric Gastroenterology and Nutrition Section, Hospital Clínico Universitario Valencia, INCLIVA, Valencia, Spain
First published on 16th November 2022
Abstract
Breast milk (BM) is important for adequate infant development, and it contains bioactive compounds, such as bacteria, cytokines and some adipokines which play a role in infant microbial, metabolic, and immunological maturation. However, little is known about its impact on growth and development in early life. The objective of this study was to evaluate the impact of milk microbiota, cytokine, and adipokine profiles on the risk of overweight at 12 months of life to find the possible mechanisms of host–microbe interactions. In this study, BM samples from 100 healthy women collected during 15 d after birth were included. BM microbiota was analysed by 16S rRNA gene sequencing, and cytokine and adipokine levels were measured by the Luminex approach. In addition, infant weight and length were recorded during the first 12 months and z-scores were obtained according to the WHO databases. Infants were classified as risk of overweight (ROW) and no-risk of overweight (NOROW) based on their body mass index z-score (BMIZ) and infant weight-for-length z-score (WLZ) at 12 months. In order to study host–microbe interactions, epithelial intestinal and mammary cell lines were exposed to milk microbes to assess the host response by interleukin (IL)-6 production as a potential inflammatory marker. BM was dominated by Staphylococcus and Streptococcus genera, and the most abundant cytokines were IL-6 and IL-18. Leptin levels were positively correlated with the pregestational body mass index (BMI). Higher relative abundance of the Streptococcus genus was associated with higher IL-10 and higher relative abundance of the Bifidobacterium genus was associated with lower IL-6 concentrations in milk. Infant WLZ at 12 months could be partially predicted by Streptococcus genus proportions and IL-10 and IL-18 levels in BM. BM microbiota significantly induced cytokine responses in mammary epithelial cells. Higher levels of IL-6 production were observed in mammary cells exposed to BM microbiota from mothers with ROW offspring compared to mothers with NOROW offspring. In conclusion, BM microbiota is related to the cytokine profile. IL-10 and IL-18 levels and the abundance of the Streptococcus genus could affect early infant development. Further research is needed to clarify the specific impact of BM microbiota and cytokines on infant growth and the risk of overweight.
Introduction
Early nutrition exerts both short- and long-term effects on human health in programming its immunological, metabolic, and microbiological development. Breastfeeding confers multiple health benefits to offspring, including protection from infections and probable protection against obesity.1 The child obesity rates are increasing worldwide, and it is estimated that on average nearly one in eight children aged 7–8 years is obese in European countries (World Health Organization (WHO) Europe 2018, Children Obesity Surveillance Initiative, highlights 2015–17, preliminary data), with Spain being one of the countries with the highest obesity rates. Thus, European health care systems are supporting strategies to reduce the prevalence of obesity.Breastfeeding has been associated with a significant reduced risk of obesity, showing a dose–response effect between the breastfeeding duration and the reduction of the risk.2 In this regard, breast milk (BM) is the best food and the first option for infant nutrition,3,4 as it has functions with implications for adequate gut microbial assembly and immune system development.5
Alterations in microbial colonisation in early life due to C-section,6–8 antibiotic exposure,6 and lack of breastfeeding have been associated with a higher prevalence of obesity in children.1,2
A distinct microbial pattern has been observed in infants fed with formula compared to breastfed infants,9 mainly due to the predominance of Bifidobacteriaceae in breastfed infants during the first months of life.10 These observations could be explained by the presence of microbes and oligosaccharides, as well as other components in BM. Thus, it has been hypothesised that shifts in milk microbiota due to some maternal disorders, such as obesity11,12 or other maternal and infant factors that shape the BM microbiota composition,13–15 could be transferred to the neonates through an unbalanced microbial colonisation. However, the mechanisms that drive this relationship have been underexplored and these relationships are still poorly understood. To our knowledge, no study has shown the role of BM microbiota in infant growth and development.
Therefore, in this scenario, our main objective was to assess the impact of milk microbiota and immune and adipokine profiles on infant growth and the risk of overweight at 12 months of life, as well as to find potential mechanisms behind the BM microbiota–immune signal–obesity risk relationship using in vitro approaches.
Materials and methods
Study design and volunteers
A subgroup of 100 healthy mother–infant dyads ≤15 d post birth following breastfeeding practices from the MAMI cohort (Maternal Microbes, Valencia, Spain)16 were included in this study based on the availability of clinical and anthropometric data including maternal age, gestational age, delivery mode, maternal body mass index (BMI), and weight gain during pregnancy.All participants received oral and written information about the study and written consent was obtained from them. This study is registered on the ClinicalTrial.gov platform with registration number NCT03552939 and it is approved by the Ethics Committees of the Hospital Clínico Universitario de Valencia (Spain).
Infant anthropometric measures
Simultaneous weight and length measurements were collected during periodical paediatric visits by clinicians during the first months of life at birth, 1, 6, and 12 months. The age and sex specific BMI z-score (BMIZ) and weight-for-length z-score (WLZ) were calculated using WHO Anthro software.17 Children were classified as at risk of overweight (ROW) and no-risk of overweight (NOROW) based on their BMIZ and WLZ at 12 months. BMIZ and WLZ greater than the 85th percentile (z-score ≥0.99) were considered as at risk of overweight as has been previously described elsewhere.18 The changes in the BMIZ and WLZ were calculated by taking the difference in the z-score between time points as described elsewhere.19BM samples
Breast milk samples were collected from mothers (n = 100) ≤15 d (average = 11.32 d) after birth following a protocol described previously.13 The samples were centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C for 15 min, and the upper fat layer was removed. BM pellets were used for DNA extraction and in vitro cell model analysis and the supernatants were used for the determination of cytokine and adipokine profiles.
000g at 4 °C for 15 min, and the upper fat layer was removed. BM pellets were used for DNA extraction and in vitro cell model analysis and the supernatants were used for the determination of cytokine and adipokine profiles.
BM microbiota profile
Total DNA was extracted from the samples using 1.5–2.0 mL of BM using the Master-Pure DNA Extraction kit (Epicentre, Madison, WI, USA) following a previously described protocol.20 Then, the total DNA was purified using the MagSi-NGS Plus kit (Amsbio, Abingdon, UK) following the manufacturer's instructions. Controls for DNA extraction and PCR amplification were also included.Milk microbiota composition was assessed by the sequencing of the V3–V4 variable region of the 16S rRNA gene following the Illumina protocols as described by García-Mantrana et al.21 using a MiSeq-Illumina platform (FISABIO sequencing service, Valencia, Spain). Briefly, a Nextera XT Index kit (Illumina, CA, USA) was used for the multiplexing step, and the libraries were sequenced using a 2 × 300 pb paired-end run (MiSeq Reagent kit v3).
Trimmomatic software22 was used to search and remove the residual adaptors and DADA2 pipeline v.1.1623 was used for quality filtering, sequence joining, and chimera removal. Taxonomic assignment was achieved using the Silva v132 database, including the species level classification.24 Additional filtering was performed in which samples with less than 1000 reads, amplicon sequence variants (ASVs) with a relative abundance less than 0.01% and those present less than 3 times in at least 20% of the samples were removed. Also, the decontam package25 in the Rstudio environment was used to identify the possible contaminants which were removed from the final analysis (n = 48 ASVs).
BM cytokine and adipokine profiles
The quantification of cytokines (GM-CSF, IFN-γ, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12p70, IL-13, IL-17A, IL-18, IL-21, IL-22, IL-23, IL-27, and TNF-α) was performed by ProcartaPlex™ Multiplex immunoassay (Thermo Fisher Scientific, Austria) using a Th1/Th2/Th9/Th17 Cytokine 18-Plex Human ProcartaPlex™ panel following the manufacturer's instructions. Assay sensitivity was as follows: 1.2 pg mL−1 for GM-CSF; 0.2 pg mL−1 for IFN-γ; 0.2 pg mL−1 for IL-1β; 0.8 pg mL−1 for IL-2; 1.5 pg mL−1 for IL-4; 0.3 pg mL−1 for IL-5; 0.4 pg mL−1 for IL-6; 0.5 pg mL−1 for IL-9; 0.1 pg mL−1 for IL-10; 0.04 pg mL−1 for IL-12p70; 0.1 pg mL−1 for IL-13; 0.1 pg mL−1 for IL-17A; 0.4 pg mL−1 for IL-18; 0.6 pg mL−1 for IL-21; 8.2 pg mL−1 for IL-22; 0.9 pg mL−1 for IL-23; 5.1 pg mL−1 for IL-27; and 0.4 pg mL−1 for TNF-α.The quantification of leptin was performed using a Quantikine® Colorimetric Sandwich ELISA kit (R&D Systems, Minneapolis, MN, USA) following the manufacturer's instructions. Assay sensitivity was 7.8 pg mL−1 for leptin. Adiponectin was analysed using the Adiponectin Human ProcartaPlex™ Simplex kit (Thermo Fisher Scientific) with an assay sensitivity of 4.6 pg mL−1.
Host–microbe interactions: in vitro approach
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C for 15 min, and the upper fat layer was removed. The samples were centrifuged again at same conditions to collect the bacterial pellet and this was resuspended in 500 μL of phosphate buffered saline (PBS) to be inactivated for 10 min at 65 °C.
000g at 4 °C for 15 min, and the upper fat layer was removed. The samples were centrifuged again at same conditions to collect the bacterial pellet and this was resuspended in 500 μL of phosphate buffered saline (PBS) to be inactivated for 10 min at 65 °C.
To explore the maternal mammary epithelia and milk microbe interactions, the MCF7 (ATCC HTB-22) mammary epithelial cell line was used. Cells were routinely maintained at 37 °C under a humidified atmosphere of 5% CO2 in DMEM high glucose with stable glutamine and sodium pyruvate culture medium (Capricorn Scientific, Ebsdorfergrund, Germany), supplemented with 10% v/v inactivated fetal bovine serum (FBS, Biowest, Nuaillé, France), 1% non-essential amino acids (Capricorn Scientific, Ebsdorfergrund, Germany), 10 mM HEPES (Capricorn Scientific, Ebsdorfergrund, Germany), and antibiotics (100 U mL−1 penicillin and 100 μg mL−1 streptomycin [Sigma-Aldrich, Missouri, USA]), according to a procedure reported elsewhere.26
MCF7 cells were seeded in 12-well plates at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in complete growth medium without antibiotics. After 24 h, the cells were exposed to bacterial pellet suspension diluted 1
000 cells per well in complete growth medium without antibiotics. After 24 h, the cells were exposed to bacterial pellet suspension diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (v/v) in the specified medium and were co-incubated with human milk bacteria for 20 h at 37 °C and under 5% CO2 in an incubator. Negative controls consisted of MCF7 cells incubated without bacteria. The experiment was performed in triplicate.
10 (v/v) in the specified medium and were co-incubated with human milk bacteria for 20 h at 37 °C and under 5% CO2 in an incubator. Negative controls consisted of MCF7 cells incubated without bacteria. The experiment was performed in triplicate.
After co-incubation, the cells and supernatants were used for both gene expression determination and cytokine quantification, respectively. IL-6 concentrations were measured by ELISA (Invitrogen, Vienna, Austria) using 100 μL of the supernatant, following the manufacturer's instructions.
RNA was extracted using the NucleoSpin RNA XS kit (Macherey-Nagel, Düren, Germany) following the manufacturer's instructions. cDNA from the total RNA was generated using a high-capacity cDNA reverse transcription kit (Applied Biosystems, CA, USA) after total RNA normalisation. RT-PCR analysis was performed using the SYBR Green PCR Master Mix (Roche), using 1 μL of cDNA and the specific primers (0.15 μM): IL-6 F (5′-GTGTGAAAGCAGCAAAGAGGC-3′), IL-6 R (5′-TGCAGGAACTGGATCAGGACT-3′),27 actin (ACTB) F (5′-TTGTTACAGGAAGTCCCTTGCC-3′), and ACTB R (5′-ATGCTATCACCTCCCCTGTGTG-3′),28 which was used as a housekeeping gene. The annealing temperature was 58 °C for both targets. LC480 Conversion version 2014.1 and LinRegPCR v. 11.0 software were used for efficiency calculation,29,30 and the relative gene expression was quantified according to the efficiency-corrected method using the REST 2009 software tool.31
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 viable cells per well and expanded at 37 °C under a humidified atmosphere of 5% CO2 in DMEM high glucose with stable glutamine and sodium pyruvate culture medium (Capricorn Scientific), supplemented with 10% v/v FBS (Biowest), antibiotics (100 U mL−1 penicillin and 100 μg mL−1 streptomycin [Sigma-Aldrich]), and zeocin (200 μg mL−1, Invivogen, CA, USA).
000 viable cells per well and expanded at 37 °C under a humidified atmosphere of 5% CO2 in DMEM high glucose with stable glutamine and sodium pyruvate culture medium (Capricorn Scientific), supplemented with 10% v/v FBS (Biowest), antibiotics (100 U mL−1 penicillin and 100 μg mL−1 streptomycin [Sigma-Aldrich]), and zeocin (200 μg mL−1, Invivogen, CA, USA).
The cells were exposed to the same BM pellets as detailed above, and the supernatants were collected after 24 h of stimulation. SEAP activity was measured using p-nitrophenyl phosphate as a substrate according to manufacturer's instructions (Thermo Fisher Scientific, Waltham, USA). The signal was quantified using a CLARIOstar microtiter plate reader (BMG Labtech, Ortenberg, Germany) at 405 nm. The cells were lysed in PBS containing 0.1% Triton, 1 mM phenylmethylsulphonyl fluoride (PMSF), and 1 mM ethylenediaminetetraacetic acid (EDTA). The protein content of each well was determined using the Bradford Protein Assay (Bio-Rad, CA, USA). SEAP activity was calculated according to the formula: SEAP activity = (A405 nm test − A405 nm initial) × the total assay volume (mL)/mM extinction coefficient of p-nitrophenol (18.5) × the cell culture supernatant employed (mL) × time (min) and normalised to the protein content of each well.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min at 4 °C, and the resulting supernatant was filtered using a 0.20 μm filter and diluted 1
000g for 10 min at 4 °C, and the resulting supernatant was filtered using a 0.20 μm filter and diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (v/v) in DMEM. HT-29-transfected cells were exposed to this supernatant as described above, and after 24 h of stimulation, the culture media were collected, and the SEAP activity was measured as detailed before. The experiment was performed in triplicate.
10 (v/v) in DMEM. HT-29-transfected cells were exposed to this supernatant as described above, and after 24 h of stimulation, the culture media were collected, and the SEAP activity was measured as detailed before. The experiment was performed in triplicate.
Statistical analysis
A T-test and Mann–Whitney analysis were used depending on the data normality assessed by the Kolmogorov–Smirnov and Shapiro–Wilk tests (GraphPad Prism V5.04). A nonmetric multidimensional scaling (NMDS) analysis based on the Bray–Curtis distance was performed to assess the effect of perinatal factors on the BM microbiota, cytokine, and adipokine compositions.33The Spearman correlations between cytokine and adipokine concentrations and the relative abundances of the bacterial genera, adjusted for mode of birth, intrapartum antibiotic (ATB), and breastfeeding practices at 15 d, were obtained using SPSS V.27 and the heatmap plot was obtained using RStudio.34–36 Multivariate linear regression (backward regression) analysis was then used to determine the ability of microbiota, cytokine, and adipokine concentrations to predict longitudinal growth outcomes at 12 months post-partum. The following software was used for analysis: SPSS V.2737 (IBM Corp., released 2020; IBM SPSS Statistics for Windows, version 27.0., Armonk, NY: IBM Corp.), GraphPad Prism v. 5.04 (GraphPad Software, San Diego, CA, USA, https://www.graphpad.com) and RStudio.34
Results
Subjects and clinical data
100 families were included in the study (Table 1). 85% of the mother–infant pairs were following exclusive breastfeeding (EBF) at the time of milk collection. Approximately half of the included infants were females (57%), 61% were born vaginally and 39% were born by C-section delivery (Table 1). From the 100 children, 17% were defined as ROW and 82% were defined as NOROW at 12 months according to the BMIZ and WLZ following the WHO growth curves (Table 1). In our cohort, we only observed significant differences in the BMIZ and WLZ between ROW and NOROW infants at 6 and 12 months of life (p < 0.001). As expected, a higher weight gain (kg) over pregnancy was observed in mothers with ROW offspring compared to mothers with NOROW offspring (p = 0.045). The changes in growth from 6 to 12 months are presented in Fig. S1.† The changes in the WLZ and BMIZ values (Fig. S1A and S1B†) were significantly lower in the NOROW offspring than in the ROW offspring.| Total (n = 100) | NOROW (n = 82)a | ROW (n = 17)a | p-value | ||
|---|---|---|---|---|---|
| Categorical variables are expressed as positive cases-prevalence and (percentage, %) and a chi-squared test was performed to assess the significance. Normally distributed data are presented as mean ± standard deviation (SD) and non-normal data as median and interquartile range [IQR]. BMI, body mass index; NW, normal weight; OW, overweight; BMIZ, body mass index z-score; WLZ, weight-for-length z-score; NOROW, no risk of overweight; ROW, risk of overweight. p < 0.05 was considered statistically significant.a One infant has not available information on weight and length at 12 months and it was not included. | |||||
| Maternal characteristics | |||||
| Maternal age (years) | 34.77 ± 3.74 | 34.76 ± 3.78 | 34.76 ± 3.78 | 0.993 | |
| Gestational age (weeks) | 40 [39–40] | 40 [39–40] | 40 [39–41] | 0.313 | |
| Pre-gestational BMI (kg m−2) | 22.8 [21.0–25.1] | 22.8 [21.0–25.4] | 23.0 [20.4–23.6] | 0.610 | |
| Weight gain (kg) during pregnancy | 12 [10–14] | 12 [9–14] | 14.5 [11–18] | 0.045 | |
| Antibiotic treatment during pregnancy (%) | 31 (31%) | 25 (30%) | 6 (35%) | 0.697 | |
| Intrapartum antibiotic exposure (%) | 43 (43%) | 33 (40%) | 10 (59%) | 0.160 | |
| Infant characteristics | |||||
| Gender: female (%) | 57 (57%) | 46 (56%) | 10 (59%) | 0.837 | |
| Birth mode: vaginal birth (%) | 61 (61%) | 52 (63%) | 8 (47%) | 0.209 | |
| Antibiotic treatment, 15 days (%) | 7 (7%) | 6 (7%) | 1 (6%) | 0.834 | |
| Breastfeeding duration (months) | 9.5 [6–12] | 9.5 [6–12] | 12 [6–12] | 0.713 | |
| Breast feeding at 15 days | |||||
| Exclusive breastfeeding (EBF) | 85 (85%) | 70 (85%) | 14 (82%) | 0.753 | |
| Mixed feeding (MF) | 15 (15%) | 12 (15%) | 3 (18%) | ||
| BMIZ | At birth | −0.11 ± 1.06 | −0.17 ± 1.02 | 0.15 ± 1.20 | 0.258 |
| 1 month | −0.49 ± 0.99 | −0.59 ± 0.96 | −0.01 ± 1.03 | 0.027 | |
| 6 months | −0.26 ± 0.87 | −0.42 ± 0.77 | 0.50 ± 0.92 | <0.001 | |
| 12 months | 0.20 ± 0.91 | −0.06 ± 0.75 | 1.48 ± 0.41 | <0.001 | |
| WLZ | At birth | −0.19 ± 1.15 | −0.24 ± 1.12 | 0.06 ± 1.30 | 0.330 |
| 1 month | −0.49 ± 1.22 | −0.56 ± 1.25 | −0.16 ± 1.01 | 0.220 | |
| 6 months | −0.13 ± 0.82 | −0.28 ± 0.72 | 0.58 ± 0.91 | <0.001 | |
| 12 months | 0.20 ± 0.86 | −0.06 ± 0.68 | 1.46 ± 0.35 | <0.001 | |
Factors shaping the BM microbiota, cytokine, and adipokine compositions at 15 d of lactation
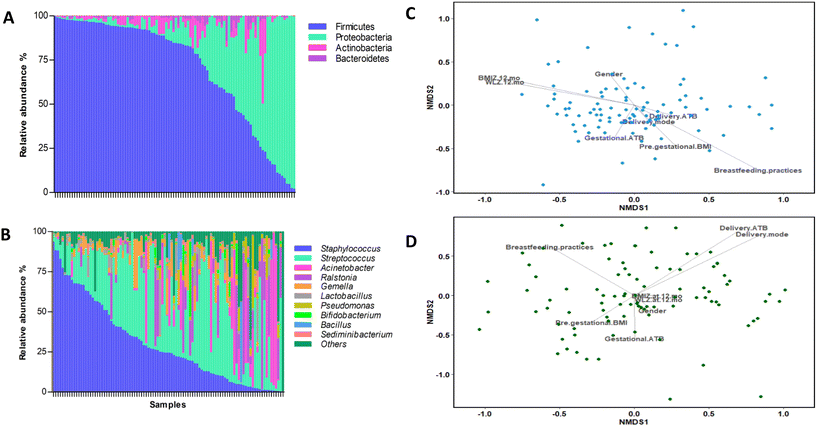
In general, BM microbial communities were characterised by a dominance of Firmicutes [84.49%; interquartile range (IQR): 48.39–94.09] and Proteobacteria [7.88%; IQR: 1.54–38.45] phyla, followed by Actinobacteria [1.82%; IQR: 0.55–5.11] and Bacteroidetes [0.66%; IQR: 0.15–1.73] phyla (Fig. 1A). At the genus level, the most abundant taxa were Streptococcus [28.10%; IQR: 8.48–46.92] and Staphylococcus [24.37%; IQR: 8.17–49.96], followed by Acinetobacter [0.18%; IQR: 0.04–1.50] and Ralstonia [0.08%; IQR: 0–6.24] (Fig. 1B).We performed a NMDS analysis to identify the main perinatal factors contributing to the BM microbiota. We found that breastfeeding practices significantly affect the NMDS ordination of the samples based on BM microbiota (envfit; breastfeeding practices: R2 = 0.088, p = 0.011) (Fig. 1C). Additionally, the BMIZ at 12 months showed a significant correlation with the NMDS ordination (envfit; BMIZ at 12 months: R2 = 0.065, p = 0.044) (Fig. 1C).
The concentration of cytokines, leptin, and adiponectin detected in BM are listed in the ESI (Table S1†). The levels obtained varied among cytokines in terms of concentration and detection, with the most abundant cytokines being IL-6, IL-18, IL-21, and IL-1β (Table S1†), even though the levels of detection of IL-6, IL-18, IL-21, TNF-α and leptin in the BM were above the assay limit of detection in more than 48% of subjects. We explored whether perinatal factors could affect the BM microbiota composition and the cytokine concentration. We found that the delivery mode and ATB significantly impacted the NMDS ordination based on the cytokine and adipokine profiles (envfit; delivery mode: R2 = 0.0615, p = 0.049; delivery ATB: R2 = 0.0671, p = 0.042) (Fig. 1D).
To assess the influence of pre-gestational BMI and breastfeeding practices on the cytokine, microbiota, and adipokine concentrations, the correlations between these parameters were also studied.
The pre-gestational maternal BMI was positively correlated with the BM microbiota: Staphylococcus genus (ρ = 0.207, p = 0.039) (Table S2†) and the BM cytokines TNF-α (ρ = 0.30, p = 0.03) and IL-22 (ρ = 0.20, p = 0.048) (Table S2†). Additionally, the pre-gestational maternal BMI was positively correlated with leptin (ρ = 0.388, p < 0.001) (Fig. S2A†), while no correlation was observed for BM adiponectin (ρ = −0.109, p = 0.28). Furthermore, mothers with mixed feeding mode showed higher leptin (p = 0.003) (Fig. S2B†) concentration compared with those with exclusive breastfeeding (p = 0.003) (Fig. S2B†), and no significant difference was observed for adiponectin (p = 0.451) (Fig. S2C†) concentration compared with those with exclusive breastfeeding.
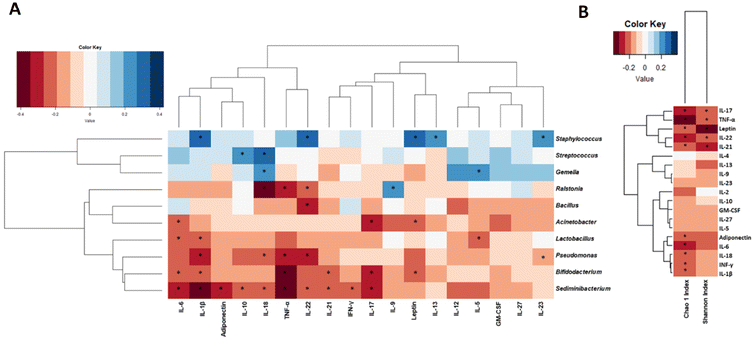
Associations between BM microbiota, milk cytokines, and adipokines
Significant associations between cytokines and BM microbial genera were identified (Fig. 2A). Higher Bifidobacterium genus was associated with lower IL-6 (ρ = −0.23, p = 0.025), TNF-α (ρ = −0.40, p < 0.001) and IL-21 (ρ = −0.21, p = 0.044). The Streptococcus genus was positively related to IL-18 (ρ = 0.28, p = 0.006) and IL-10 (ρ = 0.24, p = 0.018), while a higher abundance of the Staphylococcus genus was associated with higher IL-22 (ρ = 0.32, p = 0.002) and IL-23 (ρ = 0.30, p = 0.004).Negative correlations were also observed between Acinetobacter and IL-17 (ρ = −0.26, p = 0.011) and also between Pseudomonas and IL-18 (ρ = −0.24, p = 0.021) and IL-22 (ρ = −0.28, p = 0.007).
With regard to the associations between adipokines and BM microbiota (Fig. 2A), negative correlations were observed between leptin and the Bifidobacterium (ρ = −0.20, p = 0.048) or the Acinetobacter (ρ = −0.24, p = 0.021) genus. Similarly, adiponectin also showed a negative relationship with the relative abundance of Sediminibacterium (ρ = −0.27, p = 0.010). Associations between the Chao1 and Shannon indices and cytokines/adipokines were also observed (Fig. 2B). Both leptin (ρ = −0.21, p = 0.037) and adiponectin (ρ = −0.22, p = 0.031) showed a negative correlation with the Chao1 index, which was also negatively correlated with IL-1β (ρ = −0.21, p = 0.035). Similarly, the Shannon index was also negatively correlated with leptin (ρ = −0.37, p < 0.001) and TNF-α (ρ = −0.22, p = 0.029).
BM microbiota, cytokine, and adipokine profiles were associated with infant growth trajectories and the risk of overweight at 12 months of life
The concentrations of cytokines, leptin, and adiponectin in BM in mothers with ROW offspring compared to mothers with NOROW offspring are shown in Table 2.| Cytokines and adipokines (pg mL−1) | NOROW (n = 82) | ROW (n = 17) | ||||
|---|---|---|---|---|---|---|
| IQR | pg mL−1 | % det | IQR | pg mL−1 | % det | |
| Data shown are expressed as [IQR]. Detectability frequencies (% det). GM-CSF, granulocyte macrophage colony-stimulating factor; IFN, interferon; IL, interleukin; TNF, tumor necrosis factor; NOROW, no risk of overweight; ROW, risk of overweight.a A Mann–Whitney U test was used to determine significant differences between the concentrations and a chi-square test compared detectability. p < 0.05 was considered statistically significant. Significances are highlighted in bold. | ||||||
| IL-2 | 0.00–0.00 | 0.67 ± 2.14 | 10.98 (9) | 0.00–0.00 | 0.59 ± 1.65 | 11.76 (2) |
| IL-4 | 0.00–0.00 | 0.19 ± 1.15 | 3.66 (3) | 0.00–0.00 | 0.04 ± 0.14 | 5.88 (1) |
| IL-5 | 0.00–0.00 | 0.20 ± 0.72 | 8.54 (7) | 0.00–0.00 | 0.09 ± 0.34 | 5.88 (1) |
| IL-6 | 0.00–38.77 | 57.48 ± 162.16 | 54.88 (45) | 0.00–0.00 | 142.26 ± 418.67 | 17.65 (3) |
| IL-9 | 0.00–0.00 | 0.24 ± 1.25 | 4.88 (4) | 0.00–0.00 | 0.04 ± 0.18 | 5.88 (1) |
| IL-10 | 0.00–0.34 | 1.04 ± 4.98 | 60.98 (50) | 0.00–0.23 | 0.22 ± 0.55 | 58.82 (10) |
| IL-12 | 0.00–0.10 | 0.34 ± 2.55 | 25.61 (21) | 0.00–0.10 | 0.11 ± 0.28 | 35.29 (6) |
| IL-13 | 0.00–0.00 | 0.01 ± 0.13 | 1.22 (1) | 0.00–0.00 | 0.07 ± 0.29 | 5.88 (1) |
| IL-17 | 0.00–0.00 | 0.17 ± 0.76 | 7.32 (6) | 0.00–0.00 | 0.14 ± 0.59 | 5.88 (1) |
| IL-18 | 2.42–14.88 | 11.79 ± 18.81 | 86.59 (71) | 3.42–23.59 | 23.04 ± 39.85 | 88.24 (15) |
| IL-21 | 0.38–4.50 | 12.60 ± 41.29 | 79.27 (65) | 0.19–10.04 | 12.11 ± 28.47 | 76.47 (13) |
| IL-22 | 0.00–5.88 | 4.92 ± 8.68 | 52.44 (43) | 0.00–8.56 | 5.58 ± 8.26 | 58.82 (10) |
| IL-23 | 0.00–0.00 | 0.58 ± 1.74 | 12.20 (10) | 0.00–1.73 | 1.39 ± 2.92 | 23.53 (4) |
| IL-1β | 0.00–1.88 | 10.83 ± 55.24 | 40.24 (33) | 0.00–0.67 | 2.48 ± 8.17 | 29.41 (5) |
| IFN_γ | 0.00–0.00 | 1.33 ± 9.73 | 18.29 (15) | 0.00–0.39 | 0.90 ± 2.89 | 23.53 (4) |
| GM_CSF | 0.00–0.00 | 0.13 ± 0.90 | 2.44 (2) | 0.00–0.00 | 0.00 ± 0.00 | 0 (0) |
| TNF_α | 1.66–2.44 | 2.43 ± 2.70 | 100 (82) | 1.66–2.05 | 2.19 ± 1.39 | 100 (17) |
| Leptin | 185.90–546.34 | 446.98 ± 417.23 | 97.56 (80) | 97.64–805.37 | 521.70 ± 602.82 | 100 (17) |
| Adiponectin | 7987.05–17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 068.39 068.39 |
13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 644.08 ± 8626.07 644.08 ± 8626.07 |
98.78 (81) | 7830.93–17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 357.13 357.13 |
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 490.01 ± 28 490.01 ± 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 663.63 663.63 |
100 (17) |
The most predominant cytokine in both groups was IL-6; the concentration of this cytokine was higher in the BM from mothers with ROW offspring compared to those with NOROW offspring (p = 0.031) (Table 2).
The relationship between some taxa from the BM microbiota, cytokines and adipokines and infant development is presented in Table 3, including the linear regression β coefficients for the BM microbiota and BM cytokines and adipokines, predicting the BMIZ and WLZ at 12 months, as well as the unadjusted and adjusted models (Tables 3 and S3†).
| Unadjusted analysis | Adjusted analysisa | |||||
|---|---|---|---|---|---|---|
| Breast milk compounds | β | 95% Cl | p | β | 95% Cl | p |
| Linear regression β coefficients for the BM microbiota and cytokines predicting the WLZ at 12 months.a Adjusted for mode of birth, intrapartum antibiotics (ATB) and breastfeeding practices at 15 days. p < 0.05 was considered statistically significant. Cl: confidence interval, IL: interleukin. Significances are highlighted in bold. | ||||||
| Chao 1 index | 0.20 | 0.004 to 0.036 | 0.012 | 0.021 | 0.005 to 0.037 | 0.012 |
| Streptococcus | −0.008 | −0.015 to 0.000 | 0.040 | −0.008 | −0.016 to 0.037 | 0.048 |
| Gemella | −0.022 | −0.044 to 0.000 | 0.052 | −0.021 | −0.044 to 0.002 | 0.070 |
| IL-1β | −0.004 | −0.008 to 0.000 | 0.065 | −0.004 | −0.008 to 0.001 | 0.095 |
| IL-6 | 0.001 | 0.000 to 0.002 | 0.030 | 0.001 | 0.000 to 0.002 | 0.054 |
| IL-10 | −0.054 | −0.094 to 0.013 | 0.010 | −0.056 | −0.098 to 0.014 | 0.009 |
| IL-12 | 0.061 | −0.007 to 0.129 | 0.079 | 0.061 | −0.008 to 0.131 | 0.082 |
| IL-13 | −0.967 | −2.080 to 0.146 | 0.088 | −0.916 | −2.058 to 0.226 | 0.114 |
| IL-17 | −0.270 | −0.532 to 0.008 | 0.043 | −0.264 | −0.533 to 0.005 | 0.055 |
| IL-23 | 0.077 | −0.003 to 0.157 | 0.060 | 0.082 | −0.001 to 0.164 | 0.054 |
| IL-18 | 0.019 | 0.009 to 0.030 | <0.001 | 0.019 | 0.009 to 0.030 | 0.001 |
The unadjusted models showed that lower Streptococcus (p = 0.040) and IL-10 (p = 0.010) and IL-17 (p = 0.043) levels in the BM, and higher Chao1 index (p = 0.012) and IL-6 (p = 0.030) and IL-18 (p < 0.001) were associated with a higher WLZ at 12 months.
When these models were adjusted for the mode of birth, ATB and breastfeeding practices at 15 d, lower Streptococcus (p = 0.048) and IL-10 (p = 0.009) levels in the BM and higher Chao1 index (p = 0.012) and IL-18 (p = 0.001) were associated with a higher WLZ at 12 months (Table 3).
Interestingly, relationship between some taxa from BM microbiota, cytokines and adipokines and BMIZ at 12 months was found in unadjusted analysis. Streptococcus (p = 0.044) was a predictor of higher BMIZ at 12 months (Table S3†), and in contrast, the IL-18 (p = 0.008) concentration and Chao 1 index (p = 0.007) were significantly associated with a lower BMIZ at 12 months (Table S3†). Interestingly, in the adjusted model, lower IL-10 (p = 0.049) was associated with a higher BMIZ at 12 months.
BM microbes induce IL-6 in the mammary epithelia in a ROW-dependent manner, but do not activate the NF-κB inflammatory pathway in the intestinal epithelia
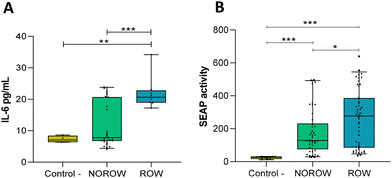
BM pellets significantly induced cytokine responses in mammary epithelial cells. Higher levels of IL-6 production were observed in mammary cells exposed to pellets from the BM of mothers with ROW offspring compared to those exposed to pellets from the BM of mothers with NOROW offspring (p = 0.001) and the control conditions (p = 0.002) (Fig. 3A). In addition, a higher gene expression of IL-6 was observed in those cells exposed to milk bacterial pellets from the mothers with ROW offspring when compared to those exposed to milk bacterial pellets from the mothers with NOROW offspring (p = 0.037) (Table S4†).However, when the intestinal epithelial cells were tested, the BM bacterial pellet samples did not induce NF-κB activation in the HT-29 reporter cell line (data not shown).
Does the faecal supernatant from ROW infants at 12 months activate the NF-κB inflammatory pathway?
When faecal supernatants from the infants at 12 months were tested, we observed NF-κB activation in the HT-29 reporter cell line. Both groups, ROW (p < 0.001) and NOROW (p = 0.006) children, induced NF-κB activation (Fig. 3B); significant differences between NOROW and ROW children were observed (Fig. 3B) (p = 0.029). Also, we observed a positive correlation between the SEAP activity induced by the faecal supernatants and the concentration of IL-6 in the BM samples (ρ = 0.38, p = 0.037).Discussion
BM contains bioactive compounds including microbiota and cytokines, which may have an impact on infant growth38,39 and health outcomes.40 Our results provide insight on the BM bioactive composition in terms of microbiota, cytokines, and adipokines and their potential link to infant growth and ROW at 12 months of life.In agreement with previous data,41 we reported that IL-6, followed by IL-18, was the most abundant cytokine in BM samples.42,43 The presence and abundance of these cytokines have been described to be relevant in the link between BM and infant development.41,44,45 Furthermore, Streptococcus and Staphylococcus were also identified as the most abundant genera.13 However, the potential interaction between these BM components and the association with the ROW are underexplored. Little is known about the factors influencing the complex association between microbial taxa, cytokines and adipokines in BM. Some studies reported the key relevance of maternal BMI and breastfeeding practices on milk microbiota and infant growth.11,46 Maternal BMI has been highlighted as one of the factors affecting other components in BM, such as lipids and human milk oligosaccharides.47,48 While its relationship to other components has been less addressed, significant associations have been described between maternal BMI with milk leptin at 1 month postpartum and milk glucose, insulin, IL-6, and TNF-α with an impact of BMI on the infant body composition.45 Similarly, our results revealed that higher leptin levels in human milk were associated with higher maternal BMI, which was in accordance with other studies.46,49,50 These results suggest a potential link between maternal signals in BM and infant growth. Interestingly, although our population was mainly of normal weight and only a few were overweight, the influence of BMI was observed.11 Indeed, our results also suggested a potential positive association between mixed feeding and leptin concentration. Different studies have shown that both formula feeding1,51 and high maternal BMI were associated with a higher risk of overweight in children between 2 and 5 years of age.52 This adipokine is related to the control of food intake and weight regulation;53 it is commonly observed in higher amounts in obese patients (compared to non-obese population) who also show a phenomenon known as leptin resistance.54 This indicates that leptin plays an important role in the control of food intake.
Thus, the association described in our study would support the role of BM cytokines as a potential route by which the maternal clinical conditions, such as BMI, could affect infant growth, since higher levels of leptin in BM could exert an effect in children. Apart from leptin, the associations that our analysis has revealed are crucial for infant development, since cytokines could impact infants’ immune system and gut epithelium via alteration of oral tolerance,55 among other potential actions.
In the present study, Streptococcus genus was positively associated with milk IL-18 and IL-10 concentrations in accordance with a previous study.12 We also found that Bifidobacterium genus abundance was negatively related to pro-inflammatory cytokines, such as IL-6 and IL1-β, and leptin in the BM. In contrast, it has been reported that IL-6 was positively associated with Staphylococcus.12 The Bifidobacterium genus has been proposed as one of the most important bacterial taxa affecting immune system development during early life,56,57 and BM would play a key role in the colonisation of the infant gut;10,58,59 differences in the gut microbiota composition in children may predict infant growth. Indeed, a study found lower Bifidobacterium and higher Staphylococcus aureus at 6 and 12 months in obese children.60 Thus, the negative relationship between this genus in maternal BM and leptin could be one of the potential links between the lower incidence of being overweight observed in breastfed infants.61
Our observations suggest that the immune signals present in BM and their relationship with the microbial components could potentially be linked and influence infant development. Higher abundances of Streptococcus and IL-10 and lower IL-6 and IL-18 concentrations were found to be predictors of lower WLZ at 12 months, and the Chao1 index and IL-18 were found to be predictors of higher BMIZ at 12 months. These relationships may be explained by the effects of cytokines in the development of the new-born. In this line, IL-10 may have immunomodulatory and anti-inflammatory effects on the alimentary tract of the new-born,62 and increased levels of IL-6 have been consistently linked to obesity.63 Contrary to our results, a study reported that BM cytokine levels did not play a substantial role in the growth of children. However, this study performed analysis with data from the first 2–3 months postpartum when the infant development might be influenced by other potential factors,41 mainly the maternal nutritional environment during pregnancy that influences growth during the initial months.64 Cytokines, as well as other BM components, have also been described to influence infant growth. Oligosaccharide composition in human milk 3 months after delivery was significantly associated with child growth throughout the first 5 years of life, since BM oligosaccharide composition may transform into a stronger and better shielded element, linked to a diminished percentage of infections and inflammation, thus allowing infants to fully invest their energy into development.65 Also, BM lipids could have an important role in the cytokine modulation in the bowels of newborns.66
To further explore the relationship between the BM signals and the risk of obesity in infants, we carried out a proof-of-concept study where BM bacteria were exposed to a mammary gland epithelial cell line to ascertain the potential effect on the maternal side and to check the potential impact on the lactating infant. BM microbes induced a response in mammary cells, which were dependent on the infant's risk of overweight at 12 months of life. We observed that milk bacteria from the mothers with ROW offspring induced a higher IL-6 release in MCF7 cells compared to those from the mothers with NOROW offspring, suggesting that the milk microbiome could contribute to the cytokine composition. Previous studies have shown that IL-6 is associated with maturation of the intestinal immune system.42 This pleiotropic cytokine has a central role in the signalling system of the organism, exerting several, sometimes conflicting, functions,67 which are also tissue-specific. In fact, the alteration of IL-6 in BM has been associated with maternal obesity;12,68 however, increased levels of IL-6 have been consistently linked to insulin resistance69 and the chronic low-grade inflammation that is commonly observed in these diseases. Contrary to our results, a study reported that higher concentrations of IL-6 in BM were also significantly associated with lower relative weight, weight gain, and fat mass in healthy term infants at 1 month of age,45 yet these previous results do not provide conclusive evidence on how these independent effects of different BM components influence the infant body composition. In this line, despite the IL-6 expression in the mammary epithelial cells due to a pro-inflammatory signal elicited by one or several bacterial species contained in breast milk, we did not find activation of the NF-κB pathway in the epithelial intestinal cells. This observation suggests that intestinal cells are unresponsive to the bacteria contained in BM at 1 month of age to elicit a pro-inflammatory response. However, at 1 year of age, we observed that the faecal supernatants of the ROW offspring showed higher pro-inflammatory activity on intestinal epithelial cells. Most interestingly, NF-κB pathway activation was associated with higher IL-6 BM concentration, suggesting that the pro-inflammatory ability of the initial BM microbiota source and cytokines, although only detected in mammary epithelial cells in our cell culture model, initiated a pro-inflammatory intestinal environment in the infants. Further studies are needed to clarify the potential impact of milk microbiota on the cytokine release by the mammary gland tissue, and how both BM microbiota and cytokines could modulate infant development. Our results suggest that the combination of milk cytokines and microbes is needed to promote a potential gut inflammatory status, leading to a potential higher risk of overweight. The critical combination and interaction of BM compounds and their inter-relations warrant further investigation.
This study has some limitations, including the sample size, which needs to be extended to fully reveal the implications of the results due to the higher intra- and inter-variability among mothers. BM is a complex fluid, and especially the immune signals among others, could be influenced by several factors that contribute to this observed variability across the population. This aspect is especially relevant in in vitro experiments, where further analysis with larger cohorts in overweight risk subjects are needed to clarify the influence of BM microbiota on the immunological active content of BM and its impact on child development. Also, the analysis of the potential relationship between the gut microbiota of infants and the BM microbiota is lacking. This would be interesting for understanding the modulation of BM on the infant gut microbiota in those at risk of overweight and those without the risk of overweight.
In conclusion, our study has shown the role of BM microbiota in infant growth and development. Our observations suggest that the association between BM cytokines and microbiota could be related to the growth of children during the first 12 months, although the potential mechanisms behind remain uncovered and this warrants further investigation. Despite that it is required to evaluate other potential environmental and host factors that may influence these associations, our results shed light on the link between BM, cytokines, adipokines, and infant growth and contribute to the knowledge that will be essential for the development of future strategies targeting infant growth modulation through breastfeeding.
Author contributions
EC-M: writing – original draft, methodology, investigation, validation, visualization, and data interpretation. MS-R, KR-A, and CB: investigation, validation, visualization, and data interpretation. MJR-L and FJP-C: supervision, writing – review and editing, visualization, and interpretation. MCC and CM-C: supervision, conceptualization, writing – review and editing, visualization, and interpretation.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank the families involved in the MAMI study as well as all the members of the MAMI cohort study. This work was supported by the research grant from the European Research Council under the European Union's Horizon 2020 research and innovation program (ERC starting grant, no. 639226) and La Marató-TV3 (DIM-2-ELI, ref. 2018-27/30-31). E. C.-M. was supported by a predoctoral fellowship from the Generalitat Valenciana—European Social Fund (GRISOLÍA2019). K. R.-A. holds a fellowship from the Spanish Ministry of Economy, Industry, and Competitiveness (FPU19/05150).References
- T. Harder, R. Bergmann, G. Kallischnigg and A. Plagemann, Duration of breastfeeding and risk of overweight: A meta-analysis, Am. J. Epidemiol., 2005, 162, 397–403 CrossRef PubMed.
- J. Yan, L. Liu, Y. Zhu, G. Huang and P. Peter Wang, The association between breastfeeding and childhood obesity: a meta-analysis, BMC Public Health, 2014, 14, 1–11 CrossRef PubMed.
- World Health Organization, WHO|Exclusive breastfeeding for six months best for babies everywhere, WHOGeneva, 2011 Search PubMed.
- J. C. K. Wells, Breast-feeding as ‘personalized nutrition’, Eur. J. Clin. Nutr., 2018, 72, 1234–1238 CrossRef PubMed.
- F. Ladomenou, J. Moschandreas, A. Kafatos, Y. Tselentis and E. Galanakis, Protective effect of exclusive breastfeeding against infections during infancy: A prospective study, Arch. Dis. Child., 2010, 95, 1004–1008 CrossRef PubMed.
- M. Selma-Royo, M. Calatayud, I. García-Mantrana, A. Parra-Llorca, R. Escuriet, C. Martínez-Costa and M. C. Collado, Perinatal environment shapes microbiota colonization and infant growth: impact on host response and intestinal function, Microbiome, 2020, 8, 1–44 CrossRef PubMed.
- M. Reyman, M. A. van Houten, D. van Baarle, A. A. T. M. Bosch, W. H. Man, M. L. J. N. Chu, K. Arp, R. L. Watson, E. A. M. Sanders, S. Fuentes and D. Bogaert, Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life, Nat. Commun., 2019, 10, 1–12 CrossRef CAS PubMed.
- P. Lif Holgerson, L. Harnevik, O. Hernell, A. C. R. Tanner and I. Johansson, Mode of birth delivery affects oral microbiota in infants, J. Dent. Res., 2011, 90, 1183–1188 CrossRef CAS PubMed.
- N. T. Ho, F. Li, K. A. Lee-sarwar, H. M. Tun, B. P. Brown, P. S. Pannaraj, J. M. Bender, M. B. Azad, A. L. Thompson, S. T. Weiss, M. A. Azcarate-peril, A. A. Litonjua, A. L. Kozyrskyj, H. B. Jaspan, G. M. Aldrovandi and L. Kuhn, Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations, Nat. Commun., 2018, 9, 1–13 CrossRef PubMed.
- G. W. Tannock, B. Lawley, K. Munro, S. G. Pathmanathan, S. J. Zhou, M. Makrides, R. A. Gibson, T. Sullivan, C. G. Prosser, D. Lowry and A. J. Hodgkinson, Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk, Appl. Environ. Microbiol., 2013, 79, 3040–3048 CrossRef CAS PubMed.
- E. Cortés-Macías, M. Selma-Royo, C. Martínez-Costa and M. C. Collado, Breastfeeding Practices Influence the Breast Milk Microbiota Depending on Pre-Gestational Maternal BMI and Weight Gain over Pregnancy, Nutrients, 2021, 13, 1–15 CrossRef PubMed.
- M. C. Collado, K. Laitinen, S. Salminen and E. Isolauri, Maternal weight and excessive weight gain during pregnancy modify the immunomodulatory potential of breast milk, Pediatr. Res., 2012, 72, 77–85 CrossRef CAS PubMed.
- E. Cortes-Macías, M. Selma-Royo, I. García-Mantrana, M. Calatayud, S. González, C. Martínez-Costa and M. C. Collado, Maternal diet shapes the breast milk microbiota composition and diversity: impact of mode of delivery and antibiotic exposure, J. Nutr., 2020, 151, 330–340 CrossRef PubMed.
- C. Gomez-Gallego, I. Garcia-Mantrana, S. Salminen and M. C. Collado, The human milk microbiome and factors influencing its composition and activity, Semin. Fetal Neonatal Med., 2016, 21, 400–405 CrossRef PubMed.
- S. Moossavi, S. Sepehri, B. Robertson, L. Bode, S. Goruk, C. J. Field, L. M. Lix, R. J. de Souza, A. B. Becker, P. J. Mandhane, S. E. Turvey, P. Subbarao, T. J. Moraes, D. L. Lefebvre, M. R. Sears, E. Khafipour and M. B. Azad, Composition and variation of the human milk microbiota are influenced by maternal and early-life factors, Cell Host Microbe, 2019, 25, 324–335 CrossRef CAS PubMed.
- I. García-Mantrana, C. Alcántara, M. Selma-Royo, A. Boix-Amorós, M. Dzidic, J. Gimeno-Alcañiz, I. Úbeda-Sansano, I. Sorribes-Monrabal, R. Escuriet, F. Gil-Raga, A. Parra-Llorca, C. Martínez-Costa and M. C. Collado, MAMI: A birth cohort focused on maternal-infant microbiota during early life, BMC Pediatr., 2019, 19, 1–8 CrossRef PubMed.
- World Health Organization, WHO | WHO Anthro Survey Analyser and other tools, WHOGeneva, 2010 Search PubMed.
- World Health Organization, WHO|Child Growth Standards, Methods and development. Length/height-for-age, weight-for-age, weight-for-length, weight-for-height and body mass index-for-age, WHO Geneva, 2006 Search PubMed.
- A. W. Kamng'ona, R. Young, C. D. Arnold, E. Kortekangas, N. Patson, J. M. Jorgensen, E. L. Prado, D. Chaima, C. Malamba, U. Ashorn, Y. M. Fan, Y. B. Cheung, P. Ashorn, K. Maleta and K. G. Dewey, The association of gut microbiota characteristics in Malawian infants with growth and inflammation, Sci. Rep., 2019, 9, 1–13 CrossRef PubMed.
- A. Boix-Amorós, M. C. Collado and A. Mira, Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation, Front. Microbiol., 2016, 7, 1–9 Search PubMed.
- I. García-Mantrana, C. Alcántara, M. Selma-Royo, A. Boix-Amorós, M. Dzidic, J. Gimeno-Alcañiz, I. Úbeda-Sansano, I. Sorribes-Monrabal, R. Escuriet, F. Gil-Raga, A. Parra-Llorca, C. Martínez-Costa and M. C. Collado, MAMI: A birth cohort focused on maternal-infant microbiota during early life, BMC Pediatr., 2019, 19, 1–8 CrossRef PubMed.
- A. M. Bolger, M. Lohse and B. Usadel, Trimmomatic: A flexible trimmer for Illumina sequence data, Bioinformatics, 2014, 30, 2114–2120 CrossRef CAS PubMed.
- B. J. Callahan, P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. A. Johnson and S. P. Holmes, DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, 13, 581–587 CrossRef CAS PubMed.
- C. Quast, E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer, P. Yarza, J. Peplies and F. O. Glöckner, The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic Acids Res., 2013, 41, 590–596 CrossRef PubMed.
- N. M. Davis, Di. Proctor, S. P. Holmes, D. A. Relman and B. J. Callahan, Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data, Microbiome, 2018, 6, 1–14 CrossRef PubMed.
- A. Boix-Amorós, M. T. Hernández-Aguilar, A. Artacho, M. C. Collado and A. Mira, Human milk microbiota in sub-acute lactational mastitis induces inflammation and undergoes changes in composition, diversity and load, Sci. Rep., 2020, 10, 1–14 CrossRef PubMed.
- Y. H. Huang, M. H. Chou, Y. Y. Du, C. C. Huang, C. L. Wu, C. L. Chen and J. H. Chuang, Expression of toll-like receptors and type 1 interferon specific protein MxA in biliary atresia, Lab. Invest., 2007, 87, 66–74 CrossRef CAS PubMed.
- F. Pattyn, P. Robbrecht, A. De Paepe, F. Speleman and J. Vandesompele, RTPrimerDB: the real-time PCR primer and probe database, major update 2006, Nucleic Acids Res., 2006, 34, 684–688 CrossRef PubMed.
- C. Ramakers, J. M. Ruijter, R. H. Lekanne Deprez and A. F. M. Moorman, Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data, Neurosci. Lett., 2003, 339, 62–66 CrossRef CAS PubMed.
- J. M. Ruijter, C. Ramakers, W. M. H. Hoogaars, Y. Karlen, O. Bakker, M. J. B. van den hoff and A. F. M. Moorman, Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data, Nucleic Acids Res., 2009, 37, 1–12 CrossRef PubMed.
- W. P. Michael, G. W. Horgan and D. Leo, Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR, Nucleic Acids Res., 2002, 30, 1–10 CrossRef PubMed.
- C. Bäuerl, J. M. Coll-Marqués, C. Tarazona-González and G. Pérez-Martínez, Lactobacillus casei extracellular vesicles stimulate EGFR pathway likely due to the presence of proteins P40 and P75 bound to their surface, Sci. Rep., 2020, 10, 1–12 CrossRef PubMed.
- D. Philip, VEGAN, a package of R functions for community ecology, J. Veg. Sci., 2003, 14, 927–930 CrossRef.
- T. R Studio, R Studio Inc., Boston, MA, USA, 2016.
- G. R. Warnes, B. Bolker, L. Bonebakker, R. Gentleman, W. Huber, A. Liaw, T. Lumley, M. Maechler, A. Magnusson, S. Moeller, M. Schwartz, V. Bill and G. Tal, gplots: Various R Programming Tools for Plotting Data. R package vesiron 3.10.3https://CRAN.R-project.org/package=gplots, 2019.
- A. Day, heatmap.plus: Heatmap with more sensible behavior. R package version 1.3, https://CRAN.R-project.org/package=heatmap.plus, 2015, pp. 1–4 Search PubMed.
- IBM Corp, Released 2020. IBM SPSS Statistics for Windows, Version 27.0, IBM Corp, Armonk, NY Search PubMed.
- H. Nuss, A. Altazan, J. Zabaleta, M. Sothern and L. Redman, Maternal pre-pregnancy weight status modifies the influence of PUFAs and inflammatory biomarkers in breastmilk on infant growth, PLoS One, 2019, 14, 1–17 CrossRef PubMed.
- R. Garofalo, Cytokines in Human Milk, J. Pediatr., 2010, 156, S36–S40 CrossRef CAS PubMed.
- K. E. Lyons, C. A. Ryan, E. M. Dempsey, R. P. Ross and C. Stanton, Breast milk, a source of beneficial microbes and associated benefits for infant health, Nutrients, 2020, 12, 1–30 CrossRef PubMed.
- A. Saso, O. Blyuss, D. Munblit, A. Faal, S. E. Moore and K. Le Doare, Breast milk cytokines and early growth in Gambian infants, Front. Pediatr., 2019, 6, 1–10 Search PubMed.
- M. F. Böttcher, M. C. Jenmalm, R. P. Garofalo and B. Björkstén, Cytokines in breast milk from allergic and nonallergic mothers, Int. Arch. Allergy Immunol., 1999, 118, 319–320 CrossRef PubMed.
- Y. Takahata, H. Takada, A. Nomura, K. Ohshima, H. Nakayama, T. Tsuda, H. Nakano and T. Hara, Interleukin-18 in Human Milk, Pediatr. Res., 2001, 50, 268–272 CrossRef CAS PubMed.
- M. Fujimori, E. L. França, T. C. Morais, V. Fiorin, L. C. de Abreu and A. C. Honório-França, Cytokine and adipokine are biofactors can act in blood and colostrum of obese mothers, BioFactors, 2017, 43, 243–250 CrossRef CAS PubMed.
- D. A. Fields and E. W. Demerath, Relationship of insulin, glucose, leptin, IL-6 and TNF-α in human breast milk with infant growth and body composition, Pediatr. Obes., 2012, 7, 304–312 CrossRef CAS PubMed.
- D. Chan, S. Goruk, A. B. Becker, P. Subbarao, P. J. Mandhane, S. E. Turvey, D. Lefebvre, M. R. Sears, C. J. Field and M. B. Azad, Adiponectin, leptin and insulin in breast milk: Associations with maternal characteristics and infant body composition in the first year of life, Int. J. Obes., 2018, 42, 36–43 CrossRef CAS PubMed.
- J. Mäkelä, K. Linderborg, H. Niinikoski, B. Yang and H. Lagström, Breast milk fatty acid composition differs between overweight and normal weight women: the STEPS Study, Eur. J. Nutr., 2013, 52, 727–735 CrossRef PubMed.
- J. L. Saben, C. R. Sims, A. Abraham, L. Bode and A. Andres, Human milk oligosaccharide concentrations and infant intakes are associated with maternal overweight and obesity and predict infant growth, Nutrients, 2021, 13, 1–16 CrossRef PubMed.
- J. Murphy, R. M. Pfeiffer, B. C. D. Lynn, A. I. Caballero, E. P. Browne, E. C. Punska, H. P. Yang, R. T. Falk, D. L. Anderton, G. L. Gierach, K. F. Arcaro and M. E. Sherman, Pro-inflammatory cytokines and growth factors in human milk: an exploratory analysis of racial differences to inform breast cancer etiology, Breast Cancer Res. Treat., 2018, 172, 209–219 CrossRef CAS PubMed.
- M. Fujimori, E. L. França, T. C. Morais, V. Fiorin, L. C. de Abreu and A. C. Honório-França, Cytokine and adipokine are biofactors can act in blood and colostrum of obese mothers, BioFactors, 2017, 43, 243–250 CrossRef CAS PubMed.
- H. J. Choi, S. K. Kang and M. R. Chung, The relationship between exclusive breastfeeding and infant development: A 6- and 12-month follow-up study, Early Hum. Dev., 2018, 127, 42–47 CrossRef PubMed.
- E. Voerman, S. Santos, B. P. Golab, P. Amiano, F. Ballester, H. Barros, A. Bergström, M. A. Charles, L. Chatzi, C. Chevrier, G. P. Chrousos, E. Corpeleijn, N. Costet, S. Crozier, G. Devereux, M. Eggesbø, S. Ekström, M. P. Fantini, S. Farchi, F. Forastiere, V. Georgiu, K. M. Godfrey, D. Gori, V. Grote, W. Hanke, I. Hertz-Picciotto, B. Heude, D. Hryhorczuk, R. C. Huang, H. Inskip, N. Iszatt, A. M. Karvonen, L. C. Kenny, B. Koletzko, L. K. Küpers, H. Lagström, I. Lehmann, P. Magnus, R. Majewska, J. Mäkelä, Y. Manios, F. M. McAuliffe, S. W. McDonald, J. Mehegan, M. Mommers, C. S. Morgen, T. A. Mori, G. Moschonis, D. Murray, C. N. Chaoimh, E. A. Nohr, A. M. N. Andersen, E. Oken, A. J. J. M. Oostvogels, A. Pac, E. Papadopoulou, J. Pekkanen, C. Pizzi, K. Polanska, D. Porta, L. Richiardi, S. L. Rifas-Shiman, L. Ronfani, A. C. Santos, M. Standl, C. Stoltenberg, E. Thiering, C. Thijs, M. Torrent, S. C. Tough, T. Trnovec, S. Turner, L. van Rossem, A. von Berg, M. Vrijheid, T. G. M. Vrijkotte, J. West, A. Wijga, J. Wright, O. Zvinchuk, T. I. A. Sørensen, D. A. Lawlor, R. Gaillard and V. W. V. Jaddoe, Maternal body mass index, gestational weight gain, and the risk of overweight and obesity across childhood: An individual participant data meta-analysis, PLoS Med., 2019, 16, 1–22 CrossRef PubMed.
- F. K. Uysal, E. E. Önal, Y. Z. Aral, B. Adam, U. Dilmen and Y. Ardiçolu, Breast milk leptin: Its relationship to maternal and infant adiposity, Clin. Nutr., 2002, 21, 157–160 CrossRef CAS PubMed.
- A. G. Izquierdo, A. B. Crujeiras, F. F. Casanueva and M. C. Carreira, Leptin, obesity, and leptin resistance: where are we 25 years later?, Nutrients, 2019, 11, 1–11 CrossRef PubMed.
- B. Dawod and J. S. Marshall, Cytokines and soluble receptors in breast milk as enhancers of oral tolerance development, Front. Immunol., 2019, 10, 1–9 CrossRef PubMed.
- L. Ruiz, S. Delgado, P. Ruas-Madiedo, B. Sánchez and A. Margolles, Bifidobacteria and their molecular communication with the immune system, Front. Microbiol., 2017, 8, 1–9 Search PubMed.
- S. Saturio, A. M. Nogacka, M. Suárez, N. Fernández, L. Mantecón, L. Mancabelli, C. Milani, M. Ventura, C. G. de los Reyes-Gavilán, G. Solís, S. Arboleya and M. Gueimonde, Early-life development of the bifidobacterial community in the infant gut, Int. J. Mol. Sci., 2021, 22, 1–12 Search PubMed.
- K. Murphy, D. Curley, T. F. O′callaghan, C. O'Shea, E. M. Dempsey, P. W. O'Toole, R. P. Ross, C. A. Ryan and C. Stanton, The composition of human milk and infant faecal microbiota over the first three months of life: a pilot study, Sci. Rep., 2017, 7, 1–10 CrossRef PubMed.
- K. Fehr, S. Moossavi, H. Sbihi, R. C. T. Boutin, L. Bode, B. Robertson, C. Yonemitsu, C. J. Field, A. B. Becker, P. J. Mandhane, M. R. Sears, E. Khafipour, T. J. Moraes, P. Subbarao, B. B. Finlay, S. E. Turvey and M. B. Azad, Breastmilk Feeding Practices Are Associated with the Co-Occurrence of Bacteria in Mothers’ Milk and the Infant Gut: the CHILD Cohort Study, Cell Host Microbe, 2020, 28, 285–297 CrossRef CAS PubMed.
- M. Kalliomäki, M. C. Collado, S. Salminen and E. Isolauri, Early differences in fecal microbiota composition in children may predict overweight, Am. J. Clin. Nutr., 2008, 87, 534–538 CrossRef PubMed.
- J. D. Forbes, M. B. Azad, L. Vehling, H. M. Tun, T. B. Konya, D. S. Guttman, C. J. Field, D. Lefebvre, M. R. Sears, A. B. Becker, P. J. Mandhane, S. E. Turvey, T. J. Moraes, P. Subbarao, J. A. Scott and A. L. Kozyrskyj, Association of exposure to formula in the hospital and subsequent infant feeding practices with gut microbiota and risk of overweight in the first year of life, J. Am. Med. Assoc., 2018, 172, 1–11 Search PubMed.
- A. R. M. A. Meki, T. H. Saleem, M. H. Al-Ghazali and A. A. Sayed, Interleukins -6, -8 and -10 and tumor necrosis factor-alpha and its soluble receptor I in human milk at different periods of lactation, Nutr. Res., 2003, 23, 845–855 CrossRef CAS.
- S. Sindhu, R. Thomas, P. Shihab and D. Sriraman, Obesity Is a Positive Modulator of IL-6R and IL-6 Expression in the Subcutaneous Adipose Tissue: Significance for Metabolic Inflammation, PLoS One, 2015, 10, 1–17 CrossRef CAS PubMed.
- C. M. Wright, K. N. Parkinson and R. F. Drewett, The influence of maternal socioeconomic and emotional factors on infant weight gain and weight faltering (failure to thrive): Data from a prospective birth cohort, Arch. Dis. Child., 2006, 91, 312–317 CrossRef CAS PubMed.
- H. Lagström, S. Rautava, H. Ollila, A. Kaljonen, O. Turta, J. Mäkelä, C. Yonemitsu, J. Gupta and L. Bode, Associations between human milk oligosaccharides and growth in infancy and early childhood, Am. J. Clin. Nutr., 2020, 111, 769–778 CrossRef PubMed.
- G. J. Barrera and G. Sánchez, Cytokine modulation (IL-6, IL-8, IL-10) by human breast milk lipids on intestinal epithelial cells (Caco-2), J. Matern.-Fetal Neonat. Med., 2016, 29, 2505–2512 Search PubMed.
- S. Wueest and D. Konrad, The controversial role of IL-6 in adipose tissue on obesity-induced dysregulation of glucose metabolism, Am. J. Physiol.: Endocrinol. Metab., 2020, 319, 607–613 CrossRef PubMed.
- U. D. Erliana and A. D. Fly, The function and alteration of immunological properties in human milk of obese mothers, Nutrients, 2019, 11, 1–25 CrossRef PubMed.
- F. B. Hu, J. B. Meigs, T. Y. Li, N. Rifai and J. E. Manson, Inflammatory Markers and Risk of Developing Type 2 Diabetes in Women, Diabetes, 2004, 53, 693–700 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2fo02060b |
| This journal is © The Royal Society of Chemistry 2023 |