Dietary citrus flavonoid extract improves lactational performance through modulating rumen microbiome and metabolites in dairy cows†
Shiqiang
Yu
a,
Liuxue
Li
a,
Huiying
Zhao
a,
Shuyue
Zhang
a,
Yan
Tu
b,
Ming
Liu
a,
Yuchao
Zhao
 *ac and
Linshu
Jiang
*a
*ac and
Linshu
Jiang
*a
aBeijing Key Laboratory of Dairy Cow Nutrition, College of Animal Science and Technology, Beijing University of Agriculture, Beijing, 102206, China. E-mail: zhaoyuchao2019@126.com; jlsbua@126.com
bBeijing Key Laboratory of Dairy Cow Nutrition, Institute of Feed Research, Chinese Academy of Agricultural Sciences, Beijing 100081, China
cBeijing Beinong Enterprise Management Co., Ltd, Beijing 102206, China
First published on 18th November 2022
Abstract
The effects of dietary supplementation with citrus flavonoid extract (CFE) on milk performance, rumen fermentation, rumen microbiome, rumen metabolome, and serum antioxidant indexes were evaluated. Eight multiparous lactating cows were allocated to a replicated 4 × 4 Latin square with 25-d periods consisting of 20 d of adaptation and 5 d of sampling. Experimental treatments included a control diet (CON) and CON supplemented with 50 g d−1 (CFE50), 100 g d−1 (CFE100), and 150 g d−1 (CFE150). Feeding CFE to dairy cows increased milk production and milk lactose. Milk somatic cell count linearly reduced with increasing CFE amount. Supplementing CFE linearly increased the ruminal concentrations of total volatile fatty acids, acetate, propionate, butyrate, and microbial crude protein. Ruminal lipopolysaccharide linearly decreased with increasing CFE amount. Compared with CON, CFE150 cows exhibited a greater abundance of Firmicutes and a low abundance of Bacteroidetes. Cellulolytic bacteria (genera Ruminococcus, Clostridium, and Butyrivibrio) and carbohydrate metabolism were enriched in the CFE150 cows. For archaea and viruses, major methanogens (genera Methanobacterium and Methanosarcina) and phylum Uroviricota were inhibited in the CFE150 cows. Compared with CON, the ruminal concentrations of tyrosine, proline, pyruvate, glucose, and glucose-6-phosphate were higher in the CFE150 cows. The metabolites of citrus flavonoids, such as hippuric acid, hesperetin, and naringenin, were increased in the CFE150 cows. Supplementing CFE significantly improved the antioxidant capacity of the dairy cows. This study highlighted that dietary supplementation with CFE led to significant changes in the rumen microbial composition and metabolites, and consequently resulted in an improved lactational performance of dairy cows.
Introduction
Many dairy cows with a high-performance level are suffering from metabolic disorders that are mainly ascribed to the high metabolic load and associated failures of immunological adaptation. Phytochemicals, as potential agents for improving animal health and maintaining metabolic homeostasis in livestock farms, have become more widespread due to the growing public concern about the side effects of antibiotics. Flavonoids are ubiquitous plant polyphenols that have various health-promoting properties in humans and animals.1 In ruminants, it has been reported that flavonoid intake is evidently associated with several biological effects, e.g., improvement of production performance,1 enhancement of animal product quality,2 immune response enhancement,3 and ruminal CH4 reduction.4 Therefore, as promising natural phytochemicals, flavonoids are now being extensively studied for applications in ruminant production. Flavonoids are present in high quantity in citrus fruits and their by-products, and therefore, citrus flavonoids have been applied in the pharmaceutical and food industries.5 Flavanones (e.g., hesperidin and naringin) and O-polymethoxylated flavones (e.g., tangeretin and nobiletin) are the main components in citrus-derived flavonoids.6 These citrus-derived flavonoids, as promising additives, exhibit diverse biological activities, such as antimicrobial, antioxidant, anti-stress, and anti-inflammatory, in ruminants.2,7,8Alhidary and Abdelrahman1 reported that serum glutathione peroxidase (GSH-Px) activity and superoxide dismutase (T-SOD) activity were greater in sheep that received naringin, and that their overall immune status was enhanced based on greater antibody titers and cell-mediated immunity. The effects of hesperidin supplementation were also evaluated by Simitzis et al.,2 who observed that plasma malonaldehyde (MDA) was reduced. The results of Ying et al.9 showed that adding citrus extract at 4.5 g d−1 did not change lactational performance in mid-lactation or early-lactation periods of dairy cows, but reduced plasma non-esterified fatty acids, which may be associated with the manipulation of adipose tissue lipolysis. These studies demonstrated that citrus flavonoids have the potential to improve the health status of dairy cows.
The bioactivities of these citrus flavonoids are determined by their structure and subsequent metabolism. In order to exert the bioactivities, naringin and hesperidin must be hydrolysed to form their active aglycone forms naringenin and hesperitin, respectively.10 It was perceived that the significant beneficial effects concurred with the relatively high bioavailability of naringin and hesperidin in ruminants, based on the capacity of the rumen microbes to deglycosylate naringin and hesperidin, promoting the absorption of the aglycones in the small intestine.11 It was documented that the interaction between gastrointestinal microbes and flavonoids plays an important role in mediating the health-promoting effects of plant flavonoids in non-ruminants.12,13 On the one hand, citrus flavonoids can affect the structure and community of gastrointestinal microbiota, such as increasing beneficial bacteria and inhibiting harmful bacteria.14 On the other hand, the gastrointestinal microorganisms further enhance the bioavailability of citrus flavonoids and promote flavonoid metabolite production.14,15 Therefore, it is likely that the action of these bioactive compounds would reduce oxidative stress and inflammatory response in the rumen, thereby improving the conditions for microbial flourishing and nutrient uptake.
About 70% of energy and 60–85% of protein requirements of lactating cows come from rumen fermentation, which is carried out by the complex symbiotic microbiota. Previous studies have revealed that citrus flavonoids reduced rumen inflammation and decreased the risk of acidosis in high-grain diet-fed Holstein bulls,3 and inhibited ruminal CH4 production and the population of hydrogenotrophic methanogenic archaea.16 However, there are few studies on the effects of citrus flavonoid extract (CFE) on milk performance, rumen fermentation, rumen microbiome, and rumen metabolism in dairy cows. A better understanding of the microbial community and their functional response to CFE will help facilitate the development of mechanisms to manipulate the rumen microbiota by plant flavonoids for improving lactational performance and the health of dairy cows. We hypothesized that dietary supplementation with CFE would improve rumen fermentation, and consequently, lactational performance by regulating the rumen microbiome and metabolomic profiling. The hypothesis was tested to analyse the effect of CFE on milk performance, rumen fermentation parameters, rumen microbial composition, rumen metabolites, and serum antioxidant indexes.
Materials and methods
Experimental animals and treatments
This study was performed in strict accordance with the China Laboratory Animal Welfare and Ethics Committee Guidelines (publication no. GB/T 35892-2018). All procedures conducted in the study were approved by the Animal Care Committee of Beijing University of Agriculture (Beijing, China). Eight multiparous Chinese Holstein cows (662 ± 57.1 kg of body weight, 160 ± 22.4 days in milk, 36.1 ± 3.79 kg d−1 of milk production) were used in a replicated 4 × 4 Latin square design. Animals were allocated randomly to four treatments, including the basal diet (CON), and the basal diet supplemented with CFE at 50 g d−1 (CPE50), 100 g d−1 (CFE100), and 150 g d−1 (CFE150). Each experimental period lasted 25 d, including 20 d of adaptation to the treatments and 5 d of sampling.Cows were housed in a tie stall and had free access to water. Diets were formulated as total mixed ration (TMR) according to the NRC guidelines.17 Feed ingredients and the chemical composition are shown in ESI Table 1.† Cows were fed ad libitum once daily at approximately 0800 h. The amounts of feed supplied were adjusted to allow for 5–10% refusals (as-fed basis). The dry matter (DM) content of the TMR and feed components was determined weekly, and the TMR was adjusted accordingly.
Citrus flavonoids were extracted and purified from Citrus reticulata Blanco according to the method of Jiang et al.18 In brief, 1 kg of citrus peel powder was extracted twice with 15 L of calcium carbonate solution (0.1%) at 100 °C for 1.5 h. The extract was then evaporated to dryness. The total flavonoids were enriched using AB-8 macroporous absorption resin columns and eluted with 80% ethanol (two-fold of the column volume). The eluents were collected and concentrated to dryness for use. The content of the total flavonoids in the CFE was 56.83% (DM basis), which was determined by aluminium nitrate spectrophotometry (510 nm) with rutin equivalents.18 The concentrations of three major flavonoids including naringin, hesperidin, and neohesperidin in the CFE were quantified using a HPLC system (1290 Infinity; Agilent Technologies, Inc.), using the standard substances (Beijing Solarbio Science & Technology Co., Ltd, Beijing, China) according to Shi et al.19 The chemical composition of the CFE is presented in ESI Table 2.† The CFE product was top-dressed daily by mixing manually with 300 g of the TMR to ensure complete consumption by the cows. The consumption of the CFE lasted for approximately 1 h daily.
Dry matter intake and milk performance
The samples of the TMR and refusals were determined for DM (method 930.15),20 crude protein (method 990.03),20 acid detergent fiber (method 973.18),20 and neutral detergent fiber.21 Daily feed provision and refusals were recorded to calculate the daily DM intake (DMI). Cows were milked three times daily at 0700, 1330, and 2000 h, and the milk yield (MY) was recorded at each milking. During the last 3 d of each sampling period, milk was collected daily from each milking, and the samples were pooled proportionately based on milk production. Preservatives were added to the milk samples and the samples were stored at 4 °C for the determination of milk composition. The milk samples were determined for fat, protein, lactose, somatic cell counts (SCC), and milk urea-N (MUN) using a Milkoscan FT 6000 (Foss Electric, Hillerød, Denmark). The amount of 3.5% fat-corrected milk (FCM) was calculated according to Sklan et al.22 and the amount of energy-corrected milk (ECM) was calculated based on the equations of Sjaunja et al.23 The feed efficiency of each cow was calculated as MY/DMI, 3.5% FCM/DMI, and ECM/DMI.Rumen fermentation and rumen bacterial cell-wall components
On the last day of each period, approximately 200 mL of rumen fluid were collected 3 h after morning feeding using an oesophageal tube and a syringe. The first 200 mL of the sample was discarded to avoid saliva contamination. After filtration with four layers of gauze, ruminal pH was immediately determined using a digital pH meter (PHS-3C; Shanghai Yueping Scientific Instrument Co., Ltd, Shanghai, China). Four aliquots of each fluid sample were kept at −80 °C until the determination of ruminal fermentation parameters, rumen bacterial cell-wall components, microbiome, and metabolome, respectively. Ruminal volatile fatty acid (VFA) contents were analysed in samples containing 25% metaphosphoric acid using a gas chromatograph (7890B; Agilent Technologies, Inc.) equipped with a capillary column (30 m × 0.250 mm × 0.25 μm; DB-FFAP; Agilent Technologies, Inc.). The temperatures of the injector and detector were 170 °C and 190 °C, respectively. The oven temperature was increased from 100 °C to 250 °C. The concentration of ammonia–nitrogen (NH3–N) in the rumen fluid was analysed using a microplate reader (Multiskan FC, Thermo Fisher Scientific, New York, USA) based on the method of Broderick and Kang.24 Microbial crude protein (MCP) in the rumen fluid was analysed using the colorimetric method as described by Makkar et al.25 The concentrations of lipopolysaccharide (LPS) in the rumen fluid were analysed using a commercial ELISA kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The lipoteichoic acid (LTA) in the rumen fluid was determined using a commercial ELISA kit (Amyjet Scientific, Wuhan, China). The optical densities were measured at 405 nm and 410 nm for LPS and LTA, respectively, using a microplate reader (Multiskan FC, Thermo Fisher Scientific, New York, USA).Quantification of specific rumen microbes by real-time qPCR
The relative abundances of 13 specific rumen microbes including fungi, methanogens, protozoa, Fibrobacter succinogenes, Megasphaera elsdenii, Prevotella brevis, Prevotella ruminicola, Ruminococcus albus, Ruminococcus flavefaciens, Ruminobacter amylophilus, Selenomonas ruminantium, Succinimonas amylolytica, and Streptococcus bovis were quantified using real-time qPCR. The primer sequences of these targeted microbes are listed in ESI Table 3.† Ruminal microbial DNA was extracted by repeated bead-beating using QIAamp columns.26 The quality of the DNA samples was evaluated using an ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA) and verified by running on 1% agarose gels. Amplification was performed using a Roche LightCycler 96 system (Roche Diagnostics Deutschland GmbH, Mannheim, Germany) in a reaction system of 20 μL consisting of 10 μL of SYBR Green PCR Master Mix (Beijing Solarbio Science & Technology Co., Ltd, Beijing, China), 8 μL of primers (0.2 μmol each), and 2 μL of the DNA sample (40 ng μL−1) as a template. The procedure consisted of one cycle of 95 °C for 1 min for initial denaturation, followed by 40 cycles of 95 °C for 30 s, and primer annealing at 60 °C for 30 s and 68 °C for 1 min for extension. The cycle threshold (Ct) was used to calculate the fold change (FC) of each rumen microbe relative to the total bacteria. The relative abundance of these target microbes was calculated using the following equation: relative abundance = 2−[Ct(target)−Ct(total bacteria)].27Rumen microbial analysis by metagenomic sequencing
The collected rumen fluid samples from CON and CFE150 were used for metagenomic sequencing. Microbial genomic DNA was extracted using the E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Norcross, GA, USA). The genomic DNA was fragmented to an average size of 400 bp using a Covaris M220 (Gene Company Limited, China). The NEXTFLEX Rapid DNA-Seq Kit (Bioo Scientific, Austin, TX, USA) was used for paired-end library construction. Pooled libraries were sequenced using an Illumina Hiseq X Ten platform (2 × 150 bp).The paired-end reads were preprocessed using Fastp (version 0.20.0; https://opengene.org/fastp/) to trim off low-quality bases and adaptors.28 To remove DNA contamination from the host, the clean data were mapped to the bovine reference genome ARSUCD1.2/bosTau9 using BWA (version 0.7.9a; https://bio-bwa.sourceforge.net).29 The filtered reads were assembled using Megahit (version 1.1.2; https://github.com/voutcn/megahit).30 The assembled contigs under 500 bp of length were excluded from further analysis. Open reading frames (ORFs) were predicted using MetaGene (https://metagene.cb.k.u-tokyo.ac.jp/),31 and the predicted ORFs with a length of 100 bp or more were translated into amino acid sequences using the NCBI translation table. All predicted genes were clustered using CD-HIT (version 4.6.1; https://www.bioinformatics.org/cd-hit/) into a non-redundant gene catalog with 90% identity and 90% coverage.32 Clean reads were mapped to the non-redundant gene catalogs to calculate the gene abundance using SOAPaligner (version 2.21; https://soap.genomics.org.cn/) with 95% identity.33
All the non-redundant sequences were aligned against the GenBank non-redundant database using Diamond (version 0.8.35; (https://www.diamondsearch.org/index.php) with an E-value cut-off of E < 1 × 10−5.34 The potential functional information and information on carbohydrate-active enzymes (CAZy) were obtained using the annotation in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (version 0.8.35; https://www.diamondsearch.org/index.php) and the CAZy database (https://www.cazy.org/) with an E-value cutoff of 1 × 10−5. Principal coordinate analysis (PCoA) was conducted based on the Bray–Curtis dissimilarity matrices. All P-values were corrected for a false discovery rate (FDR) using Benjamini and Hochberg's method. All assembled and filtered raw sequence data were deposited into the NCBI Sequence Read Archive under Bioproject PRJNA809920.
Rumen fluid metabolomics analysis by LC-MS
Only the samples of CON and CFE150 were used for LC-MS analysis. The metabolites in thawed rumen fluid samples were extracted by 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 dilution with ethanol/methanol (1
4 dilution with ethanol/methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, vol/vol) according to the method of Liu et al.35 In brief, the samples were vortexed for 60 s, and the precipitated protein was removed via centrifugation at 15
1, vol/vol) according to the method of Liu et al.35 In brief, the samples were vortexed for 60 s, and the precipitated protein was removed via centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 20 min at 4 °C. The supernatant collected was subjected to the LC-MS/MS analysis. Metabolomics analysis was conducted using an UHPLC system (1290; Agilent Technologies, USA) coupled to a Triple TOF 5600 MS (Q-TOF, AB Sciex, USA) with an Acquity UPLC BEH C18 column (2.1 × 100 mm with 1.7 μm of particles). Mobile phase A was water containing 0.1% formic acid and mobile phase B was acetonitrile containing 0.1% formic acid; the flow rate was 0.40 mL min−1. The elution gradient was 95% B for 0–1 min; 95% to 65% B for 1–14 min; 65% to 45% B for 14–16 min; 40% B for 16–18 min; 40% to 95% B for 18–18.1 min; 95% B for 18.1–23 min. The injection volume was 2 μL. The MS analysis was performed under the conditions: spray voltage, −4 kV in the negative mode and 5 kV in the positive mode; curtain gas, 30 psi; source temperature, 500 °C; mass range (m/z), 50–1000. A quality control sample was injected after every eight rumen fluid samples to evaluate the stability and repeatability of the system.
000g for 20 min at 4 °C. The supernatant collected was subjected to the LC-MS/MS analysis. Metabolomics analysis was conducted using an UHPLC system (1290; Agilent Technologies, USA) coupled to a Triple TOF 5600 MS (Q-TOF, AB Sciex, USA) with an Acquity UPLC BEH C18 column (2.1 × 100 mm with 1.7 μm of particles). Mobile phase A was water containing 0.1% formic acid and mobile phase B was acetonitrile containing 0.1% formic acid; the flow rate was 0.40 mL min−1. The elution gradient was 95% B for 0–1 min; 95% to 65% B for 1–14 min; 65% to 45% B for 14–16 min; 40% B for 16–18 min; 40% to 95% B for 18–18.1 min; 95% B for 18.1–23 min. The injection volume was 2 μL. The MS analysis was performed under the conditions: spray voltage, −4 kV in the negative mode and 5 kV in the positive mode; curtain gas, 30 psi; source temperature, 500 °C; mass range (m/z), 50–1000. A quality control sample was injected after every eight rumen fluid samples to evaluate the stability and repeatability of the system.
The raw data were then normalized, and a data matrix including ion formula, retention time, m/z values, and peak intensity was generated. Metabolites were matched and identified against the human metabolome database (https://www.hmdb.ca/) and Metlin database (https://metlin.scripps.edu/). The data were analyzed using SIMCA 15 software (Umetrics, Umea, Sweden) for principal component analysis (PCA) and orthogonal partial least squares-discriminant analysis (OPLS-DA). The compounds with FC >1.1 or <0.9, P < 0.05 of Student's t-test, and variable importance in projection (VIP) > 1 were considered differentially expressed metabolites (DEM). The online platform MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/) was used to conduct the metabolic pathway analysis based on DEM using the library of Bos taurus (cow) of the KEGG.36
Serum antioxidant indexes
Blood was taken from the tail veins of each cow into vacuum tubes without a clot activator (Vacutainer, Becton Dickinson, Franklin Lakes, NJ, USA) after the morning milking and before feeding on the last day of each period. Serum was separated by centrifugation at 2000g for 15 min at room temperature, and then stored at −20 °C until analysis. The total antioxidant capacity (T-AOC), MDA, GSH-Px, and T-SOD in the serum were analyzed using commercial kits according to the supplier's instructions (Beijing Solarbio Science & Technology Co., Ltd, Beijing, China).Statistical analysis
Statistical analysis of the DMI, milk performance, rumen fermentation parameters, ruminal specific microbiota, and serum indices was performed using SAS software (version 9.4; SAS Institute Inc.) using the PROC MIXED procedure with the following model:| Yijkl = μ + Si + Pj + Tk + C(S)l(i) + eijkl, |
Results
DMI and milk performance
The DMI and milk performance of cows fed diets added with CFE of increasing levels are shown in Table 1. Although dietary supplementation with CFE did not affect the DMI, the MY was greater (P = 0.028) in cows that received CFE compared with CON. The CFE100 cows had the greatest ECM (P = 0.040), whereas those on CFE150 were statistically similar to those on CON. Supplementing CFE tended to linearly increase (P = 0.083) the feed efficiency expressed as ECM/DMI. The milk lactose percentage linearly increased (P = 0.001) with CFE addition, whereas milk protein and fat were similar among treatments. In addition, CFE supplementation linearly reduced (P = 0.043) milk SCC.| Item | CFE supplemented,a g d−1 | SEM | P-Value | |||||
|---|---|---|---|---|---|---|---|---|
| 0 | 50 | 100 | 150 | Treatment | Linear | Quadratic | ||
| a–cMean values within a row without a common superscript differ significantly (P < 0.05).a CFE, citrus flavonoid extract; DMI, dry matter intake; MY, milk yield; FCM, fat-corrected milk; ECM, energy-corrected milk; MUN, milk urea nitrogen; SCC, somatic cell count.b Calculated as [0.4324 × milk yield (kg d−1) + 16.216 × milk fat (kg d−1)].c Calculated as milk yield (kg d−1) × {[0.3887 × milk fat (%)] + [0.2356 × milk protein (%)] + [0.1653 × milk lactose (%)]}/3.1338. | ||||||||
| DMI, kg d−1 | 21.65 | 21.95 | 22.44 | 22.01 | 0.479 | 0.662 | 0.725 | 0.710 |
| MY, kg d−1 | 31.90b | 33.67ab | 34.37a | 33.96a | 0.270 | 0.028 | 0.262 | 0.423 |
| 3.5% FCMb | 34.83 | 37.07 | 38.62 | 37.45 | 1.046 | 0.078 | 0.216 | 0.312 |
| ECMc | 32.12b | 34.83ab | 36.58a | 34.88ab | 1.055 | 0.040 | 0.174 | 0.182 |
| Feed efficiency | ||||||||
| MY/DMI | 1.48 | 1.54 | 1.54 | 1.55 | 0.036 | 0.401 | 0.157 | 0.413 |
| 3.5% FCM/DMI | 1.61 | 1.69 | 1.73 | 1.71 | 0.046 | 0.234 | 0.133 | 0.269 |
| ECM/DMI | 1.48 | 1.59 | 1.64 | 1.59 | 0.043 | 0.080 | 0.083 | 0.090 |
| Milk composition, % | ||||||||
| Fat | 4.06 | 4.12 | 4.27 | 4.13 | 0.001 | 0.420 | 0.503 | 0.416 |
| Protein | 3.59 | 3.58 | 3.58 | 3.49 | 0.106 | 0.878 | 0.548 | 0.742 |
| Lactose | 4.39c | 4.82a | 5.07a | 4.76a | 0.081 | <0.001 | <0.001 | <0.001 |
| MUN, mg dL−1 | 9.57 | 9.32 | 8.87 | 9.40 | 0.343 | 0.469 | 0.608 | 0.348 |
| SCC, log10 × 103 mL−1 | 1.25a | 1.15a | 1.02b | 1.04b | 0.040 | 0.003 | 0.043 | 0.105 |
Rumen fermentation parameters and rumen bacterial cell-wall components
There were no effects of CFE supplementation on rumen fluid pH (Table 2). The concentrations of total VFA (TVFA; P = 0.022), acetate (P = 0.035), propionate (P = 0.014), and butyrate (P = 0.040) linearly increased with increasing CFE levels. The proportions of acetate, propionate, butyrate, isobutyrate, valerate, and isovalerate were not influenced by the supplementation of CFE. Moreover, feeding CFE did not affect the NH3–N content, while the ruminal MCP concentration was linearly increased (P = 0.036) with increasing CFE amount. As shown in ESI Fig. 1,† the ruminal LPS concentration decreased linearly (P = 0.006) with increasing CFE supplementation. Supplementing the diet with increasing levels of CFE tended to linearly decrease (P = 0.075) the ruminal LTA concentration.| Item | CFE supplemented,a g d−1 | SEM | P-Value | |||||
|---|---|---|---|---|---|---|---|---|
| 0 | 50 | 100 | 150 | Treatment | Linear | Quadratic | ||
| a CFE, citrus flavonoid extract; TVFA, total volatile fatty acids; VFA, volatile fatty acids; A/P, the ratio of acetate to propionate; NH3–N, ammonia–nitrogen; MCP, microbial crude protein. | ||||||||
| pH | 6.53 | 6.55 | 6.53 | 6.59 | 0.047 | 0.970 | 0.954 | 0.795 |
| TVFA, mmol L−1 | 93.97 | 95.54 | 102.43 | 103.79 | 3.490 | 0.110 | 0.022 | 0.975 |
| Individual VFA, % | ||||||||
| Acetate | 63.66 | 63.68 | 63.07 | 63.07 | 0.547 | 0.750 | 0.357 | 0.984 |
| Propionate | 19.20 | 19.24 | 19.48 | 19.62 | 0.232 | 0.521 | 0.403 | 0.898 |
| Butyrate | 11.28 | 11.10 | 11.95 | 11.72 | 0.425 | 0.404 | 0.220 | 0.956 |
| Isobutyrate | 2.11 | 2.36 | 2.01 | 2.03 | 0.123 | 0.133 | 0.290 | 0.354 |
| Valerate | 1.54 | 1.55 | 1.47 | 1.40 | 0.102 | 0.676 | 0.357 | 0.764 |
| Isovalerate | 2.21 | 2.07 | 2.03 | 2.15 | 0.114 | 0.642 | 0.657 | 0.246 |
| A/P | 3.32 | 3.34 | 3.24 | 3.22 | 0.064 | 0.501 | 0.369 | 0.829 |
| Individual VFA, mmol L−1 | ||||||||
| Acetate | 59.86 | 60.72 | 64.61 | 65.40 | 2.083 | 0.149 | 0.035 | 0.985 |
| Propionate | 18.02 | 18.39 | 19.94 | 20.40 | 0.732 | 0.073 | 0.014 | 0.953 |
| Butyrate | 10.59 | 10.71 | 12.25 | 12.21 | 0.711 | 0.153 | 0.040 | 0.897 |
| Isobutyrate | 1.97 | 2.24 | 2.05 | 2.11 | 0.116 | 0.361 | 0.634 | 0.392 |
| Valerate | 1.45 | 1.49 | 1.50 | 1.46 | 0.112 | 0.984 | 0.984 | 0.752 |
| Isovalerate | 2.08 | 1.99 | 2.07 | 2.23 | 0.131 | 0.617 | 0.365 | 0.356 |
| NH3–N, mg dL−1 | 8.70 | 10.68 | 8.69 | 8.30 | 1.266 | 0.522 | 0.602 | 0.391 |
| MCP, mg mL−1 | 0.88b | 1.01ab | 1.13a | 1.18a | 0.093 | 0.045 | 0.036 | 0.750 |
The relative abundance of specific rumen microbes determined by real-time qPCR
The relative abundances of the target microbes are presented in Table 3. There was a quadratic effect on the ruminal fungi (P = 0.031) and methanogen (P = 0.041) abundances with the lowest values at CFE100 and CFE50, respectively. Supplementing CFE linearly decreased (P = 0.026) the relative abundance of Prevotella brevis. The relative abundance of Prevotella ruminicola tended to linearly decrease (P = 0.067) with CFE inclusion in the diet. The relative abundances of Ruminococcus albus (P = 0.037) and Ruminococcus flavefaciens (P = 0.029) linearly increased with CFE supplementation. Additionally, there was a quadratic effect of CFE on Succinimonas amylolytica abundance, with the minimum value in CFE50 cows.| Item | CFE supplemented,a g d−1 | SEM | P-Value | |||||
|---|---|---|---|---|---|---|---|---|
| 0 | 50 | 100 | 150 | Treatment | Linear | Quadratic | ||
| a–cMean values within a row without a common superscript differ significantly (P < 0.05).a CFE, citrus flavonoid extract. | ||||||||
| Fungi, ×10−4 | 3.85 | 2.77 | 1.78 | 3.08 | 0.696 | 0.074 | 0.926 | 0.031 |
| Methanogens, ×10−1 | 17.53a | 4.00c | 6.99c | 11.87b | 3.377 | 0.042 | 0.472 | 0.041 |
| Protozoa, ×10−2 | 7.15 | 5.06 | 8.81 | 5.65 | 3.333 | 0.141 | 0.343 | 0.550 |
| Fibrobacter succinogenes, ×10−2 | 5.23 | 7.18 | 3.81 | 6.42 | 1.218 | 0.222 | 0.969 | 0.799 |
| Megasphaera elsdenii, ×10−5 | 2.40 | 1.43 | 1.15 | 1.89 | 0.751 | 0.650 | 0.597 | 0.265 |
| Prevotella brevis | 9.91a | 4.33ab | 5.17ab | 2.52b | 2.746 | 0.048 | 0.026 | 0.258 |
| Prevotella ruminicola, ×10−2 | 6.57 | 5.24 | 3.10 | 2.99 | 1.518 | 0.273 | 0.067 | 0.691 |
| Ruminococcus albus, ×10−2 | 1.90 | 1.82 | 3.09 | 3.67 | 0.831 | 0.066 | 0.037 | 0.857 |
| Ruminococcus flavefaciens, ×10−2 | 2.62b | 3.26b | 3.00b | 5.33a | 1.123 | 0.048 | 0.029 | 0.427 |
| Ruminobacter amylophilus, ×10−2 | 2.74 | 5.39 | 4.58 | 4.13 | 1.690 | 0.673 | 0.628 | 0.324 |
| Selenomonas ruminantium, ×10−2 | 2.54 | 2.00 | 2.24 | 2.22 | 0.436 | 0.844 | 0.733 | 0.591 |
| Succinimonas amylolytica, ×10−2 | 9.79 | 2.72 | 3.01 | 8.07 | 2.278 | 0.054 | 0.661 | 0.019 |
| Streptococcus bovis, ×10−2 | 2.68 | 4.20 | 4.37 | 2.74 | 1.806 | 0.337 | 0.427 | 0.174 |
Rumen metagenome profiling
Metagenomic sequencing of the total DNA from 16 rumen fluid samples generated a total of 762![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 789
789![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 648 reads, with an average of 47
648 reads, with an average of 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 674
674![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 353 ± 4
353 ± 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 556
556![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 442 (mean ± SD) reads per sample. After quality control and removal of host contamination, 557
442 (mean ± SD) reads per sample. After quality control and removal of host contamination, 557![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 633
633![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 640 high-quality reads were generated, with 34
640 high-quality reads were generated, with 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 852
852![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 103 ± 2
103 ± 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 279
279![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 841 reads per sample (ESI Table 4†). A total of 12
841 reads per sample (ESI Table 4†). A total of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 097, 293 contigs were generated by the de novo assembly (the N50 length of 551 ± 17 bp), with 756
097, 293 contigs were generated by the de novo assembly (the N50 length of 551 ± 17 bp), with 756![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 081 ± 27
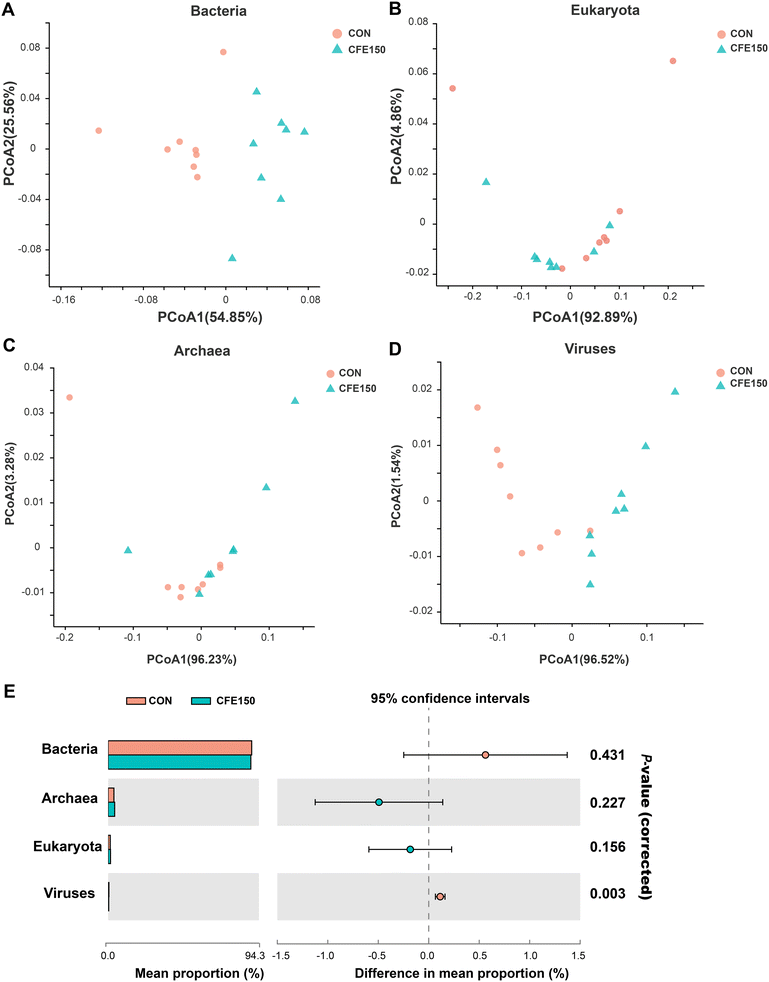
081 ± 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 721 reads for each sample. A genome assembly statistics summary is depicted in Table S3.† The rumen metagenome contains 93.95% bacteria, 1.47% eukaryota, 4.02% archaea, and 0.45% viruses (ESI Fig. 2†). As depicted in ESI Table 5,† bacteria and viruses were significantly different based on the permutational multivariate analysis of variance (PERMANOVA), but eukaryota and archaea were similar between CON and CFE150. The PCoA plot visually showed the distinct separation of bacteria and viruses between CON and CFE150 based on the Bray–Curtis distance (Fig. 1A–D). At the domain level, the relative abundance of viruses was significantly less in the rumen of CFE150 cows compared with CON (Fig. 1E).
721 reads for each sample. A genome assembly statistics summary is depicted in Table S3.† The rumen metagenome contains 93.95% bacteria, 1.47% eukaryota, 4.02% archaea, and 0.45% viruses (ESI Fig. 2†). As depicted in ESI Table 5,† bacteria and viruses were significantly different based on the permutational multivariate analysis of variance (PERMANOVA), but eukaryota and archaea were similar between CON and CFE150. The PCoA plot visually showed the distinct separation of bacteria and viruses between CON and CFE150 based on the Bray–Curtis distance (Fig. 1A–D). At the domain level, the relative abundance of viruses was significantly less in the rumen of CFE150 cows compared with CON (Fig. 1E).
The rumen microbial communities between CON and CFE150 cows at the phylum and genus levels are shown in ESI Fig. 3A–D and 4A–D.† At the phylum level, Firmicutes (44.19% ± 4.25%), Bacteroidetes (37.76% ± 3.18%), Actinobacteria (1.62%), and Proteobacteria (1.36% ± 0.20%) massively dominated the bacterial community. The phyla Ciliophora (0.75% ± 0.014%), Streptophyta (0.12% ± 0.009%), and Chytridiomycota (0.08% ± 0.007%) were the main microbiota in the eukaryotes. Regarding archaea and viruses, the phyla Euryarchaeota (3.61% ± 0.009%) and Uroviricota (0.33% ± 0.042%) dominated the community, respectively. At the genus level, the dominant bacterial microbiota were Prevotella (23.60% ± 3.92%), followed by Succiniclasticum (2.21% ± 0.18%), Bacteroides (2.10% ± 0.25%), Ruminococcus (1.88% ± 0.30%), Clostridium (1.57% ± 0.23%), and Butyrivibrio (1.55% ± 0.12%). The top genera belonging to the domains eukaryota, archaea, and viruses were Stylophora (0.002% ± 0.0001%), Methanobrevibacter (3.76% ± 0.48%), and Ithacavirus (0.03% ± 0.001%), respectively.
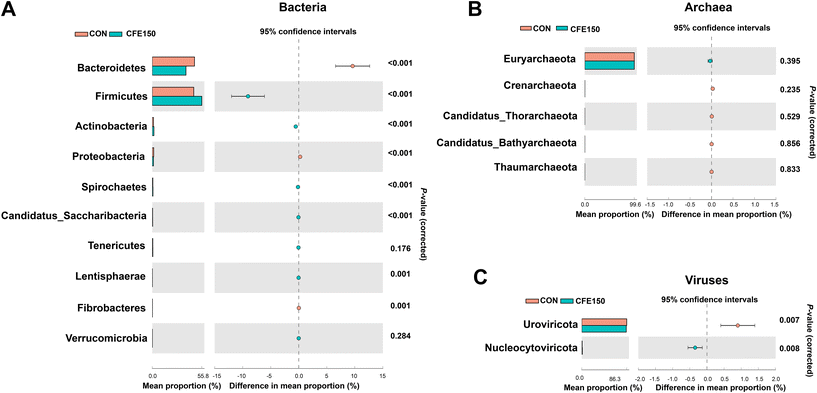
The comparison of the rumen microbial taxa at the phylum and genus levels between the CON and CFE150 groups was focused on bacteria, archaea, and viruses. The analytical results of the top 10 bacterial phyla obtained by the Wilcoxon rank-sum test are shown in Fig. 2A. The phyla Bacteroidetes, Proteobacteria, and Fibrobacteres exhibited higher abundances (P < 0.05) in the rumen of the CFE150 cows, whereas Firmicutes, Actinobacteria, Spirochaetes, Candidatus_Saccharibacteria, and Lentisphaerae exhibited higher abundances (P < 0.05) in the CFE150 cows compared to the CON cows. Compared with the CON cows, the CFE150 cows had a greater ratio of Firmicutes to Bacteroidetes (F/B) in the rumen (ESI Fig. 5†). No differences were observed in the top five phyla within archaea between two groups (Fig. 2B). For viruses, the phylum Uroviricota exhibited a higher abundance (P < 0.05) in the CON cows, whereas Nucleocytoviricota (P < 0.05) had a greater abundance (P < 0.05) in the CFE150 cows (Fig. 2C).
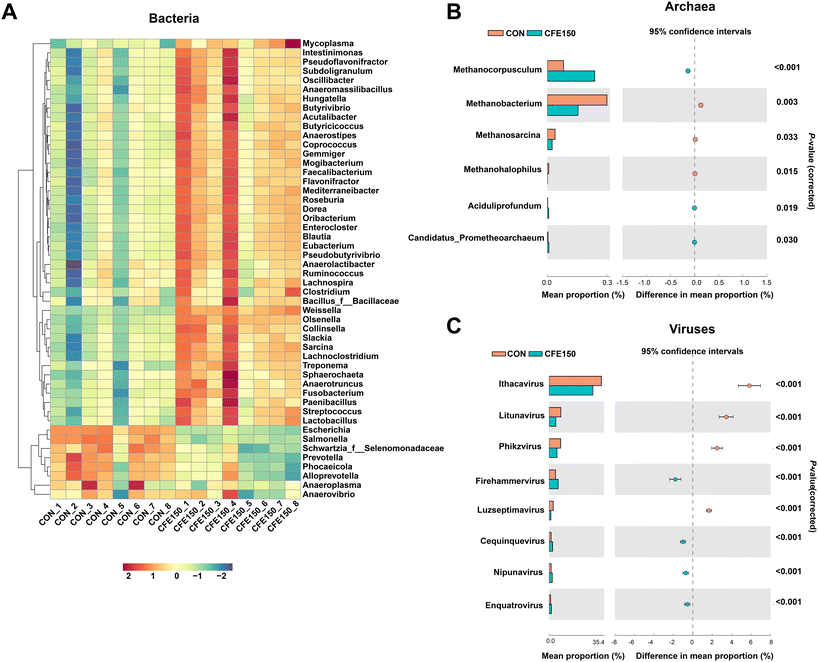
The top 50 differential bacterial genera are shown in Fig. 3A. The relative abundances of 32 genera including Ruminococcus, Clostridium, Butyrivibrio, and Pseudobutyrivibrio were greater (P < 0.05) in the CFE150 cows, whereas the relative abundances of eight genera, including Prevotella and Anaerovibrio, were greater (P < 0.05) in the CON cows. The archaea genera Methanobacterium, Methanosarcina, and Methanohalophilus showed a low abundance (P < 0.05) in the CFE150 cows (Fig. 3B), whereas the genera Methanocorpusculum, Aciduliprofundum, and Candidatus_Prometheoarchaeum were more abundant (P < 0.05) in the CON cows. The virus genera Ithacavirus, Litunavirus, Phikzvirus, and Luzseptimavirus showed a low abundance (P < 0.05) in the CFE150 cows (Fig. 3C), whereas the genera Firehammervirus, Cequinquevirus, Nipunavirus, and Enquatrovirus were more abundant (P < 0.05) in the CFE150 cows compared with the CON cows.
Comparisons of the taxa at the species level were performed using LEfSe with the non-parametric factorial Kruskal–Wallis and Wilcoxon rank-sum tests. A total of 28 species, mainly belonging to Bacteroidetes and Firmicutes, were more abundant in the CON cows compared to the CFE150 cows (LDA > 2.5 and P < 0.05; Fig. 4), whereas 17 species were significantly enriched in the CFE150 cows (LDA > 2.5 and P < 0.05).
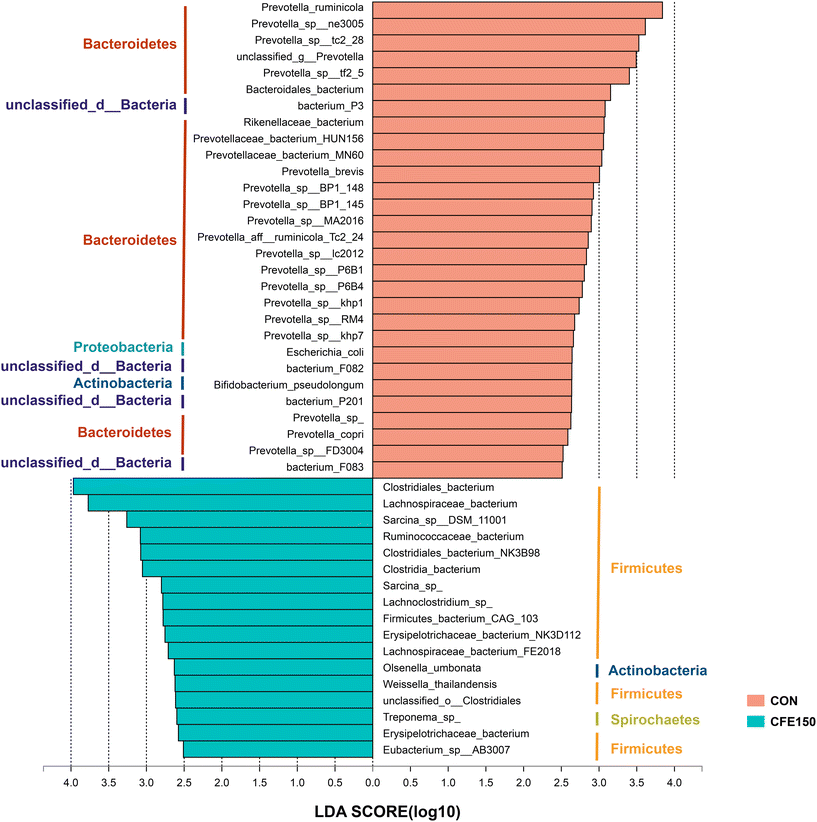 | ||
| Fig. 4 Differential rumen microbial taxa at the species level between the CON and CFE150 cows based on LEfSe. | ||
Functional analysis of the rumen microbiome
In the KEGG pathway level 2, the five most abundant pathways were “global and overview maps” (37.35% ± 4.37%), “carbohydrate metabolism” (10.58% ± 0.88%), “amino acid metabolism” (6.38% ± 0.72%), “replication and repair” (4.61% ± 0.44%), and “energy metabolism” (3.89% ± 0.50%) (ESI Fig. 6A†). The PCoA plot based on the Bray–Curtis distance showed a clear separation of two groups at pathway level 3 (ESI Fig. 6B†). As shown in ESI Fig. 6C,† the phyla Firmicutes, Bacteroidetes, Euryarchaeota, and Actinobacteria were the major contributors to the top 10 functions at pathway level 3. We considered 155 endogenous third-level metabolic pathways as rumen microbial pathways in the KEGG profiles for the further analysis. ESI Fig. 7† shows two “cellular processes” and three “environmental information processing” and eight “genetic information processing” pathways were significantly different (P < 0.05) between two groups. As shown in Fig. 5, there were 56 differential metabolic pathways (P < 0.05) at level 3 involved with “amino acid metabolism”, “biosynthesis of other secondary metabolites”, “carbohydrate metabolism”, “energy metabolism”, “glycan biosynthesis and metabolism”, “lipid metabolism”, “metabolism of cofactors and vitamins”, “metabolism of other amino acids”, “metabolism of terpenoids and polyketides”, “nucleotide metabolism”, and “xenobiotics biodegradation and metabolism”.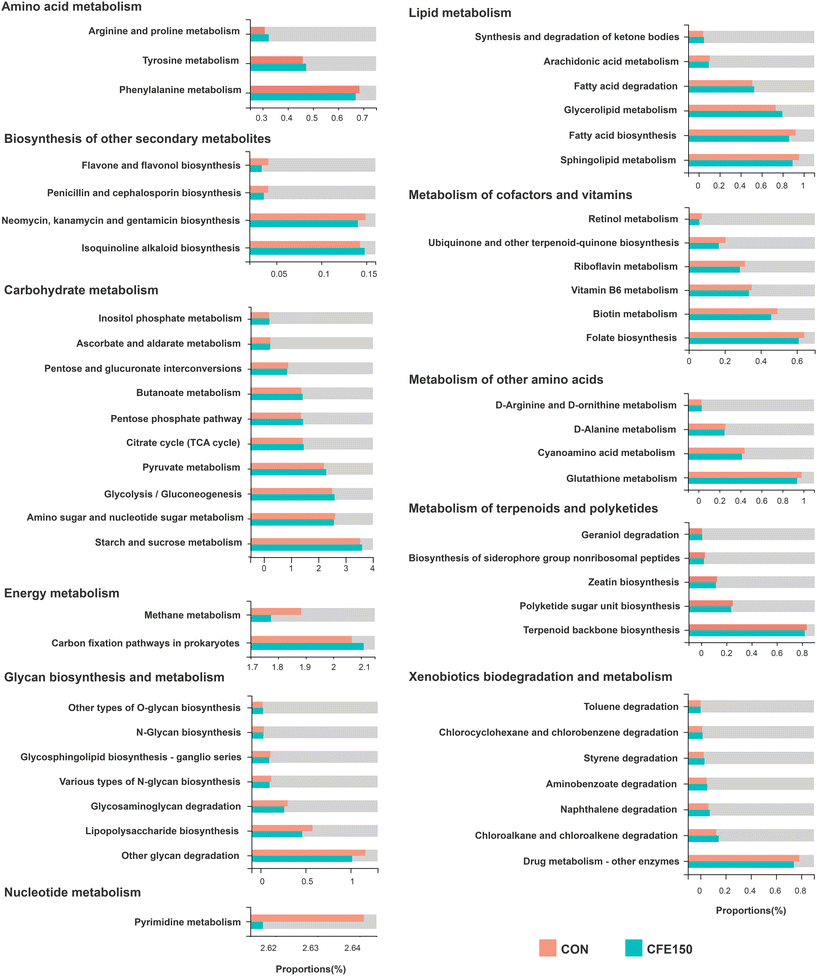 | ||
| Fig. 5 Differential KEGG functions at pathway level 3 within “Metabolism” between the CON and CFE150 cows. | ||
Due to the fact that carbohydrates are degraded by multiple enzymes, we focused on the differences in the profiles of ruminal CAZymes between the CON and CFE150 cows. As shown in ESI Fig. 8A,† the CAZymes community consisted of glycoside hydrolases (GH; 55.85% ± 2.94%), glycosyltransferases (GT; 19.77% ± 1.89%), carbohydrate esterases (CE; 13.45% ± 1.57%), carbohydrate-binding modules (CBM; 5.59% ± 0.73%), and polysaccharide lyases (PL; 2.81% ± 0.24%), and exhibited auxiliary activities (AA; 2.55% ± 0.36%). At the class level, CE and AA exhibited greater abundances (P < 0.05) in the CFE150 cows than in the CON cows (ESI Fig. 8B†), whereas PL had a greater abundance (P < 0.05) in the CON cows. At the family level, the PCoA plot revealed a separation of the two groups in CAZymes (ESI Fig. 8C†). Among the CAZymes that participated in degrading carbohydrates (including cellulose, hemicellulose, starch, protein, and lignin), 16 families (8 of GH, 7 of CE, and 1 of AA) were enriched in the CFE150 cows, whereas 14 families (11 of GH, 1 of CE and 2 of PL; ESI Table 6†) were enriched in the CON cows. For the GT class (participated in carbohydrate synthesis), GT28, GT35, and GT5 showed greater abundances in the CFE150 cows, whereas GT41 was enriched in the CON cows.
Rumen metabolomics profiling
The unsupervised PCA plots show that there was no separation between the rumen fluid metabolites of the CON and CFE150 cows (ESI Fig. 9A and B†). The OPLS-DA score plots used to verify the distinct metabolites between the CON and CFE150 cows and supervise the multivariate analysis are shown in ESI Fig. 9C and D.† All the samples fell within the 95% Hotelling T2 ellipse. The classification parameters were R2Y = 0.984 and 0.978 in the positive and negative ion modes, respectively, showing a good fit and predictive power (ESI Fig. 9E and F†). In addition, the low Q2 intercept suggested the robustness of the model, showing reliability and a low risk of overfitting.Compared with the CON cows, the relative concentrations of seven metabolites including soyasaponin A1, oleoyl glycine, lauric acid, DG(14:1(9Z)/14:1(9Z)/0:0), 3-aminoquinoline, oryzalic acid B, and isohumbertiol were low (P < 0.05) in the CFE150 cows (Table 4). Compared with the CON cows, the relative concentrations of 34 metabolites including cyclohexylamine, tetrahydrobiopterin, fructose 6-phosphate, 2-formaminobenzoylacetate, 7a-hydroxydehydroepiandrosterone, p-chlorophenylalanine, tyrosine, pyruvate, glucose, deoxycholic acid, glycyl-histidine, malonic acid, nigellic acid, 5′-carboxy-gamma-chromanol, benzylmalic acid, pyrogallin, proline, retinyl beta-glucuronide, polyethylene, trigoforin, 3-coumaric acid, urolithin C, azelaic acid, cinnamic acid, (R)-meranzin, benzyl sulfate, glucose-6-phosphate, hippuric acid, DL-benzylsuccinic acid, hesperetin, digitoxigenin, capric acid, physalin I, and naringenin were greater (P < 0.05) in the CFE150 cows.
| Metabolites | VIPa | FC | P-Value | HMDB superclass |
|---|---|---|---|---|
| a VIP, variable importance in projection; FC, fold change. | ||||
| Down-regulation | ||||
| Soyasaponin A1 | 4.59 | 0.49 | 0.029 | Lipids and lipid-like molecules |
| Oleoyl glycine | 2.47 | 0.60 | 0.003 | Organic acids and derivatives |
| 3-Aminoquinoline | 2.13 | 0.72 | 0.043 | — |
| Lauric acid | 1.99 | 0.72 | 0.046 | Lipids and lipid-like molecules |
| DG(14:1(9Z)/14:1(9Z)/0:0) | 1.89 | 0.83 | 0.013 | Lipids and lipid-like molecules |
| Oryzalic acid B | 1.36 | 0.84 | 0.037 | Organic acids and derivatives |
| Isohumbertiol | 1.24 | 0.86 | 0.026 | Lipids and lipid-like molecules |
| Up-regulation | ||||
| Cyclohexylamine | 1.02 | 1.18 | 0.037 | Organic nitrogen compounds |
| Tetrahydrobiopterin | 1.39 | 1.20 | 0.011 | Organic oxygen compounds |
| Fructose 6-phosphate | 1.24 | 1.21 | 0.029 | Organic oxygen compounds |
| 2-Formaminobenzoylacetate | 1.28 | 1.21 | 0.034 | — |
| 7a-Hydroxydehydroepiandrosterone | 1.18 | 1.24 | 0.033 | Lipids and lipid-like molecules |
| p-Chlorophenylalanine | 1.48 | 1.25 | 0.002 | Phenylpropanoids and polyketides |
| Tyrosine | 1.37 | 1.27 | 0.016 | Organic acids and derivatives |
| Pyruvate | 1.44 | 1.30 | 0.019 | Organic oxygen compounds |
| Glucose | 1.40 | 1.31 | 0.028 | Organic acids and derivatives |
| Deoxycholic acid | 1.52 | 1.32 | 0.030 | Lipids and lipid-like molecules |
| Glycyl-histidine | 1.63 | 1.39 | 0.021 | Organic acids and derivatives |
| Malonic acid | 1.69 | 1.42 | 0.019 | — |
| Nigellic acid | 2.08 | 1.44 | 0.003 | Lipids and lipid-like molecules |
| 5′-Carboxy-gamma-chromanol | 3.55 | 1.52 | <0.001 | Organoheterocyclic compounds |
| Benzylmalic acid | 3.36 | 1.53 | 0.009 | — |
| Pyrogallin | 4.31 | 1.56 | <0.001 | — |
| Proline | 4.15 | 1.57 | <0.001 | Organic acids and derivatives |
| Retinyl beta-glucuronide | 3.90 | 1.58 | 0.007 | Lipids and lipid-like molecules |
| Polyethylene | 4.82 | 1.63 | <0.001 | Organic acids and derivatives |
| Trigoforin | 4.85 | 1.65 | <0.001 | Phenylpropanoids and polyketides |
| 3-Coumaric acid | 4.82 | 1.66 | <0.001 | Phenylpropanoids and polyketides |
| Urolithin C | 4.18 | 1.70 | <0.001 | Phenylpropanoids and polyketides |
| Azelaic acid | 4.07 | 1.78 | <0.001 | Lipids and lipid-like molecules |
| Cinnamic acid | 5.72 | 1.78 | <0.001 | Phenylpropanoids and polyketides |
| (R)-Meranzin | 4.68 | 1.85 | 0.001 | Phenylpropanoids and polyketides |
| Benzyl sulfate | 4.25 | 1.89 | <0.001 | Benzenoids |
| Glucose-6-phosphate | 5.40 | 1.93 | <0.001 | Organic oxygen compounds |
| Hippuric acid | 5.86 | 1.97 | <0.001 | Benzenoids |
| DL-Benzylsuccinic acid | 5.85 | 1.98 | <0.001 | — |
| Hesperetin | 6.58 | 1.99 | <0.001 | Phenylpropanoids and polyketides |
| Digitoxigenin | 6.83 | 1.94 | <0.001 | — |
| Capric acid | 5.22 | 2.16 | 0.020 | Lipids and lipid-like molecules |
| Physalin I | 6.83 | 2.17 | <0.001 | Lipids and lipid-like molecules |
| Naringenin | 8.19 | 3.18 | <0.001 | Phenylpropanoids and polyketides |
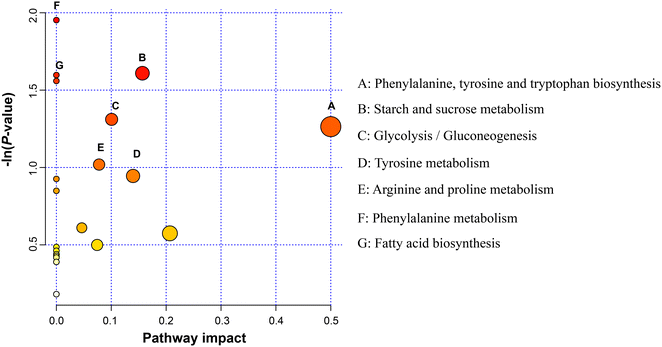
Metabolic pathway analysis based on these 41 DEM revealed the enrichment of the pathways, including “phenylalanine, tyrosine and tryptophan biosynthesis”, “starch and sucrose metabolism”, “glycolysis/gluconeogenesis”, “tyrosine metabolism”, and “arginine and proline metabolism” (Fig. 6).
Serum antioxidant indexes
As shown in ESI Fig. 10,† dietary supplementation with CFE linearly (P < 0.05) increased serum T-AOC and GSH-Px. Serum MDA was linearly reduced (P < 0.05) with increasing CFE levels. Serum T-SOD was unaffected by CFE supplementation.Discussion
Lactational performance and antioxidant capacity
Although the DMI was unaffected by treatments, dietary supplementation with CFE increased the MY and milk lactose percentage. In contrast to what was observed in our study, Ying et al.9 did not observe any effects of citrus extracts on the DMI, MY or milk composition of dairy cows. The supplementation level of citrus extract (4 g d−1) in the study by Ying et al.9 was far lower than that in our study; therefore, the difference in supplementation amounts might account for this disparity in milk performance. Similarly, several studies also reported reduced milk SCC of dairy cows that received diets supplemented with flavonoid-rich extracts from bamboo leaves,37 baicalin,38 or quercetin.39 An earlier study reported that hesperidin and naringenin administered via the intramammary route reduced milk SCC of mastitis-affected cows.40 In the present study, the antioxidant capacity of dairy cows was improved by supplementing CFE to the diet. The results confirmed the reported beneficial effects of naringin and hesperidin in improving the antioxidant defense systems in sheep1 and lambs.2 At the cellular level, the improvement of antioxidant capacity allows for an increase in energy productivity for milking and a reduction in energy loss due to accumulating reactive metabolites,41 which was one of the possible reasons for the improved milk production of the CFE-fed groups.Rumen fermentation and ruminal microbiome
Several studies have suggested that plant flavonoids might act as a carbon source for ruminal microbes, which could explain higher total VFA concentrations.42,43 It was expected that CFE improved rumen VFA production in our study. Propionate represents 60%–74% of the substrate uptake for glucose synthesis in the livers of dairy cows, and the mammary gland is one of the tissues that has an obligatory demand for glucose.44 Supplementing CFE increased the ruminal propionate concentration, which indicated that more gluconeogenic precursor would be available to the cows. Therefore, the improved lactational performance and greater lactose percentage were likely results of a greater concentration of TVFA with CFE supplementation.Firmicutes and Bacteroidetes were the most abundant ruminal phyla in dairy cows, which is in line with the previous studies on dairy cows.44,45 Bacteria within Bacteroidetes degrade non-structural carbohydrates and non-fibrous polysaccharides in the rumen,35 while bacteria belonging to Firmicutes are involved in structural carbohydrates and starch to produce acetate, propionate, and butyrate.46 Therefore, the improvement of the TVFA concentration could be attributed to the increased Firmicutes observed in our study.
The anaerobic metabolism of rumen microorganisms has a major impact on the way in which flavonoids undergo transformation in the rumen. However, only a few of the bacteria catalyzing these reactions have been identified and their degradation pathways been unraveled. Rumen bacteria within Butyrivibrio (Butyrivibrio sp. C3) and Eubacterium (Eubacterium oxidoreducens) were shown to utilize rutin and quercetin as the sources of carbon and energy.47–49 Bacteria within Butyrivibrio and Eubacterium belong to Firmicutes. Most of the identified bacterial species/strains involved in the conversion of flavanones (e.g., hesperidin and naringin) are members of different families within the Firmicutes phylum in the human gut.50 In the present study, we speculated that microbes within Bacteroidetes and Proteobacteria phyla may be inhibited by the antibacterial properties of flavonoids, whereas more members belonging to Firmicutes are favored selectively by citrus flavonoids.
Díaz et al.51 reported that the higher relative abundance of Firmicutes or an increased F/B ratio could improve production performance in bovines. Previous studies also showed that an increased F/B ratio is associated with a higher feed efficiency in animals.52,53 In the present study, we observed that the CFE150 cows displayed higher Firmicutes and low Bacteroidetes abundances compared with the CON cows. The results indicated the alteration of the top two phyla that could play an important role in the enhancement of MY and feed efficiency. Accordingly, many of the microbiota taxa alterations within the phylum Firmicutes, such as the genera Ruminococcus, Lachnospira, Clostridium, Butyrivibrio, Pseudobutyrivibrio, and the species Clostridiales bacterium, Lachnospiraceae bacterium, and Ruminococcaceae bacterium, were detected in our research. Consistently, at lower taxonomic levels, qPCR results showed that the species Ruminococcus albus and Ruminococcus flavefaciens within the genus Ruminococcus as typical cellulolytic bacteria in the rumen54 were increased in the CFE150 cows. The results revealed that CFE supplementation improved the growth of rumen cellulolytic bacteria, and consequently resulted in greater production of acetate, propionate, and butyrate. Given that cellulolytic bacteria have a high preference for NH3–N,55 as expected, the ruminal MCP concentration was increased with CFE supplementation, indicating that the utilization of the action of rumen microbes for protein synthesis was increased.
Bacteria and methanogens have a symbiotic relationship in the animal gut, and methanogens use hydrogen produced by cellulolytic bacteria as an electron donor to produce CH4.45 Flavonoids are known to have direct effects against methanogens and to suppress CH4 production.56 The major genera of rumen methanogens include Methanobrevibacter, Methanomicrobium, Methanobacterium and Methanosarcina.56 In the present study, the CFE150 cows showed a low abundance of Methanobacterium and Methanosarcina in the rumen compared to the CON cows. The qPCR results also showed that methanogen abundance was higher in the CFE150 cows than the CON cows. Unfortunately, actual CH4 emission measurements could not be conducted in our study, although it would help to explain the observed results. Corresponding to the alterations of methanogens, we observed that the “methane metabolism” in the KEGG pathway level 3 was less abundant in the CFE150 cows. Mitigating CH4 production will benefit the environment and will reduce unnecessary digested energy waste and increase energy preservation for milking.
The effects of plant flavonoids on ruminal fungi have not been much studied. In the present study, the effects of CFE on fungi were less subtle than those on bacteria, which indicated that cell-surface receptors of fungi are less sensitive to flavonoids. Hesperidin and hesperetin flavanones have different biological properties, including antiviral activity.57 Despite the growing knowledge regarding the functions and significance of viruses in other ecosystems, little to nothing is known about the roles and dynamics of rumen viruses. It was reported that rumen bacteria and viruses respond differently to dietary perturbation and have different ecological drivers.58 In the current research, CFE150 decreased virus abundance at the domain level and decreased Uroviricota abundance at the phylum level. The results could be due to the antiviral role of citrus flavonoids. However, the interaction among bacteria, fungi, archaea, and viruses under CFE supplementation remain to be further studied.
Ruminal KEGG function, CAZymes, and metabolites
Accompanied by remodeling of the ruminal taxonomic composition, the KEGG function profiles differed between the CON and CFE150 groups. We observed that the “bacterial chemotaxis” was enriched in the CFE150 cows. Bacterial chemotaxis refers to the movement of cells in response to a chemical stimulus.59 The results in the current study indicated that supplementing CFE enhanced the ability of rumen microbiota to sense and move towards favorable nutrients. The CFE150 cows showed greater abundances of gene coding for amino acid metabolism and carbohydrate metabolism. “Tyrosine metabolism” and “arginine and proline metabolism” were significantly enriched in the rumen of the CFE150 cows. Consistently, the up-regulated tyrosine and proline in the rumen were in agreement with the results. Arginine and glutamate are sources of proline synthesis. It has been reported that proline plays a vital role in protein synthesis, metabolism, and nutrition, as well as in anti-oxidative reactions and immune responses.60 Increased milk yield may be associated with an increased supply of tyrosine to the mammary gland. Since plasma tyrosine is the sole precursor of tyrosine in milk protein, an adequate amount of exogenous tyrosine appears to be essential for milk protein synthesis.61 We also found that “pyruvate metabolism” was enriched in the rumen of the CFE150 cows. The results agreed with the increased ruminal pyruvate concentration in the CFE150 cows. Pyruvate is an intermediate product between the upstream “starch and sucrose metabolism” and the downstream pathway of “glycolysis”, which were both significantly higher in the CFE150 cows, indicating a greater capacity to decompose carbohydrates. Compared with the CON cows, the CFE150 cows had a greater ruminal concentration of glucosaminic acid, which is related to the enriched pentose phosphate pathway and greater efficiency of energy harvest in the rumen.62Multiple enzymes are required for the complex degradation of carbohydrates in the rumen, including GH, PL, CE, GT, AA, and CBM. Supplementing CFE also significantly affected CAZy profiling in the rumen of dairy cows, and the most of altered CAZy belonging to GH, which are polysaccharide-degrading enzymes produced by cellulolytic bacteria.63 The increased GH abundance might be due to the increase in Firmicutes. For example, the family GH13, an amylase that degrades starch, occurs predominantly in the genus Butyrivibrio.64 The enzymes within the GH94 family exhibit cellobiose phosphorylase activity, and convert the reversible phosphorolysis of cellobiose into α-glucose 1-phosphate and glucose.65 Cellulose phosphorylases with direct or indirect involvement in cellulose degradation have been identified in a wide variety of bacteria, such as Ruminococcus, Lachnoclostridium and Oscillibacter,66 which were all enriched in the CFE150 cows. Therefore, we concluded that the abundance of GH13 and GH94 families could contribute to the increase of glucose concentration in the rumen, as observed in our study. The class CE can hydrolyze the ester bonds present in pectin and acetylated arabinoxylans.67 Regarding CE, some families with xylan esterase activity, including CE1, CE2, CE3, CE4, and CE7, showed greater abundances in the rumen of CFE150 cows, which indicated that supplementing CFE improved the microbial utilization of xylan.
Rumen bacterial cell-wall components
The LPS molecules are the characteristic components of the cell wall in Gram-negative bacteria. At the phylum level, the Gram-negative bacteria Bacteroidetes and Proteobacteria were reduced in the CFE150 cows, which might contribute to the LPS reduction and the decreased feature function “lipopolysaccharide biosynthesis” in the rumen. The bacterial LPS accumulated in the rumen can circulate into the peripheral blood stream in cows.68 Immunoactivation and systemic inflammation responses are stimulated after the interaction between LPS and lipopolysaccharide binding protein (LBP). The inflammatory responses resulting from LPS markedly disrupt the metabolic homeostasis in high-yield dairy cows with a high metabolic load.69,70 De Nardi et al.71 reported that flavonoid–essential oil mixtures did not change the ruminal LPS content, whereas the mixture reduced the ruminal concentrations of LBP and reduced the blood neutrophil concentration in heifers. Therefore, it was assumed that supplementing CFE could decrease inflammation in the mammary glands of dairy cows, highlighting that the health-promoting effects of CFE might be an additional reason for the reduced SCC.Overall, dietary supplementation with CFE significantly altered the rumen microbial structure and increased the relative abundances of structural carbohydrate-degrading bacteria and decreased the abundances of key methanogens. The main limitation of this study was that the rumen fluid samples lacked undigested solid fiber particles, which are also important to understand the microbiome profiling of liquid and solid fractions.
Conclusions
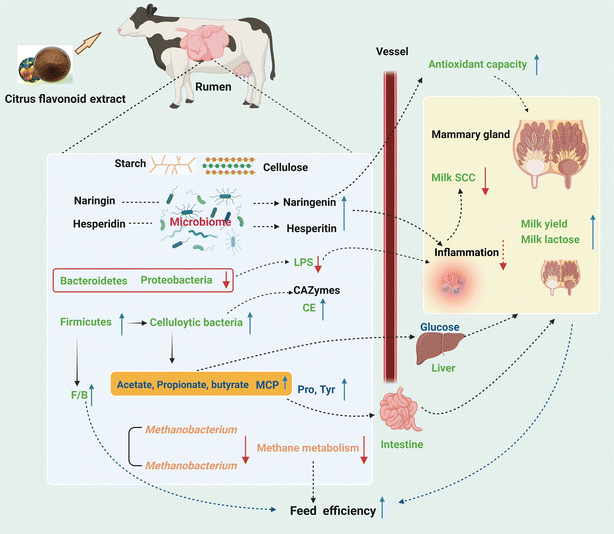
The present study provided new insights into the mode of action of CFE, such as improved MY and reduced milk SCC, increased ruminal TVFA and MCP, and decreased ruminal LPS in lactating dairy cows. Supplementation of CFE in the diet of cows in the mid-lactation period caused a significant shift in the rumen bacterial community, and also decreased the relative abundances of methanogenic archaea and Uroviricota. The greater lactational performance of dairy cows with CFE supplementation was likely a result of the increased VFA production, increased F/B in the rumen, decreased ruminal methane metabolism, and enhanced antioxidant capacity (Fig. 7), which have important implications for the dairy feeding system. The CFE serves as a promising natural supplement for supporting the lactation of dairy cows, with a positive impact on both animal health and environment. Further research is needed to elucidate the potential interaction among citrus flavonoids and rumen microbes by combining microbial culturomics, microbiome and metabolome, to facilitate the application of citrus flavonoid extracts.Author contributions
SY, LL, and HZ conducted the experiment. SZ collected the samples. YT, ML, and YZ analyzed the samples. LJ conceived and designed the experiments. SY and YZ wrote and revised the manuscript. All authors have read and approved the final manuscript.Conflicts of interest
The authors declare that they have no conflicts of interest.Acknowledgements
This study was supported by the Beijing Municipal Education Commission Classification Development Project 2022 (10066092022), the Categorized Development Project 2022 (Start-up Foundation for Talent Introduction, Beijing University of Agriculture; 5066516004/003) and the China Postdoctoral Science Foundation (2022M710181).References
- I. A. Alhidary and M. M. Abdelrahman, Effects of naringin supplementation on productive performance, antioxidant status and immune response in heat-stressed lambs, Small Ruminant Res., 2016, 138, 31–36 CrossRef.
- P. E. Simitzis, M. A. Charismiadou, M. Goliomytis, A. Charalambous, I. Ntetska, E. Giamouri and S. G. Deligeorgis, Antioxidant status, meat oxidative stability and quality characteristics of lambs fed with hesperidin, naringin or α-tocopheryl acetate supplemented diets, J. Sci. Food Agric., 2018, 99, 343–349 CrossRef PubMed.
- P. Montserrat, C. Javier, A. Anna and D. Maria, Citrus aurantium flavonoid extract improves concentrate efficiency, animal behavior, and reduces rumen inflammation of Holstein bulls fed high-concentrate diets, Anim. Feed Sci. Technol., 2019, 258, 114304 CrossRef.
- E. Oskoueian, N. Abdullah and A. Oskoueian, Effects of flavonoids on rumen fermentation activity, methane production, and microbial population, Biomed. Res. Int., 2013, 2013, 349129 Search PubMed.
- A. M. Mahmoud, B. R. J. Hernández, M. A. Sandhu and O. E. Hussein, Beneficial Effects of Citrus Flavonoids on Cardiovascular and Metabolic Health, Oxid. Med. Cell. Longevity, 2019, 2019, 5484138 Search PubMed.
- L. Yuzhi, Z. Chengying, L. Chang, Z. Shuaishua, T. Guifang, B. Yuming, F. Marie-Laure, X. Hang and Z. Jinkai, Simultaneous determination of 14 bioactive citrus flavonoids using thin-layer chromatography combined with surface enhanced Raman spectroscopy, Food Chem., 2020, 338, 128115 Search PubMed.
- C. Lenehan, A. P. Moloney, E. G. O'Riordan, A. Kelly and M. McGee, Comparison of rolled barley with citrus pulp as a supplement for growing cattle offered grass silage, Adv. Anim. Biosci., 2017, 8, s33–s37 CrossRef.
- M. Sharif, M. S. Ashraf, N. Mushtaq, H. Nawaz, M. I. Mustafa, F. Ahmad, M. Younas and A. Javaid, Influence of varying levels of dried citrus pulp on nutrient intake, growth performance and economic efficiency in lambs, J. Appl. Anim. Res., 2018, 46, 264–268 CrossRef CAS.
- Y. Ying, M. Niu, A. R. Clarke and K. J. Harvatine, Short communication: Effect of a citrus extract in lactating dairy cows, J. Dairy Sci., 2017, 100, 5468–5471 CrossRef CAS PubMed.
- I. L. F. Nielsen, W. S. S. Chee, L. Poulsen, E. Offord-Cavin, S. E. Rasmussen, H. Frederiksen, M. Enslen, D. Barron, M. Horcajada and G. Williamsone, Bioavailability is improved by enzymatic modification of the citrus flavonoid hesperidin in humans: a randomized, double-blind, crossover trial, J. Nutr., 2006, 136, 404–408 CrossRef CAS PubMed.
- G. Cécile, E. Rock, C. Morand, D. Bauchart and D. Durand, Bioavailability and antioxidant capacity of plant extracts rich in polyphenols, given as a single acute dose, in sheep made highly susceptible to lipoperoxidation, Br. J. Nutr., 2007, 98, 691–701 Search PubMed.
- L. Wang, G. Huang, R. Hou, D. Qi, Q. Wu, Y. Nie, Z. Zuo, R. Ma, W. Zhou, Y. Ma, Y. Hu, Z. Yang, Y. Li and F. Wei, Multi-omics reveals the positive leverage of plant secondary metabolites on the gut microbiota in a non-model mammal, Microbiome, 2021, 9, 192 CrossRef CAS PubMed.
- Y. Stevens, E. V. Rymenant, C. Grootaert, J. V. Camp, S. Possemiers, A. Masclee and D. Jonkers, The intestinal fate of citrus flavanones and their effects on gastrointestinal health, Nutrients, 2019, 11, 1464 CrossRef CAS PubMed.
- J. F. Lu, M. Q. Zhu, H. Zhang, H. Liu, B. Xia, Y. L. Wang, X. Shi, L. Peng and J. W. Wu, Neohesperidin attenuates obesity by altering the composition of the gut microbiota in high-fat diet-fed mice, FASEB J., 2020, 34, 12053–12071 CrossRef CAS PubMed.
- T. Unno, T. Hisada and S. Takahashi, Hesperetin Modifies the Composition of fecal microbiota and increases cecal levels of short-chain fatty acids in rats, J. Agric. Food Chem., 2015, 63, 7952–7957 CrossRef CAS.
- A. R. Seradj, L. Abecia, J. Crespo, D. Villalba, M. Fondevila and J. Balcells, The effect of Bioflavex® and its pure flavonoid components on in vitro fermentation parameters and methane production in rumen fluid from steers given high concentrate diets, Anim. Feed Sci. Technol., 2014, 197, 85–91 CrossRef.
- NRC, Nutrient Requirements of Dairy Cattle, National Academies Press, Washington, D.C., 2001 Search PubMed.
- J. Jiang, L. Yan, Z. Shi, L. Wang, L. Shan and T. Efferth, Hepatoprotective and anti-inflammatory effects of total flavonoids of Qu Zhi Ke (peel of Citrus changshan-huyou) on non-alcoholic fatty liver disease in rats via modulation of NF-kappaB and MAPKs, Phytomedicine, 2019, 64, 153082 CrossRef PubMed.
- Z. Shi, T. Li, Y. Liu, T. Cai, W. Yao, J. Jiang, Y. He and L. Shan, Hepatoprotective and anti-oxidative effects of total flavonoids from Qu Zhi Qiao (fruit of Citrus Paradisi cv.Changshanhuyou) on nonalcoholic steatohepatitis in vivo and in vitro through Nrf2-ARE signaling pathway, Front. Pharmacol., 2020, 11, 4832 Search PubMed.
- AOAC International, Official Methods of Analysis, Association of Official Analytical Chemists, Arlington, VA, 18th edn, 2006 Search PubMed.
- P. J. Van Soest, J. B. Robertson and B. A. Lewis, Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition, J. Dairy Sci., 1991, 74, 3583–3597 CrossRef PubMed.
- D. Sklan, R. Ashkenazi, A. Braun, A. Devorin and K. Tabori, Fatty acids, calcium soaps of fatty acids, and cottonseeds fed to high yielding cows, J. Dairy Sci., 1992, 75, 2463–2472 CrossRef.
- L. O. Sjaunja, I. Bævre, I. Junkkarinen, J. Pedersen and J. Setälä, A nordic proposal for an energy corrected milk (ECM) formula. Pages 156–157 in 27th Session of the International Commission for Breeding and Productivity of Milk Animals, Wageningen Academic Publishers, 1990 Search PubMed.
- G. A. Broderick and J. H. Kang, Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media, J. Dairy Sci., 1980, 63, 64–75 CrossRef CAS.
- H. P. Makkar, O. P. Sharma, R. K. Dawra and S. S. Negi, Simple determination of microbial protein in rumen liquor, J. Dairy Sci., 1982, 65, 2170–2173 CrossRef CAS.
- Z. Yu and M. Morrison, Improved extraction of PCR-quality community DNA from digesta and fecal samples, BioTechniques, 2004, 36, 808–812 CrossRef CAS.
- M. K. Abdelmegeid, A. A. Elolimy, Z. Zhou, V. Lopreiato, J. C. McCann and J. J. Loor, Rumen-protected methionine during the peripartal period in dairy cows and its effects on abundance ofmajor species of ruminal bacteria, J. Anim. Sci. Biotechnol., 2018, 9, 17 CrossRef PubMed.
- S. Chen, Y. Zhou and Y. Chen, J. Gu, fastp: an ultra-fast all-in-one FASTQ preprocessor, Bioinformatics, 2018, 34, i884–i890 CrossRef.
- H. Li and R. Durbin, Fast and accurate short read alignment with Burrows-Wheeler transform., Bioinformatics, 2009, 25, 1754–1760 CrossRef.
- D. Li, C. M. Liu, R. Luo, K. Sadakane and T. W. Lam, MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph, Bioinformatics, 2015, 31, 1674–1676 CrossRef.
- H. Noguchi, J. Park and T. Takagi, MetaGene: prokaryotic gene finding from environmental genome shotgun sequences., Nucleic Acids Res., 2006, 34, 5623–5630 CrossRef PubMed.
- L. Fu, B. Niu, Z. Zhu, S. Wu and W. Li, CD-HIT: accelerated for clustering the next-generation sequencing data., Bioinformatics, 2012, 28, 3150–3152 CrossRef.
- R. Li, Y. Li, K. Kristiansen and J. Wang, SOAP: short oligonucleotide alignment program., Bioinformatics, 2008, 24, 713–714 CrossRef PubMed.
- B. Buchfink, C. Xie and D. H. Huson, Fast and sensitive protein alignment using DIAMOND, Nat. Methods, 2015, 12, 59–60 CrossRef CAS.
- C. Liu, H. Wu, S. Liu, S. Chai, Q. Meng and Z. Zhou, Dynamic alterations in Yak rumen bacteria community and metabolome characteristics in response to feed type, Front. Microbiol., 2019, 10, 1116 CrossRef.
- Z. Pang, J. Chong, G. Zhou, D. Anderson, L. Chang, M. Barrette, C. Gauthier, J. Pierre-Étienne, L. Shuzhao and J. Xia, MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights., Nucleic Acids Res., 2021, 49, W388–W396 CrossRef CAS.
- Z. Wei, S. Yuan, L. Xin, Z. Hua, N. Hui, F. Yun, B. Xiong, J. Tong and L. Jiang, Microbiome and metabolic changes of milk in response to dietary supplementation with bamboo leaf extract in dairy cows, Front. Nutr., 2021, 8, 723446 CrossRef.
- A. Burmańczuk, B. Wojciechowska, M. GbylikSikorska, A. Gajda, W. Markiewicz, E. Sosin and T. Grabowski, Baicalin decreases somatic cell count in mastitis of dairy cows, Ann. Anim. Sci., 2021, 21, 485–496 CrossRef.
- B. Artur, H. Piotr, M. Andrzej, P. Tomasz, K. Cezary, W. Beata and G. Tomasz, Quercetin decrease somatic cells count in mastitis of dairy cows, Res. Vet. Sci., 2018, 117, 255–259 CrossRef.
- A. Burmanczuk, B. Wojciechowska, M. Gbylik-Sikorska, A. Gajda, W. Markiewicz, E. Sosin and G. Tomasz, The effect of hesperidin, chrysin, and naringenin on the number of somatic cell count in mastitis in dairy cows after multiple intramammary administration, Ann. Anim. Sci., 2022, 22, 155–172 CrossRef CAS.
- N. Argov-Argaman, M. Cohen-Zinder, H. Leibovich, M. Yishay, H. Eitam, R. Agmon, O. Hadaya, R. Mesilati-Stahy, J. Miron and A. Shabtay, Dietary pomegranate peel improves milk quality of lactating ewes: emphasis on milk fat globule membrane properties and antioxidative traits, Food Chem., 2020, 313, 125822 CrossRef.
- L. L. M. Berger, R. Blank, F. Zorn, S. Wein, C. C. Metges and S. Wolffram, Ruminal degradation of quercetin and its influence on fermentation in ruminants, J. Dairy Sci., 2015, 98, 5688–5698 CrossRef PubMed.
- T. Ma, D. Chen, Y. Tu, N. Zhang, B. Si and Q. Diao, Dietary supplementation with mulberry leaf flavonoids inhibits methanogenesis in sheep, Anim. Sci. J., 2017, 88, 72–78 CrossRef PubMed.
- M. Xue, H. Sun, X. Wu, J. Liu and L. L. Guan, Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance, Microbiome, 2020, 8, 64 CrossRef PubMed.
- B. Wang, M. P. Ma, Q. Y. Diao and Y. Tu, Saponin-induced shifts in the rumen microbiome and metabolome of young cattle, Front. Microbiol., 2019, 10, 356 CrossRef PubMed.
- A. A. Anum, Y. Chao, Z. Jianbo, K. Qudratullah, L. Zeyi, L. Chen, D. Mei, Y. Ping, L. Ruijun, H. Jianli and D. Xuezhi, Effects of dietary energy levels on rumen fermentation, microbial diversity, and feed efficiency of Yaks (Bos grunniens), Front. Microbiol., 2020, 11, 625 CrossRef.
- K. J. Cheng, G. A. Jones, F. J. Simpson and M. P. Bryant, Isolation and identification of rumen bacteria capable of anaerobic rutin degradation, Can. J. Microbiol., 1969, 15, 1365–1371 CrossRef CAS PubMed.
- H. G. Krishnamurty, K. J. Cheng, G. A. Jones, F. J. Simpson and J. E. Watkin, Identification of products produced by the anaerobic degradation of rutin and related flavonoids by Butyrivibrio sp. C3, Can. J. Microbiol., 1970, 16, 759–767 CrossRef CAS.
- L. R. Krumholz and M. P. Bryant, Eubacterium oxidoreducens sp. nov. requiring H2 or formate to degrade gallate, pyrogallol, phloroglucinol and quercetin, Arch. Microbiol., 1986, 144, 8–14 CrossRef.
- A. Braune and M. Blaut, Bacterial species involved in the conversion of dietary flavonoids in the human gut, Gut Microbes, 2016, 7, 216–234 CrossRef.
- C. J. Diaz, C. Cabral, L. M. Redondo, V. N. Pin, D. Colombatto, M. D. Farber and M. M. Fernandez, Impact of chestnut and quebracho tannins on rumen microbiota of bovines, Biomed. Res. Int., 2017, 2017, 9610810 Search PubMed.
- E. Jami, B. A. White and I. Mizrahi, Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency, PLoS One, 2014, 9, e85423 CrossRef PubMed.
- P. R. Myer, T. P. Smith, J. E. Wells, L. A. Kuehn and H. C. Freetly, Rumen microbiome from steers differing in feed efficiency, PLoS One, 2015, 10, e129174 CrossRef PubMed.
- S. Koike and Y. Kobayashi, Fibrolytic rumen bacteria: their ecology and functions, Asian-Australas. J. Anim. Sci., 2009, 22, 131–138 CrossRef.
- M. P. Bryant and I. M. Robinson, Studies on the Nitrogen requirements of some ruminal cellulolytic bacteria., Appl. Microbiol., 1961, 9, 96–103 CrossRef PubMed.
- R. Bodasa, N. Prietoa, R. García-Gonzálezb, S. Andrésa, F. J. Giráldeza and S. López, Manipulation of rumen fermentation and methane production with plant secondary metabolites, Anim. Feed Sci. Technol., 2012, 176, 78–93 CrossRef.
- R. J. Eberle, D. S. Olivier, M. S. Amaral, D. Willbold, R. K. Arni and M. A. Coronado, Promising natural compounds against flavivirus proteases: citrus flavonoids hesperetin and hesperidin, Plants, 2021, 10, 2183 CrossRef PubMed.
- C. L. Anderson, M. B. Sullivan and S. C. Fernando, Dietary energy drives the dynamic response of bovine rumen viral communities, Microbiome, 2017, 5, 155 CrossRef PubMed.
- R. M. Macnab and D. E. Koshland, The gradient-sensing mechanism in bacterial chemotaxis., Proc. Natl. Acad. Sci. U. S. A., 1972, 69, 2509–2512 CrossRef PubMed.
- G. Y. Wu, F. B. Bazer, R. C. Burghardt, G. A. Johnson, S. W. Kim, D. A. Knabe, P. Li, X. L. Li, J. R. McKnight, M. C. Satterfield and T. E. Spencer, Proline and hydroxyproline metabolism: implications for animal and human nutrition, Amino Acids, 2011, 40, 1053–1063 CrossRef.
- R. C. Rae and J. R. Ingalls, Lactational response of dairy cows to oral administration of L-tyrosine, J. Dairy Sci., 1984, 67, 1430–1438 CrossRef PubMed.
- M. S. Allen and P. Piantoni, Carbohydrate nutrition: managing energy intake and partitioning through lactation, Vet. Clin. North Am.: Large Anim. Pract., 2014, 30, 577–597 Search PubMed.
- H. J. Flint, K. P. Scott, S. H. Duncan, P. Louis and E. Forano, Microbial degradation of complex carbohydrates in the gut, Gut Microbes, 2012, 3, 289–306 CrossRef.
- W. Zhang, W. Liu, R. Hou, L. Zhang, S. Schmitz-Esser, H. Sun, J. Xie, Y. Zhang, C. Wang, L. Li, B. Yue, H. Huang, H. Wang, F. Shen and Z. Zhang, Age-associated microbiome shows the giant panda lives on hemicelluloses, not on cellulose, ISME J., 2018, 12, 1319–1328 CrossRef PubMed.
- V. L. Jose, R. P. More, T. Appoothy and A. S. Arun, In depth analysis of rumen microbial and carbohydrate-active enzymes profile in Indian crossbred cattle, Syst. Appl. Microbiol., 2017, 40, 160–170 CrossRef PubMed.
- Y. Pan, X. Zheng and Y. Xiang, Structure-function elucidation of a microbial consortium in degrading rice straw and producing acetic and butyric acids via metagenome combining 16S rDNA sequencing, Bioresour. Technol., 2021, 340, 125709 CrossRef PubMed.
- M. Armendáriz-Ruiz, J. A. Rodríguez-González, R. M. Camacho-Ruíz and J. C. Mateos-Díaz, Carbohydrate Esterases: An Overview., Methods Mol. Biol., 2018, 183, 39–68 CrossRef.
- M. S. Bilal, J. A. Abaker, Z. U. Aabdin, T. Xu, H. Dai, K. Zhang, X. Liu and X. Shen, Lipopolysaccharide derived from the digestive tract triggers an inflammatory response in the uterus of mid-lactating dairy cows during SARA, BMC Vet. Res., 2016, 12, 284 CrossRef PubMed.
- Q. Zebeli and B. N. Ametaj, Relationships between rumen lipopolysaccharide and mediators of inflammatory response with milk fat production and efficiency in dairy cows, J. Dairy Sci., 2009, 92, 3800–3809 CrossRef PubMed.
- F. F. Zhao, T. Y. Wu, H. R. Wang, L. Y. Ding, G. Ahmed, H. W. Li, W. Tian and Y. Z. Shen, Jugular arginine infusion relieves lipopolysaccharide-triggered inflammatory stress and improves immunity status of lactating dairy cows, J. Dairy Sci., 2018, 101, 5961–5970 CrossRef PubMed.
- R. De Nardi, G. Marchesini, J. C. Plaizier, S. Li, E. Khafipour, R. Ricci, I. Andrighetto and S. Segato, Use of dicarboxylic acids and polyphenols to attenuate reticular pH drop and acute phase response in dairy heifers fed a high grain diet, BMC Vet. Res., 2014, 10, 277 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2fo02751h |
| This journal is © The Royal Society of Chemistry 2023 |