Open Access Article
Open Access ArticleCopper iodide microhexagons: a potential therapeutic agent for surface microbial infection and melanoma†
Sunil Venkanna
Pogu‡
,
Dokkari Nagalaxmi
Yadav‡
,
Sri Amruthaa
Sankaranarayanan
,
Rupali
Srivastava
,
Shashidhar
Thatikonda
 and
Aravind Kumar
Rengan
and
Aravind Kumar
Rengan
 *
*
Dept of Biomedical Engineering, Indian Institute of Technology Hyderabad, India. E-mail: aravind@bme.iith.ac.in
First published on 26th June 2023
Abstract
Microbial infections are a constant threat to humans and responsible for increased death rate in most of the developing countries. Conventional use of chlorine- or alcohol-based disinfectants results in the development of drug-resistant microbial strains, which are limited in their response to most of the treatment strategies. In this perspective, several engineered materials have emerged as an affordable alternative to the existing treatment strategies. For the same, here we have synthesised CuI microparticles (CuI (WH) and CuI (H)) using chemical precipitation followed by a hydrothermal process. We further comparatively evaluated the antiproliferative activity of CuI (WH) and CuI (H) against microbial cultures (non-resistant and resistant), virus, and cancer cell lines. CuI (H) displayed better size, stability, and significantly higher inhibitory activity against microorganisms and cancer cell lines than CuI (WH). Upon preliminary evaluation, which confirmed that CuI (H) is more effective than CuI (WH), we evaluated its stability and surface sterilising property, and it was found that CuI (H) were stable for the recorded period of 30 days and displayed significant surface sterilising ability. The results suggest that CuI (H) is a potent antiproliferative material against microbial cultures, viral strains, and cancer cell lines while preserving its biocompatibility towards normal cell lines, representing a potential surface sterilising material in public places upon significant in vivo efficacy evaluation.
1. Introduction
Microbial infections are potential health hazards responsible for the major portion of the global mortality rate. These infections could arise due to an unhealthy lifestyle or may be due to accidental exposure to pathogens during post-surgical wound healing.1,2 The use of conventional antibiotics to treat microbial infections is limited in clinical outcomes due to the development of multi-drug resistance in microbial strains.3–5 Also, antibiotic use leads to various detrimental side effects, such as non-targeted accumulation, tissue toxicity, and renal damage.6 It is also reported that the use of chlorine- and alcohol-based surface disinfectants in public places such as hospitals and public transportation leads to the development of resistant strains increasing the chances of exposure to resistant microbes and their associated toxicities.7 In 2019, around 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people were killed by antibiotic-resistant E. coli (Escherichia coli) alone.8 It is also found that more than 80% of E. coli strains isolated from wounds are antibiotic resistant, making them even more difficult to treat.9 During the last three years, viral infections (Covid-19) have devastated the human lifestyle globally and have been a reason for the deaths of millions of people.10
000 people were killed by antibiotic-resistant E. coli (Escherichia coli) alone.8 It is also found that more than 80% of E. coli strains isolated from wounds are antibiotic resistant, making them even more difficult to treat.9 During the last three years, viral infections (Covid-19) have devastated the human lifestyle globally and have been a reason for the deaths of millions of people.10
Along with microbial infections, cancer is also a major disease of concern, responsible for millions of deaths annually.11 Several studies were reported on the interdependence of microbial infections with the emergence of cancer or vice versa. Melanoma, in particular, is a predominant type of malignant skin cancer, which arises from premalignant actinic keratoses under chronic inflammation. Wounded and burned areas on the skin are more prone to microbial infections, which further increases the chance of skin cancer development. Microbial infections on skin promote the secretion of inflammatory factors (IL-6, IL-8, and TNF-alpha) and generate ROS (reactive oxygen species), which in turn promote transformation of premalignant actinic keratoses into malignant squamous cell carcinoma.12,13 Hence, developing a highly efficient biocompatible material which exhibits potential antimicrobial and anti-cancer activity, and at the same time it is less prone to resistance induction in microbes and cancer cells, is warranted to treat pathogenic infections and associated complications.
Many metallic nano-/microparticles have been reported for various theranostic applications,14,15 and are known to be less susceptible to the development of microbial resistance.16 For a long time, copper vessels have been used to store and serve food and water due to copper metal's intrinsic antiproliferative property, preventing the infectivity of various diseases. Several studies have reported on copper nano-/microparticles having an inhibitory effect against microbial and cancer cell growth.17–19 Similarly, iodine-based nano-/microparticles have been shown to be effective in inhibiting microbial and tumour growth.20,21 Iodine rapidly diffuses into cells through sodium iodide symporters and oxidises proteins, nucleotides, and fatty acids, eventually causing cell death.22,23 Recently, iodine nanoparticles were found to be effective in enhancing proapoptotic genes, and these iodine-based materials are also applicable in tumour tissue diagnosis using the computed tomography technique.24 So, here, we hypothesise that forming composite particles consisting of copper and iodine will provide a synergistic effect against microbial and cancer cell growth, as iodine will enhance the antibacterial effect of copper. At the same time, copper will enhance the anticancer effect of iodine. Polymer coating on the metallic nano-/microparticles imparts excellent particle stability and dispersibility. Various polymers such as polyethylene glycol, polyvinylpyrrolidone, and poly(acrylic acid) (PAA) were reported as a coating materials onto metallic particles, which resulted in increased dispersibility, stability, and targeted tumour accumulation.25–28
In the present study, we have synthesised PAA polymer-coated CuI (copper iodide) microparticles using a chemical precipitation method (CuI (WH)) and further subjected them to hydrothermal reaction to obtain CuI (H) microhexagons. The obtained CuI (WH) and CuI (H) were characterised and evaluated for their antiproliferative property against microbial cultures (Escherichia coli (E. coli), Staphylococcus aureus (S. aureus), and Candida albicans (C. albicans)), virus (bacteriophage lambda), and MDR (multidrug-resistant) E. coli. CuI (WH) and CuI (H) were then tested for their biocompatibility in a non-cancerous cell line (L929) and their cytotoxic activity against cancer cell lines (4T1, HCT 116, A549, B16) (Scheme 1). Finally, the long-term stability of CuI (H) in aqueous suspension and its application in surface sterilisation were investigated.
2. Materials and methods
2.1 Materials
Copper dichloride (CuCl2), propidium iodide (PI), fluorescein diacetate (FDA), and PAA were purchased from Sigma-Aldrich. Dulbecco's modified Eagle's medium (DMEM), Roswell Park Memorial Institute medium (RPMI 1640), fetal bovine serum (FBS), pen-strep solution, phosphate-buffered saline (PBS), trypsin EDTA solution, LB (Luria Bertani) agar/broth, MH (Mueller and Hinton) agar/broth, and SD (Sabouraud Dextrose) were purchased from Himedia Laboratories, Mumbai (India). MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide), potassium iodide (KI), sodium sulphite (Na2SO3), and dimethyl sulfoxide (DMSO) were purchased from SRL Chemicals Ltd (Mumbai, India). All other reagents used were of analytical grade. Ultra-pure de-ionised water (Milli-Q) was used for conducting all the experiments.2.2 Synthesis and characterisation of CuI (WH) and CuI (H)
CuI particles were synthesised by modifying the procedure reported by Zhou et al.29 Briefly, for the synthesis of CuI (WH) particles, 25 mg of CuCl2 and 400 mg of PAA were dissolved in 20 mL of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (water
1 (water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethanol) solution and stirred for 10 min at room temperature. After that, an aqueous solution of Na2SO3 and KI was added dropwise under stirring, turning the cyan-blue solution into a pale brown and then to an off-white precipitate indicating the formation of CuI (WH). Similarly, for the synthesis of CuI (H), the obtained CuI (WH) precipitate solution was transferred into a Teflon-lined container and heated under pressure at 180 °C for 3.5 hours (hydrothermal process). After that, it was cooled to room temperature. CuI (WH) and CuI (H) were obtained by washing them with ethanol and water. CuI without PAA (CuI (WP)) was also synthesised like CuI (H) without the addition of PAA during the initial procedure.
ethanol) solution and stirred for 10 min at room temperature. After that, an aqueous solution of Na2SO3 and KI was added dropwise under stirring, turning the cyan-blue solution into a pale brown and then to an off-white precipitate indicating the formation of CuI (WH). Similarly, for the synthesis of CuI (H), the obtained CuI (WH) precipitate solution was transferred into a Teflon-lined container and heated under pressure at 180 °C for 3.5 hours (hydrothermal process). After that, it was cooled to room temperature. CuI (WH) and CuI (H) were obtained by washing them with ethanol and water. CuI without PAA (CuI (WP)) was also synthesised like CuI (H) without the addition of PAA during the initial procedure.
The fluorescence properties of CuI (WH) and CuI (H) were determined using a UV lamp (365 nm, Analytik Jena, CA, USA) and fluorescence spectrophotometer (RF 6000, Shimadzu, Japan). Absorption spectra were obtained using a UV-visible spectrophotometer (UV 1800, Shimadzu, Japan). The hydrodynamic diameters of the CuI (WH) and CuI (H) particles were determined using a DLS instrument (Particle Sizing Systems, Inc. Santa Barbara, California, USA) and the surface charge on the particles was determined using a Zeta sizer (Particle Sizing Systems, Inc. Santa Barbara, California, USA). The morphology and size of CuI (WH) and CuI (H) microparticles were visualised using a field emission scanning electron microscope (FE-SEM, Carl Zeiss atomic microscope, Supra40 Germany). The elemental composition of CuI (H) was analysed using EDS (energy dispersive X-ray spectroscopy).
2.3 Antibacterial efficacy of CuI (WH) and CuI (H)
The antibacterial activity of the CuI (WH) and CuI (H) particles was evaluated against Gram-negative bacteria (E. coli) and Gram-positive bacteria (S. aureus).2.4 Antifungal efficacy of CuI (WH) and CuI (H) particles
The antifungal activity of CuI (WH) and CuI (H) against C. albicans was evaluated.2.5 Antiviral efficacy of CuI (WH) and CuI (H) particles
2.6 Antibacterial efficacy of CuI (WH) and CuI (H) against MDR E. coli
The MDR E. coli (MCC 3671) was obtained from MTCC, India. All the experiments with MDR E. coli were performed under BSL-2 (Biosafety Level-2) facility. MDR E. coli was cultured using a similar protocol used to maintain E. coli and obtained 0.1 OD culture for experimental investigation.The effect of CuI (H) and CuI (WH) on MDR E. coli was determined using MTT assay. For which, 100 μL of MDR E. coli culture was treated with media containing CuI (WH) and CuI (H) at concentrations from 25 μg mL−1 to 100 μg mL−1 and control treated with broth without any particles. Both treated and control were incubated at 37 °C for 24 h. After incubation, the percentage of cell viability was measured using a microplate reader at 570 nm.
The effect was also qualitatively determined using AO staining and a fluorescence microscope. For which, 50 μg mL−1 concentration of CuI (WH) and CuI (H) treated MDR E. coli and control were stained using the aforementioned AO staining protocols and imaged under blue light using a fluorescence microscope.
2.7 Determining the effect of CuI (WH) and CuI (H) on normal and cancer cell lines
Similarly, the effect of CuI (H) compared to CuI (WH) was determined using the B16 cell line. B16 cells were treated with both types of particles at concentrations of 10 to 60 μg mL−1. FDA and PI staining was used for qualitative analysis.38
So far, several studies have reported the degradation of metallic nano-/microparticles to release ions upon interaction with cells or bacterial cultures. These released ions were reported to enter into the cells or bacteria via endocytosis or through ionic channels and commence the antiproliferative activity by damaging genetic material, active proteins or by generating reactive oxygen species.39,40 In the present study, the degradation of CuI (H) upon interaction with cells or bacterial culture was determined by measuring and observing the change in the intrinsic fluorescence of CuI (H). For the same reason, first we incubated CuI (H) with E. coli culture at 37 °C and the change in fluorescence of CuI (H) at 0 h, 12 h, and 24 h was quantified using a Tecan plate reader. CuI (H) in PBS was used as control.
Similarly, the change in fluorescence of CuI (H) on interaction with B16 cells was observed using fluorescence microscopy. For which, we incubated CuI (H) with B16 cells at 37 °C and observed the change in fluorescence at 0 h, 12 h, and 24 h. CuI (H) in PBS incubated at 37 °C was used as control.
2.8 Evaluating the effect of CuI (WH) and CuI (H) on the migration of B16 and L929 cells
The effect of CuI (WH) and CuI (H) on the migration of cells was evaluated using in vitro wound healing assay. Briefly, B16 and L929 cell lines were seeded in a 24-well plate at a count of 1 × 105 cells per well. After attachment and reaching sufficient confluency, a wound was created in the cells using a microtip. The cells were treated with CuI (WH) and CuI (H) at 50 μg mL−1 concentration. The effect on the migration of cells was observed using a microscope at 12 h and 24 h. The experiment was performed in triplicate, and the % migration was calculated using ImageJ software with macros plugin to quantify the migration as per the reported procedure.41,422.9 Determining CuI (H) stability and its application as a surface steriliser
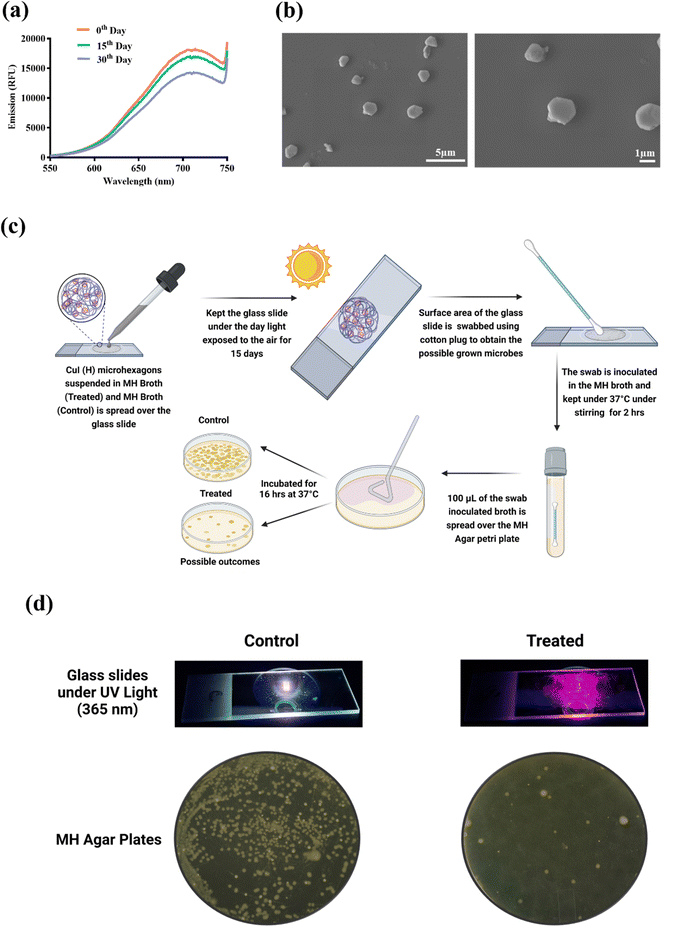
CuI (H) stability was determined by suspending CuI (H) in Milli-Q (MQ) water exposed to daylight, and its fluorescence emission was measured for the 0th, 15th, and 30th day of post-suspension. After the 30th day, suspended CuI (H) particles were drop-cast on a silicon wafer and imaged using FE-SEM. The surface sterilising property of CuI (H) was determined by spreading MH broth-suspended CuI (H) onto a glass slide and comparing it with positive control of MH broth alone spread on the glass slide. Both the glass slides were kept under daylight and exposed to air for 15 days. After 15 days, the glass slide surface was swabbed using cotton swabs, inoculated in MH broth, and incubated at 37 °C for 30 min under stirring. Then, the inoculum was spread over the MH agar Petri dish and further incubated for 16 hours at 37 °C. After the incubation, colonies formed on the agar plate were captured using camera (Schematic.2).3. Results and discussion
3.1 Synthesis and characterisation of CuI (WH) and CuI (H)
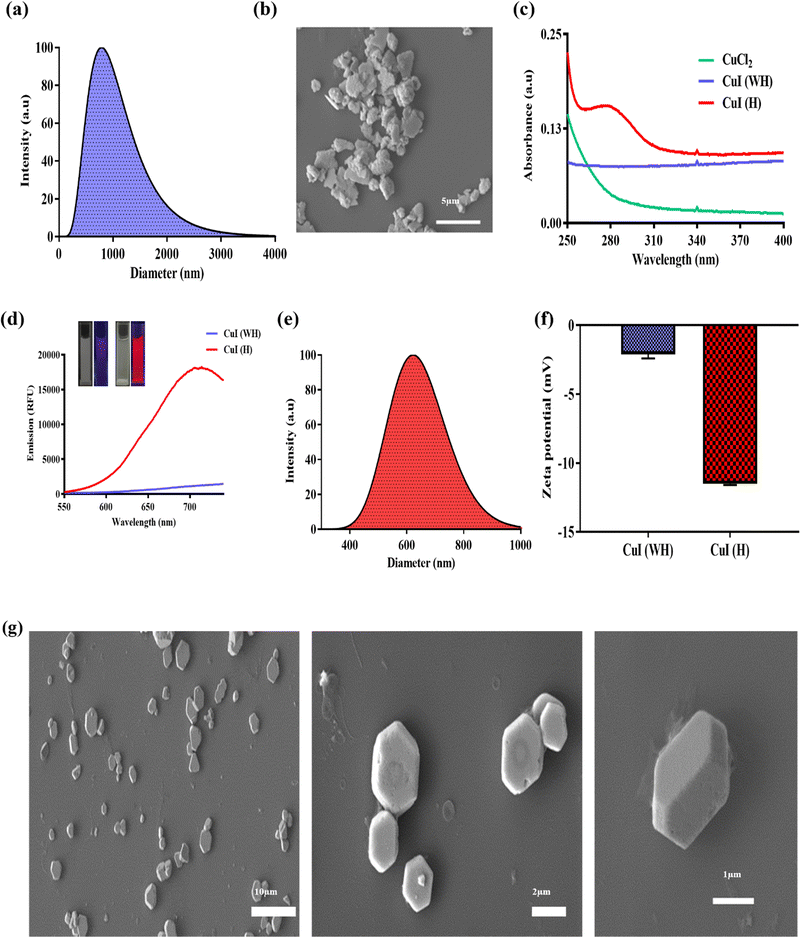
In the current study, we have synthesised PAA-coated CuI using chemical precipitation followed by hydrothermal processes. CuI (WH) synthesised without the hydrothermal method displayed large irregularly shaped particles with a size ranging from 0.8 μm to 2 μm (Fig. 1(a)). Due to the low zeta potential of −2.5 mV (Fig. 1(f)), CuI (WH) exhibited poor stability, thereby forming large clumps as seen upon FE-SEM analysis (Fig. 1(b)). Hydrothermal treatment of CuI (WH) resulted in the formation of CuI (H) microhexagons, which displayed a smaller size (0.6–0.8 μm) with a higher zeta potential forming well-dispersed microparticles (Fig. 1(e–g)). Several studies have reported the synthesis of nano-/microparticles using hydrothermal method (at higher temperature) displayed significantly better surface physio-chemical properties compared to the particles synthesized using chemical precipitation method at room temperature. In this perspective, the synthesized CuI (H) exhibited higher dispersibility and stability, which is evident from its higher zeta potential and SEM imaging compared to CuI (WH) synthesized at room temperature.43–46 EDS analysis also confirmed the presence of copper and iodine in CuI (H) (Fig. S1, ESI†). Also, CuI (H) showed peak absorption at 285 nm and emission at 715 nm when excited using 385 nm. Under UV light (365 nm), CuI (H) displayed bright red fluorescence, whereas no fluorescence was observed for CuI (WH) (Fig. 1(c) and (d)). CuI synthesised using the hydrothermal process without PAA displayed a lesser fluorescence emission intensity and poor aqueous dispersibility (Fig. S2, ESI†). Hence, we have excluded it from further experimental investigation. CuI (H) synthesised using a hydrothermal process displayed superior properties compared to CuI (WH) with respect to its size, shape, and surface charge. Hence, hydrothermal treatment and PAA coating had imparted significant characteristics to CuI, which will be helpful in various biological applications.3.2 Antibacterial efficacy of CuI (WH) and CuI (H)
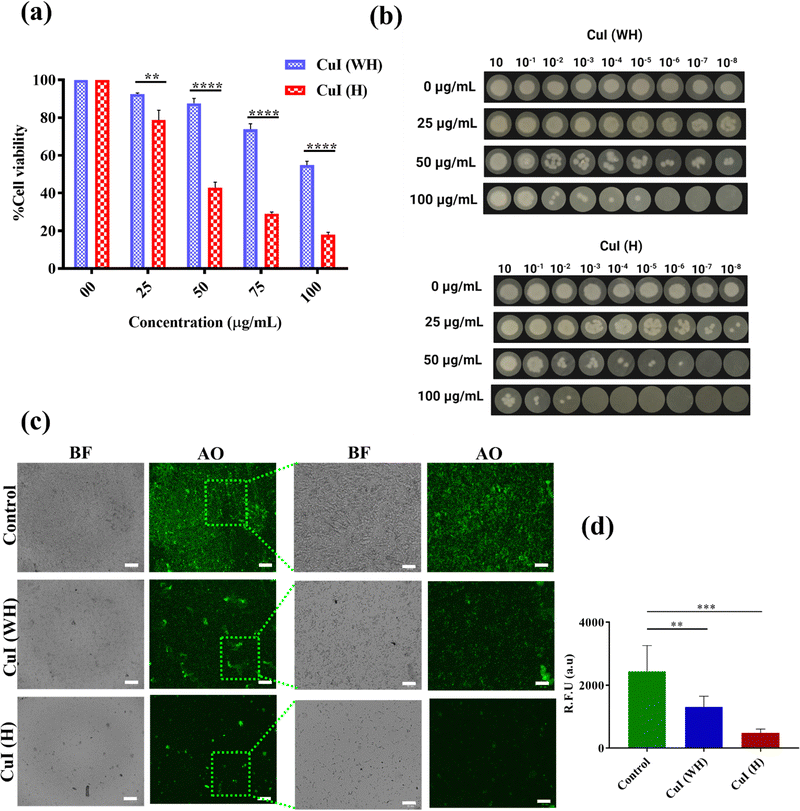
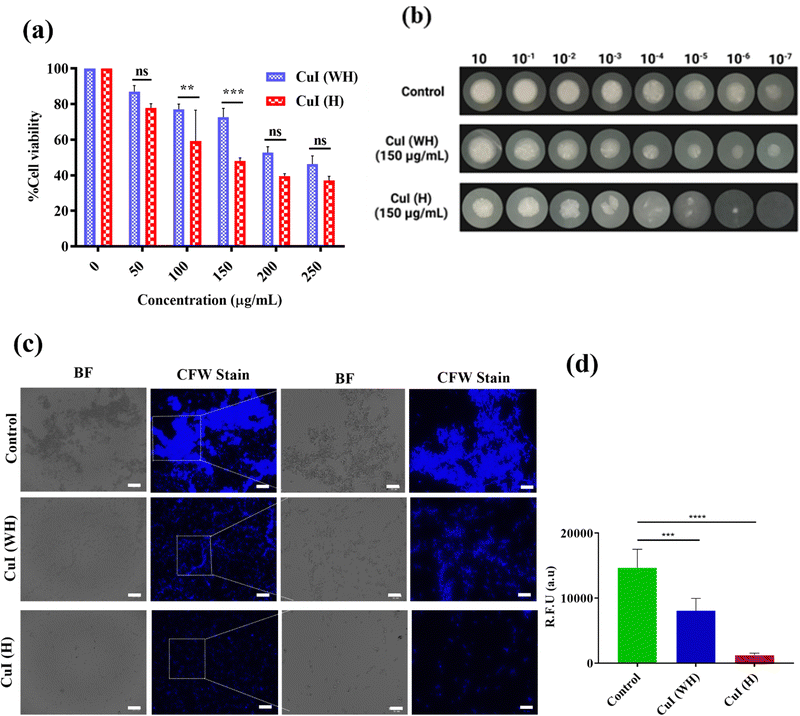
The antibacterial efficacy of CuI (H) and CuI (WH) against E. coli and S. arueus cultures was determined using MTT assay, spot formation assay, and AO staining. From the MTT assay, it was observed that the E. coli culture displayed a 50% decrease in growth at 50 μg mL−1 concentration of CuI (H), while the percentage death of E. coli at the same concentration of CuI (WH) was found to be 22% (Fig. 2(a)). The IC50 value of CuI (H) against E. coli was found to be less than that of earlier reported copper-based nanoparticles (60 μg mL−1).47Similarly, the IC50 value of CuI (H) against S. aureus was found to be 200 μg mL−1, while at the same concentration of CuI (WH), the % death of S. aureus was about 25% (Fig. 3(a)). Same results were reflected in the spot formation assay (Fig. 2(b) and 3(b)). As the concentration of CuI (H) increased, the spot size decreased in every dilution. However, in the case of CuI (WH)-treated bacteria, the decrease in spot size was insignificant compared to the control. The qualitative visualisation of the inhibitory effect of CuI (WH) and CuI (H) against both the cultures was observed using AO stain. It was found that the control and CuI (WH)-treated group displayed more highly fluorescent colonies in comparison to the CuI (H) group (Fig. 2(c) and (d) for E. coli and Fig. 3(c) and (d) for S. aureus). As AO is an intercalating agent staining DNA, the control and CuI (WH)-treated group displayed higher bacterial viability and thus DNA content, which resulted in more retention of AO and hence a higher number of fluorescent colonies under a fluorescence microscope compared to the CuI (H)-treated bacteria. Therefore, CuI (H) could be a potential antibacterial material, showing activity against both Gram-positive and Gram-negative bacterial strains.
3.3 Antifungal efficacy of CuI (H) in comparison to CuI (WH)
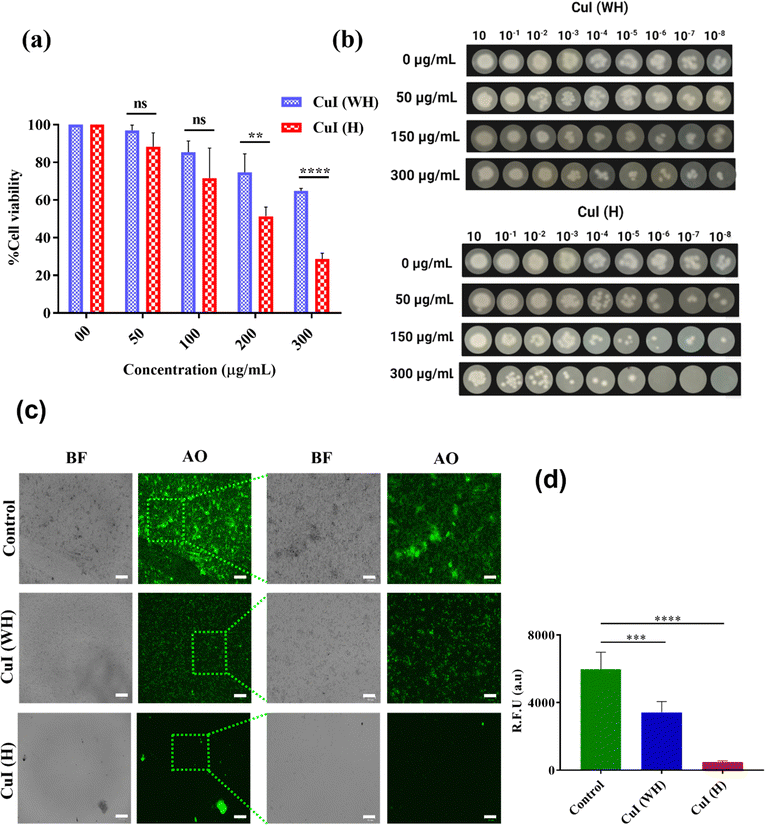
Antifungal efficacy of CuI (H) and CuI (WH) against C. albicans culture was determined using MTT assay, spot formation assay, and calcofluor white staining. After 48 h of incubation, it was observed that the C. albicans culture displayed 50% decrease in growth at 150 μg mL−1 concentration of CuI (H), while at the same concentration of CuI (WH) it was found to be 24% (Fig. 4(a)). Similar results were reflected in the spot formation assay (Fig. 4(b)). The spot size for the CuI (H)-treated sample was significantly decreased with increasing concentration at each dilution. In CuI (WH)-treated cells, the decrease in spot size was insignificant compared to the control. Similarly, the morphological effect on C. albicans after the treatment with CuI (H) and CuI (WH) was observed using a calcofluor white stain, when imaged under a fluorescence microscope. The untreated control sample displayed a very large number of colonies with well-defined hyphae stained in blue colour, whereas the CuI (WH)-treated culture showed fewer patches with improper hyphae, and the CuI (H)-treated sample showed no notable fungal patches without any hyphae (Fig. 4(c) and (d)). Therefore, it is evident that CuI (H) could qualify as a potential antifungal material.3.4 Antiviral efficacy of CuI (H) in comparison to CuI (WH)
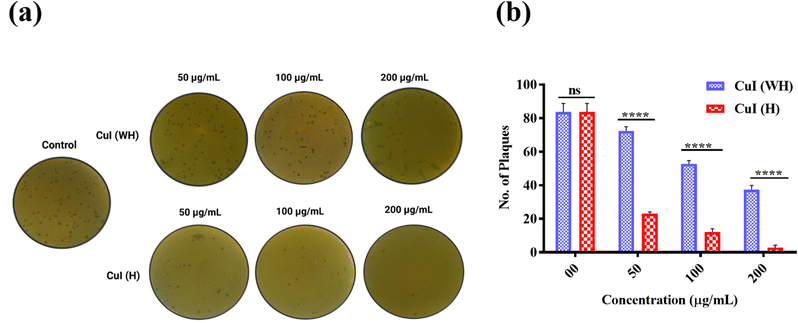
The effect of CuI (WH) and CuI (H) against bacteriophage lambda growth was evaluated using plaque formation assays. Bacteriophages were treated with same concentrations of CuI (WH) and CuI(H) microparticles and then were further allowed to infect E. coli C600.Upon 18 hours of incubation at 37 °C, a significant difference in the number of plaques formed was observed by varying the CuI (WH) and CuI (H) concentrations. At the same concentrations of CuI (WH) and CuI (H), the number of plaques formed was almost triple in the case of CuI (WH) in comparison to CuI (H) (Fig. 5(a)). Quantitative analysis was done by counting the number of plaques in triplicate. It was observed that at 50 μg mL−1 concentration of CuI (H) the number of plaques was almost 50% fewer than for the CuI (WH)-treated sample (Fig. 5(b)). CuI (H) significantly reduced the growth of bacteriophage lambda. Therefore, CuI (H) could be used as potential antiviral material.
3.5 Antibacterial efficacy of CuI (WH) and CuI (H) against MDR E. coli
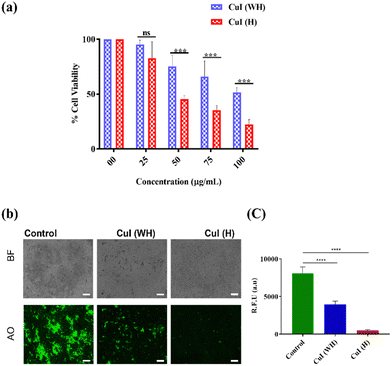
After evaluating the efficiency of CuI (H) in inhibiting non-drug-resistant bacteria, we further determined the antiproliferative efficiency of CuI (H) against MDR E. coli. CuI (H) displayed 50% inhibition of MDR E. coli at 50 μg mL−1. In comparison, at the same concentration of CuI (WH), the inhibition was around 20% (Fig. 6(a)). Similarly, we qualitatively evaluated the effect of CuI (H) using AO staining. It was found that CuI (H)-treated bacteria displayed significantly less fluorescent colonies compared to control and CuI (WH)-treated bacteria (Fig. 6(b) and (c)). CuI (H) is found to be highly effective against MDR E. coli growth. Therefore, CuI (H) could be beneficial in treating MDR strain infections.3.6 Anticancer efficacy of CuI (WH) and CuI (H)
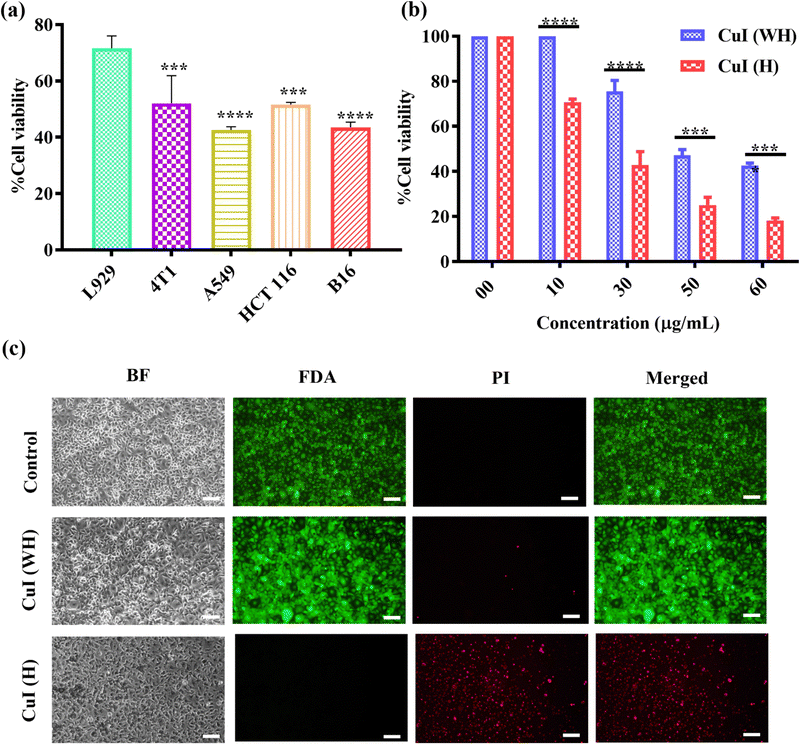
The biocompatibility and cytotoxicity of CuI (WH) and CuI (H) against non-cancerous and cancer cell lines were determined using MTT assay and qualitative analysis was done using FDI/PI staining. The IC50 value for CuI (H) against cancer cells (HCT116, 4T1, A549, and B16) was found to be 40 μg mL−1 (Fig. S3(b–d), ESI†). At the same concentration of CuI (H) used to treat L929 cells, the % of live cells was found to be more than 75% (Fig. S3(a), ESI†). This indicates that CuI (H) displays cytotoxicity against cancer cells, and, at the same, it is compatible with non-cancerous cells (Fig. 7(a)).As the size of CuI (H) is micrometric, using it within the body will have certain drawbacks, such as low tumour accumulation or it may block small blood vessels, leading to detrimental side effects. CuI (H) could be used to treat surface infections and superficial tumors, such as melanoma, to avoid such side effects. With this perspective, we have evaluated the cytotoxic efficacy of CuI (H) in comparison to CuI (WH) at different concentrations (10–60 μg mL−1). CuI (H) was found to be more effective in inhibiting the growth of B16 cells compared to CuI (WH), and IC50 values were 40 μg mL−1 and 60 μg mL−1, respectively (Fig. 7(b)).
The qualitative visualisation of the anticancer efficacy of CuI (WH) and CuI (H) against B16 cells was observed by live/dead staining using FDI/PI stains. It was found that the control and CuI (WH)-treated group displayed a higher green fluorescence (FDI) in comparison to the CuI (H) group. At the same time, more red fluorescence (PI) was observed in CuI (H)-treated cells in comparison to control and CuI (WH)-treated cells validating the quantitative results obtained using MTT assay (Fig. 7(c)). From the obtained results, it is evident that CuI (H) is more potent in inhibiting the growth of cancer cells and biocompatible towards non-cancerous cells. Therefore, CuI (H) could represent a potent anticancer material.
Metallic nanoparticles, primarily composed of gold (Au), silver (Ag), or copper (Cu), exhibit potent antimicrobial properties. The mechanism behind the antibacterial efficacy of suspended metallic particles is not well understood. According to recent reports, the antibacterial activity is associated with the release of ions from the particles upon interaction with cells or the microbes. These released ions enter plasma membranes through endocytosis or via ion channels. Upon entry, metallic ions cause a series of effects by damaging proteins and genetic materials and causing generation of reactive oxygen species leading to cell death.39,40 Previous studies have also reported CuI nanoparticles which exhibit greater efficacy against Gram-negative bacteria in comparison to Gram-positive bacteria, because of more negative charge on the surface leading to strong attraction towards the released positive ions (Cu2+).48,49
In accordance with the reported studies, we have found similar results. From the determination of change in fluorescence of CuI (H) upon incubation with B16 and E. coli culture, there was a significant loss of fluorescence intensity after 24 h, whereas in PBS no significant change in CuI (H) fluorescence was observed (Fig. S4(ac), ESI†). The obtained results validate those of previously reported studies.
3.7 Evaluating the effect of CuI (WH) and CuI (H) on the migration of B16 and L929 cells
Cell migration is a major contributing factor for transformation of benign tumours into malignant ones. Inhibition of cellular migration and subsequent metastatic growth in malignant tumours is more challenging. We have evaluated the effect of CuI (H) (50 μg mL−1) on the migration of B16 and L929 cells in comparison to CuI (WH) (50 μg mL−1) and control at 12 h and 24 h. At 12 h, it was observed that CuI (WH)-treated and untreated control cells displayed migration of 35% and 83%, respectively. In comparison, CuI (H)-treated cells displayed migration of only 19%. After 24 h, the migration in CuI (WH)-treated and untreated control cells was about 88% and 100%, respectively, while in CuI (H)-treated cells it was around 29% (Fig. 8(a) and (b)). Therefore, CuI (H) microhexagons are more potent in inhibiting cancer cell migration, simultaneously not affecting the migration of non-cancerous cells (L929) (Fig. S5, ESI†), which could be useful in preventing development of malignant squamous cell carcinoma.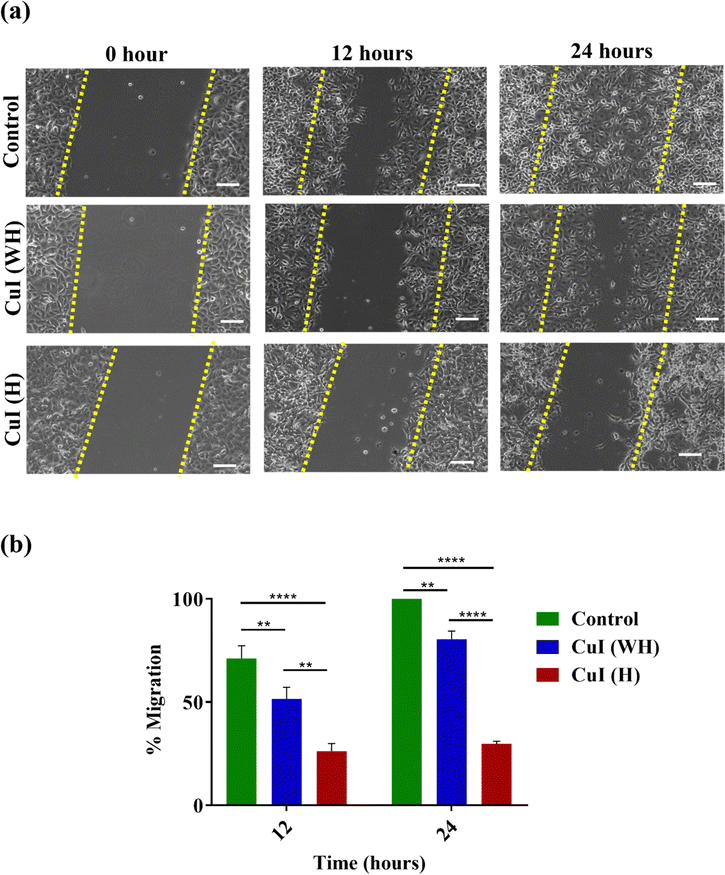 | ||
| Fig. 8 The effect of CuI (WH) and CuI (H) on cell migration of B16 cells. (a) Qualitative. (b) Quantitative. p < 0.05, ****p < 0.0001, ***p < 0.001. Scalebar: 100 μm. | ||
3.8 Determining the stability of CuI (H) and its application as a surface steriliser
The aqueous stability of CuI (H) for 30 days was evaluated using a fluorescence spectrophotometer and FE-SEM. The fluorescence intensity of 30 days aqueous suspended CuI (H) was decreased by only 18% in comparison to the 0th day, while the shape of CuI (H) was retained as same as that of the 0th day (Fig. 9(a) and (b)). Similarly, slides spread with MH broth-suspended CuI (H) and MH broth alone exposed to daylight and air for 15 days were used to determine the surface disinfectant property of CuI (H) (Fig. 9(c)). The number of colonies in the CuI (H) inoculum-spread Petri dish were significantly fewer in comparison to the MH broth inoculum-spread Petri dish (Fig. 9(d)). From the results, it is evident that CuI (H) retains its stability in an aqueous solution even under daylight and retains its antiproliferative property. Therefore, CuI (H) could be used as an efficient surface disinfectant material in microbial infection-prone areas.4. Conclusion
Microbial infections are a continuing threat to human health. The use of antibiotics to treat microbial infections is known to elicit microbial resistance, which further attenuates the efficiency of treatment. Recent reports also suggested the emergence of microbial infections in tumour regions, which further aggravates tumour-related complications. So, developing a treatment modality effective in treating microbial infections and inhibiting cancer growth without inducing resistance will be of great importance. In the present study, CuI microparticles were synthesised using the chemical precipitation method followed by hydrothermal reaction. The high temperature involved in the hydrothermal process as well as PAA coating played a significant role in forming well-dispersed and uniform-sized CuI (H) microhexagons. Studies have reported that smaller sized and higher zeta potential particles will display greater stability and cell attraction, hence significantly higher therapeutic efficacy.50–52 Similarly, the smaller size and higher zeta potential (−16 mV) of CuI (H) compared to CuI (WH) led to a higher stability and antiproliferative activity against microbial culture, including resistant strain (E. coli), viral and cancer cells in comparison to CuI (WH). CuI (H) microparticles also retained their structure and fluorescence in an aqueous solution for 30 days and found to be effective antiproliferative agent against microbial growth. From the obtained results, it can be deduced that CuI (H) could represent a potential material for treating surface microbial infections and superficial tumors, such as melanoma. CuI (H) could also be used as a surface disinfectant in microbial infection-prone areas. However, a significant in vivo analysis of the antimicrobial property of CuI (H) is required for a potential clinical translation.Author contributions
Sunil Venkanna Pogu: conceptualization, investigation, experimental studies, writing – original draft. Dokkari Nagalaxmi Yadav: investigation, experimental studies, writing – original draft. Sri Amruthaa Sankaranarayanan: experimental studies. Rupali Srivastava: experimental studies. Shashidhar Thatikonda: supervision, fund acquisition. Aravind Kumar Rengan: conceptualization, supervision, fund acquisition, project administration, review.Conflicts of interest
The authors declare that there are no conflicts of interest.Acknowledgements
The authors would like to acknowledge MHRD IMPRINT (4291), ICMR (no. 35/1/2020-GIA/Nano/BMS), ICMR-CoE grant, DST-AMT (DST/TDT/AMT/2017/227), SERB-CRG (CRG/2020/005069), IITH/BME/SOCH3 project grants, and SUPRA (SPR/2022/230). The authors gratefully acknowledge Dr Manjula Reddy, Chief Scientist, CSIR-Hyderabad for providing the bacteriophage lambda (ATCC 23724-B2) and E. coli cultures. A fellowship from the Department of Biotechnology, Ministry of Science & Technology, Government of India (DBT/2020/IIT-H/1356) is gratefully acknowledged by author S. V. P. Author D. N. Y. would like to gratefully acknowledge DST INSPIRE (DST/INSPIRE/03/2019/001517) for funding her fellowship. Author S. A. S. would like to gratefully acknowledge MoE-PMRF (ID 2000832) for funding her fellowship. The authors would also like to acknowledge their labmate Himasree Buddhiraju. Licensed version of BioRender.com was used for preparing graphical abstract & schematic.References
- A. Shakir, D. Abate, F. Tebeje and F. Weledegebreal, Infect. Drug Resist., 2021, 14, 4629–4639 CrossRef PubMed
.
- R. E. Mengesha, B. G. S. Kasa, M. Saravanan, D. F. Berhe and A. G. Wasihun, BMC Res. Notes, 2014, 7, 4–9 CrossRef
.
- A. E. Ghenea, R. Cioboată, A. I. Drocaş, E. N. Ţieranu, C. M. Vasile, A. Moroşanu, C. G. Ţieranu, A. I. Salan, M. Popescu, A. Turculeanu, V. Padureanu, A. L. Udriştoiu, D. Calina, D. Cârţu and O. M. Zlatian, Antibiotics, 2021, 10, 868 CrossRef
.
- S. Bungau, D. M. Tit, T. Behl, L. Aleya and D. C. Zaha, Curr. Opin. Environ. Sci. Health, 2021, 19, 100224 CrossRef
.
- S. Qin, W. Xiao, C. Zhou, Q. Pu, X. Deng, L. Lan, H. Liang, X. Song and M. Wu, Signal Transduction Target. Ther., 2022, 7, 1–27 CrossRef
.
- Z. Tang, B. Song, W. Zhang, L. Guo and J. Yuan, Anal. Chem., 2019, 91, 14019–14028 CrossRef CAS
.
- E. Avershina, V. Shapovalova and G. Shipulin, Front. Microbiol., 2021, 12, 2044 Search PubMed
.
- C. J. Murray, K. S. Ikuta, F. Sharara, L. Swetschinski, G. Robles Aguilar, A. Gray, C. Han, C. Bisignano, P. Rao, E. Wool, S. C. Johnson, A. J. Browne, M. G. Chipeta, F. Fell, S. Hackett, G. Haines-Woodhouse, B. H. Kashef Hamadani, E. A. P. Kumaran, B. McManigal, R. Agarwal, S. Akech, S. Albertson, J. Amuasi, J. Andrews, A. Aravkin, E. Ashley, F. Bailey, S. Baker, B. Basnyat, A. Bekker, R. Bender, A. Bethou, J. Bielicki, S. Boonkasidecha, J. Bukosia, C. Carvalheiro, C. Castañeda-Orjuela, V. Chansamouth, S. Chaurasia, S. Chiurchiù, F. Chowdhury, A. J. Cook, B. Cooper, T. R. Cressey, E. Criollo-Mora, M. Cunningham, S. Darboe, N. P. J. Day, M. De Luca, K. Dokova, A. Dramowski, S. J. Dunachie, T. Eckmanns, D. Eibach, A. Emami, N. Feasey, N. Fisher-Pearson, K. Forrest, D. Garrett, P. Gastmeier, A. Z. Giref, R. C. Greer, V. Gupta, S. Haller, A. Haselbeck, S. I. Hay, M. Holm, S. Hopkins, K. C. Iregbu, J. Jacobs, D. Jarovsky, F. Javanmardi, M. Khorana, N. Kissoon, E. Kobeissi, T. Kostyanev, F. Krapp, R. Krumkamp, A. Kumar, H. H. Kyu, C. Lim, D. Limmathurotsakul, M. J. Loftus, M. Lunn, J. Ma, N. Mturi, T. Munera-Huertas, P. Musicha, M. M. Mussi-Pinhata, T. Nakamura, R. Nanavati, S. Nangia, P. Newton, C. Ngoun, A. Novotney, D. Nwakanma, C. W. Obiero, A. Olivas-Martinez, P. Olliaro, E. Ooko, E. Ortiz-Brizuela, A. Y. Peleg, C. Perrone, N. Plakkal, A. Ponce-de-Leon, M. Raad, T. Ramdin, A. Riddell, T. Roberts, J. V. Robotham, A. Roca, K. E. Rudd, N. Russell, J. Schnall, J. A. G. Scott, M. Shivamallappa, J. Sifuentes-Osornio, N. Steenkeste, A. J. Stewardson, T. Stoeva, N. Tasak, A. Thaiprakong, G. Thwaites, C. Turner, P. Turner, H. R. van Doorn, S. Velaphi, A. Vongpradith, H. Vu, T. Walsh, S. Waner, T. Wangrangsimakul, T. Wozniak, P. Zheng, B. Sartorius, A. D. Lopez, A. Stergachis, C. Moore, C. Dolecek and M. Naghavi, Lancet, 2022, 399, 629–655 CrossRef CAS
.
- N. S. Alharbi, J. M. Khaled, S. Kadaikunnan, A. S. Alobaidi, A. H. Sharafaddin, S. A. Alyahya, T. N. Almanaa, M. A. Alsughayier and M. R. Shehu, Saudi J. Biol. Sci., 2019, 26, 1557–1562 CrossRef CAS
.
- B. W. J. H. Penninx, M. E. Benros, R. S. Klein and C. H. Vinkers, Nat. Med., 2022, 28, 2027–2037 CrossRef CAS
.
- F. Bray, M. Laversanne, E. Weiderpass and I. Soerjomataram, Cancer, 2021, 127, 3029–3030 CrossRef
.
- A. Krueger, J. Zaugg, S. Chisholm, R. Linedale, N. Lachner, S. M. Teoh, Z. K. Tuong, S. W. Lukowski, M. Morrison, H. P. Soyer, P. Hugenholtz, M. M. Hill and I. H. Frazer, Front. Microbiol., 2022, 12, 1–17 Search PubMed
.
- B. Joob and V. Wiwanitkit, Asian Pac. J. Trop. Dis., 2014, 4, 204–206 CrossRef
.
- Y. Wen, J. Hu, J. Liu and M. Li, Chem. Mater., 2021, 33, 7089–7099 CrossRef CAS
.
- H. Zhang and X. B. Yin, ACS Appl. Mater. Interfaces, 2022, 14, 26528–26535 CrossRef CAS
.
- M. M. Mamun, A. J. Sorinolu, M. Munir and E. P. Vejerano, Front. Chem., 2021, 9, 1–23 Search PubMed
.
- A. Singh, A. Ahmed, A. K. Keshri, N. Arora, F. Anjum, S. S. Rawat and A. Prasad, Bionanoscience, 2021, 11, 637–642 CrossRef
.
- J. Salvo and C. Sandoval, Burns Trauma, 2022, 10 DOI:10.1093/burnst/tkab047
.
- X. Zhang, L. Detering, D. Sultan, H. Luehmann, L. Li, G. S. Heo, X. Zhang, L. Lou, P. M. Grierson, S. Greco, M. Ruzinova, R. Laforest, F. Dehdashti, K. H. Lim and Y. Liu, ACS Nano, 2021, 15, 1186–1198 CrossRef CAS PubMed
.
- K. Kirakci, D. Bůžek, P. Peer, V. Liška, J. Mosinger, I. Křížová, M. Kloda, S. Ondrušová, K. Lang and J. Demel, ACS Appl. Nano Mater., 2022, 5, 1244–1251 CrossRef CAS
.
- W. Lu, S. Song, Y. Qian, Q. Liu, P. Li, Y. Han, J. Zhang, J. Xu, J. Sun and A. Wu, ACS Nano, 2021, 15, 2933–2946 CrossRef PubMed
.
- D. Verma Atul, Certain distance degree based Topol. indices Zeolite LTA Fram., 2018, 11–14 Search PubMed
.
- B. L. Cline, W. Jiang, C. Lee, Z. Cao, X. Yang, S. Zhan, H. Chong, T. Zhang, Z. Han, X. Wu, L. Yao, H. Wang, W. Zhang, Z. Li and J. Xie, ACS Nano, 2021, 15, 17401–17411 CrossRef CAS PubMed
.
- J. Yu, X. He, Q. Zhang, D. Zhou, Z. Wang and Y. Huang, ACS Nano, 2022, 16, 6835–6846 CrossRef CAS PubMed
.
- R. Khursheed, K. Dua, S. Vishwas, M. Gulati, N. K. Jha, G. M. Aldhafeeri, F. G. Alanazi, B. H. Goh, G. Gupta, K. R. Paudel, P. M. Hansbro, D. K. Chellappan and S. K. Singh, Biomed. Pharmacother., 2022, 150, 112951 CrossRef CAS PubMed
.
- A. Cartwright, K. Jackson, C. Morgan, A. Anderson and D. W. Britt, Agronomy, 2020, 10, 1–20 CrossRef
.
- M. C. Crisan, M. Teodora and M. Lucian, Appl. Sci., 2021, 12, 141 CrossRef
.
- T. Han, Z. Ma and D. Wang, ACS Macro Lett., 2021, 10, 354–358 CrossRef CAS
.
- Y. Zhou, M. lü, G. Zhou, S. Wang and S. Wang, Mater. Lett., 2006, 60, 2184–2186 CrossRef CAS
.
- T. Appidi, G. Ravichandran, S. V. Mudigunda, A. Thomas, A. B. Jogdand, S. Kishen, K. Subramaniyam, N. Emani, G. Prabusankar and A. K. Rengan, Mater. Today Commun., 2021, 29, 102987 CrossRef CAS
.
- S. B. Alvi, P. S. Rajalakshmi, A. Jogdand, A. Y. Sanjay, B. Veeresh, R. John and A. K. Rengan, Biomater. Sci., 2021, 9, 1421–1430 RSC
.
- S. Sharma, J. Acharya, M. R. Banjara, P. Ghimire and A. Singh, BMC Res. Notes, 2020, 13, 1–5 CrossRef
.
- F. Dharul Salam, M. Nadar Vinita, P. Puja, S. Prakash, R. Yuvakkumar and P. Kumar, Mater. Lett., 2020, 261, 126998 CrossRef CAS
.
- D. N. Yadav, S. A. Sankaranarayanan, A. M. Thanekar and A. K. Rengan, Mater. Today Nano, 2023, 100348 CrossRef
.
- P. Alekhya, C. Aruna Sunder and Prathiba, Avicenna J. Clin. Microbiol. Infect., 2020, 7, 124–128 CrossRef CAS
.
- A. Ajdidi, G. Sheehan, K. A. Elteen and K. Kavanagh, J. Med. Microbiol., 2019, 68, 1497–1506 CrossRef CAS PubMed
.
- S. Chhibber, P. Kaur and V. S. Gondil, J. Virol. Methods, 2018, 262, 1–5 CrossRef CAS
.
- T. Appidi, D. B. Pemmaraju, R. A. Khan, S. B. Alvi, R. Srivastava, M. Pal, N. Khan and A. K. Rengan, Nanoscale, 2020, 12, 2028–2039 RSC
.
- Z. Xiu, J. Ma and P. J. J. Alvarez, Environ. Sci. Technol., 2011, 45, 9003–9008 CrossRef CAS
.
- C. Gunawan, W. Y. Teoh, C. P. Marquis and R. Amal, ACS Nano, 2011, 7214–7225 CrossRef CAS PubMed
.
- J. Pijuan, C. Barceló, D. F. Moreno, O. Maiques, P. Sisó, R. M. Marti, A. Macià and A. Panosa, Front. Cell Dev. Biol., 2019, 7, 1–16 CrossRef
.
- J. P. S. Nunes and A. A. M. Dias, Biotechniques, 2017, 62, 175–179 CrossRef CAS PubMed
.
- E. N. Zare, X. Zheng, P. Makvandi, H. Gheybi, R. Sartorius, C. K. Y. Yiu, M. Adeli, A. Wu, A. Zarrabi, R. S. Varma and F. R. Tay, Small, 2021, 17, 1–31 CrossRef
.
- A. Lassenberger, T. A. Grünewald, P. D. J. Van Oostrum, H. Rennhofer, H. Amenitsch, R. Zirbs, H. C. Lichtenegger and E. Reimhult, Chem. Mater., 2017, 29, 4511–4522 CrossRef CAS
.
- H. Xie, M. Hong, E. M. Hitz, X. Wang, M. Cui, D. J. Kline, M. R. Zachariah and L. Hu, J. Am. Chem. Soc., 2020, 142, 17364–17371 CrossRef CAS PubMed
.
- H. Liu, H. Zhang, J. Wang and J. Wei, Arabian J. Chem., 2020, 13, 1011–1019 CrossRef CAS
.
- S. S. Skeeters, A. C. Rosu, Divyanshi, J. Yang and K. Zhang, ACS Appl. Mater. Interfaces, 2020, 12, 50203–50211 CrossRef CAS PubMed
.
- A. Pramanik, D. Laha, D. Bhattacharya and P. Pramanik, Colloids Surf., B, 2012, 96, 50–55 CrossRef CAS PubMed
.
- I. P. Parkin, Advances, 2021, 11, 18179–18186 Search PubMed
.
- E. Fröhlich, Int. J. Nanomed., 2012, 7, 5577–5591 CrossRef
.
- L. E. Shi, Z. H. Li, W. Zheng, Y. F. Zhao, Y. F. Jin and Z. X. Tang, Food Addit. Contam.: Part A, 2014, 31, 173–186 CrossRef CAS
.
- K. P. Steckiewicz, J. Zwara, M. Jaskiewicz, S. Kowalski, W. Kamysz, A. Zaleska-Medynska and I. Inkielewicz-Stepniak, Oxid. Med. Cell. Longevity, 2019, 2019 DOI:10.1155/2019/6740325
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ma00110e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |