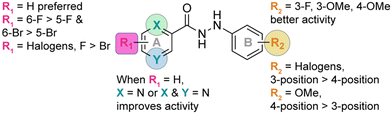Discovery and development of novel substituted monohydrazides as potent antifungal agents†
Nishad
Thamban Chandrika‡
,
Keith D.
Green‡
,
Abbygail C.
Spencer
,
Oleg V.
Tsodikov
 and
Sylvie
Garneau-Tsodikova
and
Sylvie
Garneau-Tsodikova
 *
*
Department of Pharmaceutical Sciences, College of Pharmacy, University of Kentucky, 789 South Limestone Street, Lexington, KY 40536-0596, USA. E-mail: sylviegtsodikova@uky.edu
First published on 14th June 2023
Abstract
Novel substituted monohydrazides synthesized for this study displayed broad-spectrum activity against various fungal strains, including a panel of clinically relevant Candida auris strains. The activity of these compounds was either comparable or superior to amphotericin B against most of the fungal strains tested. These compounds possessed fungistatic activity in a time-kill assay and exhibited no mammalian cell toxicity. In addition, they prevented the formation of fungal biofilms. Even after repeated exposures, the Candida albicans ATCC 10231 (strain A) fungal strain did not develop resistance to these monohydrazides.
Introduction
For the past two decades, fungal infections including superficial or mucosal and severe systemic invasive fungal infections (IFIs) have been on the rise, posing a significant threat to human health.1,2 Opportunistic fungal pathogens such as Candida species, Aspergillus species, and Cryptococcus species are responsible for the majority of IFIs. IFIs are a significant cause of morbidity and mortality, particularly for patients with compromised immune function.3,4 The increasing number of immunocompromised individuals, including cancer patients, organ transplant recipients, individuals infected with HIV, patients with diabetes and chronic obstructive pulmonary diseases, and the increasing elderly population has resulted in a surge in diseases caused by fungi in recent decades.5–8 The extensive and repeated use of the existing classes of antifungal drugs resulted in the resistance of fungal strains to available drug therapies.9–13 In addition, Candida auris, first identified in 2009 in Asia, has quickly become a cause of severe recalcitrant infections and deaths around the world.14–17C. auris is often multidrug-resistant, causing outbreaks among hospital patients and nursing home residents.18–20 In March 2023, the Centers for Disease Control and Prevention (CDC) issued a press release, which underscored a rapid spread of C. auris infections throughout healthcare facilities in the US, tripling in the numbers of clinical cases during a recent two-year span.21 It is clear that the current antifungal armamentarium is insufficient to stem the spread of C. auris infections. In order to combat the increasing cases of fungal infections, new antifungal drugs are needed that are broad spectrum and less toxic than the currently available agents.Despite the increasing threat of spread of antimicrobial-resistant fungi, treatment options remain limited.22,23 The three available classes of current antifungals: the azoles, such as fluconazole (FLC) and voriconazole (VRC), the echinocandins, such as caspofungin (CFG), and the polyenes, such as amphotericin B (AmB) are still staple treatments of IFIs. Overreliance on these agents and their excessive use have caused an increase in the percentage of fungal species resistant to these classes of drugs.24,25 IFIs have also emerged as the leading co-infection in patients hospitalized with severe COVID-19 infections.26–29 The cost of antimicrobial resistance is immense, both for the economy and the human health.30–32 The narrow spectrum of antifungal activity, limited efficacy, significant side effects, drug–drug interactions, and toxicity associated with existing antifungal drugs highlight the urgent need to develop novel antifungal therapeutic agents.33–35
In recent years, our group has been actively involved in developing novel small molecules as well aminoglycoside-based antifungal agents to treat both topical and systemic fungal infections.36–50 We previously reported the development of bis(N-amidinohydrazones), N-(amidino)-N′-aryl-bishydrazones, and N,N′-diaryl bishydrazones as potential antibacterial and antifungal agents.51,52 Next, we explored the efficacy of monohydrazones over bishydrazones.53 With the promising results we observed for monohydrazones, we now decided to explore novel substituted monohydrazides as potential antifungals. Herein, we report the synthesis and antifungal activity of 64 novel substituted monohydrazides by in vitro studies as well as by time-kill studies. We also explore their efficacy against biofilms. Finally, we investigate their toxicity profile against mammalian cell lines and their potential susceptibility to the development of resistance by C. albicans.
Results and discussion
Synthesis
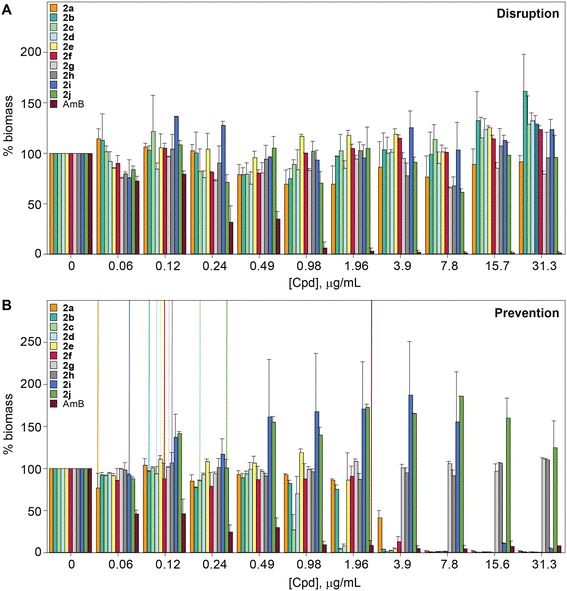
For this study, we synthesized 64 monohydrazides with different R1 and R2 substituents (Fig. 1). The coupling reaction between various commercially available acids and commercially available substituted hydrazines using coupling reagents (N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride and 1-hydroxybenzotriazole hydrate) resulted in the formation of compounds 1a–9j in 18–86% yields. The entire library of monohydrazides was separated into nine different series (1–9) based on the R1 and R2 substituents (denoted by a–j in compound numbering) on rings A and B, respectively. | ||
| Fig. 1 The synthetic scheme for the preparation of the compounds used in this study. Note: the chemical yields are provided in parentheses. | ||
Antifungal minimum inhibitory concentration (MIC) values
We tested the antifungal activity of the 64 monohydrazides (1a–9j) against a panel of seven Candida albicans strains (A–G): ATCC 10231 (A), ATCC 64124 (B), ATCC MYA-2876 (C), ATCC 90819 (D), ATCC MYA-2310 (E), ATCC MYA-1237 (F), and ATCC MYA-1003 (G) (Tables 1 and S1†). We also explored activity of the monohydrazides against a panel of five non-albicans Candida strains: Candida glabrata ATCC 2001 (H), Candida krusei ATCC 6258 (I), Candida parapsilosis ATCC 22019 (J), Candida auris strain AR Bank # 384 (K), and C. auris strain AR Bank # 390 (L) (Tables 1 and S1†). Finally, we explored the activity of these monohydrazides against seven non-Candida strains: Cryptococcus neoformans ATCC MYA-895 (M), Aspergillus terreus ATCC MYA-3633 (N), Aspergillus flavus ATCC MYA-3631 (O), Aspergillus nidulans ATCC 38163 (P), Aspergillus fumigatus NRRL 163 (Q), A. fumigatus NRRL 5109 (R), and A. fumigatus NRRL 6113 (S) (Tables 1 and S1†). We used the double dilution method with concentrations ranging from 0.06 to 31.3 μg mL−1 for monohydrazides 1a–9j, and the commercially available positive antifungal control amphotericin B (AmB). Herein, based on the MIC values (i.e., no visible growth), we defined antifungal activity as excellent (≤1.95 μg mL−1), good (3.9–7.8 μg mL−1), or poor (≥15.6 μg mL−1). Antifungal activity data for the most active compounds are presented in Table 1 and the remaining are available in Table S1 along with the entire data in Table S3.†| Fungal strain | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Candida albicans | Non-albicans Candida | Non-Candida | |||||||||||||||||
| Cpd # | A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S |
| Strains: A = C. albicans ATCC 10231, B = C. albicans ATCC 64124, C = C. albicans ATCC MYA-2876(S), D = C. albicans ATCC 90819(R), E = C. albicans ATCC MYA-2310(S), F = C. albicans ATCC MYA-1237(R), G = C. albicans ATCC MYA-1003(R), H = C. glabrata ATCC 2001, I = C. krusei ATCC 6258, J = C. parapsilosis ATCC 22019, K = C. auris AR Bank # 0384, L = C. auris AR Bank # 0390, M = C. neoformans ATCC MYA-895, N = A. terreus ATCC MYA-3633, O = A. flavus ATCC MYA-3631, P = A. nidulans ATCC 38163, Q = A. fumigatus NRRL 163, R = A. fumigatus NRRL 5109, and S = A. fumigatus NRRL 6113. Note: here, the (S) and (R) indicate that ATCC reports these strains to be susceptible (S) and resistant (R) to itraconazole (ITC) and fluconazole (FLC). | |||||||||||||||||||
| 1a | 0.24 | 1.95 | 3.9 | 7.8 | 3.9 | 0.98 | 1.95 | 7.8 | 1.95 | 3.9 | 1.95 | 0.24 | 1.95 | 1.95 | >31.3 | 3.9 | >31.3 | >31.3 | >31.3 |
| 1d | 0.49 | 7.8 | 7.8 | 1.95 | 3.9 | 1.95 | 1.95 | 7.8 | 1.95 | 15.6 | 1.95 | 1.95 | 0.49 | 3.9 | >31.3 | 3.9 | >31.3 | >31.3 | >31.3 |
| 1f | 0.98 | 1.95 | 1.95 | 1.95 | 1.95 | 1.95 | 1.95 | 3.9 | 0.98 | 7.8 | 1.95 | 1.95 | 0.98 | 31.3 | >31.3 | 1.95 | >31.3 | >31.3 | >31.3 |
| 2a | ≤0.06 | 0.98 | 0.98 | 0.49 | 1.95 | 0.98 | 0.98 | 1.95 | 0.24 | 1.95 | 1.95 | 0.24 | 0.12 | 1.95 | 7.8 | 0.49 | >31.3 | >31.3 | >31.3 |
| 2b | 0.12 | 0.98 | 0.98 | 0.49 | 0.98 | 0.98 | 0.49 | 1.95 | 0.49 | 1.95 | 0.98 | 0.98 | 0.12 | 0.98 | >31.3 | 0.24 | >31.3 | >31.3 | >31.3 |
| 2c | 0.24 | 0.98 | 0.98 | 1.95 | 1.95 | 1.95 | 0.98 | 15.6 | 0.49 | 3.9 | 0.98 | 0.98 | 0.24 | 3.9 | >31.3 | 1.95 | 7.8 | 3.9 | >31.3 |
| 2d | 0.12 | 0.98 | 3.9 | 0.98 | 1.95 | 1.95 | 0.49 | 1.95 | 0.12 | 1.95 | 3.9 | 0.98 | 0.24 | 1.95 | 31.3 | 0.49 | 31.3 | 7.8 | 15.6 |
| 2e | 0.12 | 0.98 | 1.95 | 0.24 | 0.98 | 0.98 | 0.98 | 1.95 | 1.95 | 3.9 | 1.95 | 1.95 | 0.49 | 3.9 | 31.3 | 0.49 | >31.3 | 7.8 | 15.6 |
| 2f | 0.12 | 0.98 | 1.95 | 0.49 | 0.98 | 0.98 | 0.98 | 0.98 | 0.49 | 3.9 | 3.9 | 0.12 | 0.49 | 31.3 | >31.3 | 0.49 | >31.3 | >31.3 | 31.3 |
| 2g | 0.12 | 1.95 | 0.98 | 0.98 | 31.3 | 0.49 | 0.49 | 0.98 | 0.49 | 1.95 | 0.49 | 0.12 | 0.24 | 0.98 | 15.6 | 0.49 | >31.3 | 15.6 | 31.3 |
| 2h | 0.12 | 1.95 | 3.9 | 0.98 | 1.95 | 1.95 | 0.98 | 3.9 | 0.24 | 3.9 | 0.24 | 0.98 | 0.49 | 0.98 | 3.9 | 0.49 | >31.3 | 15.6 | >31.3 |
| 2i | ≤0.06 | 0.98 | 0.98 | 0.98 | 0.98 | 1.95 | 0.49 | 7.8 | 0.24 | 1.95 | 0.24 | 0.49 | 0.24 | 0.98 | 1.95 | 0.49 | >31.3 | >31.3 | 31.3 |
| 2j | 0.24 | 1.95 | 1.95 | 0.98 | 1.95 | 3.9 | 1.95 | 7.8 | 0.98 | 1.95 | 7.8 | 1.95 | 0.49 | 1.95 | >31.3 | 0.49 | >31.3 | >31.3 | >31.3 |
| 3a | 0.49 | 0.49 | 3.9 | 3.9 | 3.9 | 3.9 | 1.95 | 31.3 | 0.24 | 1.95 | 1.95 | 1.95 | ≤0.06 | 1.95 | 7.8 | 0.98 | >31.3 | >31.3 | >31.3 |
| 3c | 0.24 | 0.98 | 0.49 | 3.9 | 1.95 | 1.95 | 0.98 | 15.6 | 0.24 | 3.9 | 1.95 | 0.98 | 0.24 | 3.9 | >31.3 | 1.95 | >31.3 | >31.3 | >31.3 |
| 3f | 0.24 | 0.98 | 0.98 | 0.98 | 0.98 | 0.49 | 0.49 | 1.95 | 0.24 | 3.9 | 3.9 | 0.49 | ≤0.06 | 31.3 | >31.3 | 1.95 | >31.3 | >31.3 | >31.3 |
| 4a | 0.49 | 0.49 | 1.95 | 3.9 | 3.9 | 3.9 | 0.98 | >31.3 | 0.24 | 7.8 | 1.95 | 1.95 | 0.06 | 0.98 | 15.6 | 1.95 | >31.3 | >31.3 | >31.3 |
| 4c | 0.98 | 3.9 | 0.98 | 1.95 | 3.9 | 0.98 | 3.9 | 3.9 | 0.24 | 1.95 | 0.98 | 1.95 | 0.49 | 1.95 | >31.3 | 3.9 | >31.3 | 15.6 | >31.3 |
| 5c | 0.98 | 0.98 | 0.49 | 3.9 | 1.95 | 1.95 | 0.98 | 15.6 | 0.24 | 3.9 | 1.95 | 0.98 | 0.49 | 3.9 | >31.3 | 1.95 | 15.6 | 7.8 | >31.3 |
| 5d | 0.24 | 0.98 | 1.95 | 1.95 | 3.9 | 1.95 | 0.98 | 15.6 | 0.49 | 3.9 | 1.95 | 0.98 | 0.49 | 1.95 | 7.8 | 0.98 | 31.3 | 15.6 | 15.6 |
| 5f | 0.24 | 1.95 | 0.98 | 1.95 | 0.98 | 0.98 | 0.98 | 0.98 | 0.49 | 7.8 | 1.95 | 1.95 | 0.24 | 15.6 | >31.3 | 1.95 | >31.3 | >31.3 | 15.6 |
| 6c | 1.95 | 3.9 | 0.49 | 1.95 | 7.8 | 0.98 | 3.9 | 7.8 | 0.24 | 0.98 | 0.98 | 1.95 | 0.98 | 1.95 | >31.3 | 3.9 | >31.3 | 31.3 | >31.3 |
| 7a | 0.24 | 0.98 | 1.95 | 3.9 | 1.95 | 3.9 | 1.95 | 31.3 | 0.24 | 1.95 | 1.95 | 1.95 | ≤0.06 | 3.9 | >31.3 | 0.98 | >31.3 | >31.3 | >31.3 |
| 7d | 0.98 | 1.95 | 1.95 | 1.95 | 3.9 | 1.95 | 0.98 | 15.6 | 0.49 | 3.9 | 1.95 | 0.98 | 0.49 | 3.9 | 7.8 | 0.49 | >31.3 | >31.3 | >31.3 |
| 7f | 1.95 | 1.95 | 1.95 | 1.95 | 1.95 | 0.98 | 0.98 | 7.8 | 0.98 | 7.8 | 3.9 | 0.98 | 1.95 | 31.3 | >31.3 | 1.95 | 31.3 | 31.3 | 31.3 |
| 8a | 0.98 | 0.98 | 1.95 | 3.9 | 1.95 | 1.95 | 0.98 | 15.6 | 0.49 | 7.8 | 1.95 | 1.95 | ≤0.06 | 3.9 | >31.3 | 1.95 | >31.3 | >31.3 | >31.3 |
| 8d | 0.24 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | 0.49 | 15.6 | 0.24 | 7.8 | 1.95 | 0.98 | 0.49 | 1.95 | 7.8 | 0.49 | 31.3 | 15.6 | >31.3 |
| 8f | 1.95 | 1.95 | 1.95 | 1.95 | 1.95 | 0.98 | 0.98 | 7.8 | 0.98 | 7.8 | 3.9 | 0.98 | 0.98 | 31.3 | >31.3 | 1.95 | 31.3 | 15.6 | 31.3 |
| 9a | 0.24 | 1.95 | 3.9 | 0.98 | 7.8 | 0.98 | 0.98 | 7.8 | 0.49 | 3.9 | 0.98 | 0.98 | 0.12 | 0.98 | 15.6 | 0.98 | >31.3 | >31.3 | >31.3 |
| 9b | 0.24 | 1.95 | 1.95 | 3.9 | 7.8 | 0.98 | 0.98 | 3.9 | 0.24 | 1.95 | 1.95 | 0.98 | 0.24 | 0.98 | >31.3 | 0.98 | >31.3 | >31.3 | >31.3 |
| 9c | 0.49 | 0.98 | 1.95 | 1.95 | 1.95 | 1.95 | 0.98 | 7.8 | 0.49 | 1.95 | 1.95 | 0.98 | 0.49 | 1.95 | >31.3 | 1.95 | 15.6 | 7.8 | >31.3 |
| 9f | 0.49 | 0.98 | 1.95 | 0.98 | 1.95 | 0.98 | 1.95 | 0.98 | 0.98 | 3.9 | 0.49 | 0.49 | 0.98 | >31.3 | >31.3 | 0.98 | 31.3 | 31.3 | 31.3 |
| 9g | 0.24 | 0.98 | 0.98 | 0.98 | 3.9 | 0.49 | 0.49 | 0.98 | 0.24 | 3.9 | 3.9 | 0.98 | 0.24 | 0.98 | 7.8 | 0.49 | >31.3 | >31.3 | >31.3 |
| AmB | 1.95 | 1.95 | 1.95 | 0.98 | 0.98 | 1.95 | 0.98 | 0.98 | 0.98 | 0.98 | 1.95 | 1.95 | 7.8 | 3.9 | 31.3 | 15.6 | 0.98 | 0.98 | 1.95 |
From the data reported in Table 1 and S1,† we observed that compounds from series 2 and series 9 performed better compared to those in the other seven series of compounds against the sixteen strains (A–P) tested. A detailed analysis of the nine series (i.e., series 1–9) led to the following conclusions. In the case of monohydrazides 1a–1j (i.e., R1 = 2,4-diF, X = C, Y = C; Fig. 1), compounds 1h, 1i, and 1j generally displayed poor activity against strains A–P, with the exception of the following combinations; 1h (MIC = 0.98 μg mL−1 against strain M), 1i (MIC = 1.95 μg mL−1 and 0.49 μg mL−1 against strains I and M, respectively), and 1j (MIC = 1.95 μg mL−1 against strain I). Among series 1, compounds 1f, 1a, 1d, and 1c performed better with excellent activity (0.24–1.95 μg mL−1) against twelve strains (A–G, I, K–M, and P), nine strains (A, B, F, G, I, and K–N), eight strains (A, D, F, G, I, and K–M), and seven strains (A, B, G, I, and K–M), respectively. Monohydrazides 2a–2j (i.e., R1 = H, X = N, Y = C; Fig. 1) displayed excellent to good activity (0.06–7.8 μg mL−1) against all fungal strains tested with the exception of compounds 2b, 2c, 2d, 2e, 2f, 2g, and 2j against strains O, H (2c only), N (2f only), and E (2g only). All the compounds from series 3 (3a, 3c, 3d, 3e, 3f, and 3g) (i.e., R1 = 5-F, X = N, Y = C; Fig. 1) displayed excellent to good activity (0.06–7.8 μg mL−1) against most of the strains A–P. However, some compounds from series 3 displayed poor activity (15.6–>31.3 μg mL−1). Those include compounds 3a, 3c, 3d, and 3g against strain H, compounds 3e and 3f against strain N, compounds 3e and 3g against strain J, compound 3e against strain P, and all “3”-compounds except 3a against strain O. Among series 3, compounds 3f, 3c, 3a, and 3g were the best in terms of their activity. In the case of monohydrazides from series 4 (i.e., R1 = 5-Br, X = N, Y = C; Fig. 1), compounds 4a, 4c, 4d, and 4f displayed excellent to good activity against strains A–G and I–N (0.24–7.8 μg mL−1). However, compound 4g performed below par with poor activity against strains B, E, G, L, and O. For series 5 (i.e., R1 = 6-F, X = N, Y = C; Fig. 1), compounds 5a, 5c, 5d, and 5f exhibited excellent to good activity (0.06–7.8 μg mL−1) against 14 to 15 (out of 16) of the non-A. fumigatus fungal strains tested, with the exception of compounds 5a, 5c, 5d, and 5f against either strains H, N, and/or O. Compounds 5e and 5g displayed excellent to good activity against strains A–I, K–M, and P (0.98–7.8 μg mL−1), and strains A–G, I, K–N, and P (0.49–7.8 μg mL−1), respectively. Amongst monohydrazides 6a, 6c, 6d, 6f, and 6g (i.e., R1 = 6-Br, X = N, Y = C; Fig. 1), compounds 6a and 6d exhibited excellent to good activity (0.06–7.8 μg mL−1) against all 16 non-A. fumigatus strains tested with the exception of strains H and O. Compounds 6c and 6f displayed excellent to good activity (0.24–7.8 μg mL−1) against all 16 non-A. fumigatus strains tested with the exception of strain O. On the other hand, compound 6g performed poorly against strains B, E, G, H, L, and O. For compounds from series 7 (i.e., R1 = 3,5-diF, X = N, Y = C; Fig. 1), compounds 7a, 7d, and 7f performed better than other members of this series. Compounds 7a, 7d, and 7f exhibited excellent to good activity (0.06–7.8 μg mL−1) against all 16 non-A. fumigatus strains tested, with the exception of compounds 7a, 7d, and 7f against strains H and O, strain H, and strains N and O, respectively. Compounds 7e, and 7g displayed excellent activity (0.49–1.98 μg mL−1) against strains E–G, I, and M, and strains A, G, I, L, M, and P, respectively. For series 8 (i.e., R1 = 3,6-diF, X = N, Y = C; Fig. 1), compound 8d exhibited excellent to good activity (0.24–7.8 μg mL−1) against all the strains tested, with the exception of strain H. Compounds 8a, 8f, and 8g displayed excellent to good activity (0.06–7.8 μg mL−1) against strains A–G, I–N, and P, strains A–M, and P, and strains A–G, I, K–M, and P, respectively. Amongst series 8, compounds 8c and 8e performed subpar when compared to other compounds in the same series. Finally, in the case of series 9 (i.e., R1 = H, X = N, Y = N; Fig. 1), compound 9c exhibited excellent activity (0.49–1.95 μg mL−1) against 14 strains A–G, I–N, and P. Among the remaining compounds in series 9, compounds 9f, 9g, 9b, 9a, 9d, 9e, and 9i displayed excellent to good activity (0.12–7.8 μg mL−1) against all 16 non-A. fumigatus strains tested with the exception of compounds 9f, 9b, 9a, 9d, 9e, and 9i against strains E (9i only was inactive), N (9f only was inactive), and O. Additionally, we explored the activity of compounds 1a, 1d, 1f, 2a–2j, 3a, 3c, 3f, 4a, 4c, 5c, 5d, 5f, 6c, 7a, 7d, 7f, 8a, 8d, 8f, 9a–9c, 9f, and 9g against clinically derived A. fumigatus strains Q, R, and S. A few compounds (2c, 2d, 2e, 5c, and 9c) exhibited good activity against strains Q and R. The highest activity against strain S was 15.6 μg mL−1 for compounds 2d, 2e, 5e and 5f. In summary, compounds 1f, 2a, 2b, 2c, 2d, 2e, 2f, 2g, 2h, 2i, 2j, 3f, 5d, 5f, 8d, 9b, 9c, 9f, and 9g exhibited a comparable or superior activity to that of the FDA-approved antifungal agent AmB against most of the tested strains, with the exception of strains Q, R and S, against which AmB was 4–8-fold more potent than the most active monohydrazides. From all of the observations made for compounds 1a–9j, we concluded that series 2 performed the best followed by series 9.
Based on the promising antifungal activities given in Table 1 and S1,† we selected the entire series 2, and some of the representative compounds from series 1 and 3–9 for further testing against a panel of ten C. auris strains (AR Bank # 381–390) (Table 2 and S2†). Using the same concentration range as above (0.06 to 31.3 μg mL−1) for all of the selected monohydrazides and using AmB as a positive control, we determined MIC values. Monohydrazides 2a, 2b, 2g, 2h, 2i, 5d, 8d, 9b, and 9f displayed excellent activity (0.06–1.95 μg mL−1) against all ten C. auris strains tested. From the remaining set of compounds tested, we found that compounds 2c, 2d, 2e, 2f, 2j, 3c, 3d, 3e, 3g, 4d, 5c, 5e, 5g, 6d, 7d, 7g, 9c, and 9g exhibited excellent to good activity (0.12–7.8 μg mL−1) against all ten C. auris strains tested. Compounds 7f, 8f, and 8g exhibited excellent activity (0.49–1.95 μg mL−1) against all of the strains tested, with the exception of compounds 7f, 8f, and 8g against C. auris strain AR Bank # 390. Similar to the result observed in Table 1, compounds 1b, 1j, 7e, and 8e displayed poor activity against most of the C. auris strains tested.
| Fungal strain (AR Bank #) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cpd # | 381 | 382 | 383 | 384 (K) | 385 | 386 | 387 | 388 | 389 | 390 (L) | 391 | 392 | 393 | 394 | 395 | 396 | 397 | 398 | 399 | 400 |
| Strains: 381–390 = Candida auris, 391, 392, and 394 = Candida duobushaemulonii, 393 and 395 = Candida haemulonii, 396 = Kodameae ohmeri, 397 = Candida krusei, 398 = Candida lusitaniae, 399 and 400 = Saccharomyces cerevisiae. | ||||||||||||||||||||
| 2a | 0.49 | 0.98 | 1.95 | 1.95 | 0.98 | 0.49 | 0.49 | 0.49 | 0.12 | 0.24 | 0.49 | 0.24 | 0.12 | 0.49 | ≤0.06 | 0.24 | 0.24 | 0.98 | 1.95 | 1.95 |
| 2b | 0.98 | 0.49 | 0.49 | 0.98 | 0.49 | 0.24 | 0.49 | 0.49 | 0.49 | 0.49 | 0.24 | 0.24 | 0.12 | 012 | ≤0.06 | 0.12 | 0.12 | 0.98 | 0.98 | 0.98 |
| 2c | 3.9 | 0.98 | 0.98 | 0.98 | 3.9 | 0.49 | 0.49 | 3.9 | 0.49 | 0.98 | 1.95 | 1.95 | 0.49 | 0.98 | 0.49 | 0.49 | 0.49 | 1.95 | 7.8 | 7.8 |
| 2d | 0.98 | 1.95 | 1.95 | 3.9 | 0.98 | 0.49 | 0.98 | 0.98 | 0.49 | 0.98 | 0.49 | 0.49 | 0.24 | 0.24 | 0.12 | 0.24 | 0.24 | 1.95 | 3.9 | 1.95 |
| 2e | 1.95 | 3.9 | 1.95 | 1.95 | 1.95 | 0.98 | 1.95 | 1.95 | 0.98 | 1.95 | 0.98 | 0.98 | 0.49 | 0.40 | 0.12 | 0.24 | 0.24 | 0.98 | 0.98 | 0.98 |
| 2f | 1.95 | 1.95 | 1.95 | 3.9 | 0.98 | 0.24 | 0.49 | 0.49 | 0.24 | 0.12 | 0.98 | 0.98 | 0.49 | 0.98 | 0.49 | 0.49 | 0.49 | 0.98 | 1.95 | 0.98 |
| 2g | 0.49 | 0.49 | 0.49 | 0.49 | 0.49 | 0.49 | 0.12 | 0.12 | 0.24 | 0.12 | 0.12 | 0.49 | 0.49 | 3.9 | 0.24 | 0.98 | ≤0.06 | 1.95 | 0.49 | 0.49 |
| 2h | 1.95 | 0.24 | 0.49 | 1.95 | 0.49 | 0.12 | 0.49 | 0.49 | 0.49 | 0.98 | 0.49 | 0.24 | 0.98 | 0.24 | 0.24 | 0.12 | 0.24 | 0.49 | 1.95 | 1.95 |
| 2i | 0.98 | 0.12 | 0.24 | 0.98 | 0.24 | ≤0.06 | 0.24 | 0.24 | 0.24 | 0.49 | 0.24 | 0.49 | 0.49 | 0.24 | 0.24 | 0.12 | 0.12 | 0.49 | 1.95 | 0.98 |
| 2j | 3.9 | 0.98 | 1.95 | 7.8 | 1.95 | 0.98 | 0.98 | 3.9 | 1.95 | 3.9 | 0.98 | 0.98 | 0.24 | 0.49 | 0.24 | 0.49 | 0.49 | 3.9 | 3.9 | 3.9 |
| 9b | 0.49 | 0.12 | 0.24 | 0.98 | 0.24 | ≤0.06 | 0.24 | 0.24 | 0.24 | 0.49 | 0.24 | 0.12 | 0.24 | 0.12 | 0.24 | 0.12 | 0.12 | 0.49 | 0.98 | 0.98 |
| 9c | 1.95 | 0.98 | 1.95 | 1.95 | 7.8 | 0.49 | 0.98 | 3.9 | 0.98 | 0.98 | 3.9 | 0.98 | 0.49 | 1.95 | 0.49 | 0.49 | 0.49 | 1.95 | 7.8 | 3.9 |
| 9e | 7.8 | 15.6 | 3.9 | 1.95 | 31.3 | 7.8 | 15.6 | 7.8 | 7.8 | 3.9 | 1.95 | 1.95 | 1.95 | 1.95 | 3.9 | 1.95 | 3.9 | 3.9 | 3.9 | 3.9 |
| 9f | 1.95 | 0.98 | 0.98 | 0.98 | 0.98 | 0.49 | 1.95 | 0.98 | 0.98 | 0.98 | 0.49 | 0.49 | 0.24 | 0.49 | 0.49 | 0.98 | 0.49 | 0.98 | 0.98 | 0.98 |
| 9g | 3.9 | 1.95 | 0.98 | 3.9 | 0.98 | 0.49 | 0.49 | 0.49 | 0.49 | 0.98 | 0.49 | 0.24 | 0.24 | 7.8 | 1.95 | 0.98 | 1.95 | 3.9 | 7.8 | 1.95 |
| AmB | 0.98 | 0.98 | 1.95 | 1.95 | 1.95 | 0.98 | 0.98 | 1.95 | 1.95 | 1.95 | 7.8 | 7.8 | 15.6 | 0.98 | 3.9 | 1.95 | 0.49 | 3.9 | 15.6 | 1.95 |
Next, we tested the compounds from the entire series 2 and selected members of series 9 against a panel of ten additional other fungal strains, including three Candida duobushaemulonii strains (AR Bank # 391, AR Bank # 392, and AR Bank # 394), two Candida haemulonii strains (AR Bank # 393 and AR Bank # 395), two Saccharomyces cerevisiae strains (AR Bank # 399 and AR Bank # 400), and one each of the following strains: Kodameae ohmeri (AR Bank # 396), Candida krusei (AR Bank # 397), and Candida lusitaniae (AR Bank # 398) (Table 2). Monohydrazides 2a, 2b, 2e, 2f, 2h, 2i, 9b, and 9f displayed excellent activity (0.06–1.95 μg mL−1) against all ten additional strains tested, whereas the remaining compounds 2c, 2d, 2g, 2j, 9c, 9e, and 9g exhibited excellent to good activity (0.06–7.8 μg mL−1) against all ten strains. Overall, as shown in Table 2 and S2,† the most active monohydrazides displayed excellent activity against a panel of ten C. auris (AR Bank # 381–390) and ten other fungal strains (AR Bank # 391–400).
Structure–activity relationship (SAR) analysis
The identity and the substitution pattern of the substituent(s) on rings A (R1) and B (R2) had a considerable influence on the activity of monohydrazides 1a–9j (summarized in Fig. 2). We first investigated the effect of the R2 substituents while keeping R1 constant (i.e., comparing the monohydrazides within each series). For series 1, we observed that when R1 = 2,4-diF (Fig. 1), the introduction of either 4-OMe (1f), 3-F (1a), or 4-F (1d) as R2 substituents resulted in better antifungal activity than other substituents (i.e., 4-Cl (1e), 3,5-diF (1i), and 3,5-diCl (1j)). In the case of series 2 (R1 = H; Fig. 1), compounds 2a, 2i, 2b, 2g, 2d, and 2e that displayed excellent activity had 3-F, 3,5-diF, 3-Cl, 2,4-diF, 4-F, and 4-Cl as R2 substituents. When comparing series 3 and 4 (R1 = 5-F and 5-Br, respectively; Fig. 1), we observed that the most active monohydrazides in each series (i.e., (3f, 3c, and 3a) and (4c, 4a, and 4f)) had (4-OMe, 3-OMe, and 3-F) and (3-OMe, 3-F, and 4-OMe) as R2 substituents, respectively. For R1 groups = 6-F and 6-Br (series 5 and series 6; Fig. 1), the most active compounds in each series had R2 substituents as 4-OMe (5f), 4-F (5d), and 3-OMe (5c) and 3-OMe (6c), 4-OMe (6f), and 3-F (6a), respectively. In the case of series 7 and 8 (R1 = 3,5-diF and 3,6-diF, respectively; Fig. 1), compounds 7d, 7a, and 7f (from series 7) and compounds 8d, 8f, and 8a (from series 8) that displayed excellent activity had 4-F, 3-F, and 4-OMe as well as 4-F, 4-OMe, and 3-F, as R2 substituents. For series 9 (R1 = H; Fig. 1), the R2 substituents 3-OMe (9c), 4-OMe (9f), 2,4-diF (9g), 3-F (9a), and 4-F (9d) resulted in better activity compared to other substituents (i.e., 3-Cl (9b), 4-Cl (9e), 2,5-diF (9h), 3,5-diF (9i), and 3,5-diCl (9j)). In sum, we observed that in each series (1–9), the most of the active monohydrazides had either 3-F (a), 3-OMe (c), or 4-OMe (f) as R2 substituents.Next, we looked into the effect of varying the R1 substituent while keeping R2 constant (i.e., comparing all “a” compounds 1a–9a, then all “b” compounds 1b–9b, etc.). We found that the monohydrazides displaying the best antifungal activity generally had H (series 2 where X = N and Y = C as well as series 9 where X = N and Y = N) as an R1 substituent. For compounds with R2 = 3-F (a), the most active compounds (from most to least active) were 2a (R1 = H, X = N, Y = C), 9a (R1 = H, X = N, Y = N), 7a (R1 = 3,5-diF), and 8a (R1 = 3,6-diF), respectively. For compounds with R2 = 3-Cl (b), the introduction of H as an R1 substituent resulted in compounds 2b and 9b with a better overall antifungal activity. The most active compounds amongst monohydrazides with R2 = 3-OMe (c), compounds 9c, 2c, 3c, and 5c, had H (X = N, Y = C), H (X = N, Y = N), 5-F, and 6-F as R1 substituents, respectively. For compounds with R2 = 4-F (d), the most active compounds 2d, 8d, 5d, and 9d possessed H (X = N, Y = C), 3,6-diF, 6-F, and H (X = N, Y = N) as R1 substituents, respectively. For compounds with R2 = 4-Cl (e), the most active compounds 2e and 9e possessed H as R1 substituents. For compounds with R2 = 4-OMe (f), the most active compounds had 6-F (5f), H (2f and 9f), 2,4-diF (1f), and 5-F (3f) as R1 substituents. For monohydrazides with R2 = 2,4-diF (g), the presence of H and 5-F as R1 substituents resulted in compounds 9g, 2g, and 3g with better overall antifungal activity than those with other R1 substituents. Similar activity profiles were observed for compounds with R2 = 2,5-diF (h), R2 = 3,5-diF (i), and R2 = 3,5-diCl (j). The introduction of H (X = N, Y = C) and H (X = N, Y = N) as R1 substituents resulted in compounds 2h, 9h, 2i, 9i, 2j, and 9j with better overall activity. In general, we observed that the monohydrazides with the best overall antifungal activity with diverse R2 groups had H (series 2 and 9) as an R1 substituent.
For further in-depth analysis of the antifungal activity, we explored the effect of regioisomers on ring A by comparing series 3 (R1 = 5-F) with series 5 (R1 = 6-F) and series 4 (R1 = 5-Br) with series 6 (R1 = 6-Br). We observed that compounds 5d (R2 = 4-F), 5e (R2 = 4-Cl), and 5f (R2 = 4-OMe) performed better than their counterparts 3d (R2 = 4-F), 3e (R2 = 4-Cl), and 3f (R2 = 4-OMe), whereas compounds 3a (R2 = 3-F) and 3g (R2 = 2,4-diF) displayed better activity compared to 5a (R2 = 3-F) and 5g (R2 = 2,4-diF). While comparing series 4 (R1 = 5-Br) with series 6 (R1 = 6-Br), we found compounds 6d (R2 = 4-F), 6f (R2 = 4-OMe), and 6g (R2 = 2,4-diF) to be better antifungals than their counterparts 4d (R2 = 4-F), 4f (R2 = 4-OMe), and 4g (R2 = 2,4-diF). From the data reported above, we were able to point to the superiority of series 5 and 6 (R1 = 6-F and R1 = 6-Br) over series 3 and 4 (R1 = 5-F and R1 = 5-Br). For disubstituted monohydrazides (series 7 and 8), compounds 7a (R2 = 3-F) and 7f (R2 = 4-OMe) exhibited similar activity to 8a (R2 = 3-F) and 8f (R2 = 4-OMe), whereas compounds 8c (R2 = 3-OMe), 8d (R2 = 4-F), and 8g (R2 = 2,4-diF) were better than their counterparts from series 7. These observations point to fact that having a substituent at the 6-position of ring A substantially increases the activity of the compounds.
We next evaluated the impact of a halogen identity on antifungal activity by comparing series 3 and 4 (5-F vs. 5-Br) as well as series 5 and 6 (6-F vs. 6-Br). For every R2 substituents, compounds from series 3 (3a, 3c, 3d, 3f, and 3g) performed significantly better than compounds from series 4 (4a, 4c, 4d, 4f, and 4g). A similar trend was observed in the case of series 5 and 6. Compounds 5a (R2 = 3-F), 5c (R2 = 3-OMe), 5d (R2 = 4-F), 5f (R2 = 4-OMe), and 5g (R2 = 2,4-diF) performed better than their counterparts 6a (R2 = 3-F), 6c (R2 = 3-OMe), 6d (R2 = 4-F), 6f (R2 = 4-OMe), and 6g (R2 = 2,4-diF). From these observations, we concluded that a fluorine is preferred over a bromine as a R1 substituent for antifungal activity.
Finally, we explored the effect of the position of the R2 substituents on ring B (i.e., 3- vs. 4-position) within each series for the entire nine series of compounds (1avs.1d, 1cvs.1f, and 1bvs.1e, etc.). For monohydrazides where R2 substituents are halogens (3-F (a) and 4-F (d) or 3-Cl (b) and 4-Cl (e)), in general we observed better antifungal activity for 3-position isomers over 4-position isomers. Indeed, for compounds with R2 = 3-F (a) and 4-F (d), 1a, 2a, 3a, 4a, and 6a performed better than their counter parts 1d, 2d, 3d, 4d, and 6d. Similarly, for compounds with R2 = 3-Cl (b) and 4-Cl (e), 1b, 2b, and 9b displayed better antifungal activity than compounds 1e, 2e, and 9e. However, for compounds with R2 = OMe (1c–9c with 3-OMe vs.1f–9f with 4-OMe), the 4-postion isomers were better antifungals when compared to the 3-position isomers. Compounds 1f, 3f, 5f, 7f, and 8f displayed better activity than their counter parts 1c, 3c, 5c, 7c, and 8c. Next, we explored the effect of a halogen identity on ring B (i.e., F (a and d) vs. Cl (b and e)) on antifungal activity. When comparing compounds with R2 = 4-F (d) and R2 = 4-Cl (e), we found that the presence of a fluorine (compounds 1d, 3d, 5d, 7d, 8d, and 9d) is preferred over that of a chlorine atom (1e, 3e, 5e, 7e, 8e, and 9e). However, when comparing compounds with R2 = 3-F and R2 = 3-Cl (1avs.1b, 2avs.2b, and 9avs.9b), we did not observe any distinct differences in antifungal activity.
In vitro cytotoxicity assay
Having established the excellent activity of our monohydrazides against most fungal strains tested, we next considered the toxicity of these compounds towards mammalian cell lines. We investigated the toxicity profile of compounds 2a, 2b, 2d, 2g, 2i, 9a, 9b, 9c, 9f, 9g, as well as controls AmB and VRC (within a concentration range of 0.12–31.3 μg mL−1) against three mammalian cell lines: J774A.1, HEK-293, and HepG2 (Fig. 3). At the highest concentration tested (31.3 μg mL−1), none of the monohydrazides used in this study displayed toxicity against the J774A.1 and HEK-293 cell lines. Against HepG2, compounds 2a, 2b, 2d, 2g, 9b, 9c, 9f, and 9g exhibited no toxicity up to 31.3 μg mL−1. However compounds 2i and 9a displayed minimal toxicity (89% and 94% cell survival, respectively) at a concentration of 31.3 μg mL−1 against HepG2. Considering the excellent antifungal MIC values for these compounds, these cytotoxicity data provides us with a reasonable therapeutic window.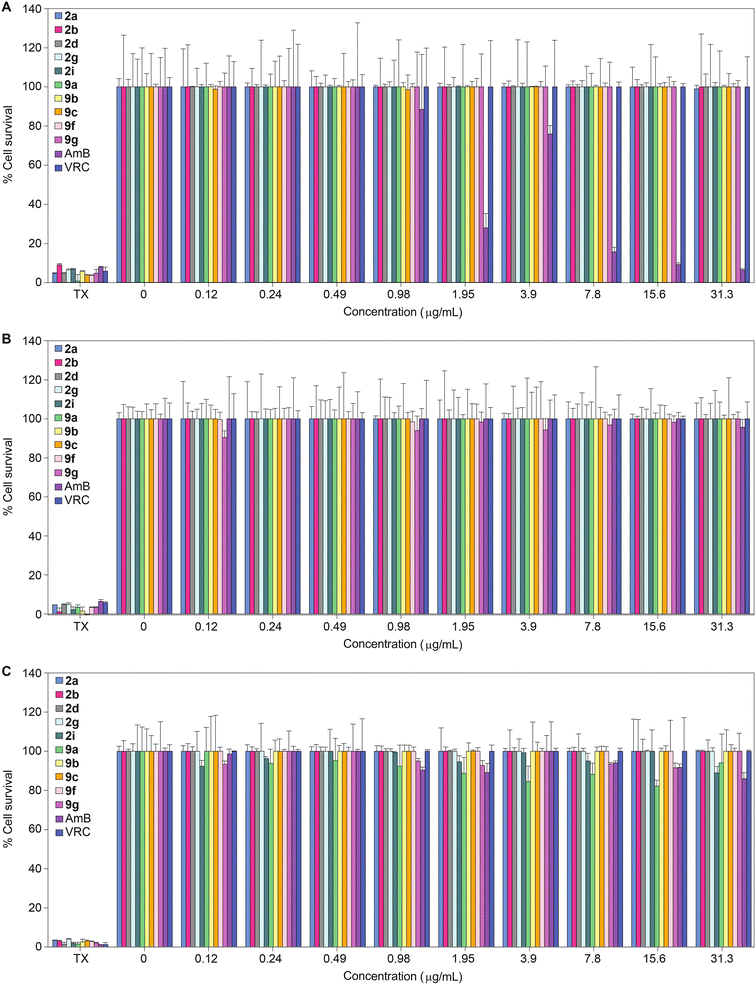 | ||
| Fig. 3 2D bar graphs normalized at 100% depicting the dose-dependent cytotoxic activity of monohydrazides 2a, 2b, 2d, 2g, 2i, 9a, 9b, 9c, 9f, and 9g, as well as AmB and VRC against A. J774A.1, B. HEK-293, and C. HepG2 mammalian cell lines. Note: For Triton X-100® (TX) the eight bars are colored differently and correspond to the colors of the respective compounds for which TX was used as a positive control. Note: The corresponding non-normalized data are presented in Fig. S173.† | ||
Physicochemical properties and in silico ADMET evaluation
A theoretical evaluation of physicochemical and absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties can provide important information regarding the drug likeness of the synthesized monohydrazides. We computed the physicochemical and ADMET properties of potent compounds 2a–2j, 9a–9c, 9f, and 9g using ADMETlab 2.0.54,55 All compounds assessed satisfied the Lipinski's rule of five and had a total polar surface area (TPSA) in the range of 50–77 Å, which suggested good cell membrane permeability. Additionally, the lipophilicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) and solubility (log
P) and solubility (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S) scores for the compounds screened were within the acceptable range (−3.6 to −1.6 for log
S) scores for the compounds screened were within the acceptable range (−3.6 to −1.6 for log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S and 1 to 2.5 for log
S and 1 to 2.5 for log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) (Table S4†). Next, we assessed the effect of these monohydrazides on the heart by virtually screening them for inhibition of the human ether-à-go-go-related (hERG) potassium ion channel. The prediction suggested that none of our compounds would be hERG blockers. When virtually analyzed for hepatotoxicity, the compounds were predicted to be high risk for drug-induced liver injury. The compounds from series 2 (2a–2j) were predicted to be moderately carcinogenic, whereas compounds 9a–9c, 9f, and 9g were predicted to be non-carcinogenic. The in silico predictions revealed that all the compounds screened had acceptable human intestinal absorption (HIA) values and, except for compounds 9a, 9c, and 9g, the remaining compounds were predicted to have higher blood–brain barrier (BBB) penetration. Interestingly, the monohydrazides screened were predicted to be neither P-glycoprotein (Pgp) inhibitors nor Pgp substrates. We also evaluated plasma protein binding (PPB), volume distribution (VD), and clearance (CL) by ADMETlab 2.0. Except for compound 2j, all compounds had a <90% PPB values, suggesting high therapeutic index for these compounds. The predicted VD values for all the compounds were within the optimal range of 0.04–20 L kg−1. As far as CL is concerned, from all the compounds virtually tested, only compounds 2e and 2j were predicted to have low clearance (Table S5†).
P) (Table S4†). Next, we assessed the effect of these monohydrazides on the heart by virtually screening them for inhibition of the human ether-à-go-go-related (hERG) potassium ion channel. The prediction suggested that none of our compounds would be hERG blockers. When virtually analyzed for hepatotoxicity, the compounds were predicted to be high risk for drug-induced liver injury. The compounds from series 2 (2a–2j) were predicted to be moderately carcinogenic, whereas compounds 9a–9c, 9f, and 9g were predicted to be non-carcinogenic. The in silico predictions revealed that all the compounds screened had acceptable human intestinal absorption (HIA) values and, except for compounds 9a, 9c, and 9g, the remaining compounds were predicted to have higher blood–brain barrier (BBB) penetration. Interestingly, the monohydrazides screened were predicted to be neither P-glycoprotein (Pgp) inhibitors nor Pgp substrates. We also evaluated plasma protein binding (PPB), volume distribution (VD), and clearance (CL) by ADMETlab 2.0. Except for compound 2j, all compounds had a <90% PPB values, suggesting high therapeutic index for these compounds. The predicted VD values for all the compounds were within the optimal range of 0.04–20 L kg−1. As far as CL is concerned, from all the compounds virtually tested, only compounds 2e and 2j were predicted to have low clearance (Table S5†).
We also investigated in silico the metabolic properties of potent compounds 2a–2j, 9a–9c, 9f, and 9g using SwissADME against the five isoforms of cytochrome P450 (CYP) monooxygenase (CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4). None of the compounds (with the exception of compound 2j against CYP2C19 and CYP2D6) were identified as possible inhibitors of CYP2C19, CYP2C9, CYP2D6, and CYP3A4. However, except for compound 2i, all compounds were predicted to inhibit CYP1A2 (Table S6†).
Time-kill studies
In order to understand whether the monohydrazides prepared are either fungistatic (inhibit or prevent fungal growth) or fungicidal (kill the fungi), we performed a time-kill assay. We tested compound 2b (one of the most active compounds from MIC studies) at 1× and 4× MIC against C. albicans ATCC 10231 (strain A) to observe the dose-dependent effect on fungal growth (Fig. 4). In addition, we evaluated the control AmB at 0.98 μg mL−1 against the same C. albicans strain A for comparison. We measured the number of colony-forming units (CFU) at 0, 3, 6, 9, 12, 18, and 24 h. We observed that at 4× MIC, compound 2b exhibited a fungistatic behavior.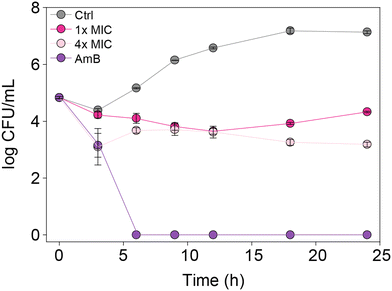 | ||
| Fig. 4 Time-kill curves for C. albicans ATCC 10231 (strain A) with AmB (0.98 μg mL−1, purple), 2b (1× MIC, fushia), 2b (4× MIC, pale pink), and untreated (grey). | ||
Antibiofilm activity
The biofilm is an important virulence contributor for pathogenic fungi, as the biofilm protects the fungi from drugs and shields the pathogens from the immune system.56–58 Biofilms are complex communities of one or more species of microorganisms surrounded by an extracellular matrix, which are not only attached to each other, but also to solid surfaces.59,60 Because of their complex structural features, antifungal agents do not penetrate the matrix well, and they cannot readily reach the pathogens embeded in these multiple layers.61 Thus, biofilms are more resistant to antifungal drugs compared to plankatonic cells.62,63 Since the compounds from series 2 behaved exceptionally well during MIC studies, the ability of compounds 2a–2j, as well as control AmB were assessed against C. albicans ATCC 10231 (strain A) in both destruction of pre-formed biofilms and prevention of biofilm formation. A large amount of fungal cells were exposed to the monohydrazides at time 0 h to evaluate the effect of these compounds in the prevention of Candida biofilm formation (Fig. 5B). Monohydrazides 2g, 2h, 2i, and 2j were not capable of preventing C. albicans ATCC 10231 (strain A) biofilm formation, whereas monohydrazides 2b, 2c, 2d, and 2e exhibited promising activity (1.96–3.9 μg mL−1) against biofilm formation of the same fungal strain. Moderate activity (7.8 μg mL−1) was observed for compounds 2a and 2f in prevention of biofilm formation assays. Amongst all the monohydrazides tested, compounds 2c and 2d were the most promising with their ability to inhibit biomass formation at 1.96 μg mL−1 (8 to 16-fold greated then MIC values). Although some of these monohydrazides were able to prevent biofilm formation, none of these compounds had any effect in terms of the disruption of already pre-formed biofilms (Fig. 5A).Resistance development studies
In order to evaluate the potential of fungi to develop resistance to monohydrazides, we repeatedly exposed C. albicans ATCC 10231 (strain A) to compounds 2a and 2b at ½× MIC to simulate fungal drug exposure in a clinical setting (Fig. 6). The fungi that grew at the highest concentration of compounds (½× MIC) were grown and used as the starting culture for a successive MIC determination. We repeated this assay for a total of 15 MIC determinations (original + 14 exposures). While normal variations in MIC values occured, no significant changes in MIC values were observed as the MIC values remained within 8-fold of the original MIC value. Considering the generally long duration of treatment with antifungal drugs, this is a promising result that suggests that a fungal strain is not likely to develop resistance to the monohydrazides, even after repeated exposures.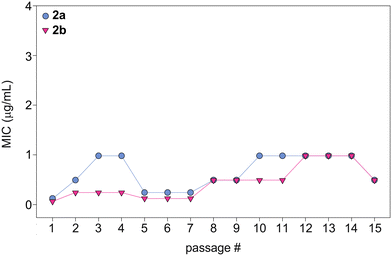 | ||
| Fig. 6 Graph showing the MIC values over 15 passages for compounds 2a (turquoise circles) and 2b (fushia inverted triangles) against C. albicans ATCC 10231 (strain A). | ||
Conclusions
In summary, we synthesized 64 novel substituted monohydrazides (1a–9j) with different R1 and R2 substituents on rings A and B. We performed a detailed antifungal activity study on these compounds against a panel of various C. albicans, non-albicans Candida, and non-Candida fungal strains. From this detailed SAR study, we established that compounds 1f, 2a, 2b, 2c, 2d, 2e, 2f, 2g, 2h, 2i, 2j, 3f, 5d, 5f, 8d, 9b, 9c, 9f, and 9g exhibited broad spectrum activity based on their MIC values. The activity of these compounds was either comparable or superior to AmB against most of the strains tested. The entire series 2, and some of the representative compounds from series 1 and 3–9 were further tested against a panel of ten C. auris and ten other fungal strains. The monohydrazides 2a, 2b, 2g, 2h, 2i, 5d, 8d, 9b, and 9g displayed excellent activity against the ten C. auris strains tested, and compounds 2a, 2b, 2e, 2f, 2h, 2i, 9b, and 9g exhibited excellent activity against the ten other fungal strains. We then explored the safety profile of our monohydrazides by investigating compounds 2a, 2b, 2d, 2g, 2i, 9a, 9b, 9c, 9f, and 9g, as well as controls AmB and VRC against three mammalian cell lines: J774A.1, HEK-293, and HepG2. The entire panel of compounds investigated exhibited minimal to no toxicity against the three different mammalian cell lines tested. We also observed compound 2b to be fungistatic at and/or above its MIC value against C. albicans ATCC 10231 (strain A) in a time-kill assay. Compounds 2a, 2b, 2c, 2d, 2e, 2f, 2g, 2h, 2i, 2j, as well as control AmB were assessed against C. albicans ATCC 10231 (strain A) in both destruction of pre-formed biofilms and prevention of biofilm formation. These monohydrazides were able to prevent the formation of biofilm against strain A. When C. albicans ATCC 10231 (strain A) was repeatedly exposed to the compounds 2a and 2b over 15 passages, no resistance was developed against these compounds. In summary, the novel substituted monohydrazides reported herein show promise as a new family of antifungal agents, and additional studies of these molecules will be reported in due course.Abbreviations
| ADMET | Absorption, distribution, metabolism, excretion, toxicity |
| AmB | Amphotericin B |
| ATCC | American Type Culture Collection |
| BBB | Blood–brain barrier |
| CFG | Caspofungin |
| CFU | Colony-forming unit |
| CL | Clearance |
| FLC | Fluconazole |
| hERG | Human ether-à-go-go-related |
| HIA | Human intestinal absorption |
| IFIs | Invasive fungal infections |
| MIC | Minimum inhibitory concentration |
| Pgp | P-glycoprotein |
| PPB | Plasma protein binding |
| SAR | Structure–activity relationship |
| TPSA | Total polar surface arear |
| TX | Triton X-100® |
| VD | Volume distribution |
| VRC | Voriconazole |
Conflicts of interest
The authors have no conflict of interest to disclose.Acknowledgements
Some NMR data reported in this publication were recorded on a Bruker AVANCE NEO 600 MHz high-performance digital NMR spectrometer supported by a NIH S10 grant S10OD28690 (to S. G.-T.). We thank the University of Kentucky PharmNMR Center (in the College of Pharmacy) for NMR support.References
- G. D. Brown, D. W. Denning and S. M. Levitz, Science, 2012, 336, 647 CrossRef CAS PubMed.
- T. Roemer and D. J. Krysan, Cold Spring Harbor Perspect. Med., 2014, 4, a019703 CrossRef PubMed.
- G. Karavalakis, E. Yannaki and A. Papadopoulou, J. Fungi, 2021, 7, 451 CrossRef CAS PubMed.
- A. Casadevall, Annu. Rev. Immunol., 2022, 40, 121–141 CrossRef PubMed.
- D. Yu and Z. Liu, Front. Microbiol., 2022, 13, 988734 CrossRef PubMed.
- M. Mei-Sheng Riley, Crit. Care Nurs. Clin. N. Am., 2021, 33, 395–405 CrossRef PubMed.
- S. Lakoh, P. S. Kamudumuli, R. O. S. Penney, S. M. Haumba, J. N. Jarvis, A. J. Hassan, N. L. E. Moudoute, B. K. Ocansey, S. Izco, S. Kipkerich, J. Sacarlal, A. T. Awopeju, N. P. Govender, C. I. M. Munyanji, K. Guyguy, E. Orefuwa and D. W. Denning, Lancet Infect. Dis., 2023, 23, 598–608 CrossRef PubMed.
- D. Forno, B. Samayoa, N. Medina, E. Arathoon, C. R. Mejia, R. Gordillo, R. Cedillos, J. Rodas, A. Ahlquist Cleveland, T. Chiller and D. H. Caceres, Mycoses, 2021, 64, 1563–1570 CrossRef CAS PubMed.
- M. C. Fisher, A. Alastruey-Izquierdo, J. Berman, T. Bicanic, E. M. Bignell, P. Bowyer, M. Bromley, R. Bruggemann, G. Garber, O. A. Cornely, S. J. Gurr, T. S. Harrison, E. Kuijper, J. Rhodes, D. C. Sheppard, A. Warris, P. L. White, J. Xu, B. Zwaan and P. E. Verweij, Nat. Rev. Microbiol., 2022, 20, 557–571 CrossRef CAS PubMed.
- A. McDermott, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2217948119 CrossRef CAS PubMed.
- N. Robbins, T. Caplan and L. E. Cowen, Annu. Rev. Microbiol., 2017, 71, 753–775 CrossRef CAS PubMed.
- D. W. Denning, Eur. J. Hosp. Pharm., 2022, 29, 109–112 CrossRef PubMed.
- J. Berman and D. J. Krysan, Nat. Rev. Microbiol., 2020, 18, 319–331 CrossRef CAS PubMed.
- H. Du, J. Bing, T. Hu, C. L. Ennis, C. J. Nobile and G. Huang, PLoS Pathog., 2020, 16, e1008921 CrossRef CAS PubMed.
- E. S. Spivak and K. E. Hanson, J. Clin. Microbiol., 2018, 56, e10588-17 CrossRef PubMed.
- M. Kordalewska and D. S. Perlin, Front. Microbiol., 2019, 10, 1918 CrossRef PubMed.
- K. Forsberg, K. Woodworth, M. Walters, E. L. Berkow, B. Jackson, T. Chiller and S. Vallabhaneni, Med. Mycol., 2019, 57, 1–12 CrossRef PubMed.
- D. W. Eyre, A. E. Sheppard, H. Madder, I. Moir, R. Moroney, T. P. Quan, D. Griffiths, S. George, L. Butcher, M. Morgan, R. Newnham, M. Sunderland, T. Clarke, D. Foster, P. Hoffman, A. M. Borman, E. M. Johnson, G. Moore, C. S. Brown, A. S. Walker, T. E. A. Peto, D. W. Crook and K. J. M. Jeffery, N. Engl. J. Med., 2018, 379, 1322–1331 CrossRef PubMed.
- A. Cortegiani, G. Misseri, A. Giarratano, M. Bassetti and D. Eyre, Crit Care., 2019, 23, 150 CrossRef PubMed.
- B. O'Brien, J. Liang, S. Chaturvedi, J. L. Jacobs and V. Chaturvedi, Lancet Microbe, 2020, 1, e193–e194 CrossRef PubMed.
- Increasing threat of spread of antimicrobial-resistant fungus in healthcare facilities. Centers for Disease Control and Prevention Press release, March 20, 2023.
- N. Robbins, G. D. Wright and L. E. Cowen, Microbiol. Spectrum, 2016, 4(5) DOI:10.1128/microbiolspec.FUNK-0002-2016.
- J. R. Perfect, Nat. Rev. Drug Discovery, 2017, 16, 603–616 CrossRef CAS PubMed.
- Y. Lee, E. Puumala, N. Robbins and L. E. Cowen, Chem. Rev., 2021, 121, 3390–3411 CrossRef CAS PubMed.
- S. E. Murphy and T. Bicanic, Front. Cell. Infect. Microbiol., 2021, 11, 759408 CrossRef CAS PubMed.
- M. Doman and K. Banyai, Front. Microbiol., 2022, 13, 919501 CrossRef PubMed.
- M. Hoenigl, D. Seidel, R. Sprute, C. Cunha, M. Oliverio, G. H. Goldman, A. S. Ibrahim and A. Carvalho, Nat. Microbiol., 2022, 7, 1127–1140 CrossRef CAS PubMed.
- A. A. Shishido, M. Mathew and J. W. Baddley, Curr. Fungal Infect. Rep., 2022, 16, 87–97 CrossRef PubMed.
- A. Amin, A. Vartanian, N. Poladian, A. Voloshko, A. Yegiazaryan, A. L. Al-Kassir and V. Venketaraman, Infect. Dis. Rep., 2021, 13, 1018–1035 CrossRef PubMed.
- C. M. Morel, R. A. Alm, C. Ardal, A. Bandera, G. M. Bruno, E. Carrara, G. L. Colombo, M. E. A. de Kraker, S. Essack, I. Frost, B. Gonzalez-Zorn, H. Goossens, L. Guardabassi, S. Harbarth, P. S. Jorgensen, S. S. Kanj, T. Kostyanev, R. Laxminarayan, F. Leonard, G. L. Hara, M. Mendelson, M. Mikulska, N. T. Mutters, K. Outterson, J. R. Bano, E. Tacconelli, L. Scudeller and G. A.-O. Network, Antimicrob. Resist. Infect. Control., 2020, 9, 187 CrossRef PubMed.
- R. Smith and J. Coast, BMJ, 2013, 346, f1493 CrossRef PubMed.
- E. Rayens and K. A. Norris, Open Forum Infect. Dis., 2022, 9, ofab593 CrossRef PubMed.
- A. M. Rauseo, A. Coler-Reilly, L. Larson and A. Spec, Open Forum Infect. Dis., 2020, 7, ofaa016 CrossRef CAS PubMed.
- J. H. Kim, L. W. Cheng and K. M. Land, Pharmaceuticals, 2022, 15, 787 CrossRef PubMed.
- X. Cui, L. Wang, Y. Lu and C. Yue, J. Infect. Public Health, 2022, 15, 986–1000 CrossRef PubMed.
- N. Thamban Chandrika and S. Garneau-Tsodikova, Chem. Soc. Rev., 2018, 47, 1189–1249 RSC.
- H. X. Ngo, S. Garneau-Tsodikova and K. D. Green, MedChemComm, 2016, 7, 1285–1306 RSC.
- N. Thamban Chandrika and S. Garneau-Tsodikova, MedChemComm, 2016, 7, 50–68 RSC.
- K. C. Howard, E. K. Dennis, D. S. Watt and S. Garneau-Tsodikova, Chem. Soc. Rev., 2020, 49, 2426–2480 RSC.
- N. Thamban Chandrika, S. K. Shrestha, H. X. Ngo and S. Garneau-Tsodikova, Bioorg. Med. Chem., 2016, 24, 3680–3686 CrossRef PubMed.
- N. Thamban Chandrika, S. K. Shrestha, H. X. Ngo, O. V. Tsodikov, K. C. Howard and S. Garneau-Tsodikova, J. Med. Chem., 2018, 61, 158–173 CrossRef CAS PubMed.
- N. Thamban Chandrika, S. K. Shrestha, H. X. Ngo, K. C. Howard and S. Garneau-Tsodikova, Bioorg. Med. Chem., 2018, 26, 573–580 CrossRef CAS PubMed.
- N. Thamban Chandrika, S. K. Shrestha, N. Ranjan, A. Sharma, D. P. Arya and S. Garneau-Tsodikova, ACS Infect. Dis., 2018, 4, 196–207 CrossRef CAS PubMed.
- M. Y. Fosso, S. K. Shrestha, K. D. Green and S. Garneau-Tsodikova, J. Med. Chem., 2015, 58, 9124–9132 CrossRef CAS PubMed.
- S. K. Shrestha, M. Y. Fosso and S. Garneau-Tsodikova, Sci. Rep., 2015, 5, 17070 CrossRef CAS PubMed.
- S. K. Shrestha, M. Y. Fosso, K. D. Green and S. Garneau-Tsodikova, Antimicrob. Agents Chemother., 2015, 59, 4861–4869 CrossRef CAS PubMed.
- S. K. Shrestha, A. Garzan and S. Garneau-Tsodikova, Eur. J. Med. Chem., 2017, 133, 309–318 CrossRef CAS PubMed.
- E. K. Dennis, J. H. Kim, S. Parkin, S. G. Awuah and S. Garneau-Tsodikova, J. Med. Chem., 2020, 63, 2455–2469 CrossRef CAS PubMed.
- H. X. Ngo, S. K. Shrestha and S. Garneau-Tsodikova, ChemMedChem, 2016, 11, 1507–1516 CrossRef CAS PubMed.
- S. Y. L. Holbrook, A. Garzan, E. K. Dennis, S. K. Shrestha and S. Garneau-Tsodikova, Eur. J. Med. Chem., 2017, 139, 12–21 CrossRef CAS PubMed.
- S. K. Shrestha, L. M. Kril, K. D. Green, S. Kwiatkowski, V. M. Sviripa, J. R. Nickell, L. P. Dwoskin, D. S. Watt and S. Garneau-Tsodikova, Bioorg. Med. Chem., 2017, 25, 58–66 CrossRef CAS PubMed.
- N. Thamban Chandrika, E. K. Dennis, S. K. Shrestha, H. X. Ngo, K. D. Green, S. Kwiatkowski, A. G. Deaciuc, L. P. Dwoskin, D. S. Watt and S. Garneau-Tsodikova, Eur. J. Med. Chem., 2019, 164, 273–281 CrossRef CAS PubMed.
- N. Thamban Chandrika, E. K. Dennis, K. R. Brubaker, S. Kwiatkowski, D. S. Watt and S. Garneau-Tsodikova, ChemMedChem, 2021, 16, 124–133 CrossRef CAS PubMed.
- A. Daina, O. Michielin and V. Zoete, Sci. Rep., 2017, 7, 42717 CrossRef PubMed.
- G. Xiong, Z. Wu, J. Yi, L. Fu, Z. Yang, C. Hsieh, M. Yin, X. Zeng, C. Wu, A. Lu, X. Chen, T. Hou and D. Cao, Nucleic Acids Res., 2021, 49, W5–W14 CrossRef CAS PubMed.
- R. Zarnowski, A. Noll, M. G. Chevrette, H. Sanchez, R. Jones, H. Anhalt, J. Fossen, A. Jaromin, C. Currie, J. E. Nett, A. Mitchell and D. R. Andes, Nat. Commun., 2021, 12, 6235 CrossRef CAS PubMed.
- C. B. Costa-Orlandi, J. C. O. Sardi, N. S. Pitangui, H. C. de Oliveira, L. Scorzoni, M. C. Galeane, K. P. Medina-Alarcon, W. Melo, M. Y. Marcelino, J. D. Braz, A. M. Fusco-Almeida and M. J. S. Mendes-Giannini, J. Fungi, 2017, 3, 22 CrossRef PubMed.
- J. F. Kernien, B. D. Snarr, D. C. Sheppard and J. E. Nett, Front. Immunol., 2017, 8, 1968 CrossRef PubMed.
- J. Chandra, D. M. Kuhn, P. K. Mukherjee, L. L. Hoyer, T. McCormick and M. A. Ghannoum, J. Bacteriol., 2001, 183, 5385–5394 CrossRef CAS PubMed.
- C. H. Kowalski, K. A. Morelli, D. Schultz, C. D. Nadell and R. A. Cramer, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 22473–22483 CrossRef CAS PubMed.
- E. Butassi, L. Svetaz, M. C. Carpinella, T. Efferth and S. Zacchino, Antibiotics, 2021, 10, 1053 CrossRef CAS PubMed.
- S. Fanning and A. P. Mitchell, PLoS Pathog., 2012, 8, e1002585 CrossRef CAS PubMed.
- E. Borghi, G. Morace, F. Borgo, R. Rajendran, L. Sherry, C. Nile and G. Ramage, Front. Microbiol., 2015, 6, 1077 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures for the preparation and characterization of compounds 1a–9j as well as MIC value determination by in vitro antifungal, in vitro cytotoxicity, time-kill, biofilm disruption, prevention of biofilm formation, and development of resistance assays. 1H and 13C NMR spectra, HPLC traces, and HR-MS figures for compounds 1a–9j. Tables for experimentally determined MIC values as well as in-silico predictions of physicochemical and ADMET properties. See DOI: https://doi.org/10.1039/d3md00167a |
| ‡ These two authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |