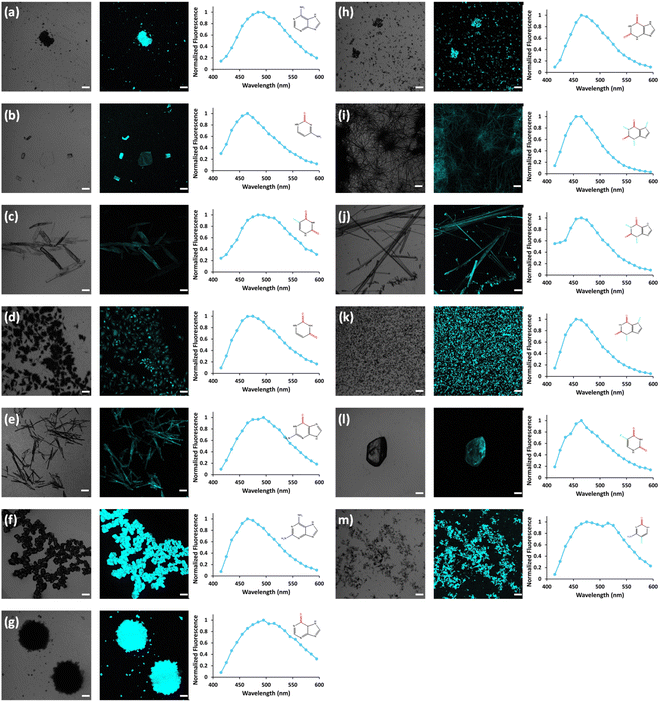
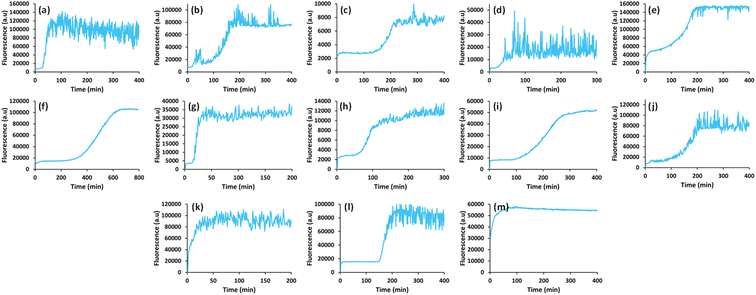
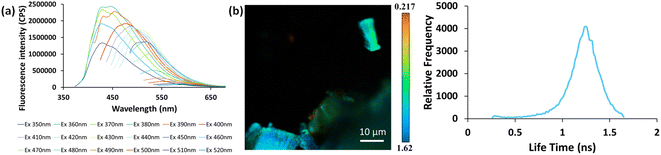
Open Access Article
Open Access ArticleIntrinsic fluorescence of nucleobase crystals†
Ruth
Aizen
 a,
Zohar A.
Arnon
a,
Zohar A.
Arnon
 a,
Or
Berger
a,
Antonella
Ruggiero
b,
Dor
Zaguri
a,
Noam
Brown
a,
Evgeny
Shirshin
cd,
Inna
Slutsky
b and
Ehud
Gazit
*ae
a,
Or
Berger
a,
Antonella
Ruggiero
b,
Dor
Zaguri
a,
Noam
Brown
a,
Evgeny
Shirshin
cd,
Inna
Slutsky
b and
Ehud
Gazit
*ae
aShmunis School of Biomedicine and Cancer Research, Tel Aviv University, Tel Aviv, 6997801, Israel. E-mail: ehudga@tauex.tau.ac.il
bDepartment of Physiology and Pharmacology, Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, 6997801, Israel
cFaculty of Physics, M. V. Lomonosov Moscow State University, Moscow, 119991 Russia
dWorld-Class Research Center “Digital Biodesign and Personalized Healthcare”, I. M. Sechenov First Moscow State Medical University (Sechenov University), 119991 Moscow, Russia
eDepartment of Materials Science and Engineering, Iby and Aladar Fleischman Faculty of Engineering, Tel Aviv University, Tel Aviv 6997801, Israel
First published on 29th November 2022
Abstract
Nucleobase crystals demonstrate unique intrinsic fluorescence properties in the visible spectral range. This is in contrast to their monomeric counterparts. Moreover, some nucleobases were found to exhibit red edge excitation shift. This behavior is uncommon in the field of organic supramolecular materials and could have implications in fields such as therapeutics of metabolic disorders and materials science.
Self-assembly of minimalistic biological and bioinspired molecules has gained tremendous interest in recent years. Ranging from short peptides1–3 to single amino acids4,5 and other metabolites,6 many ordered functional supramolecular structures with unique physical and chemical properties have been explored. In particular, the optical properties displayed by many of these building blocks when self-assembled into well-organized architectures in the solid-state have drawn major attention. Phenomena such as fluorescence, reflectance or nonlinear optics are often dictated by the supramolecular organization of the molecules.7 Photoluminescence,8 two-photon-luminescence9 and waveguiding10 of peptide nanostructures are but a few examples demonstrating the extensive research in this field.
DNA and RNA bases comprise a group of low-molecular-weight molecules with promising potential for nanotechnological applications. This is supported by density functional theory (DFT) calculations which revealed the interesting optical properties of these assemblies.11,12 For example, layered adenine and thymine were shown to have high photoemission efficiency in organic light-emitting diodes (OLEDs).13 Tubular structures composed of the caffeine non-canonical nucleobase exhibit optical wave-guiding tendency.14 Guanine crystals are widely used in the biological world in natural optically-active systems.15,16 The remarkably high refractive index of these crystals allows various organisms to use them as diffuse scatterers, reflectors and photonic crystals.17 Notable examples include the highly reflective tapetum lucidum of sharks,18 crocodiles19 and many other phyla, the silvery scales of koi fish20 and the tuneable colour-changing mechanism employed by sapphirinidae copepods and panther chameleon.21
Peptide nucleic acids (PNAs) are synthetic peptoid polymers composed of an amide backbone decorated with nucleobases as an alternative of the amino acid residues in natural peptides. Previous studies in our group have demonstrated that short PNA sequences can assemble into highly-ordered tuneable photonic crystals22 or crystalline structures showing intrinsic fluorescence and wide excitation wavelength-dependent emission.23–25 Another study demonstrated that amyloid-like structures formed by metabolite aggregates composed of the adenine nucleobase accumulated in metabolic disorders exhibit intrinsic fluorescence in the visible range which can be used to detect and monitor the corresponding disease.26 The intrinsic fluorescence of self-assembled systems has recently gained significant attention, from protein oligomers27 showing blue fluorescence, protein amyloids exhibiting structure-specific fluorescence in the visible range in the absence of aromatic amino acids,28,29 intrinsic red fluorescence in cellulose nanofibers30 and oligosaccharide-based supramolecular structures showing unique intrinsic optical properties,31 to short peptides and even single amino acids with crystallization-induced emission.32,33 Several theories were proposed to explain the possible origin of this phenomenon. Prominent examples include the formation of structure-specific supramolecular fluorophores that permit proton transfer across hydrogen bonds,28,34 electron–hole recombination due to charge transfer between charged amino acids,35 or a mechanism mediated by amino acid oxidation.36 The unique optical properties demonstrated by these materials were shown to have multiple applications ranging from metabolic disorders therapeutics and monitoring,26,36,37 to cell imaging30,32 and optoelectronics.8,13 Although nucleobases have emerged as promising candidates for optical applications and the intrinsic fluorescence of self-assembled natural peptides, amino acids, and amyloid-like metabolite assemblies were reported, as described above, no comprehensive study of this molecular family aiming to examine and compare the luminescence characteristics of each base has been performed. Here, we present an inclusive screen of the fluorescence properties of different nucleobase crystals including the primary DNA and RNA bases as well as modified building blocks. The nucleobase crystals studied here exhibit intrinsic fluorescence in the visible range and excitation wavelength-dependent intrinsic luminescence properties as observed by different characterization methods including fluorescence spectroscopy, confocal microscopy and fluorescence lifetime imaging microscopy (FLIM). We believe that these findings can be useful for various purposes ranging from materials science to biomedical applications and promote the development of new, cost-effective and biocompatible materials for optical devices.
In view of previous findings regarding the luminescent properties of self-assembling PNAs, metabolites and peptide aggregates, we employed a reductionist approach to examine the fluorescence properties of the small components that comprise some of these materials. To explore these avenues, we screened the natural nucleobases as well as non-canonical nucleobases for their optical properties. The nucleobases studied are shown in Scheme S1 (ESI†) and are classified according to their molecular structure as either purines or pyrimidines (Schemes S1a and S2a respectively, ESI†). Next, we examined the morphology and fluorescent properties of each nucleobase in the self-assembled organization. Using confocal microscopy with excitation at 405 nm, we observed that all nucleobase crystals exhibited autofluorescence signals in the visible range without the use of any extrinsic labels. This was also evident when plotting the fluorescence spectra of each nucleobase (Fig. 1). It is important to note, that for all nucleobases, the same acquisition parameters were used. To understand whether the fluorescence is aggregation-induced, we monitored the fluorescence intensity during the crystallization process (using the same conditions outlined above) over time in solution. These experiments were performed under the same set of conditions as previously reported for protein, peptide and metabolite assemblies26,27 (λex = 405 nm, λem = 450 nm) (Fig. 2a–m). Indeed, the fluorescence intensity gradually increased as the monomers self-assembled and the structures were formed. In this context, it is important to note that intrinsic fluorescence of nucleobase monomers in solution was previously reported in the UV range of the spectrum38 and intrinsic fluorescence of nucleobase crystals has never been reported. To quantitatively analyse this phenomenon, fluorescence emission spectra of the crystals were measured. Since PNA assemblies comprised of peptide–nucleobase conjugates were reported to exhibit excitation wavelength-dependent emission,23 we set out to investigate whether nucleobase crystals also present similar properties. Fluorescence emission spectra of the crystals were measured at different excitation wavelengths using solid-state fluorescence spectroscopy. In these experiments, crystals of each nucleobase were deposited on a quartz slide and placed in a desiccator to facilitate water evaporation. The fluorescence spectra of adenine showed emission peaks between 445 nm and 585 nm with the excitation wavelength ranging from 350 nm to 520 nm (Fig. 3a). Moreover, the emission peaks for adenine red-shifted with a longer excitation wavelength. Each nucleobase was found to present a different pattern of excitation-emission dependency, with several nucleobases showing a clear change in fluorescence emission spectra in response to a shift in the excitation wavelength towards the red edge of the absorption band (Fig. S3, ESI†). Fig. S4 (ESI†) shows the epifluorescence images of all nucleobases using different emission/excitation filters. This phenomenon is termed red edge excitation shift (REES)23,39,40 and was suggested to originate from inhomogeneous broadening of the spectra. Inhomogeneous broadening of the spectra (absorbance and emission) occurs when there are many interactions between the fluorophore and surrounding molecules such as in a condensed media or a rigid environment. In these cases, many different environments are possible, with a wide-ranging variation of interaction energies between them. This type of broadening is dependent on solvent polarity and the change of the fluorophore dipole moment on excitation. In a condensed and rigid systems, the relaxation dynamics and reorientation of the environment diploe around the excited state may be slower than the rate of emission and therefore when shining monochromatic light on the sample, a subset of molecules from the inhomogeneous distribution are excited and the emission spectrum is that relevant to the specific subset of photoselected molecules, which are found in a specific relaxed geometry state. Here, we assume that the crystal packing (both hydrogen bonds and pi stacking) creates a polarizable but rigid and condensed matrix which allows the slow relaxation of the dipole environment molecules.39,40 It is clear from observing the different fluorescence spectra of the different nucleobases that the nucleobase crystal 2,6-diaminopurine did not show a shift in emission when exciting with longer wavelengths. 2,6-Diaminopurine, or 2-aminoadenine has an additional amine group when compared to adenine that exhibits a clear shift in emission when exciting with longer wavelengths. This additional amine changes its ability to form hydrogen bonds and therefore the overall crystal packing. It was previously demonstrated that for a system to show the ‘red-edge’ effect’ a rigid environment or condensed lattice packing should be present.39–41 Introducing an additional amine, which is a bulky unit to the purine structure may modulate the system rigidity and therefore prevents the slow relaxation of the solvent that is required for a red shift in emission to take place. Moreover, when exciting guanine, caffeine, and theophylline with shorter wavelengths, we can see that three different peaks are present in the emission spectra. These peaks disappear upon excitation with longer wavelengths and only one peak remains and shifts. We believe that the reason for this change could be the presence of a few polymorphs and their relative abundance in the sample. As mentioned, REES is essentially a site-selective effect, which allows probing the redistribution of fluorophores between different environments. The fact that there are a few polymorphs in the samples may introduce heterogeneity in the sample. Here, it is important to note that heterogeneity can also arise from crystal defects, surface effects such as oxidation and polycrystallinity. In the case of the predominance of a few individual ‘fluorophores’ we see the different peaks. As the wavelength of excitation shift from that corresponding to band maximum further to the red edge, only a small fraction of ‘fluorophores’ is excited (photo-selected). In this case there are no dominating populations of ‘fluorophores’ and the gradual REES is observed.
Fluorescence lifetime is an intrinsic property of a ‘fluorophore’ and provides complementary information to fluorescence spectroscopy measurements.42 In order to measure the lifetime of nucleobases, Fluorescence Lifetime Imaging Microscopy (FLIM) was used to excite each nucleobase crystal with a 780 nm laser using a two-photon confocal microscope. The fluorescence lifetime image and corresponding frequency histogram for adenine are presented in Fig. 3b (left and right respectively), exhibiting unimodal distribution. We further examined the fluorescence lifetime signatures for all nucleobase crystals (Fig. S5, ESI†). Bimodal distributions for a few crystals are evident, such as in the case of theobromine, Fig. S5J (ESI†). Such bimodality can arise from polymorphism, crystallinity, defects in the crystal system, etc. The fluorescence lifetime of all nucleobases studied hereon was in the range of nanoseconds which is typical of fluorescence processes.42 This analysis supports the signal stemming from intrinsic fluorescence.
The results presented here clearly demonstrate the intrinsic fluorescence of nucleobase crystals in the visible range. In the monomeric state however, no fluorescence is detected in this region of the electromagnetic spectrum. As the assembly process progresses, the fluorescence intensity increases, reflecting the formation of supramolecular chromophores. Although the underlying mechanism is still not fully realized and further advanced experiments such as DFT calculations need to be conducted, this inclusive screen of the optical properties of nucleobases provides the starting point for more extensive research. Furthermore, the REES effect exhibited by these crystals is a unique phenomenon that is uncommon in the field of organic supramolecular materials and could have implications in fields such as nanotechnology and materials science.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in part by the Joint NSFC-ISF Grant (Research Grant Application no. 3145/19) (E. G.), and the by the Ministry of Science and Higher Education of the Russian Federation within the framework of state support for the creation and development of World-Class Research Centers “Digital biodesign and personalized healthcare” (agreement no. 075-15-2022-304) (E. S.). The authors thank Dr Sharon Gilead and Dr Sigal Rencus-Lazar for contributing discussions, knowledge and support, Dr David Levy for PXRD measurements and Dr Tal Schwartz for the use of the spectrofluorometer in his laboratory.References
- P. W. J. M. Frederix, G. G. Scott, Y. M. Abul-Haija, D. Kalafatovic, C. G. Pappas, N. Javid, N. T. Hunt, R. V Ulijn and T. Tuttle, Nat. Chem., 2015, 7, 30–37 CrossRef CAS PubMed.
- L. Adler-Abramovich and E. Gazit, Chem. Soc. Rev., 2014, 43, 6881–6893 RSC.
- C. G. Pappas, R. Shafi, I. R. Sasselli, H. Siccardi, T. Wang, V. Narang, R. Abzalimov, N. Wijerathne and R. V. Ulijn, Nat. Nanotechnol., 2016, 11, 960–967 CrossRef CAS PubMed.
- L. Adler-Abramovich, L. Vaks, O. Carny, D. Trudler, A. Magno, A. Caflisch, D. Frenkel and E. Gazit, Nat. Chem. Biol., 2012, 8, 701–706 CrossRef CAS PubMed.
- S. Shaham-Niv, P. Rehak, L. Vuković, L. Adler-Abramovich, P. Král and E. Gazit, Isr. J. Chem., 2017, 57, 729–737 CrossRef CAS.
- S. Shaham-Niv, L. Adler-Abramovich, L. Schnaider and E. Gazit, Sci. Adv., 2015, 1, e1500137 CrossRef PubMed.
- S. I. Stupp, M. U. Pralle, G. N. Tew, L. Li, M. Sayar and E. R. Zubarev, MRS Bull., 2000, 25, 42–48 CrossRef CAS.
- G. Rosenman, N. Amdursky, M. Molotskii, D. Aronov, L. Adler-Abramovich and E. Gazit, Nano Lett., 2009, 9, 3111–3115 CrossRef PubMed.
- S. Semin, A. Van Etteger, L. Cattaneo, N. Amdursky, L. Kulyuk, S. Lavrov, A. Sigov, E. Mishina, G. Rosenman and T. Rasing, Small, 2015, 11, 1156–1160 CrossRef CAS PubMed.
- X. Yan, J. Li and H. Möhwald, Adv. Mater., 2011, 23, 2796–2801 CrossRef CAS PubMed.
- A. Alparone, Chem. Phys., 2013, 410, 90–98 CrossRef CAS.
- F. F. Maia, V. N. Freire, E. W. S. Caetano, D. L. Azevedo, F. A. M. Sales and E. L. Albuquerque, J. Chem. Phys., 2011, 134, 05B601 CrossRef PubMed.
- E. F. Gomez, V. Venkatraman, J. G. Grote and A. J. Steckl, Sci. Rep., 2014, 4, 1–5 Search PubMed.
- N. Chandrasekhar, E. R. Reddy, M. D. Prasad, M. S. Rajadurai and R. Chandrasekar, CrystEngComm, 2014, 16, 4696–4700 RSC.
- R. Aizen, K. Tao, S. Rencus-Lazar and E. Gazit, J. Nanopart. Res., 2018, 20, 125 CrossRef PubMed.
- D. Gur, B. A. Palmer, S. Weiner and L. Addadi, Adv. Funct. Mater., 2017, 27, 1603514 CrossRef.
- B. A. Palmer, D. Gur, S. Weiner, L. Addadi and D. Oron, Adv. Mater., 2018, 30, 1800006 CrossRef PubMed.
- C. R. Braekevelt, Anat. Embryol., 1994, 190, 591–596 CAS.
- C. E. Dieterich and H. J. Dieterich, Graefe's Arch. Clin. Exp. Ophthalmol., 1978, 208, 159–198 CrossRef CAS PubMed.
- D. Gur, B. Leshem, D. Oron, S. Weiner and L. Addadi, J. Am. Chem. Soc., 2014, 136, 17236–17242 CrossRef CAS PubMed.
- J. Teyssier, S. V. Saenko, D. van der Marel and M. C. Milinkovitch, Nat. Commun., 2015, 6, 6368 CrossRef CAS PubMed.
- O. Berger, E. Yoskovitz, L. Adler-Abramovich and E. Gazit, Adv. Mater., 2016, 28, 2195–2200 CrossRef CAS PubMed.
- O. Berger, L. Adler-Abramovich, M. Levy-Sakin, A. Grunwald, Y. Liebes-Peer, M. Bachar, L. Buzhansky, E. Mossou, V. T. Forsyth, T. Schwartz, Y. Ebenstein, F. Frolow, L. J. W. Shimon, F. Patolsky and E. Gazit, Nat. Nanotechnol., 2015, 10, 353–360 CrossRef CAS PubMed.
- O. Berger and E. Gazit, Biopolymers, 2017, 108, e22930 CrossRef PubMed.
- Z. A. Arnon, O. Berger, R. Aizen, K. Hannes, N. Brown, L. J. W. Shimon and E. Gazit, Small Methods, 2019, 3, 1900179 CrossRef CAS.
- S. Shaham-Niv, Z. A. Arnon, D. Sade, A. Lichtenstein, E. A. Shirshin, S. Kolusheva and E. Gazit, Angew. Chem., Int. Ed., 2018, 57, 12444–12447 CrossRef CAS PubMed.
- A. Bhattacharya, S. Bhowmik, A. K. Singh, P. Kodgire, A. K. Das and T. K. Mukherjee, Langmuir, 2017, 33, 10606–10615 CrossRef CAS PubMed.
- D. Pinotsi, L. Grisanti, P. Mahou, R. Gebauer, C. F. Kaminski, A. Hassanali and G. S. Kaminski Schierle, J. Am. Chem. Soc., 2016, 138, 3046–3057 CrossRef CAS PubMed.
- L. Grisanti, M. Sapunar, A. Hassanali and N. Došlić, J. Am. Chem. Soc., 2020, 142, 18042–18049 CrossRef CAS PubMed.
- A. Khalid, L. Zhang, J. P. Tetienne, A. N. Abraham, A. Poddar, R. Shukla, W. Shen and S. Tomljenovic-Hanic, APL Photonics, 2019, 4, 020803 CrossRef.
- Y. Yu, S. Gim, D. Kim, Z. A. Arnon, E. Gazit, P. H. Seeberger and M. Delbianco, J. Am. Chem. Soc., 2019, 141, 4833–4838 CrossRef CAS PubMed.
- R. Ravanfar, C. J. Bayles and A. Abbaspourrad, Cryst. Growth Des., 2020, 20, 1673–1680 CrossRef CAS.
- Z. A. Arnon, T. Kreiser, B. Yakimov, N. Brown, R. Aizen, S. Shaham-Niv, P. Makam, M. N. Qaisrani, E. Poli, A. Ruggiero, I. Slutsky, A. Hassanali, E. Shirshin, D. Levy and E. Gazit, iScience, 2021, 24, 102695 CrossRef CAS PubMed.
- A. D. Stephens, M. N. Qaisrani, M. T. Ruggiero, G. D. Mirón, U. N. Morzan, M. C. González Lebrero, S. T. E. Jones, E. Poli, A. D. Bond, P. J. Woodhams, E. M. Kleist, L. Grisanti, R. Gebauer, J. A. Zeitler, D. Credgington, A. Hassanali and G. S. Kaminski Schierle, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2020389118 CrossRef CAS PubMed.
- S. Prasad, I. Mandal, S. Singh, A. Paul, B. Mandal, R. Venkatramani and R. Swaminathan, Chem. Sci., 2017, 8, 5416–5433 RSC.
- T. N. Tikhonova, N. R. Rovnyagina, A. Y. Zherebker, N. N. Sluchanko, A. A. Rubekina, A. S. Orekhov, E. N. Nikolaev, V. V. Fadeev, V. N. Uversky and E. A. Shirshin, Arch. Biochem. Biophys., 2018, 651, 13–20 CrossRef CAS PubMed.
- C. Wei Chung, A. D. Stephens, E. Ward, Y. Feng, M. Jo Davis, C. F. Kaminski and G. S. Kaminski Schierle, Anal. Chem., 2022, 94, 5367–5374 CrossRef PubMed.
- M. Daniels and W. Hauswirt, Science, 1971, 171, 675–677 CrossRef CAS PubMed.
- A. P. Demchenko, Luminescence, 2002, 17, 19–42 CrossRef CAS PubMed.
- W. C. Galley and R. M. Purkey, Proc. Natl. Acad. Sci. U. S. A., 1970, 67, 1116–1121 CrossRef CAS PubMed.
- C. Sissa, A. Painelli, M. Blanchard-Desce and F. Terenziani, J. Phys. Chem. B, 2011, 115, 7009–7020 CrossRef CAS PubMed.
- A. Jain, C. Blum and V. Subramaniam, Fluorescence lifetime spectroscopy and imaging of visible fluorescent proteins, Elsevier, 2009, ch. 4, pp. 147–176 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2na00551d |
| This journal is © The Royal Society of Chemistry 2023 |