Open Access Article
Open Access ArticleLarge-scale production of MXenes as nanoknives for antibacterial application†
Yuchen
Liu
ab,
Xing
Chen
c,
Jiazhi
Sun
c,
Nuo
Xu
a,
Qi
Tang
a,
Jie
Ren
a,
Cheng
Chen
*a,
Weiwei
Lei
 *b,
Chao
Zhang
a and
Dan
Liu
*b
*b,
Chao
Zhang
a and
Dan
Liu
*b
aSchool of Resources and Environment, Anhui Agricultural University, 130 Changjiang West Road, Hefei, 230036, Anhui, China. E-mail: chc@ahau.edu.cn
bInstitute for Frontier Materials, Deakin University, Locked Bag 2000, Geelong, Victoria 3220, Australia. E-mail: weiwei.lei@deakin.edu.au; dan.liu@deakin.edu.au
cKey Laboratory of Integrated Crop Pest Management of Anhui Province, School of Plant Protection, Anhui Agricultural University, Hefei 230036, China
First published on 3rd November 2023
Abstract
Antimicrobial resistance of existing antibacterial agents has become a pressing issue for human health and demands effective antimicrobials beyond conventional antibacterial mechanisms. Two-dimensional (2D) nanomaterials have attracted considerable interest for this purpose. However, obtaining a high yield of 2D nanomaterials with a designed morphology for effective antibacterial activity remains exceptionally challenging. In this study, an efficient one-step mechanical exfoliation (ECO-ME) method has been developed for rapidly preparing Ti3C2 MXenes with a concentration of up to 30 mg mL−1. This synthetic pathway involving mechanical force endows E-Ti3C2 MXene prepared by the ECO-ME method with numerous irregular sharp edges, resulting in a unique nanoknife effect that can successfully disrupt the bacterial cell wall, demonstrating better antibacterial activity than the MXenes prepared by conventional wet chemical etching methods. Overall, this study provides a simple and effective method for preparing MXenes on a large scale, and its antibacterial effects demonstrate great potential for E-Ti3C2 in environmental and biomedical applications.
Introduction
Bacteria are ubiquitous in the environment and profoundly influence human activity. Antimicrobials are the most effective tools for intervening in the consequences of bacterial infestation. However, the abuse and overuse of antimicrobials have led to antimicrobial resistance (AMR), which has seriously endangered the efficacy of antimicrobials, leading to recalcitrant bacterial infections and has emerged as one of the most pressing issues in global public health.1–4 Most conventional antimicrobials originate from natural sources and primarily act on processes essential for bacterial growth and/or survival, including the production of DNA, RNA, or essential proteins.5 Moreover, through prolonged exposure, bacteria have evolved the ability to evade the activity of existing antimicrobials, requiring the use of multiple or high doses for bacterial eradication.6 The spread of this acquired resistance will render existing antimicrobials ineffective; therefore, there is an urgent requirement for developing new antimicrobial agents.Two-dimensional (2D) nanomaterials have exhibited extraordinary potential for numerous applications since the discovery of graphene in 2004, particularly in the field of antimicrobial agents.7–14 Impressively, 2D nanomaterials represent a novel antimicrobial modality for bacteria and have been extensively explored for environmental and biomedical applications, such as water treatment, wound infection healing, and food packaging.15–22 Compared to conventional antimicrobials, 2D nanomaterials can circumvent the existing known resistance mechanisms and exhibit less susceptibility to the development of selective resistance.23 However, the antibacterial mechanisms of 2D nanomaterials vary with their size, shape, and surface chemistry, resulting in various processes and outcomes of interactions with bacteria, which significantly influence their antibacterial activity. Therefore, an in-depth investigation and understanding of the antibacterial properties and action mechanisms of different nanomaterials remain challenging.
MXene, a novel 2D nanomaterial, has been discovered with the general formula Mn+1XnTx, where M represents an early transition metal, X denotes carbon and/or nitrogen, and T indicates various terminals, typically hydroxyl, oxygen, and fluorine groups.24–29 MXenes have demonstrated outstanding biocompatibility, as the main elements (carbon and nitride) are essential while some early transition metals, such as Ti and Nb, are relatively inert in biological organisms.30–33 MXenes show flexible functionalization owing to their diverse and tunable surface, while their high hydrophilicity enables direct use in biomedical applications without complex surface hydrophilic modifications.26,34,35 However, conventional approaches to MXene preparation have some inevitable inherent limitations, such as unbearably long fabrication times, high temperatures and high pressures, as well as costly materials.36–38 Therefore, insufficient MXene production further hinders its development as a bacteriostatic agent.
Mechanical exfoliation conducted through ball milling is a well-established, efficient, and operational method for 2D nanomaterial preparation and has been successfully used to produce numerous 2D nanomaterials including graphene, boron nitride nanosheets, and transition metal dichalcogenides.39–42 Although mechanical exfoliation has been proven to be an effective approach for preparing 2D nanomaterials, the presence of strong M–A bonds in MXene precursors, which are unlike other 2D materials, makes them almost impossible to obtain by simple mechanical exfoliation. Existing strategies for MXene preparation through mechanical exfoliation require a multi-step process,43–46 which fails to improve the preparation efficiency and limits the possibility of subsequent large-scale applications. Therefore, it is necessary to develop a one-step mechanical exfoliation approach to prepare MXenes using a facile and efficient process.
Herein, an efficient one-step mechanical exfoliation (ECO-ME) method has been developed for the rapid preparation of MXenes, which significantly reduces the preparation time and energy consumption while maintaining comparable yields with conventional wet chemical etching methods. The prepared Ti3C2 MXene (E-Ti3C2) exhibited excellent hydrophilic properties, enabling E-Ti3C2 to be easily dispersed in heated agar aqueous solutions and uniformly distributed in the hydrogels. E-Ti3C2 demonstrated excellent antibacterial activity that enabled significant inhibition against Escherichia coli (E. coli), therefore, a mechanism for the antibacterial activity of E-Ti3C2 was proposed. Owing to the irregular shape and numerous sharp edges, which are attributed to the presence of mechanical forces, the nanosheets present a distinct nanoknife effect, leading to antibacterial activity by disrupting the bacterial cell wall. Such characteristics also endow E-Ti3C2 with superior antibacterial activity compared to MXenes prepared by the conventional wet chemical etching method. Furthermore, to validate the antibacterial mechanism, genetic engineering techniques were used to manipulate the overexpression of wall synthesis genes murA in E. coli to increase the cell wall thickness. For E. coli with thickened cell walls, the inhibitory effect of E-Ti3C2 was obviously reduced, thus confirming that E-Ti3C2 achieves a bactericidal effect primarily through the physical disruption of bacterial cell walls. This unique inhibitory behaviour makes E-Ti3C2 a promising candidate to combat the potential effects that may be caused by bacteria from the environment, while also providing new insights for the subsequent development of bactericides.
Experimental section
Materials
The MAX phase precursor Ti3AlC2 (400 mesh, ≥98%) was provided by 11 Technology Co., Ltd, China. Hydrochloric acid (HCl, 36–38%, AR) was purchased from Xilong Scientific Co., Ltd, China. Lithium fluoride (LiF, 99%, AR) was purchased from Shanghai Aladdin Biochemical Technology Co., Ltd, China. Sodium chloride (NaCl, AR) was purchased from Sinopharm Chemical Reagent Co., Ltd. Phosphate buffered saline (PBS, pH = 7.2–7.6) was provided by Sangon Biotech Co., Ltd (Shanghai, China). Agar was supplied by Rhawn Chemical Co., Ltd. Yeast extract (total nitrogen ≥9.0%, amino nitrogen ≥3.0%) and tryptone (total nitrogen ≥12.5%, amino nitrogen ≥2.5%) were purchased from Labgic Technology Co., Ltd. Lysozyme (BR, from chicken egg white, ≥20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 units per mg protein) was provided by Shanghai Yuanye Biotechnology Co., Ltd. All chemicals were used as received.
000 units per mg protein) was provided by Shanghai Yuanye Biotechnology Co., Ltd. All chemicals were used as received.
Synthesis of E-Ti3C2 MXene
E-Ti3C2 was obtained by the ECO-ME method. Specifically, 1 g of the pristine MAX phase precursor, Ti3AlC2, 1.6 g of lithium fluoride, and 20 mL of 9 M hydrochloric acid were added directly to a ball mill jar with a polytetrafluoroethylene (PTFE) liner. A ball-milling jar with an atmosphere protection device controlled the ball-milling atmosphere with nitrogen at atmospheric pressure. The ball mill jar was pumped to vacuum, then filled with nitrogen and purged three times; finally, the ball mill atmosphere was maintained with nitrogen. The milling ball material was chosen as silicon carbide (SiC), with a diameter of 8 mm and controlled ball-to-MAX mass ratio of 8.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. After ball milling for 2 h, the mixture was washed with deionized water using a centrifuge model H4-21KR (KeCheng, China) at 10
1. After ball milling for 2 h, the mixture was washed with deionized water using a centrifuge model H4-21KR (KeCheng, China) at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min per cycle, with a washing water volume control at 200 mL until the final pH reached approximately 7. After redispersing the sediment in water through mild manual shaking, E-Ti3C2 was collected by centrifugation at 3000 rpm for 30 min to remove the unexfoliated bulk material. Thereafter, stable aqueous solutions of E-Ti3C2 were obtained by repeated centrifugation until the final supernatant became transparent. The final yield was approximately 70%. For the high-concentration E-Ti3C2 solution, the obtained E-Ti3C2 solution was further centrifuged at 10
000 rpm for 30 min per cycle, with a washing water volume control at 200 mL until the final pH reached approximately 7. After redispersing the sediment in water through mild manual shaking, E-Ti3C2 was collected by centrifugation at 3000 rpm for 30 min to remove the unexfoliated bulk material. Thereafter, stable aqueous solutions of E-Ti3C2 were obtained by repeated centrifugation until the final supernatant became transparent. The final yield was approximately 70%. For the high-concentration E-Ti3C2 solution, the obtained E-Ti3C2 solution was further centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 1 h, the sediment was retained, and the appropriate volume of water was added and redispersed by manual shaking to obtain a high-concentration E-Ti3C2 dispersion.
000 rpm for 1 h, the sediment was retained, and the appropriate volume of water was added and redispersed by manual shaking to obtain a high-concentration E-Ti3C2 dispersion.
Synthesis of W-Ti3C2 MXene
Ti3C2 MXene (W-Ti3C2), used as a comparison, was prepared by the conventional wet chemical etching method.47 In brief, 1 g Ti3AlC2, 1.6 g lithium fluoride and 20 mL 9 M hydrochloric acid are added to a PTFE beaker, then stirred at 300 rpm for 30 h at 50 °C. The following centrifugal washing and collection steps are the same as those described for E-Ti3C2.Synthesis of agar/E-Ti3C2 MXene hydrogels
An agar/E-Ti3C2 hydrogel containing 1.5 wt% agar was obtained through a solution mixing method. Briefly, 1.5 g of agar powder was poured into 100 mL of deionized water and then heated to 95 °C and stirred at 1000 rpm for 1 h until the agar was completely dissolved. When the temperature of the agar solution was cooled to approximately 65 °C, the required amount of E-Ti3C2 solution was added to the agar solution and stirred at 150 rpm for 2 min. The resulting mixture was poured into a polymethyl methacrylate (acrylic) cube with a side length of 4.5 cm and then placed at 5 °C for rapid cooling until agar gelled to obtain the agar/E-Ti3C2 hydrogel.Hydrophilic measurements of E-Ti3C2
The hydrophilicity of E-Ti3C2 was investigated by measuring its contact angle on a nylon substrate. A certain amount of an aqueous E-Ti3C2 solution was filtered onto a nylon substrate with a pore size of 0.22 μm and a diameter of 47 mm using a facile vacuum filtration method, and the water contact angle of E-Ti3C2 was then measured.Assessment of E-Ti3C2 antibacterial activity
Wild-type E. coli and E. coli with thickened cell walls obtained by overexpressing the cell wall synthesis gene murA through genetic engineering were used in the experiments. Luria–Bertani (LB) medium was employed to provide the necessary conditions for bacterial growth, which contains 1.5 wt% agar and nutrients. Briefly, 10 g tryptone, 5 g yeast extract, 10 g sodium chloride and 15 g agar were mixed with 1 L sterilized water, and the pH was subsequently equilibrated to 7.2 ± 0.2. The mixture was then divided into appropriate volumes and autoclaved at 121 °C for 15 min. After the LB medium had cooled down to approximately 60 °C, different concentrations of E-Ti3C2 and W-Ti3C2 solutions were then added, and the mixture was subsequently poured into a 9 cm diameter Petri dish. LB medium without MXene was used as a control. 100 μL (OD600 nm = 0.6) of wild-type E. coli and E. coli with thickened cell walls were inoculated onto the medium and colonies were counted after 12 hours of incubation at 37 °C.LB liquid medium was prepared to measure bacterial growth, and the composition and preparation process were similar to those of the LB medium described above, except for the absence of agar. After autoclaving, 20 mL of the resulting LB liquid medium was dispensed into 50 mL sterile tubes. LB liquid medium containing a final concentration of 600 μg mL−1 E-Ti3C2 and an equal volume of water was added as a control. Next, 100 μL of bacterial suspension with OD600 nm = 0.6 was inoculated into the media and then placed in a shaking incubator at 180 rpm, 37 °C. The optical density at a wavelength of 600 nm was measured using a spectrophotometer. All experiments were performed in triplicate.
Confocal microscopy to investigate the interaction between bacteria and E-Ti3C2
To observe the interaction between E-Ti3C2 and E. coli, 0.25 mL of E. coli suspension at a density of 106 CFU mL−1 was withdrawn and washed thrice using 0.2 M PBS buffer at 5000 rpm for 3 min per cycle. After resuspension in PBS buffer, 100 μL of E-Ti3C2 solution (600 μg mL−1) was added and continuously shaken at 180 rpm for 1 h at 37 °C and then observed using a confocal laser scanning microscope.Evaluation of the bacterial cell wall thickness using lysozyme
100 μL bacterial suspensions of wild-type strain and E. coli strain with thickened cell walls at a concentration of 106 CFU mL−1 were spread on LB agar plates. The filter paper was then trimmed into discs with a diameter of 6 mm and placed onto the surface of LB agar plates. Subsequently, 10 μL of lysozyme (100 mg mL−1) was dropped onto the surface of the filter discs and incubated at 37 °C overnight. Equal amounts of water were also dropped onto a filter paper as a control. The diameters of the lysis halos were measured.Characterization
XRD analysis of E-Ti3C2 was performed using a Rigaku Ultima IV powder diffractometer with Cu-Kα radiation, with a step width of 0.02° at 0.6 s per step in the 2θ range from 5–65° (Rigaku Co., Japan). The morphology and structure of E-Ti3C2 were observed using a Hitachi S4800 SEM instrument with an acceleration voltage of 2 kV (Hitachi Co., Japan). A Hitachi HT7700 TEM with an acceleration voltage of 80 kV was used to further reveal the morphology of E-Ti3C2 (Hitachi Co., Japan). Chemical binding component and chemical state analyses of E-Ti3C2 were performed using a K-Alpha XPS (Thermo Fisher Scientific Inc., USA). The contact angle was measured using a Sindin SDC-200S system (Shengding Instrument Co., Ltd, China). FTIR spectra were obtained using a Nicolet IS50 FTIR spectrometer (Thermo Fisher Scientific Inc., USA). The size distribution and zeta potential were obtained from a Zetasizer Nano ZS (Malvern Panalytical, Ltd, Malvern, UK). Bacterial growth was measured at an optical density of 600 nm using a microplate spectrophotometer (Multiskan GO, Thermo Fisher Scientific Inc., USA). A Nikon TI2-S-HU confocal laser scanning microscope was used to investigate the interaction of E-Ti3C2 nanosheets with bacteria (Nikon Co., Japan).Results and discussion
Characterization of MXene prepared through the ECO-ME method
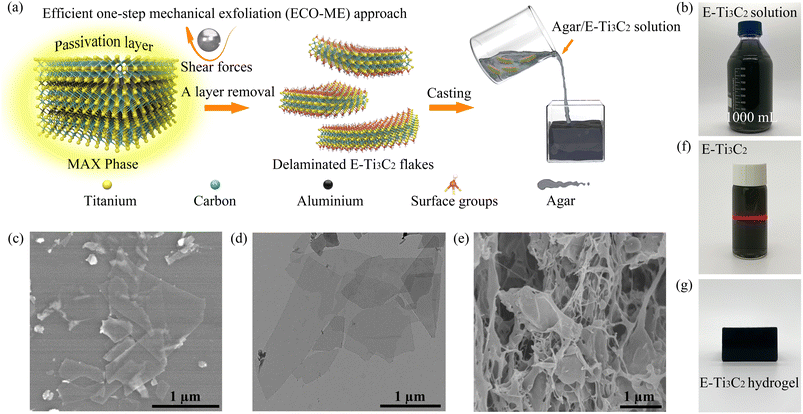
Ti3C2 MXene, the most investigated member of the MXene family, and its corresponding precursor Ti3AlC2 were selected as raw materials, and the mechanical forces provided by a ball mill were engaged in the entire preparation process for the rapid preparation of MXenes, as illustrated in Fig. 1a. In general, the presence of strong M–A bonds prevents direct acquisition of MXene from the original MAX phase by simple mechanical exfoliation, often making it necessary to divide the preparation process into a mechanical exfoliation process and an etching process.43,44 In such processes, mechanical exfoliation typically acts to reduce the size of the MAX phase particles for accelerating subsequent etching, or to peel the etched accordion-shaped MXene into individual sheets. This multistep strategy increases the operational complexity while affecting the efficiency of preparation and increasing the risk to the operator. Furthermore, the choice of etchant significantly influences the morphology and purity of the final product.45,46In the ECO-ME approach, an acid- and alkali-resistant polytetrafluoroethylene (PTFE) lining is employed to isolate the reactant mixture from direct contact with the ball mill jar, thereby avoiding damage to the equipment and preventing contamination of MXene with substances released by corrosion of the ball mill jar.48 Owing to the remarkable hardness and suitable density of silicon carbide (SiC), it was selected as the material for milling balls, which can endure impact during ball milling while offering sufficient shear forces, in addition to high acid resistance, which is essential.49 As a result of the configuration and material selection of the milling balls and jars, corrosion and potential product contamination are avoided; thus, an effective acid etchant can be used in the ECO-ME approach. Acid etchants are considerably more effective than alkaline etchants in removing the Al atomic layer from the pristine MAX phase while preventing the formation of new Al(OH)3 and AlO(OH) passivation layers.50 The liquid phase milling environment minimizes fragmentation forces during the ball milling process, thereby maintaining the lateral size and structural integrity of the product. The nitrogen milling atmosphere was employed to inhibit MXene oxidation from the edges during the preparation process.51 Furthermore, since mechanical forces are involved to accelerate the preparation process, the ECO-ME method is more energy efficient, with only 4.6% power consumption of the conventional wet chemical etching method, as shown in Table S1.†
The ECO-ME method has demonstrated the ability to prepare MXenes at high yields and with high efficiencies. The high-speed motion of the SiC milling balls first breaks the passivation layer wrapped around the MAX particles, leaving the Al atomic layer exposed to the etchant, while the shear force provided by the milling balls quickly peels off the etched E-Ti3C2, leaving the active site continuously exposed. In contrast to conventional wet chemical etching methods, the ECO-ME method maintains high yields but significantly reduces fabrication time with the possibility of obtaining aqueous MXene solutions at high concentrations (>30 mg mL−1) by subsequent centrifugation. As shown in Fig. 1b, an aqueous E-Ti3C2 solution was prepared on a large scale using the ECO-ME method. Scanning electron microscopy (SEM) revealed that the morphology of E-Ti3C2 was transformed from the bulk phase morphology of the original MAX phase (Fig. S1†) into a sheet-like structure with distinct two-dimensional features after ECO-ME treatment, while maintaining the integrity of the two-dimensional structure, as shown in Fig. 1c. Transmission electron microscopy (TEM) (Fig. 1d) further revealed the morphology of freestanding E-Ti3C2, which showed distinct two-dimensional features while possessing a small thickness. The size distribution of the individual E-Ti3C2 nanosheets prepared using the ECO-ME method has a diameter of approximately 0.5 to 1.5 μm, as illustrated in Fig. S2.†
E-Ti3C2 possesses an excellent hydrophilic nature with a water contact angle of 54.2° as measured on a nylon substrate (Fig. S3†) and the zeta potential is measured at −44.6 mV; thus, E-Ti3C2 is capable of dispersing homogeneously in water to form a stable colloid and the typical Tyndall effect of E-Ti3C2, as displayed in Fig. 1f. Meanwhile, the high hydrophilicity of E-Ti3C2 allowed it to be homogeneously dispersed in heated and completely dissolved aqueous agar solution; after sufficient stirring and subsequent drop casting in a polymethyl methacrylate (acrylic) cube, agar/MXene hydrogels (1.5 wt% agar content) with specific shapes could be obtained, as shown in Fig. 1g. The SEM image of the agar/E-Ti3C2 hydrogels after freeze-drying revealed its microstructure, wherein E-Ti3C2 was uniformly distributed throughout the hydrogel and was fully incorporated into its structure (Fig. 1e).
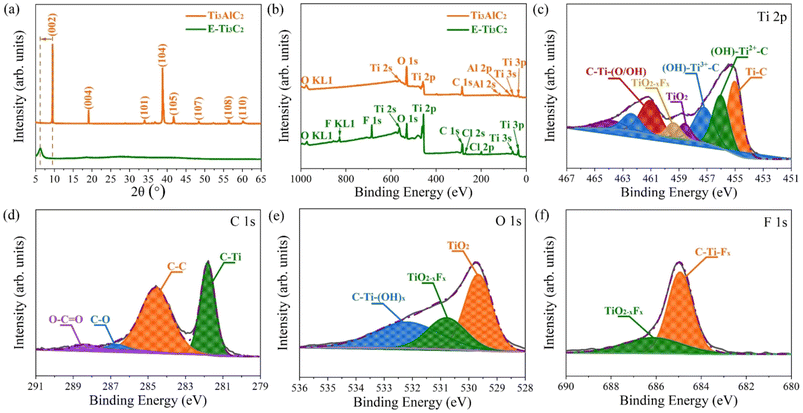
To further confirm that the MXene has been successfully prepared, the pristine MAX phase and E-Ti3C2 were investigated by X-ray diffraction (XRD), as shown in Fig. 2a. The XRD pattern of the pristine MAX phase showed several characteristic peaks including (002), (004), (101), (104), (105), (107), (108), and (110); however, after ECO-ME processing, all peaks except for (002) disappeared, indicating that the MAX phase was almost completely transformed into MXene. The characteristic peak (002) shifted from 9.49° to 6.24° with weakening and broadening, demonstrating the structural expansion and larger d-spacing caused by extraction of the Al atomic layer from the MAX phase and confirming the formation of E-Ti3C2. X-ray photoelectron spectroscopy (XPS) demonstrated the removal of the Al atomic layer. Titanium, carbon, oxygen, and aluminium were detected in the original MAX phase (Fig. 2b). After ECO-ME treatment, the peaks corresponding to aluminium were completely removed and characteristic peaks of fluorine and chlorine were observed, which could be explained by the introduction of the reactants during the preparation process; the detailed percentages of the individual elements are listed in Table S2.† For a deeper investigation of the changes in the aluminium elements through ECO-ME treatment, the high-resolution Al 2p XPS spectra are presented in Fig. S4.† The pristine MAX phase presents two distinctive peaks belonging to Al–O and Al–Ti, whereas in the E-Ti3C2 spectrum, both peaks were absent, indicating complete removal of the A-layer and confirming the successful preparation of E-Ti3C2 MXene.
High-resolution XPS spectra were obtained to understand the chemical bonding state in E-Ti3C2. In the Ti 2p region (Fig. 2c), peaks corresponding to Ti–C, (OH)–Ti2+–C, (OH)–Ti3+–C, C–Ti–(O/OH), TiO2−xFx and TiO2 were detected, where the presence of the Ti–C peak indicated successful preparation of E-Ti3C2 and the C–Ti–(O/OH) peak could be attributed to termination of the bare surface of the Ti atom by abundant surface functional groups. The (OH)–Ti2+–C and (OH)–Ti3+–C bonds were considered to be due to the formation of suboxide and/or hydroxide moieties when highly reactive E-Ti3C2 reacted with ambient moisture or oxygen. The appearance of the TiO2 and TiO2−xFx peaks can be explained by the inevitable oxidation of the sample and its subsequent termination by fluorine functional groups. Successful preparation of E-Ti3C2 was further confirmed by high-resolution XPS C 1s spectra (Fig. 2d), where a peak corresponding to C–Ti was also observed. The C–C bond can be considered to be due to the selective dissolution of Ti atoms, whereas the detection of C–O and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O is considered to be derived from the highly reactive E-Ti3C2 sheet exposed to the atmosphere. Moreover, high-resolution O 1s and F 1s spectra revealed the detailed chemical bonding situation in E-Ti3C2, as shown in Fig. 2e and f, respectively. The appearance of TiO2 and TiO2−xFx peaks in the O 1s spectra could be attributed to the inevitable oxidation of E-Ti3C2 throughout the preparation and measurement periods; furthermore, the oxidized E-Ti3C2 was terminated by a fluorine functional group. The presence of the C–Ti–(OH)x peak resulted from highly reactive E-Ti3C2 flakes exposed to moisture and oxygen under ambient conditions. The C–Ti–Fx and TiO2−xFx observed in the F 1s high-resolution XPS spectra corroborate the outcome of the C 1s and O 1s spectra, confirming that the fluorine functional group terminates E-Ti3C2.52 The binding energies of each fitted peak in the high-resolution XPS spectra are summarized in Table S3.† Fourier transform infrared (FTIR) spectroscopy further revealed functional groups on the E-Ti3C2 surface, as shown in Fig. S5.† The peak at 3423 cm−1 was attributed to the stretching vibration of the –OH group, whereas the absorption peak at 2023 cm−1 could be explained by the vibrational band of the C–O group. The bending vibrations of the C
O is considered to be derived from the highly reactive E-Ti3C2 sheet exposed to the atmosphere. Moreover, high-resolution O 1s and F 1s spectra revealed the detailed chemical bonding situation in E-Ti3C2, as shown in Fig. 2e and f, respectively. The appearance of TiO2 and TiO2−xFx peaks in the O 1s spectra could be attributed to the inevitable oxidation of E-Ti3C2 throughout the preparation and measurement periods; furthermore, the oxidized E-Ti3C2 was terminated by a fluorine functional group. The presence of the C–Ti–(OH)x peak resulted from highly reactive E-Ti3C2 flakes exposed to moisture and oxygen under ambient conditions. The C–Ti–Fx and TiO2−xFx observed in the F 1s high-resolution XPS spectra corroborate the outcome of the C 1s and O 1s spectra, confirming that the fluorine functional group terminates E-Ti3C2.52 The binding energies of each fitted peak in the high-resolution XPS spectra are summarized in Table S3.† Fourier transform infrared (FTIR) spectroscopy further revealed functional groups on the E-Ti3C2 surface, as shown in Fig. S5.† The peak at 3423 cm−1 was attributed to the stretching vibration of the –OH group, whereas the absorption peak at 2023 cm−1 could be explained by the vibrational band of the C–O group. The bending vibrations of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond contributed to a characteristic peak in the spectrum located at 1624 cm−1. The characteristic peak at 1394 cm−1 was generated by the bending vibration of the O–H bond. The peaks at 1115 cm−1 and 556 cm−1 represented the stretching vibrations of the C–F bond and Ti–O bonds, respectively.
O bond contributed to a characteristic peak in the spectrum located at 1624 cm−1. The characteristic peak at 1394 cm−1 was generated by the bending vibration of the O–H bond. The peaks at 1115 cm−1 and 556 cm−1 represented the stretching vibrations of the C–F bond and Ti–O bonds, respectively.
Antibacterial activity of prepared E-Ti3C2 MXene
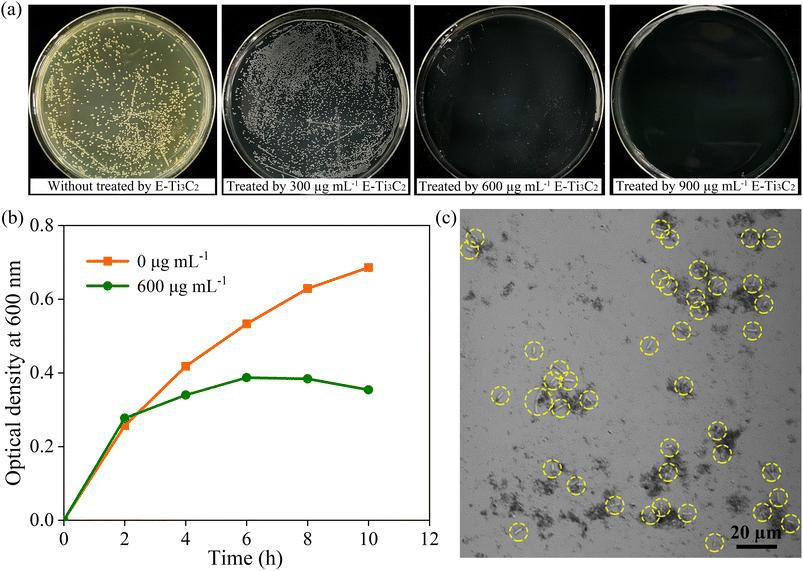
To investigate the antimicrobial activity of E-Ti3C2 in hydrogels and its inhibitory properties against bacterial growth, the model Gram-negative bacterium E. coli (wild-type) was used for the examination. To provide the necessary nutrients to sustain E. coli, Luria–Bertani (LB) agar, with the same hydrogel composition and content (1.5 wt%, agar) but with only the addition of appropriate nutrients, was employed as a medium for measuring the antibacterial effect of MXene. 100 μL of wild-type E. coli suspension at 108 CFU mL−1 was gently inoculated on the surface of the agar plates containing different concentrations of E-Ti3C2. Owing to the distinctive dark green colour of the MXene solution, the agar hydrogel plates supplemented with MXene presented a black colour, in contrast to the pure nutrient agar plates. The growth of bacterial colonies was assessed using the bacterial counting method after incubation at 37 °C for 12 h in the dark. Under the treatment of 300 μg mL−1 E-Ti3C2, the size of the bacterial colony was obviously decreased compared to those in the control sample without E-Ti3C2 MXene, indicating that E-Ti3C2 suppressed the growth of E. coli at this concentration. Besides, the inhibitory activity of E-Ti3C2 on E. coli growth became more pronounced with a further increase in the E-Ti3C2 concentration (Fig. 3a). The colonies were hardly visible at 600 μg mL−1, and with a further increase to 900 μg mL−1, the plates displayed a glossy appearance with no visible colonies, achieving 100% inhibition of the bacteria (the other two replicate experiments are shown in Fig. S6 and S7†). The growth curves of wild-type E. coli in the absence of E-Ti3C2 and in LB liquid medium containing 600 μg mL−1 E-Ti3C2 were investigated by measuring the optical density at 600 nm. The growth rate of E. coli exposed to E-Ti3C2 was significantly slower than that of the control sample after 2 h (Fig. 3b). Notably, the growth curve showed a declining trend after 6 h. Thus, E-Ti3C2 in agar hydrogels demonstrated significant antibacterial activity.To investigate the mechanism of E-Ti3C2 antibacterial activity, confocal microscopy was used to observe the interaction of E-Ti3C2 nanosheets with E. coli. The results showed that E-Ti3C2 (shown as a black fragment) is not distributed evenly in the field of vision but presents an aggregated group around E. coli (highlighted by yellow circles). At positions where more E. coli are present, more E-Ti3C2 nanosheets surround them (Fig. 3c). This can be attributed to the large specific surface area conferred by the unique two-dimensional structure of E-Ti3C2 and the abundance of functional groups and highly hydrophilic interactions with E. coli, which drive the two species to approach each other and eventually crowd together.
Antibacterial mechanism of E-Ti3C2 MXene
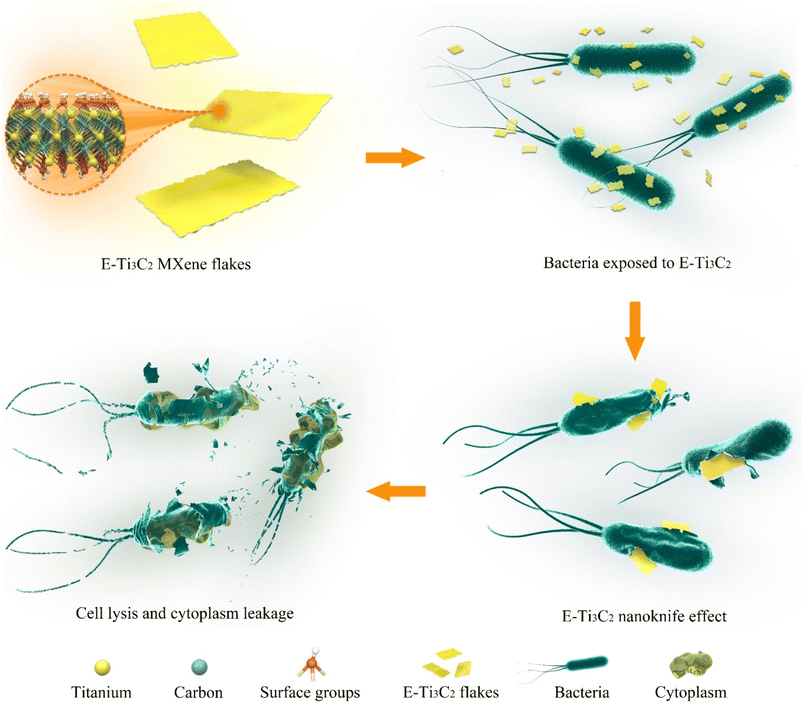
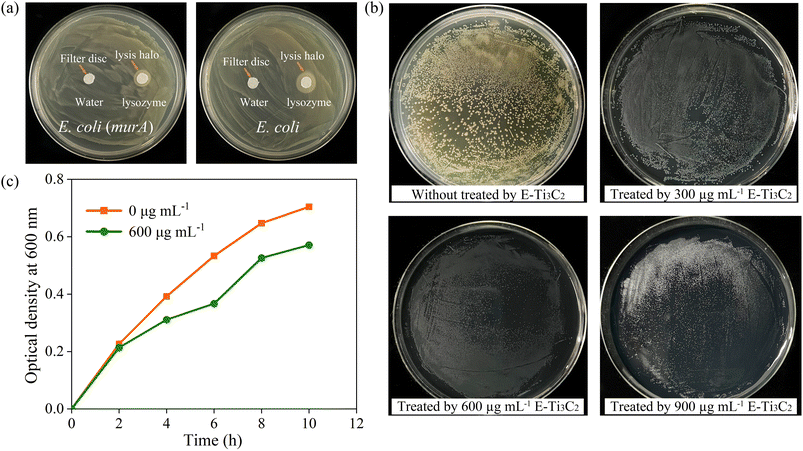
Therefore, we proposed a model for the antibacterial activity of E-Ti3C2 MXene nanosheets based on a nanoknife effect and its mechanism of action as illustrated in Fig. 4. Owing to the intensive mechanical forces involved in the preparation process, E-Ti3C2 prepared using the ECO-ME method has numerous irregular edges. The large specific surface area and abundant functional groups of E-Ti3C2 enable it to spontaneously gather around bacteria, while the high hydrophilicity of E-Ti3C2 further enhances contact with bacteria. The relative interaction between the bacteria and E-Ti3C2 sheets and the constant squeezing and movement of numerous E-Ti3C2 nanosheets result in the sharp edges of the nanosheets repeatedly injuring the bacterial cell walls, leading to their lysis and demise. Compared to other 2D materials such as graphene, MXene has a thicker monolayer, leading to higher bending rigidity.53 The ECO-ME preparation method not only accelerates preparation but also creates a large number of sharp irregular edges on MXene, which enable E-Ti3C2 to act as a nanoknife to effectively suppress or kill bacteria. As E-Ti3C2 ruptures the bacterial cell wall by repeated physical contact, leading to cytoplasmic leakage and eventual death, bacteria have a limited ability to develop resistance against the nanoknife effect; therefore, E-Ti3C2 not only kills the bacteria but also reduces the risk of bacterial resistance.To confirm the ability of the proposed E-Ti3C2 nanoknife to disrupt the E. coli cell wall, E. coli was genetically engineered to overexpress cell wall genes (murA) for increasing the cell wall thickness and to investigate its antibacterial activity. To determine whether the overexpression of murA thickens the E. coli cell wall, lysozyme, which could induce bacterial cell wall hydrolysis, was used to test the inhibition zones. The difference in cell wall thickness between wild-type and E. coli overexpressing the cell wall synthesis gene murA can be determined by distinguishing the size of the lysis halo formed by lysozyme, as shown in Fig. 5a. The lysis halo was absent in the water-added region, whereas E. coli was lysed around the filter paper to which lysozyme was added and a circular lysis halo was formed.
It can be clearly observed that lysozyme lyses the thickened cell wall of E. coli into a narrow circle, in contrast to the significantly larger lysis halo formed by lysing wild-type E. coli using the same dose of lysozyme, which produced a circle of approximately 1.35 cm in diameter on the plate (Fig. S8†). The thickened cell wall E. coli were incubated on plates containing E-Ti3C2 at a concentration of 600 μg mL−1 and although the individual size of the E. coli colonies was reduced compared to that in the control sample, no significant suppression effect was achieved as seen in the E. coli with normal cell walls. Furthermore, even with a further increase in the E-Ti3C2 concentration to 900 μg mL−1, the presence of E. coli colonies was still observed, as presented in Fig. 5b (the other two replicate experiments are shown in Fig. S9 and S10†). The weakened inhibitory effect of E-Ti3C2 on genetically modified E. coli was further confirmed by measuring the optical density at 600 nm (Fig. 5c). After overexpression of the cell wall synthesis gene, the effect of E-Ti3C2 on the growth curve of E. coli with thickened cell walls was obviously diminished and its growth inhibition rate was significantly reduced.
Thus, the antimicrobial activity of the E-Ti3C2 MXene nanosheets is attributed to their remarkable bending rigidity and sharp edges,53 which could disrupt the cell walls of E. coli. Owing to the mechanical forces involved in their preparation using the ECO-ME method, E-Ti3C2 nanosheets had irregularly shaped edges, which destroyed the bacterial cell walls, leading to their eventual rupture and death. In contrast, the inhibitory activity was significantly reduced when the E. coli cell wall was thickened through genetic engineering. This can be explained by the thickening of the cell wall, which makes it difficult for the edges of the 2D E-Ti3C2 nanosheets to puncture E. coli cells, resulting in the failure of antibacterial activity. Notably, although E-Ti3C2 was unable to kill the bacteria with the thickened cell walls, it still had an inhibitory effect on bacterial growth, as the individual E. coli colonies were significantly smaller than those in the sample without E-Ti3C2. This can be attributed to the E-Ti3C2 MXene nanosheets surrounding E. coli, which prevent the bacteria from obtaining nutrients from the environment; this results in the inhibition of bacterial growth.
Antibacterial activity of E-Ti3C2 compared with MXene prepared by conventional methods
To evaluate the antibacterial activity of E-Ti3C2 compared with the conventional wet chemical etching method prepared Ti3C2 (W-Ti3C2), a 100 μL suspension of wild-type E. coli at 108 CFU mL−1 was inoculated onto the surface of agar plates containing different concentrations of W-Ti3C2. The growth of colonies was evaluated by bacterial counting after 12 h incubation at 37 °C in darkness. It was clearly observed that in contrast to the significant antibacterial activity presented by E-Ti3C2 at a concentration of 900 μg mL−1, visible E. coli colonies were still observed on the surface of agar plates containing 900 μg mL−1 of W-Ti3C2 (Fig. S11,† the other two replicate experiments are shown in Fig. S12 and S13†). This can be attributed to the absence of mechanical forces in the preparation process of W-Ti3C2 resulting in flakes lacking sharp edges, as displayed in Fig. S14.† Furthermore, it has been demonstrated that the lateral size of the nanosheets significantly affects the cutting effect, which diminishes with increasing size of nanosheets.54,55 Therefore, the larger lateral size reduces the nanoknife effect of W-Ti3C2 and consequently impairs the antibacterial activity. As a result, E-Ti3C2 with a relatively small lateral size and numerous irregular edges produced through the ECO-ME method achieves better antibacterial activity than W-Ti3C2 prepared via conventional wet chemical etching methods.Conclusion
In summary, for the large-scale preparation of MXene, an efficient one-step mechanical exfoliation (ECO-ME) method has been developed, which dramatically reduces the preparation time and energy consumption of E-Ti3C2 MXene compared with the conventional wet chemical etching method, while maintaining a high yield and obtaining E-Ti3C2 aqueous solutions with high concentrations of more than 30 mg mL−1. Overall, E-Ti3C2, with a designed morphology, possesses excellent hydrophilicity and can be easily dispersed in agar solutions to form agar/MXene hydrogels. E-Ti3C2 nanosheets have irregular sharp edges and exhibit an extraordinary nanoknife effect that results in antibacterial activity. The antibacterial mechanism of E-Ti3C2 was confirmed using E. coli strains with thickened cell walls, indicating that the antibacterial activity of E-Ti3C2 was achieved by disrupting the bacterial cell wall. Furthermore, E-Ti3C2 demonstrated superior antibacterial activity than MXene prepared through conventional wet chemical etching approaches. These remarkable antimicrobial properties of E-Ti3C2 make it a promising candidate for combating the potential effects of bacteria and provide new insights for the development of bactericides.Author contributions
Y. C. L., C. C., D. L., and W. W. L. conceived the project. Y. C. L., J. Z. S., N. X., J. R., and X. C. carried out the experiments and characterization. Y. C. L., X. C., J. Z. S., C. Z., and C. C. worked on analysis of the results. Y. C. L., C. C., D. L., and W. W. L. wrote the manuscript. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Grants No. 42207521), Australian Research Council Discovery Program (DP220103416) and Australian Research Council Future Fellowships (FT200100730 and FT210100804). C. C. also thanks the Natural Science Foundation for the Higher Education Institutions of Anhui Province of China (KJ2020ZD10) and Anhui Provincial Returnees Innovation and Entrepreneurship Key Support Program (2022LCX003). We thank Prof. Yu Chen for his help in the bacterial experiment section.References
- C. Willyard, Nature, 2017, 543, 15 CrossRef CAS PubMed.
- N. R. Naylor, R. Atun, N. Zhu, K. Kulasabanathan, S. Silva, A. Chatterjee, G. M. Knight and J. V. Robotham, Antimicrobial Resistance & Infection Control, 2018, 7, 58 Search PubMed.
- C. A. Michael, D. Dominey-Howes and M. Labbate, Public Health, 2014, 2, 145 Search PubMed.
- B. T. Tadesse, E. A. Ashley, S. Ongarello, J. Havumaki, M. Wijegoonewardena, I. J. González and S. Dittrich, BMC Infect. Dis., 2017, 17, 616 CrossRef PubMed.
- V. M. D'Costa, C. E. King, L. Kalan, M. Morar, W. W. L. Sung, C. Schwarz, D. Froese, G. Zazula, F. Calmels, R. Debruyne, G. B. Golding, H. N. Poinar and G. D. Wright, Nature, 2011, 477, 457–461 CrossRef PubMed.
- J. M. V. Makabenta, A. Nabawy, C.-H. Li, S. Schmidt-Malan, R. Patel and V. M. Rotello, Nat. Rev. Microbiol., 2021, 19, 23–36 CrossRef CAS PubMed.
- Y. Tu, M. Lv, P. Xiu, T. Huynh, M. Zhang, M. Castelli, Z. Liu, Q. Huang, C. Fan, H. Fang and R. Zhou, Nat. Nanotechnol., 2013, 8, 594–601 CrossRef CAS PubMed.
- V. T. H. Pham, V. K. Truong, M. D. J. Quinn, S. M. Notley, Y. Guo, V. A. Baulin, M. Al Kobaisi, R. J. Crawford and E. P. Ivanova, ACS Nano, 2015, 9, 8458–8467 CrossRef CAS PubMed.
- O. Akhavan and E. Ghaderi, ACS Nano, 2010, 4, 5731–5736 CrossRef CAS PubMed.
- Y. Li, H. Yuan, A. von dem Bussche, M. Creighton, R. H. Hurt, A. B. Kane and H. Gao, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12295–12300 CrossRef CAS PubMed.
- J. D. Mangadlao, C. M. Santos, M. J. L. Felipe, A. C. C. de Leon, D. F. Rodrigues and R. C. Advincula, Chem. Commun., 2015, 51, 2886–2889 RSC.
- L. Hui, J.-G. Piao, J. Auletta, K. Hu, Y. Zhu, T. Meyer, H. Liu and L. Yang, ACS Appl. Mater. Interfaces, 2014, 6, 13183–13190 CrossRef CAS PubMed.
- X. Zou, L. Zhang, Z. Wang and Y. Luo, J. Am. Chem. Soc., 2016, 138, 2064–2077 CrossRef CAS PubMed.
- D. Liu, M. Zhang, W. Xie, L. Sun, Y. Chen and W. Lei, Appl. Catal., B, 2017, 207, 72–78 CrossRef CAS.
- Q. Li, S. Mahendra, D. Y. Lyon, L. Brunet, M. V. Liga, D. Li and P. J. J. Alvarez, Water Res., 2008, 42, 4591–4602 CrossRef CAS PubMed.
- A. Zirehpour, A. Rahimpour, A. Arabi Shamsabadi, M. Sharifian Gh and M. Soroush, Environ. Sci. Technol., 2017, 51, 5511–5522 CrossRef CAS PubMed.
- W. Yin, J. Yu, F. Lv, L. Yan, L. R. Zheng, Z. Gu and Y. Zhao, ACS Nano, 2016, 10, 11000–11011 CrossRef CAS PubMed.
- F. Cao, E. Ju, Y. Zhang, Z. Wang, C. Liu, W. Li, Y. Huang, K. Dong, J. Ren and X. Qu, ACS Nano, 2017, 11, 4651–4659 CrossRef CAS PubMed.
- W. Xu, W. Xie, X. Huang, X. Chen, N. Huang, X. Wang and J. Liu, Food Chem., 2017, 221, 267–277 CrossRef CAS PubMed.
- S. Xue, Y. Wu, M. Guo, Y. Xia, D. Liu, H. Zhou and W. Lei, Soft Matter, 2019, 15, 3680–3688 RSC.
- C. Chen, D. Liu, G. Yang, J. Wang, L. Wang and W. Lei, Adv. Energy Mater., 2020, 10, 1904098 CrossRef CAS.
- J. Wang, J. Hao, D. Liu, S. Qin, C. Chen, C. Yang, Y. Liu, T. Yang, Y. Fan, Y. Chen and W. Lei, Nanoscale, 2017, 9, 9787–9791 RSC.
- R. Y. Pelgrift and A. J. Friedman, Adv. Drug Delivery Rev., 2013, 65, 1803–1815 CrossRef CAS PubMed.
- M. Naguib, M. Kurtoglu, V. Presser, J. Lu, J. Niu, M. Heon, L. Hultman, Y. Gogotsi and M. W. Barsoum, Adv. Mater., 2011, 23, 4248–4253 CrossRef CAS PubMed.
- M. Ghidiu, M. R. Lukatskaya, M.-Q. Zhao, Y. Gogotsi and M. W. Barsoum, Nature, 2014, 516, 78 CrossRef CAS PubMed.
- M. Naguib, V. N. Mochalin, M. W. Barsoum and Y. Gogotsi, Adv. Mater., 2014, 26, 992–1005 CrossRef CAS PubMed.
- M. Naguib, O. Mashtalir, J. Carle, V. Presser, J. Lu, L. Hultman, Y. Gogotsi and M. W. Barsoum, ACS Nano, 2012, 6, 1322–1331 CrossRef CAS PubMed.
- O. Mashtalir, M. Naguib, V. N. Mochalin, Y. Dall'Agnese, M. Heon, M. W. Barsoum and Y. Gogotsi, Nat. Commun., 2013, 4, 1716 CrossRef PubMed.
- G. Yang, D. Liu, C. Chen, Y. Qian, Y. Su, S. Qin, L. Zhang, X. Wang, L. Sun and W. Lei, ACS Nano, 2021, 15, 6594–6603 CrossRef CAS PubMed.
- K. Chen, N. Qiu, Q. Deng, M.-H. Kang, H. Yang, J.-U. Baek, Y.-H. Koh, S. Du, Q. Huang and H.-E. Kim, ACS Biomater. Sci. Eng., 2017, 3, 2293–2301 CrossRef CAS PubMed.
- K. Huang, Z. Li, J. Lin, G. Han and P. Huang, Chem. Soc. Rev., 2018, 47, 5109–5124 RSC.
- A. M. Jastrzębska, A. Szuplewska, T. Wojciechowski, M. Chudy, W. Ziemkowska, L. Chlubny, A. Rozmysłowska and A. Olszyna, J. Hazard. Mater., 2017, 339, 1–8 CrossRef PubMed.
- X. Han, J. Huang, H. Lin, Z. Wang, P. Li and Y. Chen, Adv. Healthcare Mater., 2018, 7, 1701394 CrossRef PubMed.
- X. Li, Z. Huang, C. E. Shuck, G. Liang, Y. Gogotsi and C. Zhi, Nat. Rev. Chem, 2022, 6, 389–404 CrossRef PubMed.
- A. VahidMohammadi, J. Rosen and Y. Gogotsi, Science, 2021, 372, 1581 CrossRef PubMed.
- M. Naguib, R. R. Unocic, B. L. Armstrong and J. Nanda, Dalton Trans., 2015, 44, 9353–9358 RSC.
- M. Alhabeb, K. Maleski, B. Anasori, P. Lelyukh, L. Clark, S. Sin and Y. Gogotsi, Chem. Mater., 2017, 29, 7633–7644 CrossRef CAS.
- Z. Wang, V. Kochat, P. Pandey, S. Kashyap, S. Chattopadhyay, A. Samanta, S. Sarkar, P. Manimunda, X. Zhang, S. Asif, A. K. Singh, K. Chattopadhyay, C. S. Tiwary and P. M. Ajayan, Adv. Mater., 2017, 29, 1700364 CrossRef PubMed.
- C. Chen, D. Liu, L. He, S. Qin, J. Wang, J. M. Razal, N. A. Kotov and W. Lei, Joule, 2020, 4, 247–261 CrossRef CAS.
- C. Chen, J. Wang, D. Liu, C. Yang, Y. Liu, R. S. Ruoff and W. Lei, Nat. Commun., 2018, 9, 1902 CrossRef PubMed.
- W. Zhao, M. Fang, F. Wu, H. Wu, L. Wang and G. Chen, J. Mater. Chem., 2010, 20, 5817–5819 RSC.
- L. Zhang, C. Chen, J. Zhou, G. Yang, J. Wang, D. Liu, Z. Chen and W. Lei, Adv. Funct. Mater., 2020, 30, 2004139 CrossRef CAS.
- X. Su, J. Zhang, H. Mu, J. Zhao, Z. Wang, Z. Zhao, C. Han and Z. Ye, J. Alloys Compd., 2018, 752, 32–39 CrossRef CAS.
- T. Zhang, X. Jiang, G. Li, Q. Yao and J. Y. Lee, ChemNanoMat, 2018, 4, 56–60 CrossRef CAS.
- N. Xue, X. Li, M. Zhang, L. Han, Y. Liu and X. Tao, ACS Appl. Energy Mater., 2020, 3, 10234–10241 CrossRef CAS.
- M. Vakili, G. Cagnetta, J. Huang, G. Yu and J. Yuan, Molecules, 2019, 24, 2478 CrossRef CAS PubMed.
- J. Zhang, N. Kong, S. Uzun, A. Levitt, S. Seyedin, P. A. Lynch, S. Qin, M. Han, W. Yang, J. Liu, X. Wang, Y. Gogotsi and J. M. Razal, Adv. Mater., 2020, 32, 2001093 CrossRef CAS PubMed.
- P. Lakhe, E. M. Prehn, T. Habib, J. L. Lutkenhaus, M. Radovic, M. S. Mannan and M. J. Green, Ind. Eng. Chem. Res., 2019, 58, 1570–1579 CrossRef CAS.
- K. R. Mikeska, S. J. Bennison and S. L. Grise, J. Am. Ceram. Soc., 2000, 83, 1160–1164 CrossRef CAS.
- T. Li, L. Yao, Q. Liu, J. Gu, R. Luo, J. Li, X. Yan, W. Wang, P. Liu, B. Chen, W. Zhang, W. Abbas, R. Naz and D. Zhang, Angew. Chem., Int. Ed., 2018, 57, 6115–6119 CrossRef CAS PubMed.
- C. J. Zhang, S. Pinilla, N. McEvoy, C. P. Cullen, B. Anasori, E. Long, S.-H. Park, A. Seral-Ascaso, A. Shmeliov, D. Krishnan, C. Morant, X. Liu, G. S. Duesberg, Y. Gogotsi and V. Nicolosi, Chem. Mater., 2017, 29, 4848–4856 CrossRef CAS.
- J. Halim, I. Persson, P. Eklund, P. O. Å. Persson and J. Rosen, RSC Adv., 2018, 8, 36785–36790 RSC.
- V. N. Borysiuk, V. N. Mochalin and Y. Gogotsi, Comput. Mater. Sci., 2018, 143, 418–424 CrossRef CAS.
- C.-H. Yu, G.-Y. Chen, M.-Y. Xia, Y. Xie, Y.-Q. Chi, Z.-Y. He, C.-L. Zhang, T. Zhang, Q.-M. Chen and Q. Peng, Colloids Surf., B, 2020, 191, 111009 CrossRef CAS PubMed.
- S. Liu, M. Hu, T. H. Zeng, R. Wu, R. Jiang, J. Wei, L. Wang, J. Kong and Y. Chen, Langmuir, 2012, 28, 12364–12372 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3na00744h |
| This journal is © The Royal Society of Chemistry 2023 |