Open Access Article
Open Access ArticleBioactive substances inhibiting COX-2 and cancer cells isolated from the fibrous roots of Alangium chinense (Lour.) Harms
Ting Xiao†
 ab,
Xingyan Cheng†ab,
Jiaoyan Huangab,
Zhenghong Guo*c,
Ling Taoab and
Xiangchun Shen*ab
ab,
Xingyan Cheng†ab,
Jiaoyan Huangab,
Zhenghong Guo*c,
Ling Taoab and
Xiangchun Shen*ab
aThe State Key Laboratory of Functions and Applications of Medicinal Plants the Department of Pharmaceutic Preparation of Chinse Medicine, The High Educational Key Laboratory of Guizhou Province for Natural Medicinal Pharmacology and Druggability, School of Pharmaceutical Sciences, Guizhou Medical University, Guiyang 550025, China. E-mail: tingjinxiao@126.com; 3183956692@qq.com; 1134340755@qq.com; 649511230@qq.com
bThe Key Laboratory of Optimal Utilization of Natural Medicine Resources, School of Pharmaceutical Sciences, Guizhou Medical University, University Town, Guian New District, Guizhou, China. E-mail: shenxiangchun@126.com; Fax: +86-851-88416149; Tel: +86-851-88416149
cSchool of Pharmacy, Guizhou University of Traditional Chinese Medicine, University Town, Guian New District, Guiyang 550025, China. E-mail: guo_zhenghong@163.com; Fax: +86-851-88308060; Tel: +86-851-88308060
First published on 26th January 2023
Abstract
Alangium chinense has been used as a traditional folk medicine for centuries to treat rheumatism, skin diseases, and diabetes by the people of Southeast Asia. However, the bioactive constituents inhibiting COX-2 and cancer cells (HepG2, Caco-2, HeLa) remain unclear. In this study one new (14) along with twenty-four known compounds (1–13, 15–25) were isolated from the fibrous roots of Alangium chinense by chromatographic methods, and identified by NMR, and Gaussian and CD calculation. Compounds 1, 2, 13, 16, 17, 19, 20, 23, and 24 were isolated from this plant for the first time. Their inhibition effects on COX-2 enzyme and cancer cells were evaluated by MTT assay. Compounds 1–4, 13–14, and 16–18 can be used as good inhibitors against COX-2 enzyme, and compounds 1, 13, 14, and 17 were stronger than the positive control (celecoxib). In addition, molecular docking suggested that compounds 13, 17, and 18 belong to ellagic acids and have good inhibition against COX-2 enzyme. While compounds 1, 5, 13 and 21 showed cytotoxicity against HepG2 cells, compounds 2 and 21 showed cytotoxicity against Caco-2 cells, and compound 20 showed cytotoxicity against HeLa cells.
1 Introduction
Alangium chinense (Lour.) Harms belongs to the Alangium Lam. in the Alangiaceae, and it is widely distributed throughout the tropical and subtropical regions of the Eastern Hemisphere.1 A. chinense is rich in alkaloids, sugars, saponins, steroids, triterpenes, anthraquinones, and glycosides,2–4 and the roots, flowers, and leaves of A. chinense have served as ethnomedicinal treatments for rheumatic arthritis, acroanesthesia, fractures, snake bites, circulatory issues, hemostasis, toxicity, contraception, and wound healing in China.1,5,6 In addition, the fibrous roots of A. chinense named “Bai Long Xu” or “Ba Jiao Feng” in Traditional Chinese Medicine (TCM), have historically been applied as a treatment for rheumatoid arthritis and traumatic injury by the Miao people of Guizhou province, China.7 It is also the principal component of Fengshiding Capsules, which is a traditional Chinese medicine for the treatment of rheumatoid arthritis.1,8,9Cyclooxygenase-2 (COX-2) is a very important physiological enzyme playing key roles in various biological functions, especially in the mechanism of pain and inflammation. This makes it a molecule of great interest to the pharmaceutical community as a target.10 COX 2 enzyme is induced only during inflammatory processes or cancer, so selective COX-2 inhibition can significantly reduce the adverse effects, including GI tract damage and hepatotoxic effects, of traditional NSAIDs.10,11
Cancer has become a major threat to human health, with increasing mortality around the world.12 Liver cancer is the fourth leading cause of cancer-related mortality worldwide, despite the liver being the sixth most common site of primary cancer,13 and hepatocellular carcinoma (HCC) accounts for 80–90% of primary liver cancers.14 Colorectal cancer is the third most commonly diagnosed cancer and the fourth most common cause of cancer death worldwide.15 According to the latest online epidemiological database, there were more than 1.9 million new colon cancer cases in 2020, and 0.9 million deaths were recorded in the same year.16 The incidence rate and mortality rate have continuously risen in recent years. Even with the rapid development of cancer screening methods, many patients are diagnosed at an advanced stage with multiple symptoms.16 However, there are only a few effective therapeutic targets for colon cancer patients.17 Moreover, in the structure of morbidity and mortality, cervical cancer (CC) is one of the most common types of carcinomas among women in all countries of the world, in fourth place after breast cancer, colorectal cancer, and lung cancer.13 According to data from the International Agency for Research on Cancer, in 2018, the number of patients with cervical cancer was 570![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, an increase of 7.8% over the past decade, and the number of deaths from this disease amounted to 311
000, an increase of 7.8% over the past decade, and the number of deaths from this disease amounted to 311![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.13,14 In 2020, 604
000.13,14 In 2020, 604![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 new cases of cervical cancer were detected around the world, and the number of deaths was 341
100 new cases of cervical cancer were detected around the world, and the number of deaths was 341![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800.18–21
800.18–21
Therefore, despite the recommended screening programs for cancers, morbidity and mortality rates of cancers have been steadily growing in recent years worldwide. Targeted anticancer drugs, which are useful for the diagnosis of various stages of the tumor process, are very important. Due to its importance in Chinese Traditional Medicine and the necessity to identify its active constituents, we have carried out a phytochemical investigation of the fibrous roots extract of A. chinense. In the study, one new (14) and twenty-four known compounds (1–13, 15–25) were isolated and characterized from the ethyl acetate fraction. Among them, compounds 1, 2, 13, 16, 17, 19, 20, 23, and 24 were isolated from this plant for the first time. Furthermore, inhibition of COX-2 enzyme and cytotoxic activity against three human tumor cell lines (HepG2, Caco-2, and HeLa) have been evaluated with the crude extract and isolated compounds.
2 Results and discussion
2.1 Phytochemical extraction and purification
This study aimed to improve the chemical composition of A. chinense. The spectral data of compounds are basically consistent with the literature. Twenty-five compounds were isolated from ACEE using modern phytochemical techniques and were identified as cis-10-eicosaenoic acid (1), (2S)-5-hydroxy-7-methoxy-dihydroflavone (2), stigmast-4-en-3,6-dione (3), β-daucosterol (4), xanthoxylin (5), lacinilene C (6), (1S)-1-methoxylacinilene C (7), machilin-1 (8), perseal F (9), (−)-kobusin (10), (+)-fargesin (11), tephrosin (12), 3,3′,4′-tetra-O-methylellagic acid (13), (1R,6S)-6-aminocyclohex-2-ene-1-carboxylic acid (14), anabasine (15), (2S)-N-hydroxybenzylanabasine (16), 3,3′,4,4′-tetra-O-methylellagic acid (17), 3,4′-di-O-methylellagic acid (18), 3,4-methylenedioxy-3′-O-methyl-4′-O-glucoside ellagic acid (19), aralia cerebroside (20), uraci (21), chloropyrazine (22), (S)-3,4,5-trihydroxy-1H-pyrrol-2 (5H)-one (23), uvacalol k (24), and salicin (25) (Fig. 1). Among them, compound 14 was a new compound, and compound 1, 2, 13, 16, 17, 19, 20, 23, and 24 were isolated from this plant for the first time.Compound 1 (cis-10-eicosaenoic acid) was isolated as a white powder in dichloromethane. Its molecular formula was determined as C20H38O2 with an unsaturation of two. The 13C-NMR data (150 MHz, CDCl3) showed 20 carbon signals, including one carbonyl carbon, one double bond signal, one methyl signal, and sixteen methylene carbons. The NMR data of 1 (Tables 1 and 2) indicated an enoic acid skeleton consistent with 1H-NMR.22 The 1H-NMR data (600 MHz, CDCl3) showed one double bond proton confirmed at [δH 5.36 (2H, s, H-10,11)], methylene protons at [δH 2.35 (2H, t, J = 7.4, H-2), 2.03 (4H, m, H-9,12), 1.64 (2H, m, H-3), 1.25 (24H, m, H-4–8, 13–19)], and methyl protons at [δH 0.90 (3H, t, J = 6.5, H-20)].
| Position | 1 | 3 | 6 | 7 |
|---|---|---|---|---|
| 1 | 180.7 | 35.3 | 77.2 | 81.3 |
| 2 | 34.1 | 33.8 | 205.5 | 200.7 |
| 3 | 24.7 | 199.5 | 114.7 | 116.8 |
| 4 | 29.1 | 125.4 | 164.7 | 162.8 |
| 5 | 29.1 | 161.1 | 128.4 | 128.3 |
| 6 | 29.5 | 202.3 | 123.4 | 123.6 |
| 7 | 29.8 | 46.8 | 156.4 | 157.8 |
| 8 | 29.2 | 39.1 | 112.3 | 112.4 |
| 9 | 29.3 | 51.0 | 145.2 | 143.3 |
| 10 | 129.7 | 34.2 | 121.1 | 121.4 |
| 11 | 129.8 | 20.9 | 29.2 | 28.3 |
| 12 | 27.2 | 39.8 | 22.3 | 22.2 |
| 13 | 29.8 | 42.5 | 22.0 | 21.9 |
| 14 | 29.7 | 55.9 | 33.7 | 31.3 |
| 15 | 29.5 | 24.0 | 15.9 | 15.8 |
| 16 | 29.7 | 28.0 | ||
| 17 | 29.7 | 56.5 | ||
| 18 | 32.0 | 11.9 | ||
| 19 | 22.7 | 17.5 | ||
| 20 | 14.0 | 36.0 | ||
| 21 | 19.0 | |||
| 22 | 34.0 | |||
| 23 | 26.0 | |||
| 24 | 45.8 | |||
| 25 | 29.1 | |||
| 26 | 19.8 | |||
| 27 | 18.7 | |||
| 28 | 23.1 | |||
| 29 | 12.0 | |||
| OCH3 | 52.8 |
| Position | 1 | 3 | 6 | 7 |
|---|---|---|---|---|
| 1 | ||||
| 2 | 2.35 t (7.4) | |||
| 3 | 1.64 m, 1.64 m | 6.02 s | 5.84 s | |
| 4 | 1.25 m | 6.14 s | ||
| 5 | 7.41 s | 7.47 s | ||
| 6 | ||||
| 7 | ||||
| 8 | 7.22 s | 7.00 s | ||
| 9 | 2.03 m, 2.03 m | |||
| 10 | 5.36 s | |||
| 11 | 5.36 s | 3.21 m | 3.24 m | |
| 12 | 2.03 m, 2.03 m | 1.27 d (7.0) | 1.17 d (7.0) | |
| 13 | 1.25 m | 1.28 d (7.0) | 1.19 d (7.0) | |
| 14 | 1.51 s | 1.28 s | ||
| 15 | 2.27 s | 2.16 s | ||
| 16 | 2.83 s | |||
| 17 | ||||
| 18 | 0.91 d (6.5) | |||
| 19 | 1.14 s | |||
| 20 | 0.90 t (6.5) | |||
| 21 | 0.82 d (7.0) | |||
| 22 | ||||
| 23 | ||||
| 24 | ||||
| 25 | ||||
| 26 | 0.80 d (6.6) | |||
| 27 | 0.70 s | |||
| 28 | ||||
| 29 | 0.81 d (6.5) |
Compound 2 ((2S)-5-hydroxy-7-methoxy-dihydroflavone) was isolated as a yellow powder in methanol. Its molecular formula was determined as C16H14O4 with an unsaturation of ten. The 13C-NMR data (150 MHz, CD3OD) showed 15 carbon signals, including twelve aromatic carbons, one carbonyl carbon, one methylene signal, and one methyne carbon. The NMR data of 2 (Table 3 and Table 4) indicated a flavonoid skeleton consistent with 1H-NMR.23 The 1H-NMR data (600 MHz, CD3OD) showed seven aromatic protons confirmed at [δH 7.50 (2H, d, J = 7.7, H-2′, 6′), 7.42 (2H, t, J = 7.7, H-3′, 5′), 7.37 (1H, t, J = 7.7, H-4′), 6.09 (1H, d, J = 2.3, H-6), 6.06 (1H, d, J = 2.3, H-8)], methyne proton [δH 5.49(1H, dd, J = 12.8, 3.0, H-2)], and one methylene proton [δH 3.13(1H, dd, J = 17.2, 12.8, H-3α), 2.80 (1H, dd, J = 17.2, 3.13, H-3β)].
| Position | 2 | 5 | 15 | 16 | 22 | 23 | 24 |
|---|---|---|---|---|---|---|---|
| 1 | 106.9 | 129.3 | |||||
| 2 | 80.6 | 164.7 | 58.2 | 66.7 | 149.4 | 165.9 | 77.7 |
| 3 | 43.3 | 94.6 | 29.6 | 25.3 | 144.8 | 142.1 | 74.4 |
| 4 | 197.8 | 168.3 | 21.7 | 24.3 | 152.1 | 71.8 | |
| 5 | 165.3 | 91.6 | 22.5 | 35.5 | 142.8 | 100.3 | 68.7 |
| 6 | 95.9 | 167.9 | 45.7 | 53.0 | 144.7 | 128.1 | |
| 7 | 169.6 | 63.6 | |||||
| 8 | 95.0 | ||||||
| 9 | 164.5 | ||||||
| 10 | 104.1 | ||||||
| 1′ | 166.3 | ||||||
| 2′ | 149.6 | 148.5 | 128.1 | ||||
| 3′ | 136.0 | 139.5 | 128.3 | ||||
| 4′ | 133.6 | 136.1 | 129.3 | ||||
| 5′ | 124.5 | 124.2 | 128.3 | ||||
| 6′ | 148.2 | 148.0 | 128.1 | ||||
| 1′′ | 156.7 | 166.4 | |||||
| 2′′ | 121.8 | 129.4 | |||||
| 3′′ | 128.0 | 132.9 | |||||
| 4′′ | 119.0 | 133.0 | |||||
| 5′′ | 128.6 | 132.9 | |||||
| 6′′ | 115.1 | 129.4 | |||||
| 7′′ | 57.4 | ||||||
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
204.6 | ||||||
| 4-OCH3 | 56.1 | ||||||
| 6-OCH3 | 56.2 | ||||||
| 8-CH3 | 33.1 |
| Position | 2 | 5 | 15 | 16 | 22 | 23 | 24 |
|---|---|---|---|---|---|---|---|
| 1 | 2.08–1.75 m | 7.29 d (7.6) | 3.81 t (9.4, 9.5) | ||||
| 2 | 5.49 dd (12.8, 3.0) | 4.46 m | 3.35 dd (2.9, 11.2) | 4.54 m | |||
| 3 | 3.13 dd (17.2, 12.8), 2.80 dd (17.2, 3.13) | 6.04 d (2.3) | 2.08–1.75 m | 1.77 m | 5.37 t (9.3, 9.1) | ||
| 4 | 3.55 m, 3.27 m | 1.53 m, 1.87 m | 4.02 t (8.4, 7.9) | ||||
| 5 | 6.05 d (2.3) | 8.71 s | 1.83 m, 1.87 m | 8.39 s | 5.51 d (7.6) | ||
| 6 | 6.09 d (2.3) | 3.11 d (11.7), 2.17 m | 8.34 s | 6.60 d (8.3) | |||
| 7 | 5.16 d (7.5), 4.75 d (11.4) | ||||||
| 8 | 6.06 d (2.3) | ||||||
| 2′ | 7.50 d (7.7) | 8.04 d (8.0) | 8.58 s | 7.50–8.10 m | |||
| 3′ | 7.42 t (7.7) | 7.53 dd (7.9, 4.8) | |||||
| 4′ | 7.37 t (7.7) | 8.58 d (3.7) | 7.92 d (7.9) | ||||
| 5′ | 7.42 t (7.7) | 2.08–1.75 m | 7.43 dd (4.9, 7.8) | ||||
| 6′ | 7.50 d (7.7) | 4.46 m | 8.44 d (4.3) | ||||
| 1′′ | |||||||
| 2′′ | 7.50–8.10 m | ||||||
| 3′′ | 6.89 d (7.5) | ||||||
| 4′′ | 6.69 t (6.2) | ||||||
| 5′′ | 7.05 t (7.9) | ||||||
| 6′′ | 6.67 t (7.9) | ||||||
| 7′′ | 3.79 d (14.0), 3.13 d (14.0) | ||||||
| 8-CH3 | |||||||
| 4-OCH3 | 3.88 s | ||||||
| 6-OCH3 | 3.82 s |
Compound 3 (stigmast-4-en-3,6-dione) was isolated as a white crystal in dichloromethane. Its molecular formula was determined as C29H46O2 with an unsaturation of seven. The 13C-NMR data (150 MHz, CDCl3) showed 29 carbon signals, including two carbonyl carbons, one double bond signal, six methyl carbons, three quaternary carbons, ten methylene carbons, and six methyne carbons. The NMR data of 3 (Tables 1 and 2) indicated a steroid skeleton consistent with 1H-NMR.24 The 1H-NMR data (600 MHz, CDCl3) showed one double bond proton confirmed at [δH 6.14 (1H, s, H-4)], and six methyl protons [δH 1.14 (3H, s, H-19), 0.91 (3H, d, J = 6.5, H-18), 0.82 (3H, d, J = 7.0, H-21), 0.81 (3H, d, J = 6.5, H-29), 0.80 (3H, d, J = 6.6, H-26), 0.70 (3H, s, H-27)].
Compound 4 (β-daucosterol) was isolated as a white powder in pyridine. Its molecular formula was determined as C31H52O3 with an unsaturation of six. The 13C-NMR data (150 MHz, C5D5N) showed 35 carbon signals, including one double bond signal, one glucose carbon, six methyl carbons, and eleven methylene carbons. The NMR data of 4 (Table 5) indicated a steroid skeleton consistent with 1H-NMR.25 The 1H-NMR data (600 MHz, C5D5N) showed one double bond proton confirmed at [δH 5.33 (1H, s, H-5)], one glucose hydrogen proton [δH 5.07 (d, J = 7.8, H-1′)], one methyne proton [δH 4.02–3.93 (1H, m, H-3)], and six methyl protons [δH 0.94 (3H, s, H-19), 0.91 (3H, s, H-18), 0.89 (3H, s, H-21), 0.87 (3H, s, H-29), 0.67 (3H, s, H-26), 1.00 (3H, s, H-27)].
| Position | δC (150 MHz) | δH (J in Hz) (600 MHz) |
|---|---|---|
| 1 | 37.9 | |
| 2 | 30.7 | |
| 3 | 78.5 | 4.02–3.93 m |
| 4 | 40.4 | |
| 5 | 141.3 | 5.33 s |
| 6 | 122.3 | |
| 7 | 32.6 | |
| 8 | 32.5 | |
| 9 | 50.8 | |
| 10 | 37.3 | |
| 11 | 21.7 | |
| 12 | 39.7 | |
| 13 | 42.9 | |
| 14 | 57.2 | |
| 15 | 24.9 | |
| 16 | 28.9 | |
| 17 | 56.7 | |
| 18-CH3 | 12.4 | 0.91 s |
| 19-CH3 | 19.8 | 0.94 s |
| 20 | 36.8 | |
| 21-CH3 | 19.6 | 0.89 s |
| 22 | 34.6 | |
| 23 | 26.8 | |
| 24 | 46.5 | |
| 25 | 29.9 | |
| 26-CH3 | 19.4 | 0.67 s |
| 27-CH3 | 20.4 | 1.00 s |
| 28 | 23.8 | |
| 29-CH3 | 12.6 | 0.87 s |
| 1′ | 103.0 | 5.07 d (7.8) |
| 2′ | 75.7 | |
| 3′ | 78.9 | |
| 4′ | 72.1 | |
| 5′ | 79.0 | |
| 6′ | 63.2 |
Compound 5 (xanthoxylin) was isolated as a white crystal in methanol. Its molecular formula was determined as C10H12O4 with an unsaturation of five. The 13C-NMR data (150 MHz, CD3OD) showed 10 carbon signals, including one carbonyl carbon, two methoxy carbons, one methyl carbon, and six aromatic carbons. The NMR data of 5 (Tables 3 and 4) indicated an acetophenone skeleton consistent with 1H-NMR.26 The 1H-NMR data (600 MHz, CD3OD) showed two aromatic hydrogen protons confirmed at [δH 6.04 (1H, d, J = 2.3, H-3), 6.05 (1H, d, J = 2.3, H-5)], two methoxy protons [δH 3.82 (3H, s, H-6), 3.88 (3H, s, H-4)], and one methyl proton [δH 2.57 (3H, s, H-8)].
Compound 6 (lacinilene C) was isolated as a yellow powder in dichloromethane. Its molecular formula was determined as C15H18O3 with an unsaturation of six. The 13C-NMR data (150 MHz, CDCl3) showed 15 carbon signals, including six aromatic carbons, one double bond, one carbonyl carbon, four methyl carbons, one methylene carbon, and one quaternary carbon. The NMR data of 6 (Tables 1 and 2) indicated a sesquiterpene skeleton consistent with 1H-NMR.27 The 1H-NMR data (600 MHz, CDCl3) showed two aromatic hydrogen protons confirmed at [δH 7.41 (1H, s, H-5), 7.22 (1H, s, H-8)], one double bond proton [δH 6.02 (1H, s, H-3)], one methylene proton [δH 3.21 (1H, m, H-11)], and four methyl proton [δH 1.27 (1H, d, J = 7.0, H-12), 1.28 (1H, d, J = 7.0, H-13), 1.51 (1H, s, H-14), 2.27(1H, s, H-15)].
Compound 7 ((1S)-1-methoxylacinilene C) was isolated as a yellow powder in dichloromethane. Its molecular formula was determined as C16H20O3 with an unsaturation of seven. The 13C-NMR data (150 MHz, CDCl3) showed 16 carbon signals, including six aromatic carbons, one double bond, one carbonyl carbon, four methyl carbons, one methylene carbon, one methoxy carbon, and one quaternary carbon. The NMR data of 7 (Tables 1 and 2) indicated a sesquiterpene skeleton consistent with 1H-NMR.27 The 1H-NMR data (600 MHz, CDCl3) showed two aromatic hydrogen protons confirmed at [δH 7.47 (1H, s, H-5), 7.00 (1H, s, H-8)], one double bond proton [δH 5.84 (1H, s, H-3)], one methylene proton [δH 3.24 (1H, m, H-11)], four methyl proton [δH 1.17 (1H, d, J = 7.0, H-12), 1.19 (1H, d, J = 7.0, H-13), 1.28 (1H, s, H-14), 2.16 (1H, s, H-15)], and one methoxy proton [δH 2.83 (3H, s, 16-OCH3)].
Compound 8 (machilin-1) was isolated as a colourless oil in dichloromethane. Its molecular formula was determined as C20H24O5 with an unsaturation of eight. The 13C-NMR data (150 MHz, CDCl3) showed 20 carbon signals, including twelve aromatic carbons, two methyl carbons, two methoxy carbons, and four methylene carbons. The NMR data of 8 (Tables 6 and 7) indicated a lignin skeleton consistent with 1H-NMR.28 The 1H-NMR data (600 MHz, CDCl3) showed six aromatic hydrogen protons confirmed at [δH 6.94 (2H, dd, J = 9.1, H-5′′, 6′′), 6.78–6.95 (1H, m, H-2′′), 6.89 (2H, d, J = 8.0, H-2′, 3′), 6.78–6.95 (1H, m, H-6′)], two methoxy protons [δH 3.91 (3H, s, 5′-OCH3), 3.89 (3H, s, 3′′-OCH3)], four methylene protons [δH 5.45 (1H, d, J = 4.2, H-2), 4.64 (1H, d, J = 9.2, H-5), 2.43 (2H, dd, J = 6.3, 3.7, H-3, 4)], and two methyl protons [δH 1.00 (3H, d, J = 6.4, 4-CH3), 0.61 (3H, d, J = 6.9, 3-CH3)].
| Position | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|
| 1 | 131.5 | 54.2 | 50.2 | 115.4 | |
| 1a | 111.1 | ||||
| 2 | 84.8 | 114.3 | 86.0 | 82.0 | 151.1 |
| 3 | 47.6 | 145.0 | 144.0 | ||
| 4 | 43.5 | 146.2 | 71.9 | 71.0 | 101.1 |
| 4a | 148.4 | ||||
| 5 | 85.8 | 108.9 | 54.4 | 54.6 | |
| 6 | 120.0 | 85.9 | 87.7 | 63.9 | |
| 6a | 76.3 | ||||
| 7 | 95.0 | ||||
| 7a | 156.7 | ||||
| 8 | 44.9 | 71.8 | 69.7 | 108.7 | |
| 9 | 17.8 | 160.7 | |||
| 10 | 111.8 | ||||
| 11 | 128.6 | ||||
| 11a | 109.1 | ||||
| 12 | 191.4 | ||||
| 12a | 67.5 | ||||
| 1′ | 132.6 | 131.1 | 133.0 | 135.1 | |
| 2′ | 108.8 | 111.8 | 109.3 | 106.6 | |
| 3′ | 146.7 | 146.8 | 149.3 | 147.9 | |
| 4′ | 144.3 | 153.2 | 148.8 | 147.2 | 109.4 |
| 5′ | 114.0 | 133.7 | 111.1 | 108.2 | 128.8 |
| 6′ | 118.8 | 120.0 | 118.4 | 119.6 | 78.0 |
| 7′ | 28.5 | ||||
| 8′ | 28.3 | ||||
| 1′′ | 135.0 | 135.2 | 130.9 | ||
| 2′′ | 108.3 | 106.6 | 108.9 | ||
| 3′′ | 146.3 | 148.1 | 148.8 | ||
| 4′′ | 145.1 | 147.2 | 147.9 | ||
| 5′′ | 113.9 | 108.3 | 111.0 | ||
| 6′′ | 119.3 | 119.5 | 117.7 | ||
| 3-CH3 | 9.4 | ||||
| 4-CH3 | 11.9 | ||||
| CHO | 190.6 | ||||
| O–CH2–O | 101.2 | 101.0 | |||
| OCH3 | 56.0 | 56.1 | 56.0 | 55.9 | 56.4 |
| OCH3 | 55.9 | 56.0 | 56.1 | 55.9 | 55.9 |
| Position | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|
| 1 | 3.09 m | 3.27–3.36 m | 6.56 s | ||
| 2 | 5.45 d (4.2) | 6.91 s | 4.74 dd (10.5, 5.0) | 4.86 d (5.2) | |
| 3 | 2.43 dd (6.3, 3.7) | ||||
| 4 | 2.43 dd (6.3, 3.7) | 3.89 m, 4.25 dd (9.0, 4.0) | 4.11 d (9.4), 3.83 t (7.7) | 6.51–6.45 m | |
| 4-CH3 | 1.00 d (6.4) | ||||
| 5 | 4.64 d (9.2) | 6.94 s | 3.09 m | 2.87 d (7.4) | |
| 6 | 6.91 s | 4.74 dd (10.5, 5.0) | 4.42 d (7.0) | 4.63 dd (12.1, 2.4), 4.53–4.46 m | |
| 6a | 4.57 d (1.5) | ||||
| 7 | 5.25 d (9.0) | ||||
| 7a | |||||
| 8 | 3.56 m | 3.89 m, 4.25 dd (9.0, 4.0) | 3.83 t (7.7), 3.27–3.36 m | ||
| 9 | 1.45 | ||||
| 9-CH3 | 1.45 d (6.8) | ||||
| 10 | 6.51–6.45 m | ||||
| 11 | 7.73 d (8.7) | ||||
| 2′ | 6.89 d (8.0) | 7.38 s | 6.78–6.89 m | 6.93 s | |
| 3′ | 6.89 d (8.0) | ||||
| 4′ | 6.60 d (10.1) | ||||
| 5′ | 3.91 s | 6.78–6.89 m | 6.75–6.90 m | 5.56 d (10.1) | |
| 6′ | 6.78–6.95 m | 7.35 s | 6.78–6.89 m | 6.75–6.90 m | |
| 7′-CH3 | 1.45 s | ||||
| 8′-CH3 | 1.39 s | ||||
| 2′′ | 6.78–6.95 m | 6.78–6.89 m | 6.75–6.90 m | ||
| 3′′ | 3.89 s | ||||
| 4′′ | |||||
| 5′′ | 6.94 dd (9.1) | 6.78–6.89 m | 6.75–6.90 m | ||
| 6′′ | 6.94 dd (9.1) | 6.78–6.89 m | 6.75–6.90 m | ||
| 3-CH3 | 0.61 d (6.9) | ||||
| CHO | 9.85 s | ||||
| O–CH2–O | 5.95 s | 5.94 s | |||
| OCH3 | 3.95 s | 3.89 s | 3.91 s | 3.82 s | |
| OCH3 | 3.89 s | 3.88 s | 3.87 s | 3.73 s |
Compound 9 (perseal F) was isolated as a white powder in dichloromethane. Its molecular formula was determined as C18H18O5 with an unsaturation of nine. The 13C-NMR data (150 MHz, CDCl3) showed 18 carbon signals, including twelve aromatic carbons, one methyl carbon, two methoxy carbons, one aldehyde group carbon, and two methylene carbons. The NMR data of 9 (Tables 6 and 7) indicated a lignin skeleton consistent with 1H-NMR.29 The 1H-NMR data (600 MHz, CDCl3) showed one aldehyde group proton confirmed at [δH 9.85 (1H, s, 2′-CHO)], five aromatic hydrogen protons [δH 7.38 (1H, s, H-2′), 7.35 (1H, s, H-6′), 6.94 (1H, s, H-5), 6.91 (2H, s, H-2, 6)], two methylene protons [δH 5.25 (1H, d, J = 9.0, H-7), 3.56 (1H, m, H-8)], two methoxy protons [δH 3.95 (3H, s, 3′-OCH3), 3.89 (3H, s, 4-OCH3)], and one methyl proton [δH 1.45 (3H, d, J = 6.8, 9-CH3)].
Compound 10 ((−)-kobusin) was isolated as a colorless colloid in dichloromethane. Its molecular formula was determined as C21H22O6 with an unsaturation of ten. The 13C-NMR data (150 MHz, CDCl3) showed 21 carbon signals, including twelve aromatic carbons, three methylene carbons, two methoxy carbons, and four methyne carbons. The NMR data of 10 (Tables 6 and 7) indicated a furofuran lignan skeleton consistent with 1H-NMR.30 The 1H-NMR data (600 MHz, CDCl3) showed six aromatic hydrogen protons confirmed at [δH 6.78–6.89 (6H, m, H-2′, 5′, 6′, 2′′, 5′′, 6′′)], two methoxy protons [δH 3.89 (3H, s, 3′-OCH3), 3.88 (3H, s, 4′-OCH3)], three methylene protons [δH 5.95 (2H, s, O–CH2–O), 3.89 (1H, m, H-4α), 4.25 (1H, dd, J = 9.0, 4.0, H-4β), 3.89 (1H, m, H-8α), 4.25 (1H, dd, J = 9.0, 4.0, H-8β)], and four methyne protons [δH 4.74 (2H, dd, J = 10.5, 5.0, H-2, 6), 3.09 (2H, m, H-1, 5)].
Compound 11 ((+)-fargesin) was isolated as colorless colloidal in dichloromethane. Its molecular formula was determined as C21H22O6 with an unsaturation of ten. The 13C-NMR data (150 MHz, CDCl3) showed 21 carbon signals, including twelve aromatic carbons, three methylene carbons, two methoxy carbons, and four methyne carbons. The NMR data of 11 (Tables 6 and 7) indicated a furofuran lignan skeleton and consistent with 1H-NMR.31 The 1H-NMR data (600 MHz, CDCl3) showed six aromatic hydrogen protons confirmed at [δH 6.93 (1H, s, H-2′), 6.75–6.90 (5H, m, H-5′, 6′, 2′′, 5′′, 6′′)], two methoxy protons [δH 3.91 (3H, s, 3′-OCH3), 3.87 (3H, s, 4′-OCH3)], three methylene protons [δH 5.94 (2H, s, O–CH2–O), 4.11 (1H, d, J = 9.4, H-4α), 3.83 (1H, t, J = 7.7, H-4β), 3.83 (1H, t, J = 7.7, H-8α), 3.27–3.36 (1H, m, H-8β)], and four methyne protons [δH 4.86 (1H, d, J = 5.2, H-2), 4.42 (1H, d, J = 7.0, H-6), 3.27–3.36 (1H, m, H-1), 2.87 (1H, d, J = 7.4, H-5)].
Compound 12 (tephrosin) was isolated as a white powder in dichloromethane. Its molecular formula was determined as C23H22O7 with an unsaturation of twelve. The 13C-NMR data (150 MHz, CDCl3) showed 23 carbon signals, including twelve aromatic carbons, two methoxy carbons, one carbonyl carbon, two methyl carbons, one double bond carbon, one methylene carbon, one methyne carbon, and two quaternary carbons. The NMR data of 12 (Tables 6 and 7) indicated an isoflavone skeleton consistent with 1H-NMR.32 The 1H-NMR data (600 MHz, CDCl3) showed four aromatic hydrogen protons confirmed at [δH 7.73 (1H, d, J = 8.7, H-11), 6.56 (1H, s, H-1), 6.51–6.45 (2H, m, H-4, 10)], one double bond proton [δH 6.60 (1H, d, J = 10.1, H-4′), 5.56 (1H, d, J = 10.1, H-5′)], one methylene proton [δH 4.63 (1H, dd, J = 12.1, 2.4, H-6α), 4.53–4.46 (1H, m, H-6β)], one methyne proton [δH 4.57 (1H, d, J = 1.5, H-6a)], two methoxy protons [δH 3.82 (3H, s, 2-OCH3), 3.73 (3H, s, 3-OCH3)], and two methyl protons [δH 1.45 (3H, s, 7′-CH3), 1.39 (3H, s, 8′-CH3)].
Compound 13 (3,3′,4,4′-tetra-O-methylellagic acid) was isolated as a white needle crystal in methanol. Its molecular formula was determined as C18H14O8 with an unsaturation of twelve. The 13C-NMR data (150 MHz, DMSO-d6) showed 14 carbon signals, including ten quaternary carbons, two methines, two ester carbonyls, and four methoxy carbons. The NMR data of 13 (Tables 8 and 9) indicated an ellagic acid skeleton consistent with 1H-NMR.33 The 1H-NMR data (600 MHz, DMSO-d6) showed two characteristic aromatic protons confirmed at [δH 7.50 (2H, s, H-5,5′)], and four methoxy confirmed at [δH 4.05 (6H, s, OCH3), 3.96 (6H, s, OCH3)].
| Position | 13 | 17 | 18 | 20 | 21 | 25 |
|---|---|---|---|---|---|---|
| 1 | 111.8 | 111.5 | 111.9 | 70.4 | 132.0 | |
| 2 | 142.2 | 141.4 | 142.0 | 151.2 | 155.2 | |
| 3 | 140.7 | 140.7 | 141.1 | 73.9 | 115.3 | |
| 4 | 152.9 | 153.2 | 152.9 | 71.0 | 164.8 | 127.7 |
| 5 | 112.0 | 112.2 | 111.8 | 142.7 | 122.2 | |
| 6 | 112.5 | 112.9 | 113.2 | 100.7 | 128.2 | |
| 7 | 159.2 | 158.7 | 159.1 | 32.57 | 58.7 | |
| 8 | 130.7 | |||||
| 9 | 130.3 | |||||
| 10 | 34.8, 32.8, 32.6, 31.8 | |||||
| 1′ | 111.9 | 112.3 | 107.3 | 174.24 | 101.9 | |
| 2′ | 141.6 | 141.8 | 140.5 | 70.9 | 73.9 | |
| 3′ | 140.5 | 141.2 | 135.9 | 77.5 | ||
| 4′ | 150.7 | 154.2 | 150.3 | 70.2 | ||
| 5′ | 107.1 | 107.8 | 107.1 | 77.0 | ||
| 6′ | 113.3 | 113.8 | 114.0 | 61.3 | ||
| 7′ | 158.9 | 158.9 | 159.2 | |||
| 1′′ | 103.9 | |||||
| 2′′ | 71.5 | |||||
| 3′′ | 77.0 | |||||
| 4′′ | 71.0 | |||||
| 5′′ | 77.3 | |||||
| 6′′ | 61.4 | 61.4 | 61.4 | 61.5 | ||
| 4-OMe | 57.0 | |||||
| 3′-OMe | 61.4 | 61.8 | ||||
| 4′-OMe | 57.0 | 57.2 | 57.0 | |||
| CH3 | 14.23 | |||||
| (CH2)n | 29.7, 29.7, 29.6, 29.3, 29.2, 26.0, 24.9, 22.6 |
| Position | 13 | 17 | 18 | 20 | 21 | 25 |
|---|---|---|---|---|---|---|
| 1 | 4.52 dd (6.83, 10.5) | |||||
| 2 | 5.34 s | |||||
| 3 | 4.27 s | 7.09 d (7.7) | ||||
| 4 | 4.15 d (7.5) | 7.19 dt (8.2, 7.3) | ||||
| 5 | 7.48 s | 7.50 s | 7.49 s | 5.45 d (7.6) | 7.00 t (6.8, 6.0) | |
| 6 | 7.39 d (7.6) | 7.36 d (7.4) | ||||
| 7 | 4.65 dd (14.2), 4.47 dd (14.2) | |||||
| 8 | 5.34 s | |||||
| 9 | 5.60 s | |||||
| 10 | ||||||
| 1′ | 5.34 d (4.4) | |||||
| 2′ | 4.57 m | 4.47–5.06 m (H-2′–6′) | ||||
| 3′ | ||||||
| 4′ | ||||||
| 5′ | 7.50 s | 7.58 s | 7.49 s | |||
| 6′ | 3.71 m, 3.47 m | |||||
| 7′ | ||||||
| 1′′ | 4.93 d (7.86) | |||||
| 2′′ | 4.06 m | |||||
| 3′′ | 4.10 s | |||||
| 4′′ | 4.32 m | |||||
| 5′′ | 3.86 s | |||||
| 6′′ | 4.49 dd (2.4, 11.96), 4.36 dd (5.1, 11.62) | |||||
| 3-OMe | 4.05 s | 4.04 m | 4.05 s | |||
| 4-OMe | 3.96 s | |||||
| 3′-OMe | 4.05 s | 4.06 m | ||||
| 4′-OMe | 3.96 s | 3.99 s | 3.97 s | |||
| CH3 | 0.84 s | |||||
| (CH2)n | 1.48 s | |||||
| N–H | 7.57 d (9.0) | 11.01 s, 10.81 s |
Compound 14 ((1R,6S)-6-aminocyclohex-2-ene-1-carboxylic acid) is a white powder with the molecular formula C7H11NO2 and an unsaturation of three based on HR-ESI-MS ([M−H]− m/z 140.0717, calcd 140.0790) and NMR analyses (Table 10). It showed a carbonyl group at δC 176.6 in 13C NMR. The 1H NMR signals at δH 6.11 (1H, m, H-3), 5.74 (1H, m, H-2), 3.91 (1H, d, J = 4.4, H-6), 2.47 (2H, m, H-4), 2.13 (1H, m, H-5) and 1.70 (1H, m, H-5) along with the 13C NMR signals resonating at δC 48.9 (C-1), 131.3 (C-2), 138.4 (C-3), 34.1 (C-4), 25.7 (C-5) and 59.91 (C-6) were consistent with a cyclohexene skeleton.34 The cyclohexene carboxylic acid was confirmed by the HMBC cross-peaks of H-2 (δH 5.74) to C-1 (δC 48.9), C-4 (δC 34.1), and C-5 (δC 25.7), of H-3 (δH 6.11) to C-1 (δC 48.9), C-4 (δC 34.1) and C-5 (δC 25.7), of H-4 (δH 2.47) to C-2 (δC 131.3) and C-3 (δC 138.4), of H-5 (δH 2.13, 1.70) to C-1 (δC 48.9) and C-6 (δC 59.9), and of H-6 (δH 3.91) to C-1 (δC 48.9), C-2 (δC 131.3), and C-7 (δC 176.6) (Fig. 2).
| Position | δC | δH (J in Hz) | HMBC | H–H cosy | NOE |
|---|---|---|---|---|---|
| 1 | 48.9 | 3.49 br.s | H-6, H-5, H-4 | H-2, H-6 | |
| 2 | 131.3 | 5.74 m | C-1, C-4, C-5 | H-1, H-4 | H-3 |
| 3 | 138.4 | 6.11 m | C-1, C-4, C-5 | H-1, H-4 | H-4 |
| 4 | 34.1 | 2.47 m | C-2, C-3 | H-1 | H-5 |
| 5 | 25.7 | 2.13 m, 1.70 m | C-1, C-6 | ||
| 6 | 59.9 | 3.91 d (4.4) | C-1, C-2, C-7 | H-1 | H-1, H-5 |
| 7 | 176.6 |
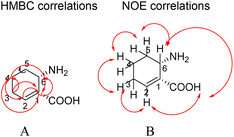 | ||
| Fig. 2 HMBC (A) and key NOE (B) correlations observed for (1R,6S)-6-aminocyclohex-2-ene-1-carboxylic acid. | ||
In addition, there were H–H cosy of H-1 (δH 5.74) to H-6 (δH 3.91), H-4 (δH 2.47), and H-5 (δH 2.13, 1.70), of H-2 (δH 5.74) to H-1 (δH 3.49) and H-4 (δH 2.47), of H-3 (δH 6.11) to H-1 (δH 3.49) and H-4 (δH 2.47), of H-4 (2.47) to H-1 (δH 3.49), and of H-6 (δH 3.91) to H-1 (δH 3.49).
The NOE cross-peaks of H-1 (δH 3.49) to H-2 (δH 5.74) and H-6 (δH 3.91), of H-6 (δH 3.91) to H-1 (δH 3.49) and H-5 (δH 1.70) revealed the relative configuration of the cyclohexene carboxylic acid (Fig. 2). NOE correlations confirmed H-1 and H-6 were β-oriented.
Compound 15 (anabasine) is a white crystal with the molecular formula C10H14N2 and an unsaturation of five. The 13C-NMR data (150 MHz, CD3OD) showed 10 carbon signals, including five aromatic carbons, four methylene carbons, and one methyne carbon. The NMR data of 15 (Tables 3 and 4) indicated an alkaloid skeleton consistent with 1H-NMR.35 The 1H-NMR data (600 MHz, CD3OD) showed four characteristic aromatic protons confirmed at [δH 8.71 (1H, s, H-2′), 8.58 (1H, d, J = 3.6 Hz, H-6′), 8.04 (1H, d, J = 7.8 Hz, H-4′), and 7.53 (1H, dd, J = 7.8, 7.8 Hz, H-5′)], two methylene protons [δH 3.17 (2H, m, H-6), 2.03 (2H, dd, J = 12.6, 6.0 Hz, H-3)], one methyne proton [δH 3.41 (1H, m, H-2)], and one omino group proton [δH 4.37 (1H, m, H-1)].
Compound 16 ((2S)-N-hydroxybenzylanabasine) was obtained as a white oil in methanol, and its molecular formula was determined as C17H20O with an unsaturation of eight. The 13C-NMR showed 17 carbon signals, including eleven aromatic carbons, five methylene carbons, and one methyne carbon. The NMR data of 16 (Tables 3 and 4) indicated an alkaloid skeleton consistent with 1H-NMR.27 The 1H-NMR data (600 MHz, CD3OD) showed eight characteristic aromatic protons confirmed at [δH 8.53 (1H, s, H-2′), 7.92 (1H, d, J = 8.0, H-4′), 7.43 (1H, dd, J = 8.0, H-5′), 8.44 (1H, d, J = 5.0, H-6′), 6.89 (1H, d, J = 7.5, H-3′′), 6.67 (1H, t, J = 7.5, H-4′′), 7.05 (1H, t, J = 8.0, H-5′′), and 6.69 (1H, t, J = 8.0, H-6′′)], five methylene protons [δH 1.69 (1H, m, H-3), 1.53, 1.77 (2H, m, H-4), 1.83, 1.87 (2H, m, H-5), 3.31, 2.09 (2H, m, H-6), and 3.79, 3.13 (2H, d, J = 14.0, H-7′′)], and one methyne proton [δH 3.35 (1H, m, H-2)].
Compound 17 (3,3′,4,4′-tetra-O-methylellagic acid) is a white amorphous powder with the molecular formula C17H12O8 and an unsaturation of twelve. The 13C-NMR showed 17 carbon signals, including twelve aromatic carbons, three methoxy groups, and two carbonyl carbons. The NMR data of 17 (Tables 8 and 9) indicated an ellagic acid skeleton consistent with 1H-NMR.36 The 1H-NMR data (600 MHz, DMSO-d6) showed two characteristic aromatic protons confirmed at [δH 7.50 (1H, s, H-5), and 7.58 (1H, s, H-5′)], and three methoxy protons [δH 4.04 (3H, m, 3-OCH3), 4.06 (3H, m, 3′-OCH3) and 3.99 (3H, s, 4′-OCH3)].
Compound 18 (3,4′-di-O-methylellagic acid) is a white amorphous powder with the molecular formula C16H16O8 and an unsaturation of nine. The 13C-NMR showed 16 carbon signals, including twelve aromatic carbons, two methoxy groups, and two carbonyl carbons. The NMR data of 18 (Tables 8 and 9) indicated an ellagic acid skeleton consistent with 1H-NMR.37 The 1H-NMR data (400 MHz, DMSO-d6) showed two characteristic aromatic protons confirmed at [δH 7.49 (2H, s, H-5, H-5′)], two methoxy protons [δH 4.05 (3H, s, 3-OCH3), and 3.97 (3H, s, 3′-OCH3)].
Compound 19 (3,4-methylenedioxy-3′-O-methyl-4′-O-glucoside ellagic acid) is a white acicular crystal with the molecular formula C22H18O13 and an unsaturation of fourteen. The 13C-NMR showed 22 carbon signals, including twelve aromatic carbons, one methoxy group, two carbonyl carbons, one methylene, and one glucose group. The NMR data of 19 (Table 11) indicated an ellagic acid skeleton consistent with 1H-NMR.38 The 1H-NMR data (600 MHz, C5D5N) showed two characteristic aromatic protons confirmed at [δH 7.73 (1H, s, H-5), and 8.44 (1H, s, H-1′)], one methylene protons [δH 6.33 (2H, m, H-3a)], one glucose terminal hydrogen proton [δH 5.88 (1H, m, H-1′′)], one methoxy group protons [δH 4.24 (3H, s, 3′-OCH3)].
| Position | δC (150 MHz) | δH (J in Hz) (600 MHz) |
|---|---|---|
| 1 | 112.5 | |
| 2 | 132.0 | |
| 3 | 138.8 | |
| 3a (CH2) | 104.6 | 6.33 m |
| 4 | 151.1 | |
| 5 | 104.6 | 7.73 s |
| 6 | 115.8 | |
| 6a | 158.0 | |
| 1′ | 113.1 | |
| 2′ | 142.0 | |
| 3′ | 142.6 | |
| 4′ | 153.0 | |
| 5′ | 113.4 | 8.44 s |
| 6′ | 114.0 | |
| 6′a | 158.6 | |
| 1′′ | 102.7 | 5.88 m |
| 2′′ | 78.3 | |
| 3′′ | 79.0 | |
| 4′′ | 74.6 | |
| 5′′ | 70.8 | |
| 6′′ | 62.1 | |
| 3′-OMe | 61.7 | 4.24 s |
Compound 20 (aralia cerebroside) is a white amorphous powder with the molecular formula C40H77NO10 and an unsaturation of three. The 13C-NMR showed 40 carbon signals, including one acylamine group, one glucose group, twenty-four methylene carbons, four methyne carbons and one double bond signals, and two methyl signals. The NMR data of 20 (Tables 8 and 9) indicated an alkaloid skeleton.39 The 1H-NMR data (600 MHz, DMSO-d6) showed one amide hydrogen proton confirmed at [δH 7.57 (1H, d, J = 9.0)], one glucose group proton [δH 4.93 (1H, d, J = 7.9, H-1′′), 4.06 (1H, m, H-2′′), 4.10 (1H, s, H-3′′), 4.32 (1H, m, H-4′′), 3.86 (1H, s, H-5′′), and 4.49, 4.36 (2H, dd, J = 2.4, 11.96, H-6′′)], four methyne protons [δH 5.24 (1H, s, H-2), 4.27 (1H, s, H-3), 4.25 (1H, d, J = 7.5, H-4), and 4.57 (1H, m, H-2′)], one double bond [δH 5.34 (1H, s, H-8), and 5.60 (1H, s, H-9)], two methyl protons [δH 0.84 (6H, s, CH3 × 2)], and twenty-four methylene protons [δH 1.48 (48H, s, CH2 × 24)].
Compound 21 (uracil) is a pale yellow needle crystal with the molecular formula C4H4N2O2 and an unsaturation of four. The 13C-NMR showed 4 carbon signals, including two carbonyl carbons and two aromatic carbons. The NMR data of 21 (Tables 8 and 9) indicated a nitrogen heterocyclic skeleton consistent with 1H-NMR.40 The 1H-NMR data (600 MHz, DMSO-d6) showed two characteristic aromatic protons confirmed at [δH 5.45 (1H, d, J = 7.6, H-5), and 7.39 (1H, d, J = 7.6, H-6)], and two imino group protons [δH 11.01 (1H, s, 1-NH), and 11.81 (1H, s, 3-NH)].
Compound 22 (chloropyrazine) is a white needle crystal with the molecular formula C4H3ClN2 and an unsaturation of four. The 13C-NMR showed 4 carbon signals, including three aromatic carbons. The NMR data of 22 (Tables 3 and 4) indicated a nitrogen heterocyclic skeleton consistent with 1H-NMR.41 The 1H-NMR data (600 MHz, CD3OD) showed two characteristic aromatic protons confirmed at [δH 8.39 (1H, s, H-5), and 8.34 (1H, s, H-6)].
Compound 23 ((S)-3,4,5-trihydroxy-1H-pyrrol-2 (5H)-one) is a white needle crystal with the molecular formula C5H7NO4 and an unsaturation of three. The 13C-NMR showed 4 carbon signals, including one carbonyl carbon, one double bond, and one methyne carbon. The NMR data of 23 (Tables 3 and 4) indicated an alkaloid skeleton consistent with 1H-NMR.42 The 1H-NMR data (600 MHz, CD3OD) showed one imino group proton confirmed at [δH 7.29 (1H, d, J = 7.6, H-1)] and one methyne hydrogen proton [δH 5.51 (1H, d, J = 7.6, H-5)].
Compound 24 (uvacalol k) is a white powder with the molecular formula C21H20O7 and an unsaturation of twelve. The 13C-NMR showed 4 carbon signals, including two aromatic rings, one double bond, one methylene carbon, and four methyne carbons. The NMR data of 24 (Tables 3 and 4) indicated a polyoxygenated cyclohexene skeleton consistent with 1H-NMR.43 The 1H-NMR data (600 MHz, CD3OD) showed ten aromatic hydrogen protons confirmed at [δH 7.50–8.10 (10H, m, H-2′–H-6′, H-2′′–H-6′′)], one double bond protons [δH 6.60 (1H, d, J = 8.3, H-6)], one methylene proton [δH 5.16 d (1H, J = 7.5, H-7a), 4.75 d (1H, J = 11.4, H-7b)], and four methyne protons [δH 5.37 (1H, t, J = 9.3, 9.1, H-3), 4.54 (1H, m, H-2), 4.02 (1H, t, J = 8.4, 7.9, H-4), 3.81 (1H, t, J = 9.4, 9.5, H-1)].
Compound 25 (salicin) is a white needle crystal with the molecular formula C13H18O7 and an unsaturation of five. The 13C-NMR showed 13 carbon signals, including six aromatic carbons, one glucose group, and one methylene carbon. The NMR data of 25 (Tables 8 and 9) indicated an aromatic hydrocarbon skeleton consistent with 1H-NMR.44 The 1H-NMR data (600 MHz, DMSO-d6) showed four aromatic hydrogen protons confirmed at [δH 7.36 (1H, d, J = 7.4, H-6), 7.19 (1H, dt, J = 8.2, 7.3, H-4), 7.09 (1H, d, J = 7.7, H-3), and 7.00 (1H, t, J = 6.8, 6.0, H-5)], one glucose group proton [δH 5.34 (1H, d, J = 4.4, H-1′), 4.47–5.06 (4H, m, H-2′–5′), 3.71 (1H, m, H-6′), and 3.47 (1H, m, H-6′)], and one methylene proton [δH 4.65 (1H, dd, J = 14.2, H-7), 4.47 (1H, dd, J = 14.2, H-7)].
2.2 Quantum chemical Gaussian and CD calculation
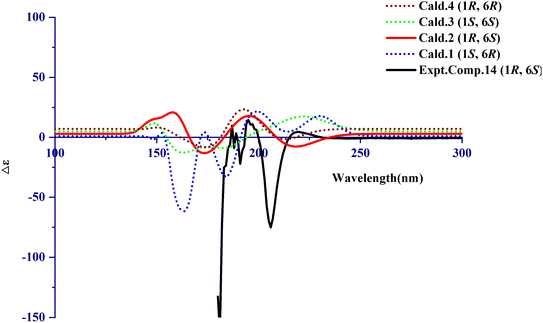
The calculated NMR of compound 14 was conducted with the Gaussian program. The DP4+ data of 1R,6S was 100% and the probabilities of isomer 1 (1S,6R), isomer 3 (1S,6S), and isomer 4 (1R,6R) were 0%. It can be inferred that 1R,6S was the correct relative configuration. In addition, the absolute configuration of compound 14 (1R,6S) was further confirmed with the ECD spectrum. Thus, the absolute configuration of compound 14 was defined as (1R,6S)-6-aminocyclohex-2-ene-1-carboxylic acid (Fig. 3 and Fig. 4).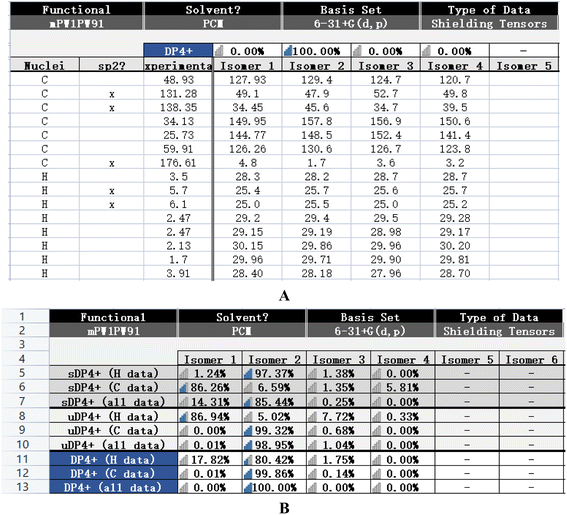 | ||
| Fig. 3 Theoretical NMR data (A and B) of compound 14 (isomer 2) and the possibility configurations of four conformations. | ||
2.3 Anti-inflammatory activity of octocarpus COX-2 inhibitors
Prostaglandins (PGs) are metabolites of arachidonic acid via the cyclooxygenase (COX) pathway. COX is a key enzyme that catalyzes the conversion of arachidonic acid to PGs and it includes COX-1 and COX-2.45 COX-1 is a structural enzyme that is essential for maintaining human physiological needs,46 whereas COX-2 is an inducible enzyme that is highly expressed during inflammation.47 COX-2 can enhance PGE2 synthesis during inflammation, which plays a significant role in the subsequent cell inflammatory responses,48 eventually producing a series of inflammatory mediators, which participate in the body's physiological and pathological processes through a cascade reaction.In addition, COX-2 is a very important physiological enzyme playing key roles in various biological functions, especially in the mechanism of pain and inflammation, making it a molecule of great interest to the pharmaceutical community as a target.49
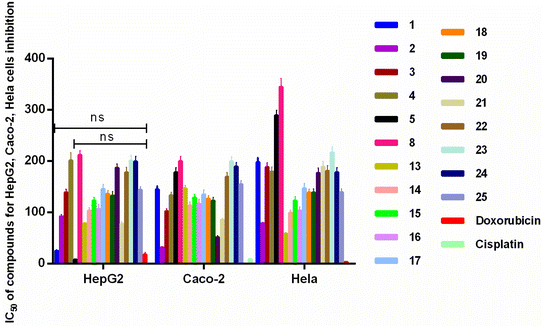
Total extract, petroleum ether extract, ethyl acetate extract, N-butanol extract and nineteen compounds were tested for the anti-inflammatory activity of octocarpus COX-2 inhibitors (Fig. 5). Among them, ethyl acetate extract (IC50 = 51.37 ± 7.89 μg mL−1), compounds 1 (IC50 = 10.80 ± 0.30 μM), 2 (IC50 = 34.07 ± 3.57 μM), 3 (IC50 = 67.90 ± 6.21 μM), 4 (IC50 = 69.70 ± 6.55 μM), 13 (IC50 = 16.49 ± 1.76 μM), 14 (IC50 = 19.32 ± 3.58 μM), 16 (IC50 = 24.20 ± 1.20 μM), 17 (IC50 = 13.93 ± 2.44 μM), and 18 (IC50 = 34.73 ± 1.53 μM) showed strong inhibitory activity against COX-2, and the inhibitory activity of compounds 1, 13, 14, and 17 was stronger than that of the positive control drug celecoxib (IC50 = 21.35 ± 5.13 μM or 8.15 ± 3.13 μg mL−1). Compounds 13, 17, and 18 belong to ellagic acids. According to reports, ellagic acids can be used as anti-inflammatory, anticancer and antioxidant agents.50,51 Moreover, ellagic acids can also inhibit the expression of COX-2 in IL-1β-induced chondrocytes.52 This is consistent with our results.
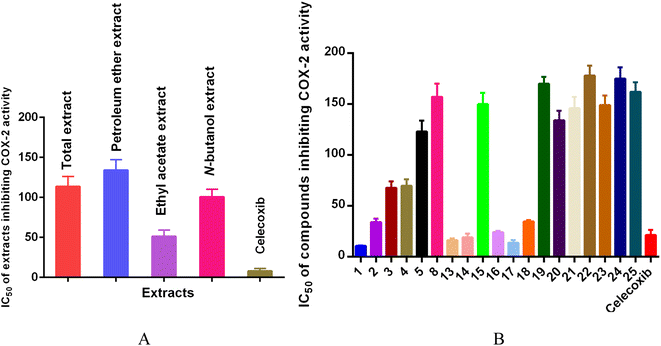 | ||
| Fig. 5 Results of extracts (A) and compounds (B) inhibiting cyclooxygenase (COX-2) activity (n = 3). | ||
In contrast, compounds 5 (IC50 = 123.14 ± 10.55 μM), 8 (IC50 = 157.33 ± 12.85 μM), 15 (IC50 = 150.00 ± 11.00 μM), 19 (IC50 = 170.00 ± 7.00 μM), 20 (IC50 = 134.00 ± 10.00 μM), 21 (IC50 = 146.00 ± 11.00 μM), 22 (IC50 = 178.00 ± 10.00 μM), 23 (IC50 = 149.00 ± 10.00 μM), 24 (IC50 = 175.00 ± 11.00 μM), and 25 (IC50 = 162.00 ± 10.00 μM) showed relatively weak inhibitory activity against COX-2.
2.4 Cytotoxic activity assay against cancer cells
Cancer refers to a group of some of the most diagnosed and deadliest pathophysiological conditions worldwide. Although cancer is a common disease worldwide, its molecular pathology is characterized by a wide spectrum of biological aggressiveness that makes its control difficult, and thus a life-threatening disease.53Nineteen compounds were tested for activity against HepG2 cells, Caco-2 cells, and HeLa cells (Fig. 6). Compounds 1, 5, 13 and 21 showed good inhibitory effect on HepG2 cells with IC50 of 25.00 ± 1.08, 8.21 ± 1.12, 78.85 ± 1.06, 78.5 ± 4.41 μM, and there were no significant differences between compounds 1 and 5 and positive control doxorubicin. Compounds 2 and 21 showed good inhibitory effect on Caco-2 cells with IC50 of 85.98 ± 2.96 μM. And compound 20 showed good inhibitory effect on HeLa cells with IC50 of 51.90 ± 2.56 μM. A. chinense showed great anticancer potential, and it could become a good anticancer drug in the near future.
2.5 Molecular docking
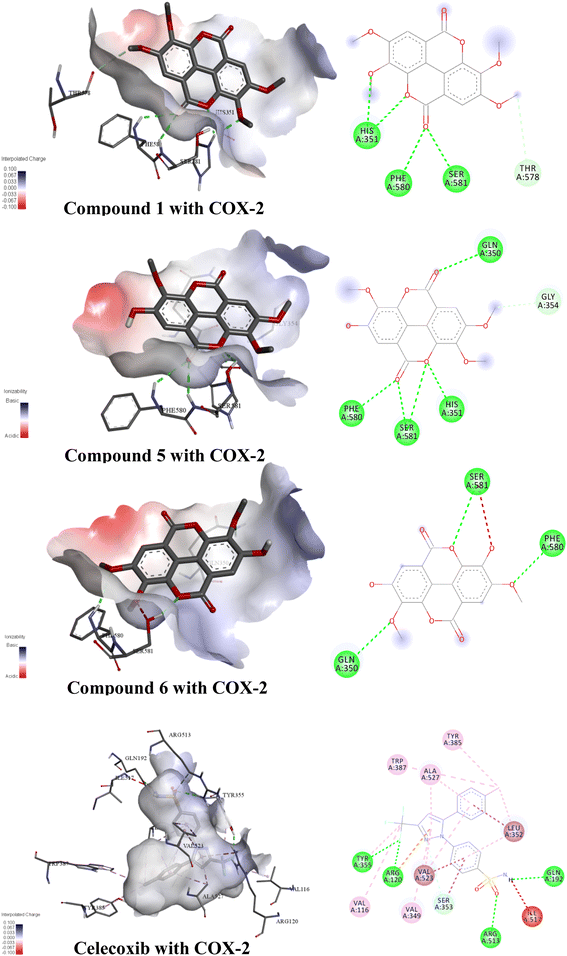
Molecular docking is an important tool in computer-aided molecular design, which can be used to predict biological experiments by simulating the interaction of small molecules and biological macromolecules.54 The binding energy and binding modes of compounds 13, 17, and 18 with COX-2 were measured (Fig. 7, Table 12). Compound 18 had the lowest binding energy of −8.1 kcal mol−1. Conversely, as the number of hydroxyl groups went up and the number of methoxy groups was reduced, ellagic acids had the lowest binding energy.| Target | Compound | Binding free energy (kcal mol−1) | Numbers of bonds | Amino acids |
|---|---|---|---|---|
| COX-2 | Compound 1 | −7.7 | 5 | His351, Phe580, Ser581, Thr578 |
| Compound 5 | −7.8 | 6 | Gln350, Gly354, Phe580, Ser581, His351 | |
| Compound 6 | −8.1 | 4 | Ser581, Phe590, Gln350 | |
| Celecoxib | −10.0 | 22 | Trp387, Ala527, Tyr385, Leu352, Gln192, Ile517, Arg513, Tyr355, Arg120, Val523, Val116, Val349, Ser353 |
3 Material and methods
3.1 Chemicals and reagents
Silica gels with 100–200 and 200–300 meshes were purchased from Qingdao Ocean Chemical Co., Ltd (Shandong, China), Toyopearl HW-40F was purchased from Tosoh (Tokyo, Japan), acetonitrile of HPLC grade was purchased from J&K Scientific Inc (Beijing, China). Dichloromethane, ethyl acetate, petroleum ether (Fuyu Fine Chemical Inc, Tianjin, China), a high performance liquid chromatograph (Shimadzu Corporation, Japan), a circulating water multipurpose vacuum pump (Lichen Bangxi Instrument Inc, Shanghai, China), COX-2 LM0168 (LMAI Bio, Shanghai, China), DMEM medium, FBS (Fetal Bovine Serum), and penicillin-streptomycin were purchased from Thermo Fisher Scientific Co., Ltd. MTT reagent, doxorubicin, and cisplatin were acquired from Sigma Aldrich (St. Louis, MO, USA).3.2 Plant material
The fibrous root of Alangium chinense (Lour.) Harms was purchased from Nayong City, Guizhou Province, China. The morphological characteristics of the plant were compared with those recorded in the Flora of China (Editorial Committee of Flora of China, Chinese Academy of Sciences, 1983) and identified by Qingde Long, an associate professor of Guizhou Medical University, with voucher specimen number 20191010.3.3 Extraction and isolation
The 30 kg of dried fibrous root of A. chinense were cut into segments, then extracted with 75% ethanol at 25 °C three times, once a week. The obtained extract was filtered through a Brinell funnel. The filtrate was collected and further concentrated with a rotary evaporator. The extract was re-suspended in water with a separation funnel, and then the same amounts of petroleum ether, ethyl acetate, and N-butanol were added for continuous extraction. Then the extraction layer was evaporated using a rotary evaporator to remove the organic reagent to obtain 70.400 g, 159.800 g, and 203.200 g, respectively.A total of 70.400 g of petroleum ether extract (APEE) was dissolved in dichloromethane and then mixed with silica gel at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, and then loaded onto a chromatograph silica gel column (75 × 1000 mm) using a gradient of petroleum ether–ethyl acetate solvent system as eluant to obtain fractions Fr. 1 to Fr. 9 based on the TLC analysis. Eluents from the resin column were dried under vacuum. Among them, Fr. 1 (27.900 g) was separated with an aqueous elution of petroleum ether
3, and then loaded onto a chromatograph silica gel column (75 × 1000 mm) using a gradient of petroleum ether–ethyl acetate solvent system as eluant to obtain fractions Fr. 1 to Fr. 9 based on the TLC analysis. Eluents from the resin column were dried under vacuum. Among them, Fr. 1 (27.900 g) was separated with an aqueous elution of petroleum ether![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate (v/v = 1
ethyl acetate (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0–1
0–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with a silica gel column to obtain fractions Fr. 1.1 to Fr. 1.6 based on the TLC analysis. Fr. 1.1 (0.800 g) was separated with an aqueous elution of petroleum ether
1) with a silica gel column to obtain fractions Fr. 1.1 to Fr. 1.6 based on the TLC analysis. Fr. 1.1 (0.800 g) was separated with an aqueous elution of petroleum ether![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ether ethyl acetate (v/v = 50
ether ethyl acetate (v/v = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with a silica gel column to obtain compound 1 (0.100 g). Fr. 1.2 (1.2 g) was separated with an aqueous elution of dichloromethane
1) with a silica gel column to obtain compound 1 (0.100 g). Fr. 1.2 (1.2 g) was separated with an aqueous elution of dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 1
methanol (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with a silica gel column and further purified by semi-preparative high performance liquid chromatography with methanol
1) with a silica gel column and further purified by semi-preparative high performance liquid chromatography with methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (v/v = 80
water (v/v = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) as the mobile phase to obtain compound 2 (0.016 g, tR 8.8 min). The precipitated white crystals from Fr. 2 (18.490 g) were recrystallized with methanol and dichloromethane to obtain compound 3 (0.065 g). Fr. 2.3 (1.080 g) was separated with an aqueous elution of petroleum ether
20) as the mobile phase to obtain compound 2 (0.016 g, tR 8.8 min). The precipitated white crystals from Fr. 2 (18.490 g) were recrystallized with methanol and dichloromethane to obtain compound 3 (0.065 g). Fr. 2.3 (1.080 g) was separated with an aqueous elution of petroleum ether![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate (v/v = 100
ethyl acetate (v/v = 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with a silica gel column and further recrystallized with methanol to obtain compound 4 (0.100 g). Fr. 2.5 (1.080 g) was separated with an aqueous elution of dichloromethane
1) with a silica gel column and further recrystallized with methanol to obtain compound 4 (0.100 g). Fr. 2.5 (1.080 g) was separated with an aqueous elution of dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 1
methanol (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with a silica gel column and further purified by semi-preparative high performance liquid chromatography with methanol
1) with a silica gel column and further purified by semi-preparative high performance liquid chromatography with methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (v/v = 80
water (v/v = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) as the mobile phase to obtain compound 5 (0.016 g, tR 8.8 min).
20) as the mobile phase to obtain compound 5 (0.016 g, tR 8.8 min).
Fr. 3 (22.700 g) was separated with an aqueous elution of dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 1
methanol (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with a silica gel column and further purified by semi-preparative high performance liquid chromatography with methanol
1) with a silica gel column and further purified by semi-preparative high performance liquid chromatography with methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (v/v = 7525) as the mobile phase to obtain compound 6 (0.008 g, tR 9.2 min) and compound 7 (0.008 g, tR 9.7 min). Fr. 3.3 (1.900 g) was separated with an aqueous elution of dichloromethane
water (v/v = 7525) as the mobile phase to obtain compound 6 (0.008 g, tR 9.2 min) and compound 7 (0.008 g, tR 9.7 min). Fr. 3.3 (1.900 g) was separated with an aqueous elution of dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 1
methanol (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) with a silica gel column and further purified by semi-preparative high performance liquid chromatography with methanol
2) with a silica gel column and further purified by semi-preparative high performance liquid chromatography with methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (v/v = 75
water (v/v = 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25) as the mobile phase to obtain compound 8 (0.015 g, tR 7.5 min). Fr. 4 (17.300 g) was separated with an aqueous elution of dichloromethane
25) as the mobile phase to obtain compound 8 (0.015 g, tR 7.5 min). Fr. 4 (17.300 g) was separated with an aqueous elution of dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 1
methanol (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with a silica gel column and further purified by semi-preparative high performance liquid chromatography with methanol
1) with a silica gel column and further purified by semi-preparative high performance liquid chromatography with methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (v/v = 75
water (v/v = 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25) as the mobile phase to obtain compound 9 (0.010 g, tR 8.5 min). Fr. 4.3 (3.910 g) was separated with an aqueous elution of dichloromethane
25) as the mobile phase to obtain compound 9 (0.010 g, tR 8.5 min). Fr. 4.3 (3.910 g) was separated with an aqueous elution of dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 1
methanol (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) with a silica gel column and further purified by semi-preparative high performance liquid chromatography with methanol
2) with a silica gel column and further purified by semi-preparative high performance liquid chromatography with methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (v/v = 73
water (v/v = 73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 28) as the mobile phase to obtain compound 10 (0.010 g, tR 8.6 min) and compound 11 (0.008 g, tR 9.1 min). Fr. 5 (15.330 g) was separated with an aqueous elution of dichloromethane
28) as the mobile phase to obtain compound 10 (0.010 g, tR 8.6 min) and compound 11 (0.008 g, tR 9.1 min). Fr. 5 (15.330 g) was separated with an aqueous elution of dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 1
methanol (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) with a silica gel column and further purified by semi-preparative high performance liquid chromatography with methanol
2) with a silica gel column and further purified by semi-preparative high performance liquid chromatography with methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (v/v = 70
water (v/v = 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) as the mobile phase to obtain compound 12 (0.012 g, tR 8.5 min).
30) as the mobile phase to obtain compound 12 (0.012 g, tR 8.5 min).
In addition, a total of 159.800 g of ethyl acetate extract (ACEE) was dissolved in methanol and then mixed with silica gel at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, and then loaded onto a chromatograph over a silica gel column (85 × 1200 mm) using a gradient of petroleum ether–ethyl acetate solvent system as eluant to obtain fractions Fr. 1 to Fr. 12 based on the TLC analysis. Eluents from the resin column were dried under vacuum. Among them, Fr. 3 (0.450 g) was separated with an aqueous elution of dichloromethane
3, and then loaded onto a chromatograph over a silica gel column (85 × 1200 mm) using a gradient of petroleum ether–ethyl acetate solvent system as eluant to obtain fractions Fr. 1 to Fr. 12 based on the TLC analysis. Eluents from the resin column were dried under vacuum. Among them, Fr. 3 (0.450 g) was separated with an aqueous elution of dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 1
methanol (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with Toyopearl HW-40F and further purified by petroleum ether
1) with Toyopearl HW-40F and further purified by petroleum ether![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate (v/v = 25
ethyl acetate (v/v = 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with silica gel to obtain compound 13 (0.004 g) and compound 14 (1.000 g). Fr. 5 (0.430 g) was separated with an aqueous elution of petroleum ether
1) with silica gel to obtain compound 13 (0.004 g) and compound 14 (1.000 g). Fr. 5 (0.430 g) was separated with an aqueous elution of petroleum ether![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate (v/v = 1
ethyl acetate (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with Toyopearl HW-40F and further purified by silica gel with petroleum ether
1) with Toyopearl HW-40F and further purified by silica gel with petroleum ether![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate (v/v = 15
ethyl acetate (v/v = 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and then compound 15 (0.004 g) was obtained. Fr. 7 (0.5 g) was separated with an aqueous elution of dichloromethane
1), and then compound 15 (0.004 g) was obtained. Fr. 7 (0.5 g) was separated with an aqueous elution of dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 1
methanol (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with Toyopearl HW-40F to get five fractions and Fr. 7.3 was further purified by silica gel with dichloromethane
1) with Toyopearl HW-40F to get five fractions and Fr. 7.3 was further purified by silica gel with dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 5
methanol (v/v = 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to obtain compound 16 (0.005 g). Fr. 7.4 (0.250 g) was separated with an aqueous elution of petroleum ether
1) to obtain compound 16 (0.005 g). Fr. 7.4 (0.250 g) was separated with an aqueous elution of petroleum ether![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate (v/v = 35
ethyl acetate (v/v = 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with silica gel and then recrystallized in methanol to obtain compound 17 (0.004 g). Fr. 10 (0.980 g) was separated with an aqueous elution of petroleum ether
1) with silica gel and then recrystallized in methanol to obtain compound 17 (0.004 g). Fr. 10 (0.980 g) was separated with an aqueous elution of petroleum ether![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dichloromethane acetate (v/v = 50
dichloromethane acetate (v/v = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to get five fractions, and then Fr. 10.1 was further purified by semi-preparative high performance liquid chromatography with methanol
1) to get five fractions, and then Fr. 10.1 was further purified by semi-preparative high performance liquid chromatography with methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (v/v = 35
water (v/v = 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65) as the mobile phase to obtain compound 18 (0.003 g, tR 15 min). Fr. 10.3 (0.205 g) was separated with an aqueous elution of petroleum ether
65) as the mobile phase to obtain compound 18 (0.003 g, tR 15 min). Fr. 10.3 (0.205 g) was separated with an aqueous elution of petroleum ether![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dichloromethane (v/v = 3
dichloromethane (v/v = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with silica gel and further purified by Toyopearl HW-40F with methanol to obtain compound 19 (0.003 g). Fr. 10.4 (0.245 g) was purified with an aqueous elution of dichloromethane
1) with silica gel and further purified by Toyopearl HW-40F with methanol to obtain compound 19 (0.003 g). Fr. 10.4 (0.245 g) was purified with an aqueous elution of dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 1
methanol (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with Toyopearl HW-40F to obtain compound 20 (0.003 g). Fr. 10.5 (0.220 g) was separated with an aqueous elution of petroleum ether
1) with Toyopearl HW-40F to obtain compound 20 (0.003 g). Fr. 10.5 (0.220 g) was separated with an aqueous elution of petroleum ether![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dichloromethane (v/v = 15
dichloromethane (v/v = 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with silica gel to obtain compound 21 (0.005 g).
1) with silica gel to obtain compound 21 (0.005 g).
A. chinense samples were dissolved in n-butanol and mixed with silica gel at a ratio of 13, and then loaded onto a chromatograph over a silica gel column (85 × 1200 mm, 203.200 g) using a gradient of dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol acetate solvent system as eluant to obtain fractions Fr. 1 to Fr. 8 based on the TLC analysis, eluents from the resin column were dried under vacuum. Among them, Fr. 2 (0.647 g) was separated with an aqueous elution of dichloromethane
methanol acetate solvent system as eluant to obtain fractions Fr. 1 to Fr. 8 based on the TLC analysis, eluents from the resin column were dried under vacuum. Among them, Fr. 2 (0.647 g) was separated with an aqueous elution of dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 1
methanol (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with Toyopearl HW-40F to get seven fractions. Fr. 2.3 was recrystallized with petroleum ether and ethyl acetate to obtain compound 22 (0.004 g). Fr. 2.5 (0.500 g) was separated with an aqueous elution of dichloromethane
1) with Toyopearl HW-40F to get seven fractions. Fr. 2.3 was recrystallized with petroleum ether and ethyl acetate to obtain compound 22 (0.004 g). Fr. 2.5 (0.500 g) was separated with an aqueous elution of dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 1
methanol (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with Toyopearl HW-40F to obtain compound 23 (0.003 g). Fr. 3 (0.470 g) was separated using an aqueous elution of dichloromethane
1) with Toyopearl HW-40F to obtain compound 23 (0.003 g). Fr. 3 (0.470 g) was separated using an aqueous elution of dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 1
methanol (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with Toyopearl HW-40F to get four fractions. Fr. 3.3 was further separated with dichloromethane
1) with Toyopearl HW-40F to get four fractions. Fr. 3.3 was further separated with dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 5
methanol (v/v = 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with silica gel to obtain compound 24 (0.007 g). Fr. 4 (0.400 g) was separated using an aqueous elution of dichloromethane
1) with silica gel to obtain compound 24 (0.007 g). Fr. 4 (0.400 g) was separated using an aqueous elution of dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (v/v = 1
methanol (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with Toyopearl HW-40F to get five fractions. Fr. 4.3 was further separated by petroleum ether
1) with Toyopearl HW-40F to get five fractions. Fr. 4.3 was further separated by petroleum ether![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate (v/v = 15
ethyl acetate (v/v = 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with silica gel to obtain compound 25 (0.005 g).
1) with silica gel to obtain compound 25 (0.005 g).
The compounds isolated from A. chinense were identified with NMR data.
3.4 Compound characterization
Compound (2): amorphous white powder. For 1H NMR (600 MHz, D2O) and (150 MHz, D2O). HR-ESI-MS m/z 141.0790, [M−H]− (calcd for C7H11NO2, 140.0718).3.5 Quantum chemical Gaussian calculation
Quantum chemical Gaussian calculation was conducted according to a previous report with slight modification.55 The relative configuration of compound 14 in mol 2 format in Spartan was opened, and the conformation searched for at the molecular force field level of MMFF to obtain the dominant conformation of each compound. Then, under density functional theory (DFT), at B3LYP/6-31G(d) level, these conformations were put into an optimization and vibration analysis calculation, and no virtual frequency was checked after optimization. On this basis, the Boltzmann distributions of each dominant conformation were calculated, and then the non-dominant conformation with Gibbs free energy greater than 1.5 kcal mol−1 was removed, and the optimized dominant conformation was obtained.In addition, these dominant conformations were put into NMR calculations, and using DFT and the Gaussian default GIAO method, the magnetic shielding value of each carbon atom and hydrogen atom were calculated under gas-phase conditions at rmpw1pw91/6-31+G(d, p) level. According to the proportion of Boltzmann distribution and their respective phases of Gibbs free energy (ΔG), the average values of magnetic shielding values of all conformations of one compound can be obtained. Meanwhile, using tetramethylsilane (TMS) as a reference, the magnetic shielding values of four carbon atoms and 12 hydrogen atoms were calculated. Then all conformations of one compound were subtracted from the average value of these hydrocarbon atoms and the magnetic shielding values of the hydrocarbon atoms to get the theoretical chemical shift value of each hydrocarbon atom we need.
Finally, the calculated chemical displacement value of each hydrocarbon atom calculated by NMR and the actually measured chemical displacement value of each hydrocarbon atom adopted the feasibility analysis of DP4+, which will generate the corresponding probability data.
3.6 Analysis of CD calculation
Circular dichroism (CD) spectra were calculated according to reported methods.56 Four kinds of geometric optimization of compound 14 were calculated with the density functional theory (DFT) method and time-dependent DFT (TDDFT) via the Gaussian 09 program. The ECD spectra were optimized at the APFD/6-311+G(2d, p) level in MeOH. The computational data were fitted in OriginPro 2019b (OriginLab Inc., USA).3.7 Study of the anti-inflammatory activity of octocarpus COX-2 inhibitors
The study of the COX-2 enzyme inhibitory activity of the samples was performed according to the instructions for the kit. Compounds to be tested with 6.25, 25, 50, 100, 200, 400, 800 μM were prepared and celecoxib was used a positive control drug, and the extracts were 5, 25, 50, 100, 200, 400, 800 μg mL−1.Each sample was provided three times. The procedure was to add samples to wells separately, set up 2 replicate wells for each sample, incubate at 37 °C for 10 min, add 5 μL of COX-2 Probe and 5 μL of COX-2 substrate working solution to each well, centrifuge and mix well. Performed on a culture plate shaker, the incubation was timed for 5 min at 37 °C protected from light and then the fluorometric assay was performed. The excitation wavelength was 560 nm and the emission wavelength was 590 nm. The average fluorescence values were calculated for each sample well and a blank control well, which were recorded as RFU Blank Control, RFU 100% Enzyme Activity Control, RFU Positive Inhibitor Control and RFU Sample, RFU Relative Fluorescence Unit, respectively.
The percent inhibition for each sample was calculated with the following formula: Inhibition (%) = (RFU 100% enzyme activity control − RFU sample)/(RFU 100% enzyme activity control − RFU blank control) × 100% and the IC50 of the inhibitor was calculated.
3.8 Cytotoxic activity assay
The cytotoxic activities of all isolated compounds were tested for their anti-proliferative activity against HepG2, Caco-2, and HeLa cell lines with a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay.57 Cells were seeded at a density of 5 × 103 per well into 96-well plates for 24 h. Then the cells were treated with compounds for 48 h. Control cells were exposed to 0.1% DMSO. The test cells were treated with different concentrations of isolated compounds. Adriamycin (10 μM) was used as a positive control. Then, 20 μL of MTT (5 mg mL−1) was added to each well and incubated for 4 h, after which 150 μL of DMSO was added to each well. The reduction in cell viability was determined at 490 nm by using a microplate reader (Bio-Rad iMARK, Berkeley, CA, USA). Three replicates were obtained for each treatment.3.9 Molecular docking
To investigate the relationship between isolated ellagic acid compounds and COX-2, compounds 13, 17, and 18 were used as ligands in an in silico molecular docking study. The three-dimensional structure of COX-2 (PDB ID 5IKR) was obtained from the online Protein Data Bank (PDB). Optimization of ligand structures was processed with ChemBioDraw (PerkinElmer), and the molecular docking was implemented using AutoDock Tools (version 1.5.6), AutoDock 4.2 package (Autodock 4.2 and Autogrid 4.2), with the help of the Discovery Studio Visualizer (version 16.1.0.15350), which can be used to better visualize the interactions between ligands and receptors. X, Y, and Z were set to 38.889, 1.917, and 61.611 Å in the cubic grid box as the central dimensions of COX-2. Water molecules and persistent inhibitors were removed from the crystal structure of 5IKR. Blind docking over the whole receptor was carried out using the Genetic Algorithm method, and the resultant complex structures were investigated by using the conformations with the most favorable binding energy.4 Conclusions
It has been reported that in clinical practice, A. chinense is mostly used to treat rheumatoid arthritis, numbness and paralysis, heart failure, strain, low back pain, traumatic injury and other diseases, and its curative effect is accurate. A. chinense is widely used in Miao medicine. Among the prescriptions of Miao medicine for the treatment of rheumatic diseases, A. chinense has been used 44 times, with a frequency of 12.6%, ranking fourth.58 Although the star anise maple has good pharmacological activity and rich medication experience in China, there are still many problems that need further research. Modern pharmacology has proved that A. chinense and its extracts have many pharmacological functions. This study enriched the understanding of more compounds in the fibrous roots of A. chinense through the separation of compounds from the fibrous roots of A. chinense, especially using ethyl acetate.In this study, one new (14) and twenty-four known compounds (1–13, 15–25) were isolated, and seven compounds were isolated from this plant for the first time. After screening the anti-inflammatory and antitumor activities of the isolated compounds in vitro, it was found that compounds 1, 2, 3, 4, 13, 14, 16, 17 and 18 may have potential anti-inflammatory effects and compound 18 showed the lowest binding energy with COX-2 enzyme with molecular docking technology. It is preliminarily speculated that it is one of the main active components with anti-inflammatory effects. The above two components may be the anti-inflammatory active components of the fibrous root of A. chinense. The specific mechanism research group will carry out the next research step to provide a basis for follow-up basic research on active substances. While compounds 1, 5, 13, 20, and 21 showed cytotoxicity against HepG2, Caco-2 and HeLa cells, so they may be at least partly responsible for the antitumor activity of A. chinense. Those compounds can be used as the basis for pharmacological activity research in later stages for anti-inflammatory and anticancer effects.
Author contributions
T. X. and Z. G.: conceptualization, Gaussian and ECD calculations, writing, reviewing and editing; J. H.: purification, isolation of compounds, and COX-2, cancer cells activity assays; X. C.: formal analysis, data curation; L. T.: reviewing and editing; X. S.: conceptualization, reviewing and editing.Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
This research was supported by grant from the Basic Scientific Research Project of Guizhou Province (grant number [2021] 535, [2021] 540), the Guizhou Medical University Breeding programs of National Natural Science Foundation of China (grant number 19NSP074), the PhD Start-up Fund of Guizhou University of Traditional Chinese Medicine (grant number 2019-10), the projects of State Key Laboratory of Function and Applications of Medicinal Plants, Guizhou Medical University (grant number FAMP201908K).References
- K. Zhai, H. Duan, G. J. Khan, H. Xu, F. Han, W. Cao, G. Gao, L. Shan and Z. Wei, Salicin from Alangium chinense ameliorates rheumatoid arthritis by modulating the Nrf2-HO-1-ROS pathways, J. Agric. Food Chem., 2018, 66, 6073–6082 CrossRef CAS PubMed.
- X. Wei, J. Yang, Z. Dai, H. Yu, C. Ding, A. Khan, Y. Zhao, Y. Liu and X. Luo, Antitumor pyridine alkaloids hybrid with diverse units from Alangium chinense, Tetrahedron Lett., 2020, 61, 1–7 Search PubMed.
- X. Hu, X. Wei, Y. Zhou, X. Liu, J. Li, W. Zhang, C. Wang, L. Zhang and Y. Zhou, Genus Alangium - a review on its traditional uses, phytochemistry and pharmacological activities, Fitoterapia, 2020, 147, 104773 CrossRef CAS PubMed.
- F. Gao and S. Zhang, Salicin inhibits AGE-induced degradation of type II collagen and aggrecan in human SW1353 chondrocytes: therapeutic potential in osteoarthritis, Artif. Cells, Nanomed., Biotechnol., 2019, 47(1), 1043–1049 CrossRef CAS PubMed.
- Y. Zhang, Y. B. Liu, Y. Li, L. Li, S. G. Ma, J. Qu, J. D. Jiang, X. G. Chen, D. Zhang and S. S. Yu, Terpenoids from the roots of Alangium chinense, J. Asian Nat. Prod. Res., 2015, 17, 1025–1038 CrossRef CAS PubMed.
- Y. Zhang, Y. B. Liu, Y. Li, L. Li, S. G. Ma, J. Qu, J. D. Jiang, X. G. Chen, D. Zhang and S. S. Yu, Terpenoids from the roots of Alangium chinense, J. Asian Nat. Prod. Res., 2015, 17, 1025–1038 CrossRef CAS PubMed.
- H. H Xing, K. Zhou, Y. Yang, L. Zhou, W. Dong, Y. D. Wang, H. Y. Ma, M. Zhou, Y. Q. Ye and Q. F. Hu, A new cytotoxic alkaloid from roots of Alangium chinense, China J. Chin. Mater. Med., 2017, 42, 303–306 Search PubMed.
- Y. Liu, Z. T. Xu, B. Zhao, T. Wu and X. Q. Cai, Advances in pharmacy research of Miao medicine Alangium chinense, Stud. Trace Elem. Health, 2012, 29, 57–60 CAS.
- Y. F. Wu, J. Gong, T. Zhao, W. J. Fan, C. G. Fan, X. Li and S. F. Ni, Overview of pharmaceutical research on Alangium Lam, J. Anhui Agric. Sci., 2010, 38, 10676–10677 Search PubMed.
- V. Sharma, P. Bhatia, O. Alam, M. J. Naim, F. Nawaz, A. A. Sheikh and M. Jha, Recent advancement in the discovery and development of COX-2 inhibitors: Insight into biological activities and SAR studies (2008-2019), Bioorg. Chem., 2019, 89, 103007 CrossRef CAS PubMed.
- B. Hinz and K. Brune, Cyclooxygenase-2-10 years later, J. Pharmacol. Exp. Ther., 2002, 300, 367–375 CrossRef CAS PubMed.
- W. Zhao, B. Shan, D. He, Y. Cheng, B. Li, C. Zhang and C. Duan, Recent progress in characterizing long noncoding RNAs in cancer drug resistance, J. Cancer., 2019, 10, 6693–6702 CrossRef CAS PubMed.
- J. M Llovet, R. K. Kelley, A. Villanueva, A. G. Singal, E. Pikarsky, S. Roayaie, R. Lencioni, K. Koike, J. Zucman-Rossi and R. S. Finn, Hepatocellular carcinoma, Nat. Rev. Dis. Prim., 2021, 7, 6 CrossRef PubMed.
- M. Ringelhan, D. Pfister, T. O'Connor, E. Pikarsky and M. Heikenwalder, The immunology of hepatocellular carcinoma, Nat. Immunol., 2018, 19, 222–232 CrossRef CAS PubMed.
- P. Favoriti, G. Carbone, M. Greco, F. Pirozzi, R. E. M. Pirozzi and F. Corcione, Worldwide burden of colorectal cancer: a review, Updates Surg., 2016, 68, 7–11 CrossRef PubMed.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- X. Wang and P. A. Ward, Opportunities and challenges of disease biomarkers: a new section in the journal of translational medicine, J. Transl. Med., 2012, 10, 240 CrossRef PubMed.
- F. G. Tumours, WHO classification of tumors, WHO press, 5th edn, Geneva, Switzerland, 2020 Search PubMed.
- World Health Organization, International agency for research on cancer (IARC); global cancer observatory (GCO), Available online: https://gco.iarc.fr.
- J. Ferlay, M. Colombet, I. Soerjomataram, D. M. Parkin, M. Piñeros, A. Znaor and F. Bray, Cancer statistics for the year 2020: An overview, Int. J. Cancer, 2021, 149, 778–789 CrossRef CAS PubMed.
- A. D. Kaprin, V. V. Starinskiy and A. O. Shakhzadova, The state of cancer care for the population of Russia in 2019, M.: MNIOI named after P.A. Herzen-Branch of the Federal State Budgetary Institution “National Medical Research Center of Radiology” of the Minzdrav of Russia, 2020 Search PubMed.
- J. Wei, Y. Li, H. Mo, N. Chen and R. Lu, Study on chemical constituents of Zhuang medicine Sauropus rostratus, Lishizhen Med. Mater. Med. Res., 2017, 28, 289–291 Search PubMed.
- S. Que, Y. Zhang and Y. Zhao, Chemical constituents of Tibetan medicine Oxytropis falcata, Chin. Tradit. Herb. Drugs, 2007, 38, 1458–1460 CAS.
- R. Wang, D. Wu, H. Gao, B. Sun, J. Huang and L. Wu, Isolation and identification of steroids from the leaves of the Magnolia sieboldii K. Koch, J. Shenyang Pharm. Univ., 2009, 26, 874–877 CAS.
- Y. Yang, L. He, A. Yang, G. Shi and R. Lu, Studies on Chemical Constituents of Tibetan Medicine Rabdosia breviflora, China J. Chin. Mater. Med., 2005, 30, 153–154 Search PubMed.
- X. Tian, G. Ding, C. Peng, Y. Hu, Z. Ma and Z. Zou, Liposoluble Constituents from Dichrocephala benthamii C. B. Clarke, Chin. Pharm. J., 2012, 47, 1366–1369 CAS.
- Y. Zhang, Y. Liu, Y. Li, S. Ma, L. Li, J. Qu, D. Zhang, X. Chen, J. Jiang and S. Yu, Sesquiterpenes and Alkaloids from the Roots of Alangium chinense, J. Nat. Prod., 2013, 76, 1058–1063 CrossRef CAS PubMed.
- H. Shimomura, Y. Sashida and M. Oohara, Lignans from Machilus Thunbergii, Phytochemistry, 1988, 27, 634–636 CrossRef CAS.
- L. Tsai, J. Chen, C. Duh and I. Chen, Cytotoxic Neolignans and Butanolides from Machilus obovatifolia, Planta Med., 2001, 67, 559–561 CrossRef PubMed.
- S. W. Chang, K. H. Kim, I. K. Lee, S. U. Choi and K. R. Lee, Phytochemical Constituents of Geranium eriostemon, Nat. Prod. Sci., 2009, 15, 151–155 CAS.
- C. Kong, J. I. Lee, J. Kim and Y. Seo, In Vitro Evaluation on the Antiobesity Effect of Lignans from the Flower Buds of Magnolia denudata, J. Agric. Food Chem., 2011, 59, 5665–5670 CrossRef CAS PubMed.
- H. Ye, L. Chen, Y. Li, A. Peng, A. Fu, H. Song, M. Tang, H. Luo, Y. Luo, Y. Xu, J. Shi and Y. Wei, Preparative isolation and purification of three rotenoids and one isoflavone from the seeds of Millettia pachycarpa Benth by high-speed counter-current chromatography, J Chromatogr A, 2008, 1178, 101–107 CrossRef CAS PubMed.
- S. Numonov, S. Edirs, K. Bobakulov, K. Bozorov, F. Sharopov, W. Setzer, H. Zhao, M. Habasi, M. Sharofova and H. Aisa, Evaluation of the antidiabetic activity and chemical composition of geranium collinum root extracts-computational and experimental investigations, Molecules, 2017, 22, 983 CrossRef PubMed.
- S. Choi and R. B. Silverman, In activation and inhibition of γ-aminobutyric acid aminotransferase by conformation ally restricted vigabatrin an alogues, J. Med. Chem., 2002, 45, 4531–4539 CrossRef CAS PubMed.
- C. Yang and D. D. Tanner, A simple synthesis of (±)-1,2,3,6-tetrahydro-2,3′-bipyridine (anatabine) and (±)-3-(2-piperidinyl)pyridine (anabasine) from lithium aluminum hydride and pyridine, Can. J. Chem., 1997, 75, 616–620 CrossRef CAS.
- J. Liu, M. Yu, S. Wang and G. Zhang, Study on chemical constituents from Euphorbia thymifolia, China J. Chin. Mater. Med., 2020, 45, 5226–5231 Search PubMed.
- Q. Zhang, A. Wang, S. Fan, X. Ma, C. Wang and J. Jia, Phenolic acid derivatives of Euphorbia ebracteolata, China J. Chin. Mater. Med., 2017, 42, 2995–2998 Search PubMed.
- A. U. Rahman, F. N. Ngounou, M. I. Choudhary, S. Malik, T. Makhmoor, M. Nur-E-Alam, S. Zareen, D. Lontsi, I. F. Ayafor and B. L. Sondengam, New antioxidant and antimicrobial ellagic acid derivatives from Pteleopsis hylodendron, Planta Med., 2001, 67, 335–339 CrossRef PubMed.
- H. J. Jung, H. A. Jung, S. S. Kang, J. Lee, Y. S. Cho, K. H. Moon and J. S. Choi, Inhibitory activity of Aralia continentalis roots on protein tyrosine phosphatase 1B and rat lens aldose reductase, Arch. Pharmacal Res., 2012, 35, 1771–1777 CrossRef CAS PubMed.
- J. Gao, J. Sheng, X. Yang and J. Liu, The constituents of Russula ochroleuca basidiomycetes, Acta Bot. Yunnan., 2001, 23, 385–393 CAS.
- W. Holzer, G. A. Eller, B. Datterl and D. Habicht, Derivatives of pyrazinecarboxylic acid: 1H, 13C and 15N NMR spectroscopic investigations, Magn. Reson. Chem., 2010, 47, 617–624 CrossRef PubMed.
- S. K. EI-Desouky, K. H. Kim, S. Y. Ryu, A. F. Eweas, A. M. GamaI-Eldeen and Y. Kim, A new pyrrole alkaloid isolated from Arum palaestinum Boiss. and its biological activities, Arch. Pharmacal Res., 2007, 30, 927–931 CrossRef PubMed.
- S. Mondal and K. M. Sureshan, Total syntheses and structural validation of lincitol A, lincitol B, uvacalol I, uvacalol J, and uvacalol K, Org. Biomol. Chem., 2014, 12, 7279–7289 RSC.
- X. Cui, Z. Zheng, Y. Li, K. Yu, Y. Wang and Q. Yao, Chemical constituents from whole herbs of Viola yedoensis, Chin. Tradit. Herb. Drugs, 2021, 52, 917–924 Search PubMed.
- X. Li, L. L. Mazaleuskaya, L. L. Ballantyne, H. Meng, G. A. FitzGerald and C. D. Funk, Genomic and lipidomic analyses differentiate the compensatory roles of two COX isoforms during systemic inflammation in mice, J. Lipid Res., 2018, 59, 102–112 CrossRef CAS PubMed.
- K. Subbaramaiah and A. J. Dannenberg, Cyclooxygenase-2 transcription is regulated by human papillomavirus 16 E6 and E7 oncoproteins: evidence of a corepressor/coactivator exchange, Cancer Res., 2007, 67, 3976–3985 CrossRef CAS PubMed.
- R. J. Flower, The development of COX-2 inhibitors, Nat. Rev. Drug Discovery, 2003, 2, 179–191 CrossRef CAS PubMed.
- R. G. Kurumbail, A. M. Stevens, J. K. Gierse, J. J. McDonald, R. A. Stegeman, J. Y. Pak, D. Gildehaus, J. M. Miyashiro, T. D. Penning, K. Seibert, P. C. Isakson and W. C. Stallings, Structural basis for selective inhibition of cyclooxygenase-2 by anti-inflammatory agents, Nature, 1996, 384, 644–648 CrossRef CAS PubMed.
- V. Sharma, P. Bhatia, O. Alam, M. J. Naim, J. Nawaz, A. A. Sheikh and M. Jha, Recent advancement in the discovery and development of COX-2 inhibitors: insight into biological activities and SAR studies (2008-2019), Bioorg. Chem., 2019, 89, 103007 CrossRef CAS PubMed.
- V. B. Rahimi, M. Ghadiri, M. Ramezani and V. R. Askari, Anti-inflammatory and anti-cancer activities of pomegranate and its constituent, ellagic acid: evidence from cellular, animal, and clinical studies, Phytother. Res., 2020, 34, 685–720 CrossRef PubMed.
- J. Ríos, R. M. Giner, M. Marín and M. C. Recio, A pharmacological update of ellagic acid, Planta Med., 2018, 84, 1068–1093 CrossRef PubMed.
- Z. Lin, C. Lin, C. Fu, H. Lu, H. Jin, Q. Chen and J. Pan, The protective effect of ellagic acid (EA) in osteoarthritis: an in vitro and in vivo study, Biomed. Pharmacother., 2020, 125, 109845 CrossRef CAS PubMed.
- I. D. Karidio and S. H. Sanlier, Reviewing cancer's biology: an eclectic approach, J. Egypt. Natl. Cancer Inst., 2021, 33, 32 CrossRef PubMed.
- T. Xiao, Z. Luo, Z. Guo, X. Wang, M. Ding, W. Wang, X. Shen and Y. Zhao, Multiple roles of black raspberry anthocyanins protecting against alcoholic liver disease, Molecules, 2021, 26, 2313 CrossRef CAS PubMed.
- F. Yuan, F. Xu, R. Fan, W. Li, D. Huang, G. Tang, T. Yuan, L. Gan and S. Yin, Structural elucidation of three 9,11-Seco tetracyclic triterpenoids enables the structural revision of euphorol J, J. Org. Chem., 2021, 86, 7588–7593 CrossRef CAS PubMed.
- X. Luo, J. Cai, Z. Yin, P. Luo, C. Li, H. Ma, N. P. Seeram, Q. Gu and J. Xu, Fluvirosaones A. and B., two indolizidine alkaloids with a pentacyclic skeleton from Flueggea virosa, Org. Lett., 2018, 20, 991–994 CrossRef CAS PubMed.
- X. Luo, C. Li, P. Luo, X. Lin, H. Ma, N. P. Seeram, C. Song, J. Xu and Q. Gu, Pterosin sesquiterpenoids from Pteris cretica as hypolipidemic agents via activating liver X receptors, J. Nat. Prod., 2016, 79, 3014–3021 CrossRef CAS PubMed.
- G. Wu, Z. H. Tian, Z. L. Xu and Y. M. Zhang, Study on the application of association rules to the use of medicine in the treatment of rheumatic diseases in Miao medicine, Lishizhen Med. Mater. Med. Res., 2013, 24, 447–449 Search PubMed.
Footnote |
| † These authors contributed equally to this article. |
| This journal is © The Royal Society of Chemistry 2023 |