DOI:
10.1039/D2RA08103B
(Paper)
RSC Adv., 2023,
13, 9186-9194
A universal green approach for the synthesis of NPS-codoped carbon quantum dots with enhanced broad-spectrum antibacterial and antioxidant activities†
Received
19th December 2022
, Accepted 5th March 2023
First published on 20th March 2023
Abstract
Bio-inspired quantum dots have received widespread attention in recent years due to their great potential for biological applications. Herein, we report a one pot hydrothermal synthesis of nitrogen–phosphorus–sulphur (NPS)-codoped carbon quantum dots from endophytic bacteria without using any additional doping precursor. The synthesized CQDs were thoroughly characterized and interestingly found to have a graphene like structure. The synthesized CQDs were then utilized in bactericidal activities against Gram-negative bacteria like Salmonella typhi, Pseudomonas aeruginosa and Gram-positive bacteria like the Bacillus subtilis strain. The strains were treated with different concentrations ranging from 5–100 µg ml−1. The 5 µg ml−1 concentration appeared to be the MIC (minimum inhibitory concentration) and 100 µg ml−1 is the MBC (minimum bactericidal concentration) maintaining a short incubation period of one hour. A simple, cost-effective and eco-friendly approach to synthesize multi-elemental doped CQDs would certainly cause the method to be used in future for diverse biological applications. As compared to the broadly used antibiotics, the developed CQDs have some added advantages including lower cytotoxicity, excellent photo-stability and high selectivity.
1 Introduction
Since their discovery in 2004, carbon quantum dots (CQDs) have been widely utilized in various fields owing to their very attractive properties, e.g. chemical inertness, high water solubility, low toxicity, ease of functionalization and photo-stability.1–3 At present, CQDs are utilized in every possible field including catalysis, sensing, bio-imaging, optoelectronic devices, energy storage devices, drug and gene delivery, bactericidal agents, etc.4–6 In contrast to conventional metal/semiconductor quantum dots, CQDs have less toxicity and higher biocompatibility, which make them an ideal material to be utilized in biomedical fields.7,8 Studies have shown that heteroatom doping can significantly improve the properties of CDs. In this area, currently, extensive research is going on to establish easy and economic methods to synthesize heteroatom-doped CQDs. Heteroatoms (e.g. N, P, S etc.) with different size and electronegativities than carbon create defects in the crystal structure of CQDs and thus introduce high density of active sites, which will make them suitable for various applications.9,10 In addition, N, S and P co-doping may also provide better carrier separation for enhanced activities. Therefore, defects mediated reactive oxygen species and enhanced charge separation will make NPS-doped-CQDs an ideal candidate for bactericidal activities.11
In literature, mainly chemical methods have given to synthesize doped-CQDs utilizing the heteroatom containing compounds as precursor.12 However, biosynthesis of any nanoparticles has been proposed as the cost-effective and environmentally friendly alternative to chemical and physical methods.13,14 Bio-synthesized nanoparticles are highly recommended especially for biological applications. Recently, different biological precursors containing different heteroatoms (mainly N, P and S) are used to fabricate doped-CQDs.15 Wang et al. used cucumber juice to synthesize NPS co-doped CQDs by hydrothermal method.16 However, for large-scale synthesis, plant source-mediated synthesis is not encouraging for environmental imbalance. In this perspective, use of bacteria, which are easy to grow on laboratory on mass scale, could be an alternate option to use as carbon precursor in CQDS. Very recently, Zhang et al. used bacteria isolated from the industrial sewage of a Kunming steel factory to synthesize CQDs.17 Again, although CQDs itself is non-hazardous, their precursors are not always safe. Therefore, use of endophytic bacteria as carbon precursors could be the best alternate option, which is yet to be explored.18
Microbial contaminations are one of the major threat to public health throughout the world. Microorganisms now have developed various tactics to bypass the antimicrobial compounds. Major concern in this area is the multidrug resistance (MDR) developed by the microorganism against the antibiotic drugs.19 With the advancement in technology, currently, different nanomaterials are used to replace the conventional antibiotics and fight with the problem of MDRs. However, majority of the materials possess threats related to environmental hazards, poor biocompatibility and high cost. Therefore, cheap, durable, sustainable and effective antimicrobial agents for long-term applications are in high demand. In this context, utilization of CQDs could provide an effective solution. Due to their small size CQDs interact with the bacterial species making them an excellent nanomaterial to be used for bactericidal properties.20,21 Maintaining the cell membrane integrity is one of the crucial factors required for the survival of bacteria. The main mechanism behind the bactericidal activity of the carbon quantum dots is related to their ability to disrupt the cell-membrane integrity.22–24 Even though, there are few reports on the utilization of CQDs for antibacterial activities, most of them exhibit their activities at acidic pH but have poor activity under physiological conditions of around neutral pH.
Current work has established a general green method for the synthesis of NPS co-doped CQDs which are highly active under neutral pH. We have isolated endophytic bacteria from the plant urtica dioica and used as the starting precursor material for the synthesis of CQDs via one pot hydrothermal synthesis method. The urtica dioica is a commonly available plant and known to have different therapeutic and medicinal properties. Among the isolated endophytic bacteria, Bacillus megatarium known to be an important industrial bacterium since ages was further used. Bacteria-derived CQDs displays excellent antimicrobial activities against all the tested pathogenic strains viz. Salmonella typhi, Pseudomonas aeruginosa and Bacillus subtilis at neutral pH. To the best of our knowledge, this is the first report of any endophytic bacteria-derived indigenous NPS co-doped CQDs which are synthesised in one-step process without any foreign doping precursors.
2 Experimental
2.1 Chemicals for isolation of endophytic bacteria and synthesis of CQDs
Tween 20, sodium hypochlorite, ethanol, hydrogen peroxide and mercuric chloride was purchased from Sisco Research Laboratories Pvt. Ltd. Dialysis membrane MWCO of >1 kDa. was obtained from Sigma-Aldrich.
2.2 Chemicals for antibacterial assay
Luria-Bertani broth and agar powders were obtained from Hi-Media Laboratories. The Gram-negative and Gram-positive bacterial strains MTCC 3224, MTCC 424 and MTCC 441 were purchased from MTCC Chandigarh, India.
2.3 Isolation of endophytic bacteria
The leaf and stem samples of Urtica dioica L. were collected using a sterile scalpel from DBT campus, Bhimtal, K. U. Uttarakhand. The samples were kept in sterile plastic bags and were used within 24 hours for further studies. To remove the surface epiphytes, the samples were thoroughly washed followed by washing in Tween 20 for 1 minute in laminar air flow. It is then given treatment of 1–5% sodium hypochlorite for 2–10 minutes then Ethanol 70–95% for 30 seconds finally 0.2% hydrogen peroxide and mercuric chloride for 2–5 minutes. In laminar air flow after proper drying of the sample the surface of the stem was removed and the leaves were cut into small pieces and placed on N. A. medium on the sterile plates. The plates were then incubated at 37 °C for 24 hours.25 The colonies obtained were differentiated on the basis of morphology. Single colonies were picked from plates and streaked to obtain pure bacterial colonies. The inoculation process repeated till monocultures were obtained for further experimentation.
2.4 Synthesis of CQDs
CQDs were prepared through one pot hydrothermal carbonization of B-1 bacterial strain. The isolated bacterial strain was grown overnight with shaking in L. B. broth. The culture was then centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g, 15 min, 4 °C). The cells were finally washed with double distilled water for at-least three times to remove the supernatant. The pellet obtained was then re-suspended in 30 ml of distilled water (30 ml). Subsequently, the bacterial suspension was transferred into a Teflon-lined stainless-steel autoclave and heated up to 200 °C for 24 hours. A light brownish solution was obtained. The obtained solution was then filtered through a syringe filter (0.22 µm) diameter to remove large particles. Finally, after dialyzing using dialysis membrane of (MWCO > 1 kDa) against deionized water for 72 hours, the CQD solution was obtained and stored at 4 °C for further experiments.26 Repeatability was further checked by repeating the same experiment thrice.
000 g, 15 min, 4 °C). The cells were finally washed with double distilled water for at-least three times to remove the supernatant. The pellet obtained was then re-suspended in 30 ml of distilled water (30 ml). Subsequently, the bacterial suspension was transferred into a Teflon-lined stainless-steel autoclave and heated up to 200 °C for 24 hours. A light brownish solution was obtained. The obtained solution was then filtered through a syringe filter (0.22 µm) diameter to remove large particles. Finally, after dialyzing using dialysis membrane of (MWCO > 1 kDa) against deionized water for 72 hours, the CQD solution was obtained and stored at 4 °C for further experiments.26 Repeatability was further checked by repeating the same experiment thrice.
2.5 Quantum yield measurement
The quantum yield (QY) was determined using the standard procedure27 with quinine sulphate in 0.1 M H2SO4 as a reference (QY = 54 percent excited at 338 nm = 1.33). The following equation can be used to calculate QY:
| QCQDs = QR(ICQDs/IR) (AR/ACQDs)(ηCQDs2/ηR2) |
where QCQDs is the quantum yield of the as-prepared CQDs, I is the integrated emission intensity, η is the refractive index of the solvent utilized, and A is the absorbance at the excitation wavelength. The abbreviations CQDs and R stand for “prepared CQDs” and “reference,” respectively.
2.6 Characterization of endophytic bacteria
The endophytic bacterial strain was identified via molecular identification. The kit obtained from Hi-media was used to isolate genomic DNA. To identify the strain, PCR amplification of extracted DNA with 16S universal primers was used. After completion of PCR, the PCR results were verified on a 1% agarose gel electrophoresis using the reference ladder and observed under UV light by using ethidium bromide. By comparing the PCR product to a ladder, the DNA product was identified. To investigate phylogenetic relationships, the isolate's 16s sequences were matched with sequencing data of bacterial strains which are taxonomically identical (retrieved from the NCBI) and a phylogram was created using the MEGA X software. To obtain the more accurate evolutionary history of our sequence we performed bootstrap to create 1000 replicas of the phylogenetic tree generated using neighbor joining method.
2.7 Characterization of CQDs
Size determination of CQDs were determined by using a Transmission Electron Microscope (JEM-2100F, 200 kV, JEOL, Fresing, Germany). The average size diameter was calculated using image J software. Fluorescence spectra were recorded on the LS 55 (PerkinElmer) fluorescence spectrophotometer with excitation range of 200–800 nm. UV-vis spectra from were obtained by Thermo Scientific Evolution 260 B10. FT-IR analysis was recorded using PerkinElmer Spectrum 2 using ATR method. X-ray Photoelectron Spectrum (XPS) was obtained using ESCA+, (omicron nanotechnology, Oxford Instrument Germany) outfitted with a monochromator aluminium source (Al Kα radiation, ħν = 1486.7 eV).
2.8 Antibacterial assay
Using the standard colony forming unit (CFU) counting method antibacterial assay was performed. Overnight grown cultures of Gram-negative bacteria Salmonella typhi, Pseudomonas aeruginosa and Gram-positive bacteria Bacillus subtilis were used. These cultures were grown in L. B. broth at 37 °C for 24 h with shaking at 150 rpm. The cultures were then diluted with a dilution factor of 10−7. Different concentration of CQDs starting from 5 µg ml−1, 25 µg ml−1, 50 µg ml−1 and 100 µg ml−1 was taken and incubated at 37 °C for 1 hour with the diluted bacterial culture. After an hour interval the subsequent bacterial culture with CQDs were plated on L. B agar plates and incubated overnight at 37 °C. The experiment was performed in triplicates. Percentage survival rate was calculated using the following equation.
| % survival rate = [((number of colonies)control − (number of colonies)sample)/(number of colonies)control] × 100 |
2.9 DPPH assay for antioxidant evaluation
DPPH assay was done to evaluate the antioxidant potential of the synthesized CQDs. DPPH (1,1-diphenylpicrylhydrazyl) over the course of time it changes its color from deep violet to colorless in the presence of antioxidants. The assay was performed using the biosynthesized CQDs using Ruiz et al. (2017) method with some modifications.28 For the assay different concentrations (10, 20, 30, 40, 50 and 60 µg ml−1) of CQDs was taken. 1 ml of methanolic DPPH (4 mg/100 ml) was mixed with 1 ml of different methanolic concentrations. The samples were then loaded onto the wells of 96 micro-titer plate and the plate was kept in dark for 30 minutes. The absorbance was recorded at 517 nm in Elisa plate reader. Ascorbic acid (1 mg ml−1) as positive and methanol as negative control was taken. The experiments were done in triplicates. The percentage scavenging activity was calculated as:
| % scavenging activity = [(Acontrol − Asample)/(Acontrol)] × 100 |
where A is the absorbance.
3 ABTS radical scavenging activity
ABTS 2,2′-azino-bis (3- ethylbenzothiazoline-6-sulphonic acid) assay is the widely used assay to determine the antioxidant potential of the compound. The assay is mainly based on quenching of ABTS˙ radical. The antioxidant capacity was determined according to Re et al. (1999) with slight modifications.29 A 7 mM ABTS stock solution was prepared. The solution was then reacted with a stock solution of 2.45 mM potassium persulfate to generate the ABTS radical cation (ABTS+). Before use the final solution was kept in dark for 12–16 h and the absorbance of solution was adjusted to 0.70 O. D value at 734 nm. Different concentration of CQDs were then treated with ABTS to a final concentration of 3 ml. The samples were then incubated for 6 min after which the absorbance was determined at 734 nm. The percentage inhibition was calculated by:
| % inhibition activity = [(Acontrol − Asample)/(Acontrol)] × 100 |
4 Results and discussion
The nucleotide sequence was submitted in the Genbank (Accession No. OK303397). The phylogenetic analysis of 16S DNA sequence of the isolated bacterial culture was obtained using the neighbour-joining method which determines the branch length as well as topology of the phylogenetic tree. The results show 100% similarity with Bacillus megaterium strain of our identified bacterial strain of Bacillus megaterium (Fig. 1). Subsequently the isolated endophytic bacteria were used as precursor for hydrothermal synthesis of doped CQDs.
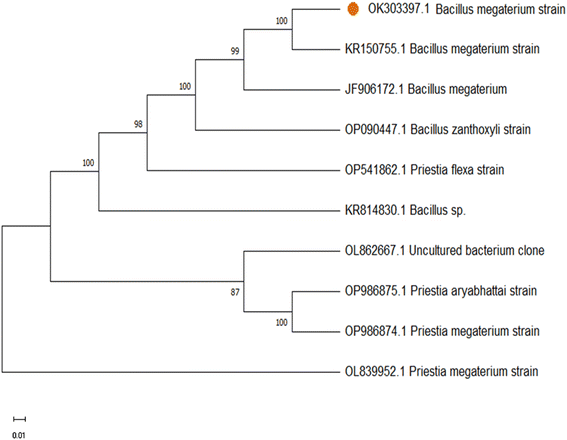 |
| | Fig. 1 Phylogenetic tree of the endophytic bacterial isolate identified as Bacillus megaterium strain. | |
One of the fascinating feature of C-dots is its optical properties. The optical image of CQDs displays brown colour under normal light, whereas it shows bluish-green luminescence under UV light of wavelength 365 nm (inset of Fig. 2a). UV-visible absorption spectrum (Fig. 2a) shows peaks at ∼270 nm and a broad shoulder at ∼330 nm. The former peak arrives due to the π–π* transitions of sp2 carbon atoms, representing the formation of graphitized carbon particles.30 The later peak refers to the n–π* transition mainly due to the presence of different functional groups (especially C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O).31 Upon excitation (wavelength ranges of ∼340–480 nm) it shows broad and strong emission peaks as shown in Fig. 2b. Highest intensity peak observed at 420 nm upon excitation with 360 nm is the effect of different surface states of C-dots.32 The excitation-dependent spectra show a shift in photoluminescence (PL) peak from 420 to 520 nm under excitation range of 340–480 nm as shown in Fig. 2b. Several hypotheses have been proposed to explain the wavelength – dependent emission spectra of CQDs.33 As the carbon particle decreases in size, quantum confinement takes place meaning that the particle is smaller than the de Broglie wavelength of the electron in the dot, creating a deviation from bulk properties. This confinement creates a quantization of the energy into discrete levels in the conduction and valence bands, so the C-dot can be understood as “virtual atom”. The emission energy depends on the radius of the particle so that as the particle gets smaller, both the excitation and emission spectra shift to shorter wavelengths. In a distribution of C-dots with varying sizes, different subsets get preferentially excited as the excitation wavelength varies. As CQDs of different size are excited, the emission spectrum shifts with excitation wavelength.34 Another potential explanation for the PL behaviour of carbon dots is the existence of different emissive sites on the surface of the dot. These sites are related to different defects on the surface, which are selectively excited depending on the wavelength used. With quinine sulphate as the reference, the quantum yield of the synthesized CQDs was measured to be ∼30% under the excitation of 360 nm, which is a relatively high degree in comparison to the previous literature and proves the material's capability in fluorescence based sensing and bio-imaging applications. FTIR spectrum of the synthesized CQDs was analysed to determine their surface functional groups. The very broad peak at 3307 cm−1 is attributed to the asymmetric stretching vibrations of –OH and/or N–H groups. Another weak band at 2131 cm−1 may be attributed to C–N vibration.35 One of the intense characteristic peak at 1639 cm−1 assigned to aromatic C
O).31 Upon excitation (wavelength ranges of ∼340–480 nm) it shows broad and strong emission peaks as shown in Fig. 2b. Highest intensity peak observed at 420 nm upon excitation with 360 nm is the effect of different surface states of C-dots.32 The excitation-dependent spectra show a shift in photoluminescence (PL) peak from 420 to 520 nm under excitation range of 340–480 nm as shown in Fig. 2b. Several hypotheses have been proposed to explain the wavelength – dependent emission spectra of CQDs.33 As the carbon particle decreases in size, quantum confinement takes place meaning that the particle is smaller than the de Broglie wavelength of the electron in the dot, creating a deviation from bulk properties. This confinement creates a quantization of the energy into discrete levels in the conduction and valence bands, so the C-dot can be understood as “virtual atom”. The emission energy depends on the radius of the particle so that as the particle gets smaller, both the excitation and emission spectra shift to shorter wavelengths. In a distribution of C-dots with varying sizes, different subsets get preferentially excited as the excitation wavelength varies. As CQDs of different size are excited, the emission spectrum shifts with excitation wavelength.34 Another potential explanation for the PL behaviour of carbon dots is the existence of different emissive sites on the surface of the dot. These sites are related to different defects on the surface, which are selectively excited depending on the wavelength used. With quinine sulphate as the reference, the quantum yield of the synthesized CQDs was measured to be ∼30% under the excitation of 360 nm, which is a relatively high degree in comparison to the previous literature and proves the material's capability in fluorescence based sensing and bio-imaging applications. FTIR spectrum of the synthesized CQDs was analysed to determine their surface functional groups. The very broad peak at 3307 cm−1 is attributed to the asymmetric stretching vibrations of –OH and/or N–H groups. Another weak band at 2131 cm−1 may be attributed to C–N vibration.35 One of the intense characteristic peak at 1639 cm−1 assigned to aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibrations.36 The Raman spectra for the synthesized CQDs as seen in Fig. 2d shows the characteristic G band at 1615 cm−1 and D band at 1330 cm−1 with an intensity ratio ID/IG ∼1.2.37 XRD pattern (ESI, Fig. S1†) of the developed materials shows a broad peak appeared at ∼23°, which can be indexed to the (002) crystal plane of graphene QDs. Therefore, characterization of the synthesized carbon quantum dots supports the formation of graphene like structure.
C stretching vibrations.36 The Raman spectra for the synthesized CQDs as seen in Fig. 2d shows the characteristic G band at 1615 cm−1 and D band at 1330 cm−1 with an intensity ratio ID/IG ∼1.2.37 XRD pattern (ESI, Fig. S1†) of the developed materials shows a broad peak appeared at ∼23°, which can be indexed to the (002) crystal plane of graphene QDs. Therefore, characterization of the synthesized carbon quantum dots supports the formation of graphene like structure.
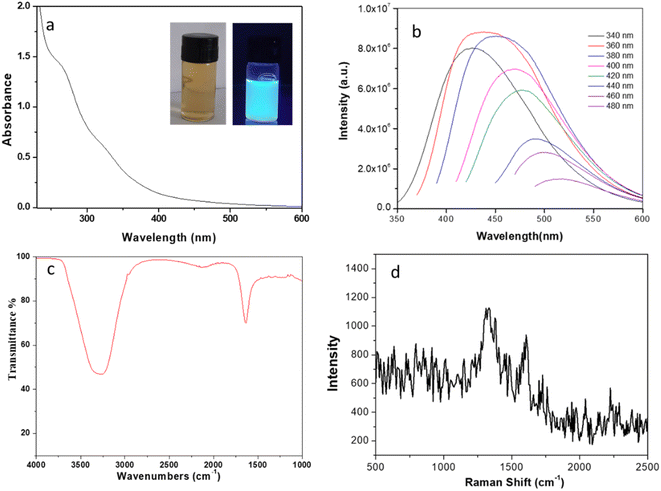 |
| | Fig. 2 (a) UV-vis absorption spectra of CQDs; inset shows the photograph under visible light and UV light (365 nm) irradiation. (b) Fluorescence spectra of CQDs under different wavelengths (340–480 nm) (c) FTIR spectrum of as synthesized CQDs (d) Raman spectra of CQDs. | |
The dispersion of the CQDs was verified by the TEM (Transmission electron microscopy) as shown in Fig. 3a. The TEM image revealed that the synthesized CQDs have a narrow size distribution of ∼2–5 nm (average size 3.5 ± 0.4 nm). High-resolution TEM image (Fig. 3b) shows the lattice fringes of 0.32 nm corresponding to (002) plane of graphene38 which reveal that CQDs display typical to graphitic carbon lattice parameters.
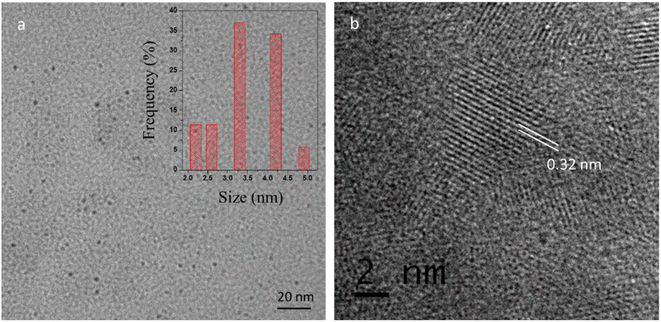 |
| | Fig. 3 (a) TEM image of CQDs (inset shows the particle size distribution of corresponding CQDs) (b) HRTEM image of CQDs showing the lattice fringes. | |
Elemental composition and speciation within the developed CQDs were determined by XPS. The XPS survey scan spectrum as shown in Fig. 4a indicates the presence of intense peaks at ∼284.6 eV, ∼399.0 eV, ∼531.0 eV, ∼133.1 eV and ∼169.6 eV attributed to characteristic peaks of carbon (C 1s), nitrogen (N 1s), oxygen (O 1s), phosphorous (P 2p) and sulphur (S 2p), respectively. Fig. 4b–f describes the high-resolution short scanned XPS spectra of C 1s, N 1s, O 1s, P 2p and S 2p, respectively. The three fitted peaks in the high-resolution C 1s attributed to sp2 graphitic carbon, carbonyl and carboxylic groups, as depicted in the figure. The N 1s peaks indicate the existence of nitrogen-containing groups, C–N–C and N–H, at 399.4 and 401.1 eV, respectively, which are assigned to amides and/or amino groups. The oxygen element in the forms of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–O, respectively, are assigned to the two fitted peaks at 531.6 and 532 eV in the O 1s spectra.39,40 The peaks of P 2p at 132.8 eV and 133.7 eV can be assigned to P–O and P
O and C–O, respectively, are assigned to the two fitted peaks at 531.6 and 532 eV in the O 1s spectra.39,40 The peaks of P 2p at 132.8 eV and 133.7 eV can be assigned to P–O and P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds, respectively. For S 2p spectra, the contribution at 167.6 eV can be assigned to sulfonic acids, and the peak at 168.8 eV is assigned to sulphate groups.41 This conforms the doping of N, P, S within the network of graphene like carbon dots during the synthesis. The contents of C, O, N, P, and S were ∼50.48%, 35.09%, 9.93% 0.96 and 3.35% respectively. The total doping content (especially nitrogen) was higher as compared to previous reports of CQDs synthesized using different plant sources.42–44 The synergic effect of doping, composition, and functional groups could be their utility in various fields. Herein, we checked the antimicrobial and antioxidant activities of the developed CQDs.
O bonds, respectively. For S 2p spectra, the contribution at 167.6 eV can be assigned to sulfonic acids, and the peak at 168.8 eV is assigned to sulphate groups.41 This conforms the doping of N, P, S within the network of graphene like carbon dots during the synthesis. The contents of C, O, N, P, and S were ∼50.48%, 35.09%, 9.93% 0.96 and 3.35% respectively. The total doping content (especially nitrogen) was higher as compared to previous reports of CQDs synthesized using different plant sources.42–44 The synergic effect of doping, composition, and functional groups could be their utility in various fields. Herein, we checked the antimicrobial and antioxidant activities of the developed CQDs.
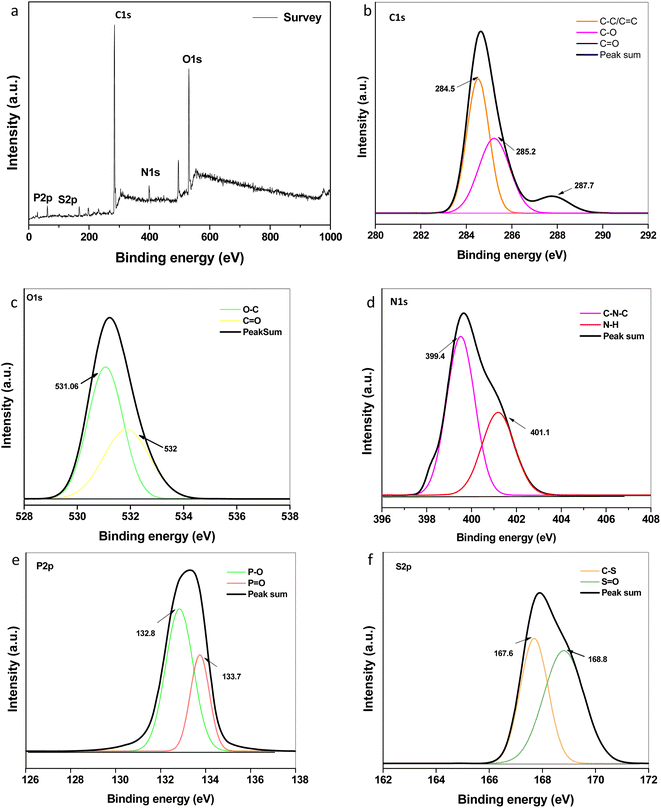 |
| | Fig. 4 High-resolution XPS spectrum: (a) survey scan (b) C 1s (c) O 1s (d) N 1s (e) P 2p and (f) S 2p spectra of the synthesized CQDs. | |
Antibacterial activities of the developed CQDs was investigated against both Gram-positive and Gram-negative bacteria viz. Salmonella typhi, Pseudomonas aeruginosa and Bacillus subtilis. As shown in Fig. 5, different concentrations (5–100 µg ml−1) of CQDs were used and the number of viable colonies were found to be significantly decreased with increasing the concentration of CQDs. Upon reaching the highest concentration i.e. 100 µg ml−1 no bacterial colonial growth was observed on the L. B. agar plates against Salmonella typhi and Pseudomonas aeruginosa. Whereas, 75 µg ml−1 of CQDs was sufficient for 100% inhibition of bacterial growth against the Bacillus subtilis.
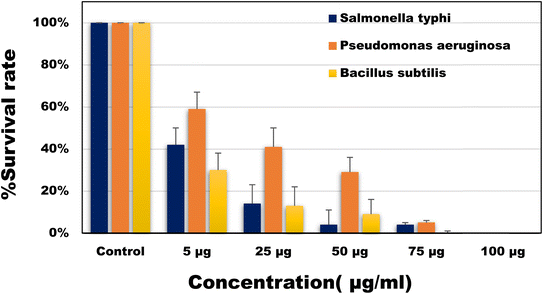 |
| | Fig. 5 Percentage survival rate of the bacterial species (Salmonella typhi, Pseudomonas aeruginosa and Bacillus subtilis) against different concentrations of CQDs. | |
The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) was also determined. MIC of CQDs against Salmonella typhi, Pseudomonas aeruginosa and Bacillus subtilis was calculated to be around 5 µg ml−1. The antibacterial properties of CQDs are mainly influenced by their extremely small size and surface functional groups. In a study45 it was seen that the role of surface charges presents on CQDs plays a crucial role in interacting with the bacterial cell wall thus also helping in the labelling of the bacterial cells.46 The bactericidal property of CQDs can be described as the destruction of the bacterial membrane (ESI; Fig. S2†), leaking of cytoplasmic material thus finally resulting in cell apoptosis.47 Comparative studies showed that the superior activities of developed CQDs as compared to most of the previously reported CQDs (Table 1).
Table 1 Comparative study of antibacterial activity of different CQDs synthesized from natural precursors
| Materials |
Size |
Method of synthesis |
MIC values |
Strain |
References |
| Cu doped NCQDs |
7.2 nm |
Hydrothermal |
600 µg ml−1 |
E. coli |
48 |
| S. epidermidis |
| Ag@S-GQDs nanocomposite |
4.0 nm |
Pyrolysis |
70 µg ml |
P. aeruginosa |
49 |
| 35 µg ml |
S. aureus |
| Sugarcane pulp derived CQDs |
2.27 nm |
Hydrothermal |
— |
Benthesicymus cereus |
50 |
| S. aureus |
| Pseudomonas aeruginosa |
| Vibrio cholera |
| E. coli |
| Pomegranate (P-Cdots) and watermelon peels (WC-dots) derived CDs |
5.0 nm |
Microwave |
40 µg ml−1 |
Pseudomonas aeruginosa |
51 |
| Bacillus Subtilis |
| S. aureus |
| NSP-CQDs |
4.0–5.0 nm |
Microwave |
100 µg ml−1 |
E. coli |
52 |
| S. aureus |
| P-doped CQDs |
3.4 nm |
Hydrothermal |
1.23 mg ml−1 |
E. coli |
53 |
| 1.44 mg ml−1 |
S. aureus |
| Curcuma longa derived CDs |
2.6 nm |
Hydrothermal |
0.25 mg ml−1 |
E. coli |
54 |
| S. aureus |
| 0.5 mg ml−1 |
K. pneumoniae |
| S. epidermidis |
| Mushroom-derived CDs |
8.0 nm |
Hydrothermal |
30.0 µg ml−1 |
S. aureus |
55 |
| K. pneumoniae |
| Pseudomonas aeruginosa |
| Endophytic bacteria derived CDs |
3.5 nm |
Hydrothermal |
5.0 µg ml−1 |
Salmonella typhi |
Present work |
| Pseudomonas aeruginosa |
| Bacillus subtilis |
4.1 Radical scavenging (DPPH) assay
The non-toxic nature of CQDs made them exceptional materials suitable for different biological applications. Antioxidant capacity of the synthesized CQDs was analysed using the well-established DPPH assay.56 The bacterial derived CQDs showed excellent free radical scavenging activity. This rise in scavenging property can be attributed to the different functional groups carboxyl, hydroxyl and amide groups present on the surface of the CQDs. At concentration of 60 µg ml−1, it shows the highest scavenging activity (∼75%) (Fig. 6). The EC50 of the CQDs was calculated to be around 24.78 µg ml−1 as compared to the EC50 of the standard ascorbic acid of 5.36 µg ml−1. Compared to previous reports of CQDs (ESI; Table S1†), the present material shows much higher activity except a recent report where similar activities were reported.57
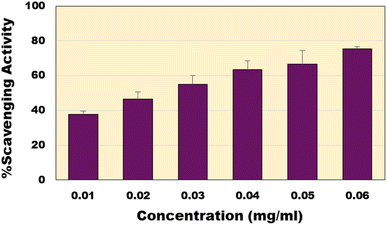 |
| | Fig. 6 Percentage scavenging activity of CQDs at various concentrations. | |
4.2 ABTS assay
In addition to DPPH assay, ABTS assay also known as Trilox equivalent antioxidant-capacity assay was also performed. The assay provides an easy, rapid and more sensitive means to evaluate antioxidant potential of any material. In this assay, EC50 value was estimated to be ∼20.65 µg ml−1 (Fig. 7), which is close to the EC50 value obtained during DPPH assay. Under similar conditions, EC50 of the ascorbic acid was estimated to be ∼4.27 µg ml−1. These results confirm the synthesis of highly efficient graphene-like carbon quantum dots which have shown very high antimicrobial and antioxidant activities.
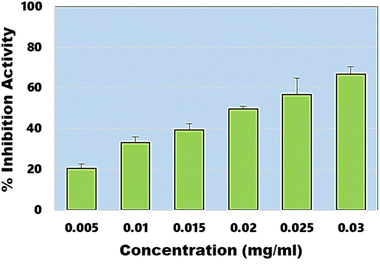 |
| | Fig. 7 Percentage inhibition activity of synthesized CQDs calculated against different concentration ranges. | |
5 Conclusion
Present study describes an easy and facile approach for the synthesis of NPS co-doped CQDs from endophytic bacteria without using any other chemicals for doping. The synthesized CQDs have mainly graphene-like structure. They possess exceptional antioxidant and broad spectrum of bactericidal activities due to their tiny size (∼3 nm) and surface functional groups. The small size of the CQDs help them to penetrate the bacterial membrane and thus helps in reactive oxygen species generation thus leading to cell apoptosis. This material does not require any further doping or functionalization as it was abundant source of carbon, nitrogen, sulphur and oxygen. Moreover, our approach can further broaden the use of endophytic bacteria in more biological applications.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
The authors would like to express deep gratitude for the support of DST, Govt. of India for financial assistance through the INSPIRE Faculty Award (IFA16-MS81). Authors wish to express sincere gratitude to the administration of GGSIP University for providing the academic environment to pursue research activities.
References
- X. Chen, Z. Song, S. Li, N. T. Thang, X. Gao, X. Gonga and M. Guo, Green Chem., 2020, 22, 3296–3308 RSC.
- C. He, X. Li, G. Fenga and W. Long, Green Chem., 2014, 16, 2566–2570 RSC.
- M. Shamsipur, A. Barati and S. Karami, Carbon, 2017, 124, 429–472 CrossRef CAS.
- B. Geng, Y. Li, J. Hu, Y. Chen, J. Huang, L. Shen, D. Pan and P. Li, J. Mater. Chem. B, 2022, 10, 3357–3365 RSC.
- M. L. Liu, B. B. Chen, C. M. Li and C. Z. Huang, Green Chem., 2019, 21, 449–471 RSC.
- Z. X. Liu, B. B. Chen, M. L. Liu, H. Y. Zou and C. Z. Huang, Green Chem., 2017, 19, 1494–1498 RSC.
- B. Geng, J. Hu, Y. Li, S. Feng, D. Pan, L. Feng and L. Shen, Nat. Commun., 2022, 13, 5735 CrossRef CAS PubMed.
- B. Geng, S. Xu, P. Li, X. Li, F. Fang, D. Pan and L. Shen, Small, 2022, 18, 2103528 CrossRef CAS PubMed.
- S. Miao, K. Liang, J. Zhu, B. Yang, D. Zhao and B. Kong, Nano Today, 2020, 33, 100879 CrossRef CAS.
- F. Li, D. Yang and H. Xu, Chem. – Eur. J., 2019, 25, 1165–1176 CrossRef CAS PubMed.
- X. Gon, Y. Liu, Z. Yang, S. Shuang, Z. Zhang and C. Dong, Anal. Chim. Acta, 2017, 968, 85–96 CrossRef PubMed.
- M. Zan, L. Rao, H. Huang, W. Xie, D. Zhu, L. Li, X. Qie, S. S. Guo, X. Z. Zhao, W. Liu and W. F. Dong, Sens. Actuators, B, 2018, 262, 555–561 CrossRef CAS.
- C. He, X. Li, G. Fenga and W. Long, Green Chem., 2022, 24, 5842–5855 RSC.
- S. Chahal, J. Macairan, R. Yousefi, N. Tufenkji and R. Naccache, RSC Adv., 2021, 11, 25354–25363 RSC.
- X. Kou, S. Jiang, S. J. Park and L. Y. Meng, Dalton Trans., 2020, 49, 6915–6938 RSC.
- C. Wang, D. Sun, K. Zhuo, H. Zhang and J. Wang, RSC Adv., 2014, 4, 54060–54065 RSC.
- S. Zhang, D. Zhang, Y. Ding, J. Hua, B. Tang, X. Ji, Q. Zhang, Y. Wei, K. Qin and B. Li, Analyst, 2019, 144, 5497–5503 RSC.
- A. Kumar, S. Kumar, K. Kiran, S. Banerjee, V. Pande and A. Dandapat, Colloids Surf., B, 2021, 206, 111948 CrossRef CAS PubMed.
- F. Prestinaci, P. Pezzotti and A. Pantosti, Pathog. Global Health, 2015, 109, 309–318 CrossRef PubMed.
- M. Alavi, E. Jabari and E. Jabbari, Expert Rev. Anti-Infect. Ther., 2021, 19, 35–44 CrossRef CAS PubMed.
- Y. Sun, M. Zhang, B. Bhandari and C. Yang, Food Rev. Int., 2020, 7, 1–20 Search PubMed.
- N. S. Bisht, A. H Tripathi, M. Pant, S. K. Upadhyay, N. G. Sahoo, S. P. S. Mehta and A. Dandapat, Colloids Surf., B, 2022, 217, 112640 CrossRef CAS PubMed.
- H. Wang, Z. Song, J. Gu, S. Li, Y. Wu and H. Han, ACS Biomater. Sci. Eng., 2019, 5, 4739–4749 CrossRef CAS PubMed.
- D. Pancholi, N. S. Bisht, V. Pande and A. Dandapat, Colloids Surf., B, 2021, 199, 111558 CrossRef CAS PubMed.
- N. Anjum and R. Chandra, Asian J. Pharm. Clin. Res., 2015, 8, 233–238 Search PubMed.
- F. Lin, C. Li and Z. Chen, Front. Microbiol., 2018, 9, 259 CrossRef PubMed.
- C. Zhao, Y. Jiao, J. Hua, J. Yang and Y. Yang, J. Fluoresc., 2018, 28, 269–276 CrossRef CAS PubMed.
- V. Ruiz, L. Yate, I. García, G. Cabanero and H. J. Grande, Carbon, 2017, 116, 366–374 CrossRef CAS.
- R. Re, N. Pellegrini, A. Proteggente, A. Pannala, M. Yang and C. Rice-Evans, Free Radical Biol. Med., 1999, 26, 1231–1237 CrossRef CAS PubMed.
- Y. Li, H. Shu, X. Niu and J. Wang, J. Phys. Chem. C, 2015, 119, 24950–24957 CrossRef CAS.
- S. Zhang, D. Zhang, Y. Ding, J. Hua, B. Tang, X. Ji and B. Li, Analyst, 2019, 144, 5497–5503 RSC.
- Y. S. Lin, Y. Lin, A. P. Periasamy, J. Cang and H. T. Chang, Nanoscale Adv., 2019, 1, 2553–2561 RSC.
- L. Guo, J. Ge, W. Liu, G. Niu, Q. Jia, H. Wang and P. Wang, Nanoscale, 2016, 8, 729–734 RSC.
- S. Zhu, Y. Song, X. Zhao, J. Shao, J. Zhang and B. Yang, Nano Res., 2015, 8, 355–381 CrossRef CAS.
- M. Shariati-Rad, T. Mohseninasab and F. Parno, RSC Adv., 2018, 8, 2173–2180 RSC.
- W. Yang, J. Ni, F. Luo, W. Weng, Q. W. Z. Lin and G. Chen, Anal. Chem., 2017, 89, 8384–8390 CrossRef CAS PubMed.
- Y. Fang, S. Guo, D. Li, C. Zhu, W. Ren, S. Dong and E. Wang, ACS Nano, 2012, 6, 400–409 CrossRef CAS PubMed.
- J. Kim and J. S. Suh, ACS Nano, 2014, 8, 4190–4196 CrossRef CAS PubMed.
- F. Yan, Z. Bai, F. Zu, Y. Zhang, X. Sun, T. Ma and L. Chen, Microchim. Acta, 2019, 186, 1–11 CrossRef PubMed.
- X. W. Hua, Y. W. Bao, H. Y. Wang, Z. Chen and F. G. Wu, Nanoscale, 2017, 9, 2150–2161 RSC.
- N. A. Travlou, D. A. Giannakoudakis, M. Algarra, A. M. Labella, E. Rodríguez-Castellón and T. J. Bandosz, Carbon, 2018, 135, 104–111 CrossRef CAS.
- J. Zhong, X. Chen, M. Zhang, C. Xiao, L. Cai, W. A. Khan, K. Yu, J. Cui and L. He, Chin. Chem. Lett., 2020, 31, 769–773 CrossRef CAS.
- H. Wang, M. Zhang, Y. Ma, B. Wang, M. Shao, H. Huang, Y. Liu and Z. Kang, J. Mater. Chem. B, 2020, 8, 2666–2672 RSC.
- C. Wang, D. Sun, K. Zhuo, H. Zhang and J. Wang, RSC Adv., 2014, 4, 54060–54065 RSC.
- M. Gagic, S. Kociova, K. Smerkova, H. Michalkova, M. Setka, P. Svec and V. Milosavljevic, J. Colloid Interface Sci., 2020, 580, 30–48 CrossRef CAS PubMed.
- C. I. Weng, H. T. Chang, C. H. Lin, Y. W. Shen, B. Unnikrishnan, Y. J. Li and C. C. Huang, Biosens. Bioelectron., 2015, 68, 1–6 CrossRef CAS PubMed.
- M. A. Quinteros, C. A. Viviana, R. Onnainty, V. S. Mary, M. G. Theumer, G. E. Granero and P. L. Páez, Int. J. Biochem. Cell Biol., 2018, 104, 87–93 CrossRef CAS PubMed.
- F. Nichols, J. E. Lu, R. Mercado, M. D. Rojas-Andrade, S. Ning, Z. Azhar and S. Chen, Langmuir, 2020, 36, 11629–11636 CrossRef CAS PubMed.
- S. Kadian, G. Manik, N. Das, P. Nehra, R. P. Chauhan and P. Roy, J. Mater. Chem. B, 2020, 8, 3028–3037 RSC.
- S. Pandiyan, L. Arumugam, S. P. Srirengan, R. Pitchan, P. Sevugan, K. Kannan and V. Gandhirajan, ACS Omega, 2020, 5, 30363–30372 CrossRef CAS PubMed.
- W. A. Qureshi, B. Vivekanandan, J. A. Jayaprasath, D. Ali, S. Alarifi and K. Deshmukh, J. Nanomater., 2021, 2021, 9096838 Search PubMed.
- K. M. Tripathi, H. T. Ahn, M. Chung, X. A. Le, D. Saini, A. Bhati and T. Kim, ACS Biomater. Sci. Eng., 2020, 6, 5527–5537 CrossRef CAS PubMed.
- S. Chai, L. Zhou, S. Pei, Z. Zhu and B. Chen, Micromachines, 2021, 12, 1116 CrossRef PubMed.
- A. Saravanan, M. Maruthapandi, P. Das, J. H. Luong and A. Gedanken, Nanomaterials, 2021, 11, 369 CrossRef CAS PubMed.
- T. Boobalan, M. Sethupathi, N. Sengottuvelan, P. Kumar, P. Balaji, B. Gulyás and A. Arun, ACS Appl. Nano Mater., 2020, 3, 5910–5919 CrossRef CAS.
- H. M. Chandra and S. Ramalingam, Food Sci. Biotechnol., 2011, 20, 15–21 CrossRef CAS.
- A. Sachdev and P. Gopinath, Analyst, 2015, 140, 4260–4269 RSC.
|
| This journal is © The Royal Society of Chemistry 2023 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article b,
Veena Pandea and
Anirban Dandapat
b,
Veena Pandea and
Anirban Dandapat *c
*c
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g, 15 min, 4 °C). The cells were finally washed with double distilled water for at-least three times to remove the supernatant. The pellet obtained was then re-suspended in 30 ml of distilled water (30 ml). Subsequently, the bacterial suspension was transferred into a Teflon-lined stainless-steel autoclave and heated up to 200 °C for 24 hours. A light brownish solution was obtained. The obtained solution was then filtered through a syringe filter (0.22 µm) diameter to remove large particles. Finally, after dialyzing using dialysis membrane of (MWCO > 1 kDa) against deionized water for 72 hours, the CQD solution was obtained and stored at 4 °C for further experiments.26 Repeatability was further checked by repeating the same experiment thrice.
000 g, 15 min, 4 °C). The cells were finally washed with double distilled water for at-least three times to remove the supernatant. The pellet obtained was then re-suspended in 30 ml of distilled water (30 ml). Subsequently, the bacterial suspension was transferred into a Teflon-lined stainless-steel autoclave and heated up to 200 °C for 24 hours. A light brownish solution was obtained. The obtained solution was then filtered through a syringe filter (0.22 µm) diameter to remove large particles. Finally, after dialyzing using dialysis membrane of (MWCO > 1 kDa) against deionized water for 72 hours, the CQD solution was obtained and stored at 4 °C for further experiments.26 Repeatability was further checked by repeating the same experiment thrice.

![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O).31 Upon excitation (wavelength ranges of ∼340–480 nm) it shows broad and strong emission peaks as shown in Fig. 2b. Highest intensity peak observed at 420 nm upon excitation with 360 nm is the effect of different surface states of C-dots.32 The excitation-dependent spectra show a shift in photoluminescence (PL) peak from 420 to 520 nm under excitation range of 340–480 nm as shown in Fig. 2b. Several hypotheses have been proposed to explain the wavelength – dependent emission spectra of CQDs.33 As the carbon particle decreases in size, quantum confinement takes place meaning that the particle is smaller than the de Broglie wavelength of the electron in the dot, creating a deviation from bulk properties. This confinement creates a quantization of the energy into discrete levels in the conduction and valence bands, so the C-dot can be understood as “virtual atom”. The emission energy depends on the radius of the particle so that as the particle gets smaller, both the excitation and emission spectra shift to shorter wavelengths. In a distribution of C-dots with varying sizes, different subsets get preferentially excited as the excitation wavelength varies. As CQDs of different size are excited, the emission spectrum shifts with excitation wavelength.34 Another potential explanation for the PL behaviour of carbon dots is the existence of different emissive sites on the surface of the dot. These sites are related to different defects on the surface, which are selectively excited depending on the wavelength used. With quinine sulphate as the reference, the quantum yield of the synthesized CQDs was measured to be ∼30% under the excitation of 360 nm, which is a relatively high degree in comparison to the previous literature and proves the material's capability in fluorescence based sensing and bio-imaging applications. FTIR spectrum of the synthesized CQDs was analysed to determine their surface functional groups. The very broad peak at 3307 cm−1 is attributed to the asymmetric stretching vibrations of –OH and/or N–H groups. Another weak band at 2131 cm−1 may be attributed to C–N vibration.35 One of the intense characteristic peak at 1639 cm−1 assigned to aromatic C
O).31 Upon excitation (wavelength ranges of ∼340–480 nm) it shows broad and strong emission peaks as shown in Fig. 2b. Highest intensity peak observed at 420 nm upon excitation with 360 nm is the effect of different surface states of C-dots.32 The excitation-dependent spectra show a shift in photoluminescence (PL) peak from 420 to 520 nm under excitation range of 340–480 nm as shown in Fig. 2b. Several hypotheses have been proposed to explain the wavelength – dependent emission spectra of CQDs.33 As the carbon particle decreases in size, quantum confinement takes place meaning that the particle is smaller than the de Broglie wavelength of the electron in the dot, creating a deviation from bulk properties. This confinement creates a quantization of the energy into discrete levels in the conduction and valence bands, so the C-dot can be understood as “virtual atom”. The emission energy depends on the radius of the particle so that as the particle gets smaller, both the excitation and emission spectra shift to shorter wavelengths. In a distribution of C-dots with varying sizes, different subsets get preferentially excited as the excitation wavelength varies. As CQDs of different size are excited, the emission spectrum shifts with excitation wavelength.34 Another potential explanation for the PL behaviour of carbon dots is the existence of different emissive sites on the surface of the dot. These sites are related to different defects on the surface, which are selectively excited depending on the wavelength used. With quinine sulphate as the reference, the quantum yield of the synthesized CQDs was measured to be ∼30% under the excitation of 360 nm, which is a relatively high degree in comparison to the previous literature and proves the material's capability in fluorescence based sensing and bio-imaging applications. FTIR spectrum of the synthesized CQDs was analysed to determine their surface functional groups. The very broad peak at 3307 cm−1 is attributed to the asymmetric stretching vibrations of –OH and/or N–H groups. Another weak band at 2131 cm−1 may be attributed to C–N vibration.35 One of the intense characteristic peak at 1639 cm−1 assigned to aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibrations.36 The Raman spectra for the synthesized CQDs as seen in Fig. 2d shows the characteristic G band at 1615 cm−1 and D band at 1330 cm−1 with an intensity ratio ID/IG ∼1.2.37 XRD pattern (ESI, Fig. S1†) of the developed materials shows a broad peak appeared at ∼23°, which can be indexed to the (002) crystal plane of graphene QDs. Therefore, characterization of the synthesized carbon quantum dots supports the formation of graphene like structure.
C stretching vibrations.36 The Raman spectra for the synthesized CQDs as seen in Fig. 2d shows the characteristic G band at 1615 cm−1 and D band at 1330 cm−1 with an intensity ratio ID/IG ∼1.2.37 XRD pattern (ESI, Fig. S1†) of the developed materials shows a broad peak appeared at ∼23°, which can be indexed to the (002) crystal plane of graphene QDs. Therefore, characterization of the synthesized carbon quantum dots supports the formation of graphene like structure.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–O, respectively, are assigned to the two fitted peaks at 531.6 and 532 eV in the O 1s spectra.39,40 The peaks of P 2p at 132.8 eV and 133.7 eV can be assigned to P–O and P
O and C–O, respectively, are assigned to the two fitted peaks at 531.6 and 532 eV in the O 1s spectra.39,40 The peaks of P 2p at 132.8 eV and 133.7 eV can be assigned to P–O and P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds, respectively. For S 2p spectra, the contribution at 167.6 eV can be assigned to sulfonic acids, and the peak at 168.8 eV is assigned to sulphate groups.41 This conforms the doping of N, P, S within the network of graphene like carbon dots during the synthesis. The contents of C, O, N, P, and S were ∼50.48%, 35.09%, 9.93% 0.96 and 3.35% respectively. The total doping content (especially nitrogen) was higher as compared to previous reports of CQDs synthesized using different plant sources.42–44 The synergic effect of doping, composition, and functional groups could be their utility in various fields. Herein, we checked the antimicrobial and antioxidant activities of the developed CQDs.
O bonds, respectively. For S 2p spectra, the contribution at 167.6 eV can be assigned to sulfonic acids, and the peak at 168.8 eV is assigned to sulphate groups.41 This conforms the doping of N, P, S within the network of graphene like carbon dots during the synthesis. The contents of C, O, N, P, and S were ∼50.48%, 35.09%, 9.93% 0.96 and 3.35% respectively. The total doping content (especially nitrogen) was higher as compared to previous reports of CQDs synthesized using different plant sources.42–44 The synergic effect of doping, composition, and functional groups could be their utility in various fields. Herein, we checked the antimicrobial and antioxidant activities of the developed CQDs.




