Open Access Article
Open Access ArticleElicitation for activation of the actinomycete genome's cryptic secondary metabolite gene clusters
Seham S. El-Hawary†
a,
Marwa H. A. Hassan† b,
Ahmed O. Hudhudc,
Usama Ramadan Abdelmohsen
b,
Ahmed O. Hudhudc,
Usama Ramadan Abdelmohsen *de and
Rabab Mohammed*b
*de and
Rabab Mohammed*b
aDepartment of Pharmacognosy, Faculty of Pharmacy, Cairo University, Cairo, Egypt
bDepartment of Pharmacognosy, Faculty of Pharmacy, Beni-Suef University, Beni-Suef 62511, Egypt. E-mail: rmwork06@yahoo.com; rababmohammed@bsu.pharm.edu.eg
cDepartment of Pharmacognosy, Faculty of Pharmacy, Merit University, Sohag 82511, Egypt
dDepartment of Pharmacognosy, Faculty of Pharmacy, Minia University, Minia 61519, Egypt. E-mail: usama.ramadan@mu.edu.eg
eDepartment of Pharmacognosy, Faculty of Pharmacy, Deraya University, New Minia 61111, Egypt
First published on 16th February 2023
Abstract
This review summarizes the recent advances in the elicitation approaches used to activate the actinomycete genome's cryptic secondary metabolite gene clusters and shows the diversity of natural products obtained by various elicitation methods up to June 2022, such as co-cultivation of actinomycetes with actinomycetes, other non-actinomycete bacteria, fungi, cell-derived components, and/or algae. Chemical elicitation and molecular elicitation as transcription factor decoys, engineering regulatory genes, the promoter replacement strategy, global regulatory genes, and reporter-guided mutant selection were also reported. For researchers interested in this field, this review serves as a valuable resource for the latest studies and references.
1. Introduction
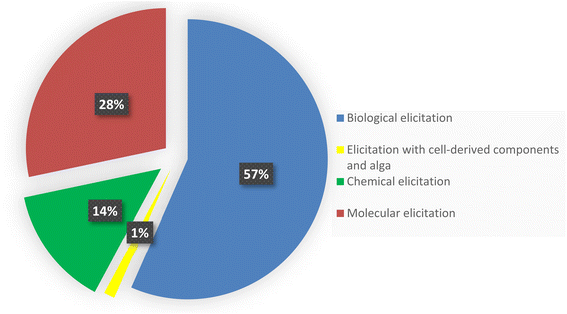
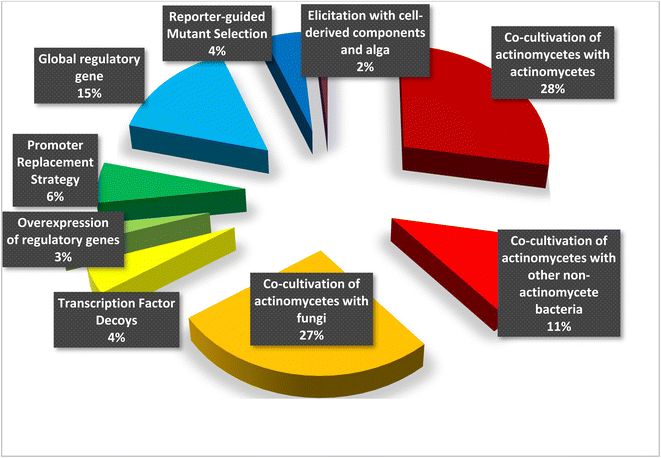
Regular protocols used for the discovery of bacterial-based natural products often involve microorganism fermentation on different media followed by chemical and biological screening of the extracts, seeking any interesting bioactive compounds. The advent of genome-sequencing technology has enabled us to scan the substantial amount of published genome data for gene clusters encrypting the previously recognized natural products, bringing in a second golden age in drug discovery. Recently, numerous new approaches were published unlocking bacterial cryptic pathways, including studies that focus on raising transcription levels and enlivening dead genes.1–3 Actinomycetes are unicellular, Gram-positive filamentous bacteria, belonging to the order Actinomycetales. They are broadly dispersed in nature and can be found in diverse habitats across the world. Actinomycetes are notable for producing a diverse variety of natural products with a wide range of biological activities and structurally diverse specialized metabolites.1 Over 200 drugs based on metabolites isolated from actinomycetes are currently in clinical trials or have been approved by the FDA for the treatment of infections, immune system abnormalities and cancer chemotherapeutic and to treat emerging health threats.2 Under conventional culture conditions, the majority of bioactive metabolites encrypted in actinomycete genomes are not activated and tendency to rediscover known secondary metabolites increases.3 Therefore, various approaches has been devoted to eliciting the underlying gene clusters in order to discover and isolate novel lead natural products and molecular scaffolds;4–6 such as: mixed fermentation or co-cultivation,7 fermentation conditions alteration which known as “one strain many compounds” (OSMAC)8 and eliciting the bacterial cells with external signals either chemically or biologically.9,10 This review summarizes notable and successful examples of elicitation methods based on literature search from 2015 until June 2022, including changing culture conditions, modifying the medium, co-cultivating with different strains, and adding a biosynthetic precursors or epigenetic modifiers (Fig. 1). This review is complementary to the first part that was previously published.112. Biological elicitation
Biological elicitation is one of the most actively pursued techniques to trigger the synthesis of natural products in actinomycetes.2.1. By microbial co-cultivation
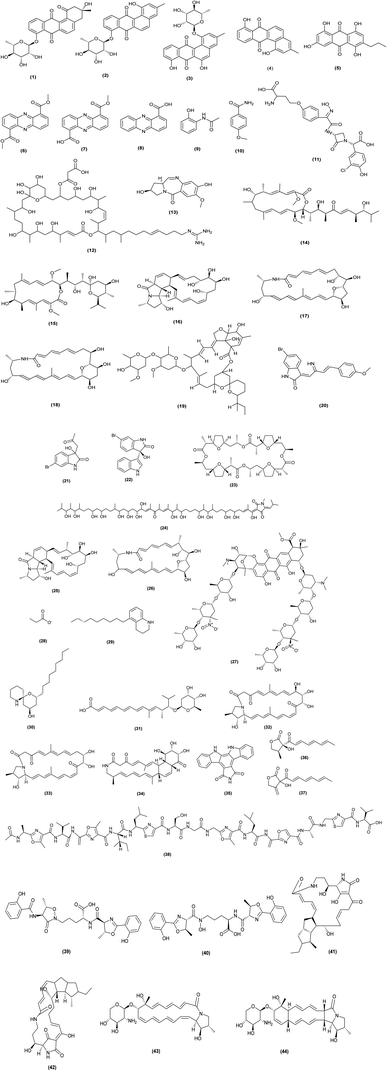
The confrontation of microorganisms in various habitat may activate cryptic gene clusters and trigger silent pathways to produce new secondary metabolites. These outcomes are based on the effective interactions between co-cultivated microorganisms.12 For example, horizontal gene transfer, physical cell–cell interactions and production of enzymes or quorum sensing molecules.11 Co-cultivation-based elicitation can thus lead to the generation of molecules that are not synthesized in monoculture, thus aiding in the discovery of cryptic and weakly expressed secondary metabolites. However, finding suitable partners for co-cultivation remains challenging. Co-cultivation can be further categorized as (a) co-cultivation of actinomycetes with other actinomycetes, (b) co-cultivation of actinomycetes with other non-actinomycete bacteria, and (c) co-cultivation of actinomycetes with fungi. The following section describe the process, the differences and the results on each of the three categories mentioned above.| Interaction partners | Induced secondary | Metabolites activity | Reference |
|---|---|---|---|
| Actinokineospora spheciospongiae strain EG49 and Rhodococcus sp. UR59 | Actinosporins E (1) | Anti-malarial | 13 |
| Actinosporins H (2) | |||
| Actinosporins G (3) | |||
| Tetragulol (4) | |||
| Capillasterquinone B (5) | |||
| Micromonospora sp. UR56 and Actinokineospora sp. EG49 | 1,6-Dicarboxylate (6) | Antibacterial | 14 |
| Phencomycin (7) | |||
| Tubermycin (8) | Cytotoxic antibiofilm | ||
| N-(2-hydroxyphenyl)-acetamide (9) p-anisamide (10) | |||
| Micromonospora sp. UA17, Gordonia sp. UA19, and Nocardia sp. UA 23 | Chlorocardicin (11) | Antibacterial | 15 |
| Neocopiamycin A (12) | Antifungal | ||
| Chicamycin B (13) | Antiparasitic | ||
| Streptomyces sp. UR23 and Nocardia sp. UR27 | Bafilomycin D (14) | Antitrypanosomal agents | 16 |
| Bafilomycin A1 (15) | |||
| Actinosynnema mirum NBRC 14064 | Mirilactams C–E, 1–3 (16, 17, 18) | Macrolactams | 17 |
| Tsukamurella pulmonis TP-B0596 | |||
| Streptomyces avermitilis and Streptomyces albus J1074 | Avermectin (19) | Pesticides | 18 |
| Saccharomonospora sp. UR22 and Dietzia sp. UR66 | Saccharomonosporine A (20) | Antiproliferative activity | 19 |
| Convolutamydine F (21) | |||
| (S) 6-Bromo-3-hydroxy-3-(1H-indol-3-yl) indolin-2-one (22) | |||
| Streptomyces coelicolor M145 | Amycomycin (24) | Antibiotic activity | 20 |
| Amycolatopsis sp. AA4 | |||
| Micromonospora wenchangensis HEK-797 | Dracolactams A (25) | Antifungal antibiotics | 21 |
| Tsukamurella pulmonis TPB0596 | Dracolactams B (26) | ||
| Rhodococcus sp. WMMA-185 | Keyicin (27) | Selectively active against gram-positive bacteria | 22 |
| Micromonospora sp. WMMB-235 | |||
| Cutibacterium avidum | Propionate (28) | It is a conjugate base of a propionic acid | 23 |
| Bifidobacterium longum subsp. infantis and Bifidobacterium bifidum | |||
| Streptomyces nigrescens HEK616 | 5-Alkyl-1,2,3,4-tetrahydroquinolines (29) | Novel antifungals | 24 |
| Tsukamurella pulmonis TP-B0596 | |||
| Streptomyces nigrescens HEK616 | [5,5]-Spirohemiaminals (30) | Broad antimicrobial activities | 25 |
| Tsukamurella pulmonis TP-B0596 | |||
| Streptomyces sp. ANAM-5 and AIAH-10 | Ethyl acetate extract | Antimicrobial and anticancer activities | 26 |
| Streptomyces tendae KMC006 | Gordonic acid (31) | Weak antibacterial activity | 27 |
| Gordonia sp. KMC005 | |||
| Streptomyces sp. NZ-6 | Niizalactams A–C (32–34) | Antifungal antibiotic | 28 |
| Tsukamurella pulmonis TP-B0596 | |||
| Streptomyces cinnamoneus NBRC 13823 | Arcyriaflavin E (35) | Cytotoxic | 29 |
| Tsukamurella pulmonis | |||
| Streptomyces sp. CJ-5 | Chojalactones A–C (36–37) | Cytotoxic | 30 |
| Tsukamurella pulmonis TP-B0596 | |||
| Streptomyces lividans TK23/pGSBC1 | Goadsporin C (38) | Antibiotic | 31 |
| Tsukamurella pulmonis TP-B0596 | |||
| Catenuloplanes sp. RD067331 | Catenulobactins A and B (39, 40) | Siderphore | 32 |
| Tsukamurella pulmonis TP-B0596 | |||
| Umezawaea sp. RD066910 | Umezawamides A and B (41, 42) | Anti-leukemia and antifungal activity | 33 |
| Tsukamurella pulmonis TP-B0596 | |||
| Nocardiopsis sp. FU40 (ΔApoS) | Ciromicin A (43) | Cytotoxicity (acute myelogenous leukemia cells) | 34 |
| Rhodococcus wratislaviensis | Ciromicin B (44) | Cytotoxicity (stem-like myeloid progenitor cells) | 35 |
Co-cultivation of two actinomycetes associated with marine sponge, Actinokineospora sp. EG49 and Micromonospora sp. UR56 resulted in the accumulation of five metabolites that were not traced in their independent cultures. These metabolites were isolated and identified as dimethyl phenazine-1,6-dicarboxylate (6), phencomycin (7), tubermycin (8), N-(2-hydroxyphenyl)-acetamide (9), p-anisamide (10). Compounds 6–8 and 10 showed high antibacterial, antibiofilm and cytotoxic activities.14
A change in natural product biosynthesis occurred in Micromonospora sp. UA17 isolated from the Red Sea Sponge Coscinoderma mathewsi, when co-cultivated with the mycolic acid containing Nocardia sp. UA 23 and Gordonia sp. UA19. Chlorocardicin (11), neocopiamycin A (12), and chicamycin B (13) were identified in the co-culture extracts, but not in the corresponding monocultures. This suggests a role for mycolic acid in the stimulation of encoded natural product biosynthesis pathways. Co-culture extracts showed antibacterial, antiparasitic and antifungal activities higher than monocultures.15 Streptomyces sp. UR23 co-fermentation with a strain containing mycolic acid Nocardia sp. UR27 stimulating the production of a possible antitrypanosomal agents bafilomycin D (14) and bafilomycin A1 (15).16 In addition, three new compounds identified as mirilactams (C–E) (16–18) were isolated in the co-culture of Actinosynnema mirum NBRC 14064 with Tsukamurella pulmonis TP-B0596.17
Nguyen and his colleagues found that the butenolide compounds in Streptomyces albus J1074 stimulated the production of avermectin (19) in Streptomyces avermitilis in an in vivo metabolic profiling assay with imaging mass spectrometry (IMS). This demonstrated the complex chemical interactions in streptomycetes through interspecies signals.18
Furthermore, the combined culture of a Dietzia sp. UR66 with Saccharomonospora sp. UR22 resulted in the production of a novel brominated oxo-indole alkaloid Saccharomonosporine A (20), convolutamydine F (21) and (S) 6-bromo-3-hydroxy-3-(1H-indol-3-yl) indolin-2-one (22) with a promising Pim-1 kinase inhibitors that mediates tumor cell growth inhibitory effect, along with other known induced metabolites; (S) 6-bromo-3-hydroxy-3-(1H-indol-3-yl) indolin-2-one (22) and nonactin (23).19 Amycomicin (24), a modified fatty acid with an epoxide isonitrile head, was discovered to be a selective and potent inhibitor of Staphylococcus aureus after co-culture with Amycolatopsis sp. AA4 as the producing strain and Streptomyces coelicolor M145 as the inducing strain.20
In the co-culture broth of Tsukamurella pulmonis TPB0596 with a rare actinomycete Micromonospora wenchangensis HEK-797, only two new macrolactams (dracolactams A and B) were detected, whereas the axenic medium of strain HEK-797 yielded a 26-membered polyene macrolactam, which was probably the precursor of dracolactams A (25) and B (26).21
From another co-culture of Rhodococcus sp. WMMA-185 and Micromonospora sp. WMMB-235, a two-marine invertebrate-associated bacteria, a novel antibiotic, keyicin (27), was isolated. It had specific inhibitory effect against Gram-positive bacteria Methicillin Sensitive Staphylococcus aureus (MSSA) and B. subtilis, with MIC values of 2.5 μM and 9.9 μM, respectively. Unlike many anthracyclines, keyicin may alter fatty acid metabolism and showing antibacterial activity without causing nucleic acid damage.22
Cutibacterium avidum growth and production of propionate (28) enhanced by co-cultivation with Bifidobacterium longum subsp. infantis and Bifidobacterium bifidum which confirmed the metabolic cross-feeding strategy.23
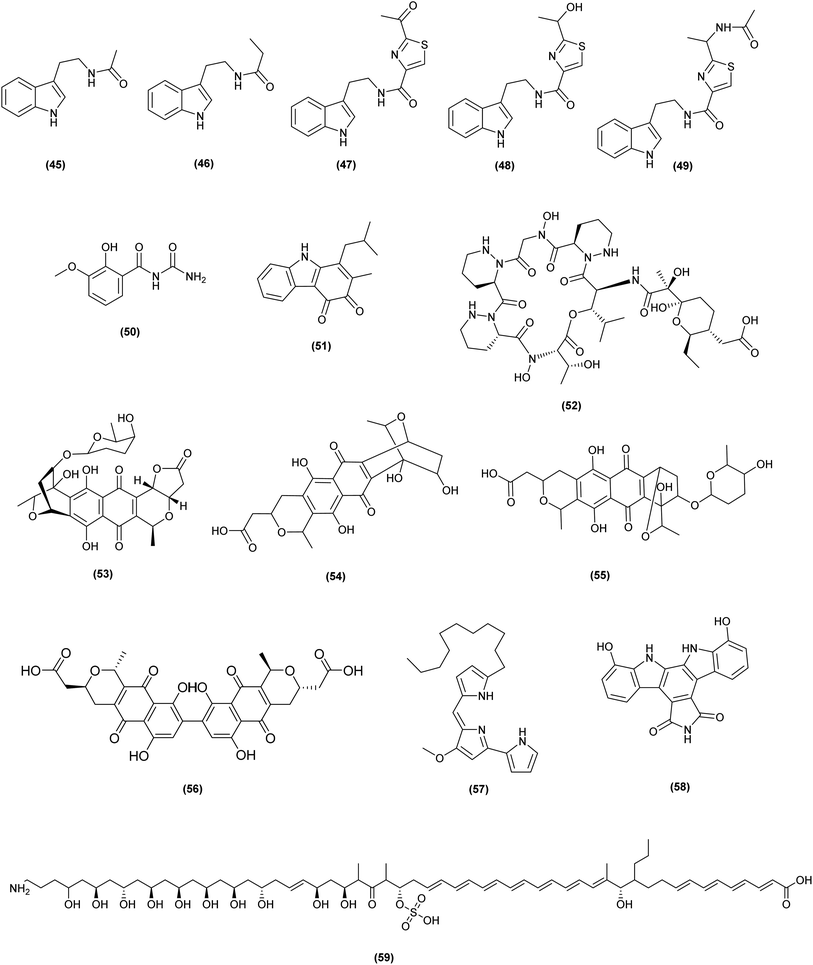
| Interaction partners | Induced secondary | Metabolites activity | Reference |
|---|---|---|---|
| Streptomyces sp. CGMCC4.7185 | Tryptamine (45, 46) | Algicidal | 36 |
| Bacillus mycoides | Bacillamides (47, 48, 49) | ||
| Streptomyces sp. RKND-216 | N-Carbamoyl-2-hydroxy-3-methoxybenzamide (50) | Antimicrobial | 37 |
| Alteromonas sp. RKMC-009 | Cytotoxic activity | ||
| Streptomyces sp. RKND-216 | Carbazoquinocin G (51) | Antimicrobial | 37 |
| M. smegmatis ATCC 120515 | Cytotoxic activity | ||
| Streptomyces albogriseolus strain B24 | Dentigerumycin E (52) | Antiproliferative | 38 |
| Bacillus cereus | Antimetastatic | ||
| Streptomyces sp. Strain PTY087I2 methicillin-resistant Staphylococcus aureus (MRSA) | Granaticin (53) | Increased antibacterial activity | 39 |
| Granatomycin D (54) | |||
| Dihydrogranaticin B (55) | |||
| Streptomyces coelicolor | Actinorhodin (56) | A benzoisochromanequinone dimer polyketide antibiotic | 40 |
| Myxococcus xanthus DK1622 | |||
| Streptomyces coelicolor A3 | Undecylprodigiosin (57) | Immunosuppressive | 41 |
| E. coli C600 | Antitumor | ||
| Streptomyces sp. MA37 | Be-13793c (58) | Anti-cancer activity | 42 |
| Pseudomonas sp. | |||
| Streptosporangium sp. KDCAGE35 | Funisamine (59) | 43 | |
| Bacillus sp. Strain KDCAGE13 |
Liang et al. stated that co-cultivation method induced more mass characteristics than the heat-killed inducer cultures, and both approaches resulted in the initiation of major characteristics not seen with any other induction method. N-Carbamoyl-2-hydroxy-3-methoxybenzamide (50) and carbazoquinocin G (51), two new secondary metabolites produced through co-culturing of producer Streptomyces sp. RKND-216 with inducers Alteromonas sp. RKMC-009 and M. smegmatis ATCC 120515, respectively.37
Dentigerumycin E (52), a new piperazic acid-bearing cyclic peptide isolated from the co-culture of marine Streptomyces albogriseolus strain B24 and Bacillus cereus and shown antimetastatic and antiproliferative activities against: liver, lung, stomach, breast and colorectal cancer.38
Co-cultivation of Streptomyces sp. PTY087I2 with human pathogens such as: Bacillus subtilis, Pseudomonas aeruginosa, methicillin-sensitive and methicillin-resistant Staphylococcus aureus, resulted in increased production of the three antibiotics, granaticin (53), granatomycin D (54), and dihydrogranaticin B (55). Furthermore, biological activities against the Gram-positive human pathogens used in these study was significantly increased.39 Increasing the use of co-culture studies to enable competitive interactions could boost metabolite synthesis and help to learn more about these microbial relationships.
| Interaction partners | Induced secondary | Metabolites activity | Reference |
|---|---|---|---|
| Mycobacterium smegmatis | Malformin C (60) | Increase in cytotoxic activity | 44 |
| Aspergillus niger | |||
| Streptomyces natalensis | Natamycin (61) | Polyene macrolide polyketide antibiotics | 45 |
| Penicillium chrysogenum | |||
| Streptomyces lividans | Bionectriamines A (62) | 46 | |
| Bionectriamines B (63) | |||
| Bionectria sp. | Tris(2,4-di-tert-butylphenyl) phosphate (64) | ||
| 6,8-Dihydroxyisocoumarin-3-carboxylic acid (65) | |||
| Streptomyces albospinus RLe7 | Amphotericin B (66) | Antifungal activity | 47 |
| Coniochaeta sp. FLe4 | Induction of red pigmented fungal phenotype | ||
| Aspergillus flavipes | Cytochalasans (67–72) | Cytotoxic effects against Streptomyces sp. | 48 |
| Streptomyces sp. | |||
| Streptomyces coelicolor | Cyclo (Phe–Tyr) (73) | 49 | |
| Aspergillus niger | Cyclo (Phe–Phe) (74) | ||
| Phenylacetic acid (75) | |||
| Furan-2-carboxylic acid (76) | |||
| 2-Hydroxyphenylacetic acid (77) | |||
| Streptomyces rochei MB037 | Borrelidin (78) | Antibacterial activity | 50 |
| Rhinocladiella similis 35 | Borrelidins F (79), J (80) and K (81) | ||
| 7-Methoxy-2,3-dimethylchromone-4-one (82) | |||
| Streptomyces leeuwenhoekii C34 | Luteoride D (83) | 51 | |
| Aspergillus fumigatus MR2012 | Pseurotin G (84) | ||
| Terezine D (85) | |||
| 11-O-methylpseurotin A (86) | |||
| Streptomyces leeuwenhoekii C58 | Chaxapeptin | Inhibit the human lung cancer cell line A549 | 51 |
| Aspergillus fumigatus MR2012 | Pentalenic acid (87) | ||
| Streptomyces albospinus RLe7 | Djalonensone (88) | 52 | |
| Phomopsis sp. FLe6 | 2,5-Furandimethanol (89) | ||
| Streptomyces piomogenus AS63D | Penicisteroid C (90) | Antimicrobial | 53 |
| Aspergillus niger | Cytotoxic activities | ||
| Streptomyces sp. CMB-M0423 | Heronapyrrole B (91) | Fungistatic | 54 |
| Aspergillus sp. CMB-StM0423 | |||
| Aspergillus fumigatus | Fumigermin (92) | A reversible germination inhibitor | 55 |
| Streptomyces rapamycinicus | |||
| Trichoderma sp. (307) | (3R, 7R)-7-hydroxy-de-O-methyllasiodiplodin (93) | Potent α-glucosidase inhibitory activity | 6 |
| Acinetobacter johnsonii (B2) | (3R)-5-oxo-de-O-Methyllasiodiplodin (94) | ||
| Streptomyces coelicolor A3 (2) | Gtri-02 (95) | 56 | |
| Aspergillus niger N402 | |||
| Actinomycete WAC 2288 | Ibomycin (96) | Anticryptococcus activity | 57 |
| Cryptococcus neoformans |
In Streptomyces natalensis HW-2 fermentation, a fungal elicitor from Penicillium chrysogenum AS 3.5163 showed an inductive effect on the production of natamycin (61). It was first observed that the fungal elicitor altered transcriptional levels in S. natalensis HW-2. The main conclusion is that the fungal elicitor increases the amount of precursor and changes the expression of natamycin related genes and secondary metabolic regulators.45
Co-cultivation of Bionectria sp., obtained from the seeds of the tropical plant Raphia taedigera with Streptomyces lividans yielded two novel o-aminobenzoic acid derivatives, bionectriamines A (62) and B (63), along with two other previously reported tris(2,4-di-tert-butylphenyl) phosphate (64) and 6,8-dihydroxyisocoumarin-3-carboxylic acid (65). None of the reported compounds were found in axenic fungal or bacterial cultures, showing that co-cultivation with bacteria activated silent biogenetic gene clusters.46 Amphotericin B (66), a well-known antifungal compound resulted from the co-cultivation of Streptomyces albospinus RLe7 and Coniochaeta sp. FLe4, endophytes isolated from the roots of the Brazilian medicinal plant Lychnophora ericoides. Amphotericin B was identified as one of the substances responsible for antifungal action as well as the induction of a red-pigmented fungal phenotype.47
A series of cytochalasans (rosellichalasin (67) and five aspochalasins (68–72)) were produced as a result of the co-culture of Aspergillus flavipes and Streptomyces sp. which isolated from the same marine sediment. These cytochalasans were reported to be formed by A. flavipes, and they were able to have cytotoxic activity against Streptomyces sp. at a wide range of concentrations while having no effect on the producer, favoring the producer in competition.48 The secondary metabolism of the filamentous model microorganisms Streptomyces coelicolor and Aspergillus niger is greatly influenced by their co-cultivation. A. niger formed the cyclic dipeptide cyclo (Phe–Tyr) (73), cyclo (Phe–Phe) (74), phenylacetic acid (75), furan-2-carboxylic acid (76) and 2-hydroxyphenylacetic acid (77) in reaction to S. coelicolor, according to NMR-based metabolomics combined with multivariate data analysis.49
3. Elicitation with cell-derived components and alga (Fig. 4)
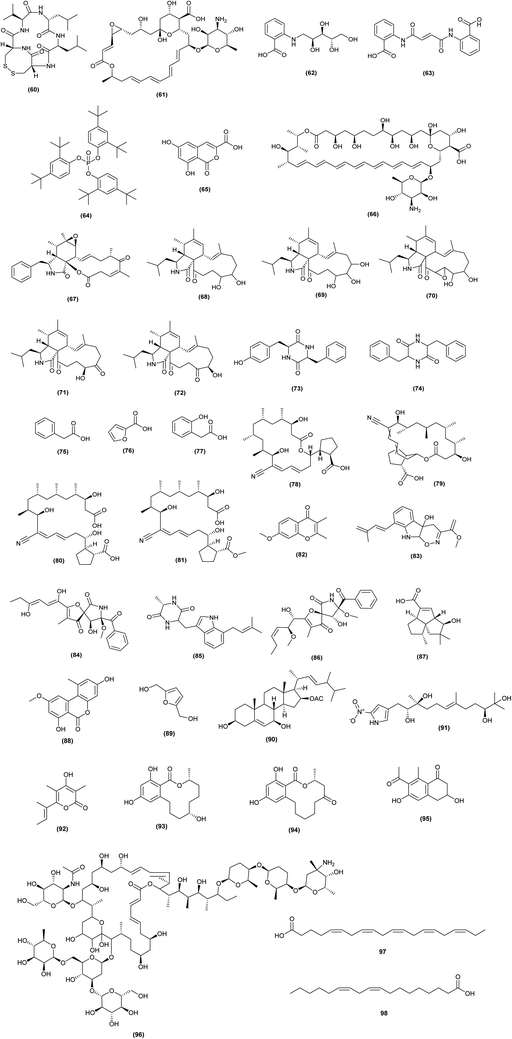
Many studies have been conducted to induce antibiotic encoding biosynthetic genes in actinomycetes by chemical and biological elicitors, which lead to production of numerous valuable bioactive metabolites based on the whole genome results. Mohammadipanah et al. revealed that the supernatant of Pseudomonas aeruginosa UTMC 1404 culture could act as stimuli to induce antibacterial synthetic pathways in Promicromonospora kermanensis DSM 45485. The eliciting agents in P. aeruginosa culture cell filtrate were resistant to detergent, acidic, and basic conditions and had an amphipathic character.58Mixed fermentation of Nocardia bhagyanarayanae I-27 and green microalga Tetradesmus obliquus AARL G022 yielded essential Ω3 fatty acids, including eicosapentaenoic acid (97) and α-linolenic acid (98).59 Eicosapentaenoic acid granted by the European Commission as Orphan designation for the treatment of familial adenomatous polyposis.
4. Chemical elicitation (Table 4 and Fig. 5)
Antibiotics at sub-inhibitory concentrations (SIC) have been demonstrated to alter gene expression at the transcriptional level, influencing 5–10% of all transcripts. Many phenotypic changes have been seen in the presence of sub-inhibitory antibiotic concentrations, including biofilm formation and enhanced bacterial motility. Antibiotic at SICs are being employed to trigger the production of cryptic natural compounds.60 Lincomycin at 1/10 of its MIC significantly boosted the expression of the pathway-specific regulatory gene actII-ORF4 in the blue-pigmented antibiotic actinorhodin (ACT) (99) biosynthetic gene cluster, resulting in ACT overproduction in Streptomyces coelicolor A3(2).61| Elicitor | Strain | Secondary metabolites Induced | Mechanism of elicitation | References |
|---|---|---|---|---|
| Lincomycin | Streptomyces coelicolor A3 | Actinorhodin (99) antibiotic | At 1/10 of its MIC, lincomycin significantly boosted the expression of the pathway-specific regulatory gene actII-ORF4 in the blue-pigmented antibiotic actinorhodin (ACT) biosynthetic gene cluster, resulting in ACT overproduction | 61 |
| Triclosan | S. coelicolor A3(2) strain 1147 (wild-type) | Actinorhodin (99) | 62 | |
| Triclosan | Streptomyces albus industrial strain KO-606 | Salinomycin (100) | ||
| Chloramphenicol | S. antibioticus strain KO-1164 | Nonribosomal peptide (NRP) antibiotics | Chloramphenicol raised amino acid pool sizes 1.5 to 6-fold, boosting CDA and actinomycin production | |
| Actinomycin D (101), calcium-dependent antibiotic (CDA), and piperidamycin) | ||||
| Streptomycin and paromomycin | Streptomyces diastatochromogenes 1628 | Toyocamycin (102) | Ribosome engineering strategies for activating genes for toyocamycin biosynthesis | 63 |
| Tetramycin A (103) | ||||
| Tetramycin P (104) | ||||
| Tetrin B (105) | ||||
| Furosemide fenofibrate | Saccharopolyspora cebuensis | Cebulantin (106) | ceb, a class I lanthipeptide BGC | 64 |
| (Inhibitory effect on vibrio strains) | ||||
| Etoposide and ivermectin | Streptomyces albus J1074 | Surugamide I (107) inhibit a cysteine protease implicated in cancer | Chemogenetic high-throughput screening approach (“HiTES”) to discover small molecule elicitors of silent biosynthetic gene clusters | 65 |
| Acyl-surugamide A (108) a novel antifungal | ||||
| Scandium | Micromonospora sp. BBHARD22 | Aloesaponarin II (109) | ||
| Nonomuraea sp. BBHARD23 | Hypogeamicin B (110) | |||
| Streptomycin | Microbispora sp. BCCAGE54 | Tetarimycin B (111) | Identified through the heterologous production of an environmental-derived type II polyketide synthase (PKS) gene cluster | 43 |
| Histone deacetylase inhibitors (hdais)(valproic acid) | Promicromonospora kermanensis DSM 45485 | Antibacterial activity | 50 | |
| Trimethoprim | Burkholderia thailandensis | Acybolins A–I and bactobolin (112-115) | Antibiotic | 66 |
| Ferric ion | Streptomyces sp. FXJ1.172 | Antibacterial cyclodepsipeptide, named NC-1 (116) | Lead to the activation of an uncharacterized cyclodepsipeptide, NC-1 | 67 |
| Rifampicin | Streptomyces somaliensis SCSIO ZH66 | Fredericamycin A (117) | Using ribosome engineering and response surface methodology | 68 |
| Cl-ARC | Streptomyces ghanaensis ATCC 14672 | Oxohygrolidin (118) | Elevate the expression of cryptic biosynthetic genes | 69 |
| Streptomyces hygroscopicus ATCC 53653 | 9-Methylstreptimidone (119) | |||
| Streptomyces sp. WAC0256 | Cyclotetralactone dynactin | |||
| Nonactin (23) |
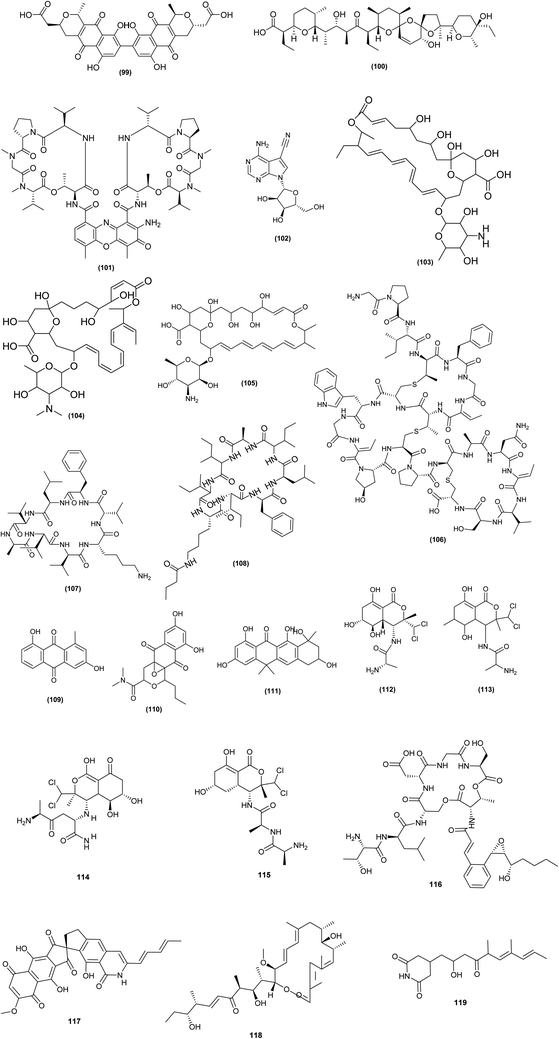 | ||
| Fig. 5 Chemical structures of compounds 99–119 reported upon chemical elicitation. Bactobolins: bactobolin (106), bactobolin A (107), bactobolin B (108), bactobolin C (109). | ||
Interestingly, it was found that addition of a SIC (0.1 μM) of triclosan to the culture media of S. coelicolor A3 (2) strain 1147 (wild-type) increased actinorhodin (99) synthesis by 5.2-fold. Triclosan had a positive effect on actinorhodin even in the high-producing S. coelicolor strain KO-1130 (rpoB), elevating titers by 82%. Moreover, addition of a SIC (1 μM) of triclosan to Streptomyces albus industrial strain KO-606 increased salinomycin (100) production by 40%, achieving 30.4 g L−1 of salinomycin titer. At subinhibitory concentrations of 0.3 g ml−1 (1/30 of its MIC), chloramphenicol (ribosome-targeting drug) increased the production of nonribosomal peptide antibiotic (actinomycin D) (101) in S. antibioticus strain KO-1164 by 1.6-fold CDA by S. coelicolor A3(2), presumably via increasing the intracellular amino acid pool size.62 From GYM agar plates containing various concentrations of streptomycin and paromomycin at doses of 1.5- to 10-fold greater than their respective MICs, 307 spontaneous antibiotic-resistant mutants of Streptomyces diastatochromogenes strain 1628 were identified. When streptomycin and paromomycin were used as screening antibiotics, the yields of toyocamycin (102), tetramycin A (103), tetramycin P (104) and tetrin B (105) were 244, 680, 583, and 569 mg L−1, respectively, which were 4.1-, 7.8-, 5.1-, and 13- fold higher than the yields of the wild-type strain.63
To uncover cryptic, bioactive metabolites, bioactivity assays are coupled with high-throughput elicitor screening (HiTES). The use of this technique in Saccharopolyspora cebuensis, with inhibition of Escherichia coli growth as a read-out, resulted in the production of cebulantin (106), a new lanthipeptide, induced by the clinical drugs furosemide and fenofibrate.64
HiTES was used successfully to awaken a cryptic gene cluster in Streptomyces albus J1074. The cytotoxins etoposide and ivermectin were shown to be powerful inducers, allowing identifying and isolating 14 new small chemical products from the chosen cluster. One of these metabolites is surugamide I (107), a strong cathepsin B inhibitor implicated in cancer, while a novel acyl-surugamide A (108) had good antifungal activity.65 The production of the anthraquinones aloesaponarin II (109) and hypogeamicin B (110) from the actinomycetes Micromonospora sp. BBHARD22 and Nonomuraea sp. BBHARD23, isolated from hypogean cave, increased six-fold under scandium metal exposure conditions. Tetarimycins (111) production from Microbispora sp. BCCAGE54 was increased thirty-threefold when exposed to streptomycin.43
5. Molecular elicitation
In addition to the cultivation-dependent elicitation techniques previously discussed, a review of metabolic product identification techniques for microorganisms with silent gene clusters discovered through genome mining has been undertaken. Another way to explore the actinomycetes' hidden potential is to mine microbial genomes for silent gene clusters, characterize the cryptic products using gene-knockout experiments, and combine these processes with analytical techniques like MALDI-TOF imaging and HPLC-MS.705.1. Transcription factor decoys (TFD) (Table 5 and Fig. 6)
Many methods have been devised to fully activate the untapped biosynthetic gene cluster (BGC), either by cloning and expressing the whole biosynthetic gene cluster (BGCs) or by directly modifying regulators in native hosts. Transcription factor decoys are DNA molecules that are created to interfere with gene regulation and may cause both the deactivation of a target BGC that is naturally active as well as the de-repression of a target BGC that is silent.71| Elicitor | Strain | Secondary metabolites Induced | Mechanism of elicitation | References |
|---|---|---|---|---|
| Phosphopantheteine transferases (PPtase) | Streptomyces alboniger NRRL B-1832 | Puromycin A (120) | Overexpression of PPtases affects secondary metabolism since it regulates primary metabolism, which causes physiological changes and changes in metabolic flux | 72 |
| Puromycin B (121) | ||||
| Puromycin C (122) | ||||
| Avenolide | Streptomyces avermitilis | Avermectin (pesticides) (123) | 73 | |
| TFDs, Rssp8M28I can activate butyrolactol A and Rssp7M24I can activate oxazolepoxidomycin A | Streptomyces sp. F-5635 (ssp8) | Butyrolactol A (124) | The presence of TFDs in the cell can bind to repressors, reducing the interaction between repressors and promoters and resulting in the activation of the silenced genes | 74 |
| Streptomyces sp. F-4335 (ssp7) | Oxazolepoxidomycin A (125) | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Overexpression of regulatory genes | ||||
| SARP-type regulator gene papR2 | Streptomyces sp. SHP22-7 | Amicetin (126) | Amicetin/plicacetin gene cluster activation induced by PapR2 | 75 |
| Plicacetin (127) | ||||
| SigA | Corynebacterium glutamicum | β-carotene (128) | In the stationary development phase, overexpression of sigA caused a 2-fold increase in the accumulation of the native carotenoid decaprenoxanthin | 76 |
| Bisanhydrobacterioruberin (BABR) (129) | ||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Promoter replacement strategy | ||||
| pCRISPR-Cas9-LigD and pCRISPomyces-2 sgRNA | Streptomyces sp. WAC5374, WAC8241 and 6273 | Thiolactomycin (130) | ligD, an enzyme necessary for NHEJ, and a homology template are both present in the plasmid | 4 |
| Amicetin (126) | ||||
| Phenanthroviridin (131) | ||||
| 5-Chloro-3-formylindole (132) | ||||
| CRISPR-Cas9 technique | S. roseosporus NRRL 15998 | Auroramycin (133) | Utilising a CRISPR-Cas9-based knock-in of KasOp* promoter's silent type I PKS BGC | 78 |
| Streptomyces viridochromogenes | Pentangular type II polyketide (134) | Effective and precise promoter cassette introduction using CRISPR-Cas9 promotes expression of biosynthetic genes and causes the creation of distinctive metabolites that are not seen in the parent strain | 83 | |
| S. roseosporus | Macrolactam (135) | 83 | ||
| Photocyclized alteramide A (136) | ||||
| FR-90098 9 (137) | ||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Global regulatory genes | ||||
| whiB-like (wbl) regulatory genes | Streptomyces somaliensis SCSIO ZH66 | Violapyrone B (138) | Play significant roles in secondary metabolism and morphological differentiation | 79 |
| Violapyrone A (139) | ||||
| Violapyrone J (140) | ||||
| Violapyrone C (141) | ||||
| Violapyrone H (142) | ||||
| ΔadpA | Streptomyces ansochromogene | Oviedomycin (143) | adpa, a global regulatory gene, is disrupted to activate a cryptic oviedomycin gene cluster (pks7) | 5 |
| adpAa gene inactivation or abrC3 global regulatory gene overexpression | Streptomyces argillaceus ATCC 12956 | Antimycin A1a (144) | Regulatory elements that respond to butyrolactone hormone | 80 |
| Antimycin A1b (145) | ||||
| Antimycin A2a (146) | ||||
| Antimycin A2b (147) | ||||
| Antimycin A3a (148) | ||||
| Antimycin A3b (149) | ||||
| Antimycin A4a (150) | ||||
| Antimycin A4b (151) | ||||
| Carotenoids (leprotene) (152) | ||||
| 3-Hydroxyleprotene (153) 3,3′-dihydroxyleprotene (154) | ||||
| β-isorenieratene (155) | ||||
| Germicidins B (156) | ||||
| Germicidins C (157) | ||||
| Desferrioxamine B (158) | ||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Reporter-guided mutant selection | ||||
XylE-neo cassette + the promoter of ccaR gene (XylE, a catecholase that dyes colonies brown when catechol is present, was fused to neo, a gene that codes for kanamycin resistance + ccaR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) gene, a transcriptional activator of clavulanic acid) gene, a transcriptional activator of clavulanic acid) |
Streptomyces venezuelae ISP5230 and Streptomyces sp. PGA64 | Jadomycin B (159) | Streptomyces spp. reporter-guided mutant selection. The strain is mutagenized using UV light after a double reporter construct called xylE-neo is put under the control of a selected quiet promoter | 81 |
| Gaudimycin D (160) | ||||
| Gaudimycin E (161) | ||||
| Mutagenesis by Tn insertion in conjunction with genetic reporters | Streptomyces albus J1074 | Antimycins (144–151) and candicidins (162) | The XNR 3174 gene, which encodes an unidentified transcriptional regulator, was shown to be the site of transposon insertion in the most well-known strain of S. albus, ATGSal2P2:TN14. Avenolide-like substance butenolide 4 was discovered to be produced by the mutant strain | 82 |
| Transposon (Tn) mutagenesis | Streptomyces coelicolor | Tripyrrole antibiotic: Undecylprodigiosin (163) and streptorubin B (RED (164)) | A hyperactive transposase-based Tn5 transposition system trigger's 724 mutants involving 365 genes, includes 17 genes in the RED biosynthetic gene cluster | 84 |
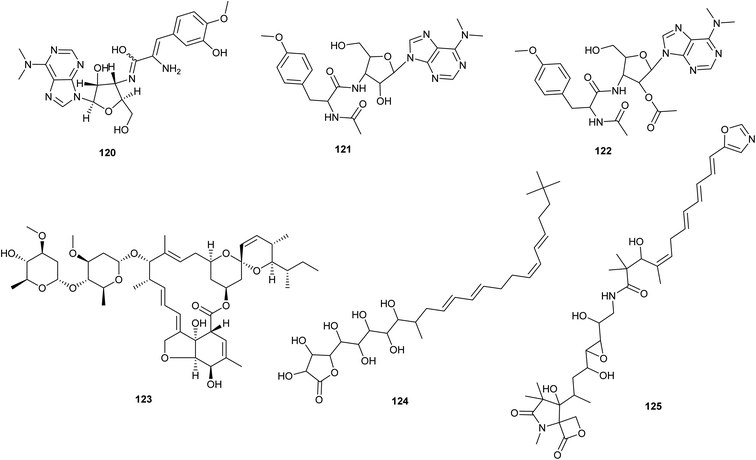 | ||
| Fig. 6 Chemical structures of compounds 120–125 reported upon molecular elicitation with transcription factor decoys. | ||
Puromycin A (120), a potent cytotoxic agent, along with two new derivatives (puromycin B and C) (121,122) were identified from Streptomyces alboniger NRRL B-1832 by enhancing the phosphopantetheinylation of carrier proteins. To activate the puromycin cryptic/silenced biosynthetic pathway, two broad-selective phosphopantetheinyl transferase (PPtase) genes from Bacillus subtilis and S. verticillus were conjugated into the S. alboniger NRRL B-1832 strain.72 By altering the expression of a novel TetR-family regulator and its target gene product, it was possible to increase the production of the anthelmintic avermectin in Streptomyces avermitilis. In order to promote the production of avermectin (123) and morphological differentiation in S. avermitilis, Niu et al. 2016 found SAV3619, a TetR-family transcriptional regulator named AveT, to be an activator.
Transcription Factor Decoys (TFD) activation efficiently activated BGCs from Streptomyces sp. F-5635 (ssp8) (a) and Streptomyces sp. F-4335 (ssp7), resulting in the isolation of butyrolactol A (124), a wide antifungal, and a new chemical, oxazolepoxidomycin A (125).74
5.2. Overexpression of regulatory genes (Table 5 and Fig. 7)
The streptomyces antibiotic regulatory protein (SARP) that regulates PapR2 expression in Streptomyces lividans or Streptomyces sp. SHP22-7 leads to a significantly improved antibiotic BGC such as: red amicetin (126) and plicacetin (127) (broad-spectrum antibacterial and antiviral disaccharide pyrimidine nucleoside antibiotics), respectively.75 β-carotene (128) and the non-native bisanhydrobacterioruberin (129) production were stimulated by overexpression of the principal sigma factor gene sigA from Corynebacterium glutamicum.76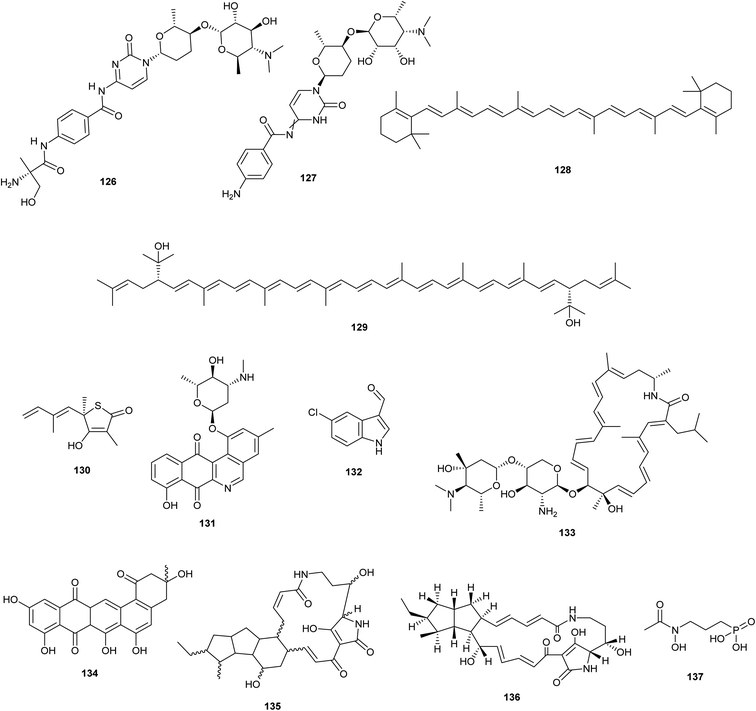 | ||
| Fig. 7 Chemical structures of compounds reported upon molecular elicitation with overexpression of regulatory genes (126–129) and Promoter Replacement Strategy (130–137). | ||
5.3. Promoter replacement strategy (Fig. 7)
As a selective and effective method for gene editing, CRISPR/Cas9 knocks in promoter cassettes to cause the expression of biosynthetic genes. As a result, more strain collections were mined in search of new antibiotics.77 (Culp et al. 2019). employed CRISPR-Cas9 genome engineering to inactivate the genes producing two of the most prevalent antibiotics, streptothricin and streptomycin, in 11 actinomycete strains. As a result, the modified strain WAC6273 orf17 was used to isolate the uncommon antibiotic amicetin and its new derivatives. Besides, the antibacterial substances thiolactomycin (130), phenanthroviridin (131), and 5-chloro-3-formylindole (132) were discovered utilizing activity-guided purification from two distinct WAC5374 and WAC8241 ΔstrI altered strains.4In Streptomyces roseosporus NRRL 15998, Lim et al. 2018 found auroramycin (133), a strong glycosylated polyene macrolactam antibiotic, by inducing the expression of a 95 kb BGC utilizing a 97 bp kasO* promoter.
5.4. Global regulatory gene (Table 5 and Fig. 8)
Violapyrone B (VLP B) (138), a pyrone chemical, was identified when the whiB-like (wbl A50) regulatory gene from S. somaliensis SCSIO ZH66, a deep-sea-derived strain, was rendered inactive. Further investigation led to the characterization of the VLP biosynthetic gene cluster and isolation of the VLPs analogues named A (139), J (140), C (141) and H (142) as a result of inactivation of vioB.79Global regulatory genes, such as adpa, which is the global regulatory gene in several streptomyces species, are activated by genetic manipulation, which results in the activation of cryptic gene clusters. Adpa was disrupted in S. ansochromogenes, resulting in the development of a mutant whose fermented extract displayed inhibitory zones against Gram-positive bacteria (S. aureus, B. subtilis and B. cereus) in addition to cytotoxic activities which done not occur in the wild type strain which indicate that disruption of adpa gene resulted in new metabolites. Chromatographic analysis revealed the presence of the new metabolite, oviedomycin (143) in Δadpa fermented extract and absence of nikkomycin traditional produced by the wild type strain adpa.5
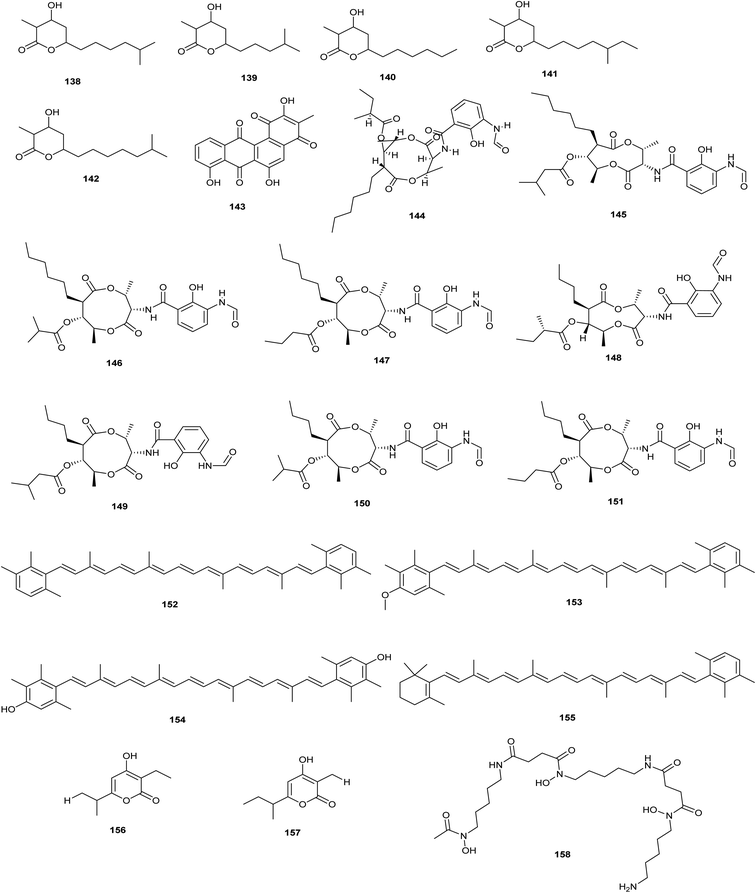 | ||
| Fig. 8 Chemical structures of compounds 138–158 reported upon molecular elicitation with global regulatory genes. | ||
Becerril et al. adopted four different approaches to activate BGCs in S. argillaceus and isolate encoded compounds; first, they changed culture conditions of S. argillaceus which lead to the production of antimycins (144–151) as a result of the encoded gene cluster 27 being activated. Additionally, they activated the cryptic gene cluster 26 when they expressed it in a heterologous host, which led to the isolation of three new carotenoids (152–155). Furthermore, the discovery of novel secondary metabolites is greatly aided by the global regulatory genes. Herein, the authors inactivated the global regulatory gene adqa resulted in identification of two compounds; germicidins B (156) and C (157) isolated as mixture and decreased production of mithromycin. On the other hand, they overexpressed the global regulatory gene abrc3 in S. argillaceus resulted in the isolation of desferrioxamine (158) (Becerril et al. 2018).
5.5. Reporter-guided mutant selection (Table 5 and Fig. 9)
Reporter-guided mutant selection is one of the most recent approaches (RGMS) for activating silent gene clusters in native producers. It is an efficient and generally applicable technique, in which reporters are employed to aid in the selection of target over-expressing mutants that are then tested for the desired phenotype utilizing reporter gene expression.81 This approach is predicated on the straightforward idea that the yield of the final metabolite is proportional to the degree of expression of the concerned biosynthetic gene cluster.82 Guo et al. adopted the RGMS approach in activating two angucycline-type gene clusters that were silent in two strains of Streptomyces in standard laboratory conditions. In this report, authors reactivated jadomycin biosynthesis in Streptomyces venezuelae ISP5230 lead to isolation of jadomycin B (159). In addition, they activated a silenced pga gene cluster, (accessionno. AY034378) in Streptomyces sp. PGA64 resulting in the isolation of two novel anthraquinone aminoglycosides named, gaudimycin D (160) and E (161). This report validated RGMS as a promising strategy in activating cryptic genes especially after activation of the pga gene cluster which has remained dormant despite more than ten years of continuous efforts using alternative ways.81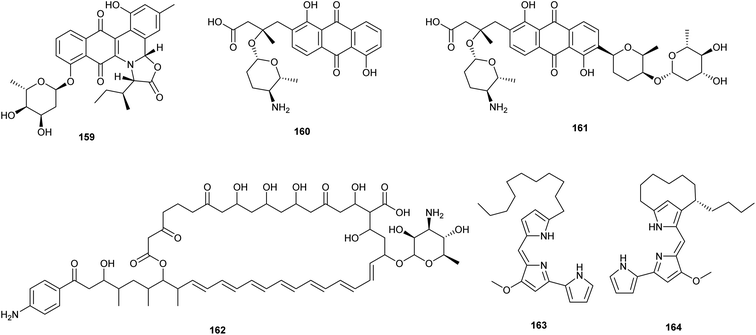 | ||
| Fig. 9 Chemical structures of compounds 159–164 reported upon molecular elicitation with reporter-guided mutant selection. | ||
Ahmed et al. applied a reporter-guided screening method to activate cryptic polycyclic tetramate macrolactam gene clusters in Streptomyces albus J1074. Herein, the low expressed S. albus J1074 PTM gene cluster was awakened using a combination of reporter-guided screening and transposon mutagenesis with gusA's selection as a reporter resulting in the production of antimycins (144–151) and candicidins (162).82
6. Conclusions
Recent advances in the whole-genome sequencing of Actinomycetes has uncovered a wide new field of microbial biology and chemistry. Under ordinary growth conditions, most biosynthetic gene clusters found in genomes were found to be “silent”. The scientific world has acknowledged that inactive gene clusters contain a significant source of therapeutic compounds. Several techniques for activating silent BGC have been developed. These methods enable the discovery of new metabolite scaffolds and improve the biosynthesis of minor secondary metabolites. The use of cross-species co-cultures, molecular and chemical elicitation for the biosynthesis of novel natural products have emerged as successful techniques in recent years (Fig. 10). In addition, appropriate dereplication methods, such as molecular networking and metabolomics, are needed to analyze the potential uniqueness of the evoked metabolites and prevent isolating existing compounds. Finally, novel chemical scaffolds from actinomycetes will be found through structural identification of the new hidden products utilizing LC-MS, MS/MS, and NMR.Author contributions
A. O. H. gathered a complete survey of all isolated compounds; A. O. H. and M. H. A. H. prepared the manuscript; U. R. A. interpreted and revised the results, and wrote the manuscript; S. S. E., R. M. and U. R. A. scientifically discussed the results and contributed to editing of the manuscript.Conflicts of interest
The authors declare that they have no conflict of interest.References
- U. Ramadan Abdelmohsen, Kristina Bayer and Ute Hentschel, Nat. Prod. Rep., 2014, 31, 381–399 RSC.
- N. J. Barrows, R. K. Campos, S. T. Powell, K. R. Prasanth, G. Schott-Lerner, R. Soto-Acosta, G. Galarza-Muñoz, E. L. McGrath, R. Urrabaz-Garza, J. Gao, P. Wu, R. Menon, G. Saade, I. Fernandez-Salas, S. L. Rossi, N. Vasilakis, A. Routh, S. S. Bradrick and M. A. Garcia-Blanco, Cell Host Microbe, 2016, 20, 259–270 CrossRef CAS PubMed.
- K. Ochi and T. Hosaka, Appl. Microbiol. Biotechnol., 2012, 971, 87–98 Search PubMed.
- E. J. Culp, G. Yim, N. Waglechner, W. Wang, A. C. Pawlowski and G. D. Wright, Nat. Biotechnol., 2019, 37, 1149–1154 CrossRef CAS PubMed.
- J. Xu, J. Zhang, J. Zhuo, Y. Li, Y. Tian and H. Tan, J. Biol. Chem., 2017, 292, 19708–19720 CrossRef CAS PubMed.
- L. Zhang, S. Niaz, D. Khan, Z. Wang, Y. Zhu, H. Zhou, Y. Lin, J. Li and L. Liu, Mar. Drugs, 2017, 15, 35 CrossRef PubMed.
- J. Chen, P. Zhang, X. Ye, B. Wei, M. Emam, H. Zhang and H. Wang, Mar. Drugs, 2020, 18, 2009–2019 Search PubMed.
- R. Pan, X. Bai, J. Chen, H. Zhang and H. Wang, Front. Microbiol., 2019, 10, 1–20 CrossRef CAS PubMed.
- P. R. Jensen, K. L. Chavarria, W. Fenical, B. S. Moore and N. Ziemert, J. Ind. Microbiol. Biotechnol., 2014, 41, 203–209 CrossRef CAS PubMed.
- Y. M. Chiang, S. L. Chang, B. R. Oakley and C. C. C. Wang, Curr. Opin. Chem. Biol., 2011, 15, 137–143 CrossRef CAS PubMed.
- U. R. Abdelmohsen, T. Grkovic, S. Balasubramanian, M. S. Kamel, R. J. Quinn and U. Hentschel, Biotechnol. Adv., 2015, 33, 798–811 CrossRef CAS PubMed.
- F. Canon, T. Nidelet, E. Guédon, A. Thierry and V. Gagnaire, Front. Microbiol., 2020, 11, 2088 CrossRef PubMed.
- H. A. Alhadrami, B. Thissera, M. H. A. Hassan, F. A. Behery, C. J. Ngwa, H. M. Hassan, G. Pradel, U. R. Abdelmohsen and M. E. Rateb, Mar. Drugs, 2021, 19, 109 CrossRef CAS PubMed.
- M. S. Hifnawy, H. M. Hassan, R. Mohammed, M. M. Fouda, A. M. Sayed, A. A. Hamed, S. F. Abouzid, M. E. Rateb, H. A. Alhadrami and U. R. Abdelmohsen, Mar. Drugs, 2020, 18, 243 CrossRef CAS PubMed.
- Y. I. Shamikh, A. A. El Shamy, Y. Gaber, U. R. Abdelmohsen, H. A. Madkour, H. Horn, H. M. Hassan, A. H. Elmaidomy, D. H. M. Alkhalifah and W. N. Hozzein, Microorganisms, 2020, 8, 783 CrossRef CAS PubMed.
- N. M. Gamaleldin, W. Bakeer, A. M. Sayed, Y. I. Shamikh, A. O. El-Gendy, H. M. Hassan, H. Horn, U. R. Abdelmohsen and W. N. Hozzein, Antibiotics, 2020, 9, 629 CrossRef CAS PubMed.
- S. Hoshino, M. Ozeki, C. P. Wong, H. Zhang, F. Hayashi, T. Awakawa, H. Morita, H. Onaka and I. Abe, Chem. Pharm. Bull., 2018, 66, 660–667 CrossRef CAS PubMed.
- T. B. Nguyen, S. Kitani, S. Shimma and T. Nihira, Appl. Environ. Microbiol., 2018, 84, 2791–2808 Search PubMed.
- S. S. El-Hawary, A. M. Sayed, R. Mohammed, M. A. Khanfar, M. E. Rateb, T. A. Mohammed, D. Hajjar, H. M. Hassan, T. A. M. Gulder and U. R. Abdelmohsen, Front. Chem., 2018, 6, 538 CrossRef CAS PubMed.
- G. Pishchany, E. Mevers, S. Ndousse-Fetter, D. J. Horvath, C. R. Paludo, E. A. Silva-Junior, S. Koren, E. P. Skaar, J. Clardy and R. Kolter, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 10124–10129 CrossRef CAS PubMed.
- S. Hoshino, M. Okada, T. Awakawa, S. Asamizu, H. Onaka and I. Abe, Org. Lett., 2017, 19, 4992–4995 CrossRef CAS PubMed.
- N. Adnani, M. G. Chevrette, S. N. Adibhatla, F. Zhang, Q. Yu, D. R. Braun, J. Nelson, S. W. Simpkins, B. R. McDonald, C. L. Myers, J. S. Piotrowski, C. J. Thompson, C. R. Currie, L. Li, S. R. Rajski and T. S. Bugni, ACS Chem. Biol., 2017, 12, 3093–3102 CrossRef CAS PubMed.
- V. N. Rocha Martin, C. Lacroix, J. Killer, V. Bunesova, E. Voney, C. Braegger and C. Schwab, Syst. Appl. Microbiol., 2019, 42, 506–516 CrossRef PubMed.
- R. Sugiyama, S. Nishimura, T. Ozaki, S. Asamizu, H. Onaka and H. Kakeya, Org. Lett., 2015, 17, 1918–1921 CrossRef CAS PubMed.
- R. Sugiyama, S. Nishimura, T. Ozaki, S. Asamizu, H. Onaka and H. Kakeya, Angew. Chemie, 2016, 128, 10434–10438 CrossRef.
- M. Uzzal Haque, M. Ajijur Rahman, M. Anwarul Haque, A. Kumar Sarker and M. Anwar Ul Islam, J. Appl. Pharm. Sci., 2016, 6, 51–055 CrossRef.
- H. B. Park, J. S. Park, S. Il Lee, B. Shin, D. C. Oh and H. C. Kwon, J. Nat. Prod., 2017, 80, 2542–2546 CrossRef CAS PubMed.
- S. Hoshino, M. Okada, T. Wakimoto, H. Zhang, F. Hayashi, H. Onaka and I. Abe, J. Nat. Prod., 2015, 78, 3011–3017 CrossRef CAS PubMed.
- S. Hoshino, L. Zhang, T. Awakawa, T. Wakimoto, H. Onaka and I. Abe, J. Antibiot., 2015, 68, 342–344 CrossRef CAS PubMed.
- S. Hoshino, T. Wakimoto, H. Onaka and I. Abe, Org. Lett., 2015, 17, 1501–1504 CrossRef CAS PubMed.
- T. Ozaki, Y. Kurokawa, S. Hayashi, N. Oku, S. Asamizu, Y. Igarashi and H. Onaka, ChemBioChem, 2016, 17, 218–223 CrossRef CAS PubMed.
- S. Hoshino, M. Ozeki, T. Awakawa, H. Morita, H. Onaka and I. Abe, J. Nat. Prod., 2018, 81, 2106–2110 CrossRef CAS PubMed.
- S. Hoshino, C. P. Wong, M. Ozeki, H. Zhang, F. Hayashi, T. Awakawa, S. Asamizu, H. Onaka and I. Abe, J. Antibiot., 2018, 71, 653–657 CrossRef CAS PubMed.
- D. K. Derewacz, B. C. Covington, J. A. McLean and B. O. Bachmann, ACS Chem. Biol., 2015, 10, 1998–2006 CrossRef CAS PubMed.
- D. C. Earl, P. B. Ferrell, N. Leelatian, J. T. Froese, B. J. Reisman, J. M. Irish and B. O. Bachmann, Nat. Commun., 2018, 9, 1–12 CrossRef CAS PubMed.
- L. Yu, Z. Hu and Z. Ma, Nat. Prod. Res., 2015, 29, 2087–2091 CrossRef CAS PubMed.
- L. Liang, G. Wang, B. Haltli, D. H. Marchbank, H. Stryhn, H. Correa and R. G. Kerr, J. Nat. Prod., 2020, 83, 2696–2705 CrossRef CAS PubMed.
- D. Shin, W. S. Byun, K. Moon, Y. Kwon, M. Bae, S. Um, S. K. Lee and D.-C. Oh, Front. Chem., 2018, 6, 498 CrossRef CAS PubMed.
- A. Sung, S. Gromek and M. Balunas, Mar. Drugs, 2017, 15, 250 CrossRef PubMed.
- N. Lee, W. Kim, J. Chung, Y. Lee, S. Cho, K. S. Jang, S. C. Kim, B. Palsson and B. K. Cho, ISME J., 2020, 14, 1111–1124 CrossRef CAS PubMed.
- F. Mavituna, K. J. K. Luti and L. Gu, Enzyme Microb. Technol., 2016, 90, 45–52 CrossRef CAS PubMed.
- F. Maglangit, Q. Fang, K. Kyeremeh, J. M. Sternberg, R. Ebel and H. Deng, Molecules, 2020, 25, 256 CrossRef CAS PubMed.
- B. C. Covington, J. M. Spraggins, A. E. Ynigez-Gutierrez, Z. B. Hylton and B. O. Bachmann, Appl. Environ. Microbiol., 2018, 84, 1125–1143 CrossRef PubMed.
- T. Jomori, Y. Hara, M. Sasaoka, K. Harada, A. Setiawan, K. Hirata, A. Kimishima and M. Arai, J. Nat. Med., 2020, 74, 76–82 CrossRef CAS PubMed.
- D. Wang, W. Shen, J. Yuan, L. Wei, Y. Zhang, PREPRINT (Version 1), 2019, available at Research Square, DOI:10.21203/rs.2.18475/v1.
- R. S. T. Kamdem, H. Wang, P. Wafo, W. Ebrahim, F. C. Özkaya, G. Makhloufi, C. Janiak, P. Sureechatchaiyan, M. U. Kassack, W. Lin, Z. Liu and P. Proksch, Fitoterapia, 2018, 124, 132–136 CrossRef CAS PubMed.
- F. O. Chagas, A. M. Caraballo-Rodríguez, P. C. Dorrestein and M. T. Pupo, J. Nat. Prod., 2017, 80, 1302–1309 CrossRef CAS PubMed.
- L. Yu, W. Ding and Z. Ma, Nat. Prod. Res., 2016, 30, 1718–1723 CrossRef CAS PubMed.
- C. Wu, B. Zacchetti, A. F. J. Ram, G. P. Van Wezel, D. Claessen and Y. H. Choi, Sci. Rep., 2015, 5, 1–10 Search PubMed.
- M. Yu, Y. Li, S. P. Banakar, L. Liu, C. Shao, Z. Li and C. Wang, Front. Microbiol., 2019, 10, 915 CrossRef PubMed.
- J. Wakefield, H. M. Hassan, M. Jaspars, R. Ebel and M. E. Rateb, Front. Microbiol., 2017, 8, 1284 CrossRef PubMed.
- F. O. Chagas and M. T. Pupo, Microbiol. Res., 2018, 212–213, 10–16 CrossRef CAS PubMed.
- A. S. Abdel-Razek, A. Hamed, M. Frese, N. Sewald and M. Shaaban, Steroids, 2018, 138, 21–25 CrossRef CAS PubMed.
- Z. G. Khalil, P. Cruz-Morales, C. Licona-Cassani, E. Marcellin and R. J. Capon, ISME J., 2019, 13, 147–158 CrossRef CAS PubMed.
- M. Cristina Stroe, T. Netzker, K. Scherlach, T. Krüger, C. Hertweck, V. Valiante and A. A Brakhage, Targeted induction of a silent fungal gene cluster encoding the bacteria-specific germination inhibitor fumigermin, elife, 2020, 9, e52541 CrossRef PubMed.
- C. Wu, K. Ichinose, Y. H. Choi and G. P. van Wezel, ChemBioChem, 2017, 18, 1428–1434 CrossRef CAS PubMed.
- N. Robbins, M. Spitzer, W. Wang, N. Waglechner, D. J. Patel, J. S. O'Brien, L. Ejim, O. Ejim, M. Tyers and G. D. Wright, Cell Chem. Biol., 2016, 23, 1383–1394 CrossRef CAS PubMed.
- F. Mohammadipanah, F. Kermani and F. Salimi, Appl. Biochem. Biotechnol., 2020, 192, 1224–1237 CrossRef CAS PubMed.
- B. Kumsiri, J. Pekkoh, W. Pathom-aree, S. Lumyong and C. Pumas, Algal Res., 2018, 33, 239–247 CrossRef.
- W. C. Ratcliff and R. F. Denison, Science, 2011, 332, 547–548 CrossRef CAS PubMed.
- Y. Imai, S. Sato, Y. Tanaka, K. Ochi and T. Hosaka, Appl. Environ. Microbiol., 2015, 81, 3869–3879 CrossRef CAS PubMed.
- Y. Tanaka, M. Izawa, Y. Hiraga, Y. Misaki, T. Watanabe and K. Ochi, Appl. Microbiol. Biotechnol., 2017, 101, 4417–4431 CrossRef CAS PubMed.
- X. Shentu, N. Liu, G. Tang, Y. Tanaka, K. Ochi, J. Xu and X. Yu, J. Antibiot., 2016, 69, 406–410 CrossRef CAS PubMed.
- K. Moon, F. Xu and M. R. Seyedsayamdost, Angew. Chemie, 2019, 131, 6034–6038 CrossRef.
- F. Xu, B. Nazari, K. Moon, L. B. Bushin and M. R. Seyedsayamdost, J. Am. Chem. Soc., 2017, 139, 9203–9212 CrossRef CAS PubMed.
- B. K. Okada, Y. Wu, D. Mao, L. B. Bushin and M. R. Seyedsayamdost, ACS Chem. Biol., 2016, 11, 2124–2130 CrossRef CAS PubMed.
- M. Liu, N. Liu, F. Shang and Y. Huang, Eur. J. Org. Chem., 2016, 2016, 3943–3948 CrossRef CAS.
- Y. Zhang, H. Huang, S. Xu, B. Wang, J. Ju, H. Tan and W. Li, Microb. Cell Fact., 2015, 14, 1–11 CrossRef PubMed.
- S. M. Pimentel-Elardo, D. Sørensen, L. Ho, M. Ziko, S. A. Bueler, S. Lu, J. Tao, A. Moser, R. Lee, D. Agard, G. Fairn, J. L. Rubinstein, B. K. Shoichet and J. R. Nodwell, ACS Chem. Biol., 2015, 10, 2616–2623 CrossRef CAS PubMed.
- G. L. Challis, Microbiology, 2008, 154, 1555–1569 CrossRef CAS PubMed.
- H. Ren, B. Wang and H. Zhao, Curr. Opin. Biotechnol., 2017, 48, 21–27 CrossRef CAS PubMed.
- X. Yan, B. Zhang, W. Tian, Q. Dai, X. Zheng, K. Hu, X. Liu, Z. Deng and X. Qu, Synth. Syst. Biotechnol., 2018, 3, 76–80 CrossRef PubMed.
- G. Niu, K. F. Chater, Y. Tian, J. Zhang and H. Tan, FEMS Microbiol. Rev., 2016, 40, 554–573 CrossRef CAS PubMed.
- B. Wang, F. Guo, S. H. Dong and H. Zhao, Nat. Chem. Biol., 2019, 15, 111–114 CrossRef CAS PubMed.
- J. Krause, I. Handayani, K. Blin, A. Kulik and Y. Mast, Front. Microbiol., 2020, 11, 225 CrossRef PubMed.
- H. Taniguchi, N. A. Henke, S. A. E. Heider and V. F. Wendisch, Metab. Eng. Commun., 2017, 4, 1–11 CrossRef PubMed.
- J. A. Doudna and E. Charpentier, The new frontier of genome engineering with CRISPR-Cas9, Science, 2014, 346(6213), 1258096–1258099 CrossRef PubMed.
- Y. H. Lim, F. T. Wong, W. L. Yeo, K. C. Ching, Y. W. Lim, E. Heng, S. Chen, D.-J. Tsai, T.-L. Lauderdale, K.-S. Shia, Y. S. Ho, S. Hoon, E. L. Ang, M. M. Zhang and H. Zhao, ChemBioChem, 2018, 19, 1716–1719 CrossRef CAS PubMed.
- H. Huang, L. Hou, H. Li, Y. Qiu, J. Ju and W. Li, Microb. Cell Fact., 2016, 15, 1–9 CrossRef PubMed.
- A. Becerril, S. Álvarez, A. F. Braña, S. Rico, M. Díaz, R. I. Santamaría, J. A. Salas and C. Méndez, PLos One, 2018, 13, 1–14 CrossRef PubMed.
- F. Guo, S. Xiang, L. Li, B. Wang, J. Rajasärkkä, K. Gröndahl-Yli-Hannuksela, G. Ai, M. Metsä-Ketelä and K. Yang, Metab. Eng., 2015, 28, 134–142 CrossRef CAS PubMed.
- Y. Ahmed, Y. Rebets, B. Tokovenko, E. Brötz and A. Luzhetskyy, Sci. Rep., 2017, 7, 1–11 CrossRef PubMed.
- M. M. Zhang, F. T. Wong, Y. Wang, S. Luo, Y. H. Lim, E. Heng, W. L. Yeo, R. E. Cobb, B. Enghiad, E. L. Ang and H. Zhao, Nat. Chem. Biol., 2017, 13, 607–609 CrossRef CAS PubMed.
- Z. Xu, Y. Wang, K. F. Chater, H. Y. Ou, H. H. Xu, Z. Deng and M. Tao, Appl. Environ. Microbiol., 2017, 83, 2889–2905 Search PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |