Open Access Article
Open Access ArticleQuantitative analysis of phenanthrene in soil by fluorescence spectroscopy coupled with the CARS-PLS model†
Haonan Lia,
Maogang Lia,
Hongsheng Tanga,
Hua Li ab,
Tianlong Zhang
ab,
Tianlong Zhang *a and
Xiao-Feng Yang
*a and
Xiao-Feng Yang *a
*a
aKey Laboratory of Synthetic and Natural Functional Molecule of the Ministry of Education, College of Chemistry & Materials Science, Northwest University, Xi'an, 710127, China. E-mail: tlzhang@nwu.edu.cn; xfyang@nwu.edu.cn
bCollege of Chemistry and Chemical Engineering, Xi'an Shiyou University, Xi'an, 710065, China
First published on 22nd March 2023
Abstract
Polycyclic aromatic hydrocarbons (PAHs) are typical organic pollutants in soil and are teratogenic and carcinogenic. Therefore, rapid and accurate analysis of PAHs in soil can provide a theoretical basis and data support for soil contamination risk assessment. In this work, a fluorescence spectroscopy technique combined with partial least squares (PLS) was proposed for rapid quantitative analysis of phenanthrene (PHE) in soil. At first, the fluorescence spectra of 29 soil samples with different concentrations (0.3–10 mg g−1) of PHE were collected by RF-5301 PC fluorescence spectrophotometer. Secondly, the effects of different spectral preprocessing methods were investigated on the prediction performance of the PLS calibration model. And then, the influence of competitive adaptive reweighted sampling (CARS) wavelength points on the prediction performance of PLS calibration model was discussed. Finally, according to the selected wavelength points, a quantitative analytical model for PHE content in soil was constructed using the PLS calibration method. To further explore the predictive performance of the CARS-PLS calibration model, the predictive results were compared with those of the RAW spectrum-partial least squares calibration model (RAW-PLS) and the wavelet transform-standard normal variation (WT-SNV) calibration model. The CARS-PLS calibration model showed the optimal predictive performance and its coefficient of determination of cross-validation (Rcv2) and root mean square error of 10-fold cross-validation (RMSEcv) were 0.9957 and 18.98%, respectively. The coefficient of determination of prediction set (Rp2) and root mean square error of prediction set (RMSEp) were 0.9963 and 16.13%, respectively. Hence, the CARS algorithm based on fluorescence spectrum coupled with PLS can give a rapid and accurate quantitative analysis of the PHE content in soil.
1 Introduction
PAHs are a class of highly stable and toxic persistent organic pollutants produced by the incomplete combustion of organic substances such as biomass and fossil fuels, and have been listed as one of the priority pollutants by the US Environmental Protection Agency.1,2 PAHs are hydrocarbons containing two or more benzene rings, mainly including non-concentrated hydrocarbons represented by biphenyl and terphenyl, and concentrated hydrocarbons represented by naphthalene and PHE.3 Naphthalene is thickened by two benzene rings sharing two adjacent carbon atoms, and is the most abundant compound in coal tar.4 PHE is the simplest triple benzene ring nonlinear PAHs and its unique chemical structure is closely related to the carcinogenicity of PAHs.5 Derivatives of PHE are PAHs with significant carcinogenicity and PHE has become a representative compound in PAHs research.6 Soil, as an important medium, is responsible for more than 90% of the environmental load of PHE, which is difficult to degrade after it enters the soil, and its toxicity becomes stronger and stronger with the accumulation of time.7–9 The pollution caused by PHE in the soil would not only hinder its normal function, but also cause crop yield reduction and agricultural product safety problems, which will eventually cause extremely serious harm to the human body through the food chain.10–12 The International Cancer Research Institute of the World Health Organization has declared PHE a class of carcinogen that has been shown to its presence in the human body leads to the damage of monotonous cells by high concentration of free radicals and even risks of amplifying the damage.13–15 In view of the great threats to human health and soil environment caused by PHE, it is of great significance to study the monitoring of PHE pollutants on soil ecosystem pollution.Currently, the contamination of PAHs in soil is moving toward increasing severity, and therefore the detection methods for PAHs in soil are being widely studied worldwide.16 The traditional analytical methods for laboratory detection of PAHs in soil are mainly based on chromatography, which is an effective separation and analysis technique that separates and analyzes substances based on the difference in partition coefficients between the stationary and mobile phases. Gas chromatography (GC), gas chromatography-mass spectrometry (GC-MS) and high performance liquid chromatography (HPLC) have been widely used for the detection of PAHs in soil.17–20 Temerdasheva et al.21 proposed a method for the preparation of PAHs samples in soil (substrate) by dispersive liquid–liquid microextraction and successfully determined the contents of 20 PAHs in soil (substrate) by GC-MS with the limits of quantification (LOQs) of 0.2–0.5 μg kg−1. Nevertheless, these methods have some disadvantages, such as complicated sample preprocessing process, labor- and material-intensive, can not ensure the complete extraction of PAHs, and can not meet the need for dynamic monitoring of soil pollution. Therefore, there is a need to use a convenient, rapid and straightforward method for the detection and assessment of PHE in soils.
Fluorescence is the emission of the excited molecule from the zeroth vibrational level of excited singlet state.22 The high fluorescence characteristics of PAHs has made molecular fluorescence spectrometry (MWS) used to detect PAHs in the environment.23,24 Fluorescence spectroscopy is a field portable technology with the advantages of high sensitivity and good selectivity. Although the broad emission spectrum and background fluorescence from impurities in some of the biological and environmental samples make fluorescence analysis complicated, compared with traditional methods, fluorescence spectroscopy has the best performance in terms of operation time, analysis cost, accuracy, and operator health and safety.25 Besides, fluorescence technology can simultaneously measure a variety of fluorescence parameters, such as emission wavelength, excitation wavelength, intensity, polarization and fluorescence lifetime.26 The fluorescence lifetime can be measured by time-resolved spectrum or phase-resolved spectrum. In a time-resolved fluorescence spectrum, fluorescence is excited by a series of short laser pulses, measuring the delay time between the detection of a fluorescent photon and the excitation of a pulsed laser.27 Fluorescence lifetimes at different excitation and emission wavelengths have been used to simultaneously analyze PAHs mixtures. Gu et al.28 successfully developed a prior knowledge integration method based on time-resolved fluorescence to determine PAHs in edible vegetable oils with the RMSEp was less than 2%. Nevertheless, the fluorescence lifetime is extremely sensitive to micro-environment, which makes the analysis of PAHs mixtures complicated. Excitation–emission matrix (EEM) fluorescence spectroscopy can provide complete fluorescence information of PAHs in complex environment by covering different excitation and emission wavelengths.29,30 EEM fluorescence spectroscopy also has some shortcomings, so it is difficult to extract characteristic fluorescence information from the peaks with serious overlap among PAHs. Thus, it is impossible to quantitatively analyze them by a single factor.
In recent years, chemometrics and fluorescence spectroscopy have been combined to realize the qualitative and quantitative analysis of complex systems under the interference of unknown components. Yang et al.31 proposed a quantitative method based on two-dimensional (2D) fluorescence correlation spectroscopy combined with multivariate method for quantitative determination of PAHs in the environment. Huang et al.32 combined the three-dimensional (3D) fluorescence spectrum with non-smooth non-negative matrix decomposition algorithm, the fluorescence spectrum information of a single polycyclic aromatic hydrocarbon was extracted from the aliasing spectrum, and the rapid identification of polycyclic aromatic hydrocarbons was successfully realized. Li et al.33 proposed a combination of 3D fluorescence spectroscopy and multidimensional partial least squares (N-PLS) model to achieve quantitative analysis of mixed anthracene and PHE samples in soil with RMSEp of 8.04 × 10−4 g g−1 and 5.15 × 10−4 g g−1, respectively. In the above research, when analyzing the spectral data, all the measured wavelengths are used to establish calibration models. However, chemically, only part of the wavelength is related to the chemical properties being modeled, so the remaining variables are interfering. Statistically speaking, the wavelength contains redundant information, and the high linearity of col will make the model more complicated and take longer to build. Therefore, wavelength selection is of great significance for establishing a model which can reliably predict new samples. The extraction methods of spectral variables include uninformed variable elimination (UVE), successive projections algorithm (SPA), variable importance projection (VIP) etc.34,35 Nevertheless, these algorithms are either greedy algorithms or can only find local optimal solutions, so it is difficult to choose the combination with better prediction performance from many candidate variables. CARS algorithm is the law of “survival of the fittest” in the biological evolution theory.36 CARS technique is used to filter variables and obtain the absolute values of PLS regression coefficients, retain the points with larger absolute values and remove the points with smaller absolute values to obtain a series of optimal subset.37,38 Then, cross-validation (CV) method is used to select the subset of the minimum RMSECV of the model, and finally the subset is determined as the optimal wavelength combination related to the measurement elements. This method has the advantages of fast calculation speed, high variable screening efficiency, and the ability to screen out better variable combinations.
In this work, the feasibility of a calibration model for quantitative analysis of PHE fluorescence spectrum based on CARS-PLS algorithm was investigated. The fluorescence spectra of 29 soil samples with different concentrations (0.3–10 mg g−1) of PHE were collected with a RF-5301 PC fluorescence spectrophotometer and the spectra were analyzed. The effects of different spectral pretreatment methods were explored. The effects of SNV and WT combined pretreatment was emphatically explored, and the spectra pretreated by the combination of SNV and WT was screened by CARS, and 12 wavelength points closely related to PHE content were selected. Based on the selected wavelength points, a quantitative analysis model of PHE content in soil was developed using the PLS calibration model. In order to further verify the prediction performance of CARS-PLS calibration model, it was compared with the RAW-PLS and WT-SNV-PLS models. Finally, a rapid quantitative analysis model for PHE content in soil was obtained.
2 Materials and methods
2.1 Sample preparation
In order to ensure the uniformity of the prepared samples, different proportions of PHE (Macklin, analytically pure) and soil (the Institute of Geophysical and Geochemical Exploration (IGGE), Chinese Academy of Geological Sciences) were first fully ground in agate mortar for 20 min, and then thoroughly mixed for 1 h with vortex mixer. Finally, a total of 29 soil samples with different PHE contents were obtained. The content of PHE in the samples is shown in Table 1. As can be seen from the Table 1, the mass fraction range of PHE in the soil samples ranged from 0.3 to 10 mg g−1. The prepared samples were stored in powder form for testing.| No. of sample | Concentration (mg g−1) | No. of sample | Concentration (mg g−1) |
|---|---|---|---|
| a The samples numbered with * in Table 1 was randomly divided into prediction set, and the remaining samples were calibration set. | |||
| 1 | 9.867 | 16 | 4.700 |
| 2 | 9.500 | 17 | 4.300 |
| 3 | 8.967 | 18 | 4.100 |
| 4 | 8.600 | 19 | 3.750 |
| 5* | 8.330 | 20 | 3.450 |
| 6 | 7.967 | 21 | 3.200 |
| 7* | 7.700 | 22* | 2.600 |
| 8* | 7.300 | 23 | 2.350 |
| 9 | 6.950 | 24 | 2.100 |
| 10 | 6.650 | 25* | 1.750 |
| 11* | 6.400 | 26 | 1.300 |
| 12* | 5.950 | 27 | 0.650 |
| 13* | 5.750 | 28* | 0.450 |
| 14 | 5.400 | 29 | 0.300 |
| 15 | 5.150 | ||
2.2 Spectral acquisition
The fluorescence spectrum of 29 samples with different PHE contents were obtained with a fluorescence spectrophotometer (RF-5301PC, SHIMADZU, Japan) under the optimized detection condition (excitation wavelength: 350.0 nm, excitation slit width: 3 nm, spectral range: 360.0 nm–650.0 nm, scanning speed: 1200 nm min−1, the emission slit width: 10 nm, PMT voltage: 700 V). The spectra of samples were recorded adopting a solid powder installed in the testing room of the instrument. After preheating the instrument with optimized parameters (about 5 min), the spectra collection of samples was started to be acquired, and the air-related background spectra were detected and subtracted.To reduce the experimental error, the same sample was repeatedly detected for 5 times and the average spectrum was taken as the spectrum of the sample. 145 spectra were obtained from a total of 29 samples (5 spectra per sample). According to the principle that the model building samples should cover the spectral characteristics and property range of the samples to be tested and the ratio of prediction set to calibration set was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 20 samples were randomly selected as the calibration set to build the model and 9 samples were selected as test set to verify the prediction performance of the model. The delineation results are demonstrated in Table 1.
2, 20 samples were randomly selected as the calibration set to build the model and 9 samples were selected as test set to verify the prediction performance of the model. The delineation results are demonstrated in Table 1.
2.3 Competitive adaptive weighted sampling-partial least squares
PLS calibration models were constructed based on different spectra of samples in this work. PLS is a classic regression algorithm in multivariate correction technology, and it is commonly used in various fields because of its advantages including few optimization parameters, fast modeling speed, and stable prediction performance.39–41 The procedure of the construction of CARS-PLS calibration model for the quantitative analysis PHE content in soil is depicted in Fig. 1. Firstly, five different pretreatment methods SNV,42 multivariate scatterincorrection (MSC),43 first derivative (D1st),44 wavelet transform (WT)45 and WT-SNV were used for preprocessing raw spectral data to reduce the interference of instrument noise, environmental noise, and experimental error on raw spectra. In data processing, 10-fold cross-validation (10-fold CV) and R2 were applied to optimize the parameter of preprocessing methods. Then, characteristic wavelength extraction was performed based on the processed data, in which CARS was used. CARS selects a subset of N variables by sampling N times in an iterative manner and finally chooses the subset with the lowest RMSECV value as the optimal subset. Finally, PLS calibration model was constructed based on the selected wavelength points. All calculation in the work was completed by MATLAB (Version 2022a).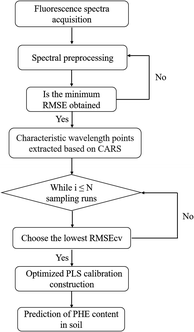 | ||
| Fig. 1 The construction of CARS-PLS calibration model for the quantitative analysis PHE content in soil. | ||
3 Results and discussion
3.1 Fluorescence spectra analysis of soil
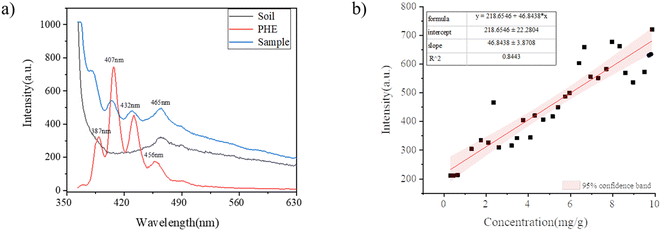
Fig. 2(a) shows the fluorescence spectra of soil, PHE and samples with different concentrations of PHE. The black line in Fig. 2(a) indicates the emission fluorescence map of the original soil sample with excitation at 350.0 nm. The characteristic peak of soil observed in fluorescence spectra was located at 465.0 nm, which may be caused by the superposition of minerals or other trace fluorescent substances in the soil. The red line in Fig. 2(a) represents the fluorescence emission map of PHE with the excitation wavelength of 350.0 nm. The characteristic peaks of PHE were located at 387.0 nm, 407.0 nm, 432.0 nm and 456.0 nm, and the fluorescence intensity was the strongest at 407.0 nm. The blue line in Fig. 2(a) denotes the spectrogram of the mixed sample of soil and PHE. The emission spectrum of the samples also consistd of four spectral bands. The characteristic peaks of the samples were located at 380.0 nm, 405.0 nm, 429.0 nm, and 465.0 nm, which were essentially the same as those of pure PHE, with no significant shift differences. In this section, a standard curve was established between the fluorescence intensity of the sample at a single wavelength and the concentration of PHE by using traditional regression analysis. As can be seen from Fig. 2(a) the sample fluorescence intensity at 380.0 nm is the main peak position of the fluorescence emission peak of the sample, so the fluorescence intensity at 380.0 nm was selected as the basis for the quantification of PHE content in soil. Fig. 2(b) shows the linear fit (with 95% confidence band) between the fluorescence peak intensity at the characteristic peak 380.0 nm and the PHE concentration in soil, with a correlation coefficient of 0.8443, and almost half of the points fall outside this 95% confidence interval, which indicates that the error between the fluorescence intensity and the PHE concentration in soil is large. This is mainly because the background noise, stray light and spectral inhomogeneity can lead to the deviation between the PHE concentration and the characteristic peak intensity during the spectral data acquisition. Hence, the use of the quantitative analysis of PHE concentration in soil could not be accurately achieved by using the standard curve. Next, the quantitative analysis of PHE concentration in soil was explored by using fluorescence spectroscopy combined with chemometric methods.3.2 Pretreatment methods selection
When collecting spectra, it is not difficult to be affected by uncontrollable factors (e.g., the state of the sample itself (particle size), the test environment (temperature and humidity) and the conditions of the instrument itself), which would make the original fluorescence spectrum difficult to analyze. Besides, this reduced the accuracy and stability of PLS calibration model. Spectral preprocessing method can effectively distinguish the correction background, improve the signal-to-noise ratio and other factors that affect the analysis precision. Therefore, when establishing the PLS calibration model, it is essential to consider appropriate spectral preprocessing methods and their integration, so as to improve the shortcomings in the original spectrum.In this work, SNV, MSC, WT, D1st and their combination on the PLS calibration model were compared to obtain the best prediction results. Among them, SNV can be used for errors caused by surface scattering, sample particle size and changes in the optical path during testing. MSC and SNV have similar functions to reduce the influence of uneven solid particle size and the resulting sample surface scattering. D1st is of used to eliminate the constant shift of the background, which is beneficial to improve the spectral resolution and realize the baseline correction of the spectrum. For D1st, the optimization range of smoothing points is from 3 to 15 (each odd point), and the optimized smoothing points for D1st is 7. WT has multi-resolution characteristics, and choosing an appropriate wavelet basis function can realize the simultaneous characterization of high-frequency unstable signals in time domain and frequency domain. The WT and SNV preprocessing methods were used to jointly process the spectra. For the WT, the range of wavelet basis function is db1–db8 and decomposition layers is 1–5. Finally, db6 and 2 were invoked as the optimal values of the wavelet basis function and decomposition layers, respectively.
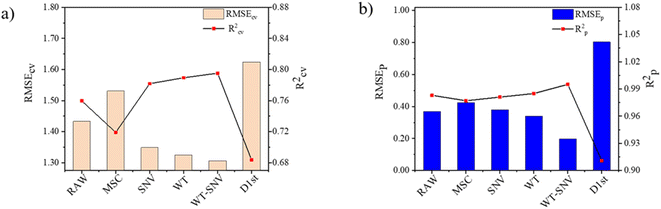
Then, a total of 5 PLS models were constructed based on raw fluorescence spectra and the variables processed by the methods mentioned above respectively. Taking R2 and RMSE as evaluation indexes, the prediction performance of PLS calibration models based on different input variables was compared to evaluate the pretreatment effect of the above spectral pretreatment methods. Prediction performance of PLS calibration models based on discrete spectral preprocessing methods is depicted in Fig. 3. (The specific parameters and performance of PLS model based on different preprocessing methods are shown in Fig. S1 in the ESI.†) As can be observed in Fig. 3, the raw fluorescence spectra of soil samples were applied for the construction of PLS calibration model, the Rcv2 and RMSECV are 0.7598 and 1.433, respectively. Moreover, the number of input variables, the determination coefficient Rp2 of the prediction set and the RMSEp are 288, 0.9834 and 0.3675, respectively. The prediction performance of the PLS calibration model based on WT-SNV preprocessed variables can obtain a better Rp2 than the RAW-PLS calibration model. Compared with the raw spectrum PLS calibration model, its Rcv2 improved from 0.7598 to 0.7953 while RMSECV reduced from 1.433 to 1.306. Meanwhile, Rp2 improved from 0.9834 to 0.9954 while RMSEp reduced from 0.3675 to 0.1984. This stems from that the combination of these two pretreatment methods not only eliminated the influence of sample inhomogeneity and surface scattering, but also reduced the interference of fluorescence background to a certain extent. Consequently, WT-SNV was selected as the preprocessing method for fluorescence spectral data of soil samples.
3.3 Feature selection based on CARS
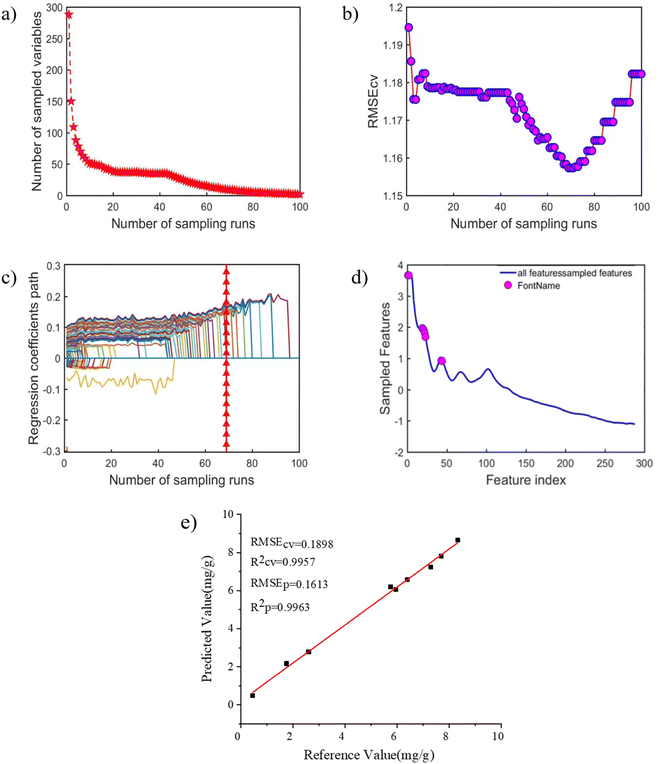
When fluorescence spectrum is combined with WT-SNV-PLS algorithm for quantitative analysis of PHE pollutants in soil, there will be redundant variables when the PLS calibration model is built with broad spectra as input variables, thus reducing the accuracy of prediction ability and increasing calculation time. Therefore, variable selection is an indispensable step in modeling. Based on spectral analysis, the PLS calibration model of soil PHE established by WT and SNV pretreatment spectra had the best prediction effect. And then, CARS algorithm was adopted in the optimal pretreatment spectrum to screen the spectral wavelength points associated with PHE in the sample spectrum. The screening results are shown in Fig. 4. Wherein, the change trend of the number of wavelength points in the process of optimizing variables is described in Fig. 4(a). It can be seen from Fig. 4(a) that the number of selected wavelength points decreased with the increase of sampling time. The trend is faster and then slower, suggesting that the wavelength point underwent a rough selection process before being selected. The change trend of RMSECV in the process of optimizing variables is demonstrated in Fig. 4(b). As the sampling times increased, the RMSECV value decreased first and increased subsequently, i.e., the number of selected wavelength points decreased gradually and the RMSECV value also decreased, indicating that the redundant wavelength points unrelated to PHE were preferably eliminated during CARS variable screening. Afterwards, the RMSECV value increased, verifying that it was caused by eliminating the wavelength point related to PHE. The trend chart of regression coefficients of each wavelength variable in the process of selecting variables is shown in Fig. 4(c), in which “*” represents the position with the smallest RMSECV value. Finally, the number of variables selected by CARS method is 12 and the combination of selected wavelength reaches the optimal level. The distribution of 12 wavelength selected by CARS method is demonstrated in Fig. 4(d). It is obvious that the characteristic variables screened by CARS corresponds to the emission characteristic wavelength of PHE.After CARS variable screening, the PLS calibration method was applied to prepare a model for the PHE content in soil. The modeling results are presented in Fig. 4(e). As can be noted in Fig. 4(e), after CARS screening, the wavelength point of PHE in CARS-PLS calibration model were reduced from 288 to 12 and the calibration model was the best. The obtained R2 and RMSECV of the calibration set are 0.9957 and 0.1898, respectively. At the same time, the R2 and RMSEp of the prediction set are 0.9963 and 0.1613, respectively. It revealed that the CARS algorithm was successfully applied to screen the wavelength points of fluorescence spectra of soil samples and construct a PLS-CARS calibration model to achieve fast, simple, accurate and environmentally friendly in situ determination of PHE content in soil.
3.4 Comparison of predicted performance based on different calibration models
To further prove the predictive performance and stability of the PLS calibration model based on soil PHE fluorescence spectrum data, RAW-PLS calibration model, WT-SNV-PLS calibration model and CARS-PLS calibration model were constructed, respectively. The predictive performance of PLS calibration models based on different methods is described in Table 2. According to Table 2, the prediction performance of the WT-PLS calibration model and CARS-PLS calibration model was improved to some extent compared with the RAW-PLS calibration model. With regard to the PLS calibration model based on WT-SNV pretreatment, the Rcv2 was enhanced from 0.7598 to 0.7953 while the RMSECV was decreased from 1.433 to 1.306. Besides, the Rp2 was enhanced from 0.9834 to 0.9954 while the RMSEp was decreased from 0.3675 to 0.1984. In terms of the PLS calibration model based on CARS, the Rcv2 was enhanced from 0.7953 to 0.9957 but the RMSECV was decreased from 1.306 to 0.1898. Furthermore, the Rp2 was enhanced from 0.9954 to 0.9963 whereas the RMSEp was decreased from 0.1984 to 0.1613. The predictive performance of PLS-CARS calibration model reaches the best level in the three PLS calibration models based on three kinds of method. The further reveals that CARS algorithm can effectively screen wavelength points and establish a better quantitative analysis model of PHE with fewer variables.| Calibration model | Latent variable | Number of variables | 10-fold-cv | Prediction set | ||
|---|---|---|---|---|---|---|
| RCV2 | RMSECV | Rp2 | RMSEp | |||
| RAW-PLS | 7 | 288 | 0.7598 | 1.433 | 0.9834 | 0.3675 |
| WT–SNV-PLS | 8 | 288 | 0.7953 | 1.306 | 0.9954 | 0.1984 |
| CARS-PLS | 8 | 12 | 0.9957 | 0.1898 | 0.9963 | 0.1613 |
4 Conclusion
In the paper, the fluorescence spectroscopy combined with CARS-PLS calibration model was successfully applied to the detection of PHE content in soil. The fluorescence spectra of 29 samples were collected. First of all, the influence of five preprocessing methods on the prediction performance of PLS calibration model was explored. Afterwards, PLS calibration models based on full spectrum, pretreatment and wavelength variable screening were established to achieve rapid determination of PHE in soil. The results show that the PLS model with key variables obtained based on the CARS algorithm had better performance than the full-spectrum PLS calibration model, and its Rcv2 and RMSEcv are 0.9957 and 0.1898 respectively. Additionally, Rp2 and RMSEp are 0.9963 and 0.1613 respectively. The obtained results sufficiently demonstrate that the CARS algorithm could be effectively utilized to extract the key variables of fluorescence spectra, and the established CARS-PLS calibration model can be employed for effective quantitative analysis of PHE content in soil. Compared with the traditional laboratory chromatographic method, the proposed method has many advantages, such as simple sample preparation, small consumption of organic reagents, highly sensitive detection, fast analysis, and low-cost, which provide theoretical basis and technical support for the analysis of other indicators of PHE in soil. In addition, the method has some shortcomings, such as the small number of samples and the single form of samples in this experiment, and we will examine the prediction performance of the model in terms of increasing the number of samples and preparing samples of different physical forms in our subsequent study.Author contributions
Haonan Li: sample, experiment, investigation, data collection, data curation, and writing – original draft preparation. Maogang Li: methodology, software, and investigation. Hongsheng Tang: writing – review & editing. Hua Li: supervision. Tianlong Zhang: funding acquisition, supervision, and writing – review & editing. Xiao-Feng Yang: funding acquisition, supervision, and project administration.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 22173071, 22073074, and 21873076).References
- N. D. Dat and M. B. Chang, Sci. Total Environ., 2017, 609, 682–693 CrossRef CAS PubMed.
- H. Zhang, J. F. Wang, H. Y. Bao and J. Li, Bull. Environ. Contam. Toxicol., 2020, 105, 446–452 CrossRef CAS PubMed.
- Z. H. Xue, X. Zheng, W. C. Yu, A. Li, S. H. Li, Y. M. Wang and X. H. Kou, J. Electrochem. Soc., 2021, 168, 057528 CrossRef CAS.
- S. T. Wang, Y. Y. Yuan, C. Y. Zhu, D. M. Kong and Y. T. Wang, Measurement, 2019, 139, 475–481 CrossRef.
- T. Doudnikova, T. Minkina, S. Sushkova, A. Barbashev, E. Antonenko, G. Bakoeva, E. Shuvaev, S. Mandzhieva, Y. Litvinov, V. Chaplygin and I. Deryabkina, Environ. Geochem. Health, 2022 DOI:10.1007/s10653-022-01281-1.
- J. W. Li, X. Shang, Z. X. Zhao, R. L. Tanguay, Q. X. Dong and C. J. Huang, J. Hazard. Mater., 2010, 173, 75–81 CrossRef CAS PubMed.
- L. Kong, Y. Gao, Q. X. Zhou, X. Y. Zhao and Z. W. Sun, J. Hazard. Mater., 2017, 343, 276–284 CrossRef PubMed.
- S. H. Wu, S. L. Zhou, H. J. Bao, D. X. Chen, C. H. Wang, B. j. Li, G. J. Tong, Y. J. Yuan and B. G. Xu, J. Hazard. Mater., 2019, 364, 108–116 CrossRef CAS PubMed.
- C. H. Wang, S. H. Wu, S. L. Zhou, Y. X. Shi and J. Song, Pedosphere, 2017, 27, 19–28 Search PubMed.
- R. P. Tong, X. Y. Yang, H. R. Su, Y. Pan, Q. Z. Zhang, J. Wang and M. Long, Sci. Total Environ., 2018, 616/617, 1365–1373 CrossRef PubMed.
- H. I. Abdel-Shafy and M. S. M. Mansour, Egypt. J. Pet., 2016, 25, 107–123 CrossRef.
- M. A. Mallah, C. X. Li, M. A. Mallah, S. Noreen, Y. Liu, M. Saeed, X. He, B. Ahmed, F. F. Feng, A. A. Mirjat, W. Wang, A. Jabar, M. Naveed, J. H. Li and Q. Zhang, Chemosphere, 2022, 296, 133948 CrossRef CAS PubMed.
- S. Huijghebaert, L. Hoste and G. Vanham, Eur. J. Clin. Pharmacol., 2021, 77, 1295 CrossRef CAS PubMed.
- B. Wu, S. H. Guo, X. J. Li and J. N. Wang, Sci. Total Environ., 2017, 11(613/614), 513–520 Search PubMed.
- J. T. Sun, L. I. Pan, D. C. W. Tsang, Y. Zhan, L. Z. Zhu and X. D. Li, Sci. Total Environ., 2018, 615, 724–740 CrossRef CAS PubMed.
- F. N. Serenjeh, P. Hashemi, A. R. Ghiasvand, F. Rasolzadeh, N. Heydari and A. Badiei, Anal. Chim. Acta, 2020, 1125, 128–134 CrossRef CAS PubMed.
- B. A. P. Agus, K. Rajentran, J. Selamat, S. D. Lestari, N. B. Umar and N. Hussain, J. Food Compos. Anal., 2023, 116, 105038 CrossRef CAS.
- J. C. W. Cheung, M. Ni, A. Y. C. Tam, T. T. C. Chan, A. K. Y. Cheunga, O. Y. H. Tsange, C. B. Yip, W. K. Lam and D. W. C. Wong, Eng. Regener., 2022, 3, 121–130 Search PubMed.
- B. Wang, L. N. Sierad, J. J. Mercuri, A. Simionescu, D. T. Simionescu, L. N. Williams, R. Velaf, P. Bajona, M. Peltz, S. Ramaswamy, Y. Hong and J. Liao, Eng. Regener., 2022, 3, 374–386 Search PubMed.
- C. M. Shao, Y. R. Yu, Q. H. Fan, X. C. Wang and F. F. Ye, Smart Med., 2022, 1, e20220008 Search PubMed.
- Z. A. Temerdasheva, T. N. Musorinaa and T. A. Chervonnaya, J. Anal. Chem., 2020, 75, 1000–1010 CrossRef.
- Y. I. Pikovskii, L. A. Korotkov, M. A. Smirnova and R. G. Kovach, Eurasian Soil Sci., 2017, 50, 1125–1137 CrossRef CAS.
- M. Tommasini, A. Lucotti, M. Alfè, A. Ciajolo and G. Zerbi, Spectrochim. Acta, Part A, 2016, 152, 134–148 CrossRef CAS PubMed.
- X. Wang, W. M. Hao, H. Zhang, Y. C. Pan, Y. Kang, X. F. Zhang, M. Q. Zuo, P. J. Tong and Y. P. Du, Spectrochim. Acta, Part A, 2015, 139, 214–221 CrossRef CAS PubMed.
- R. N. Okparanma and A. M. Mouazen, Appl. Spectrosc. Rev., 2013, 48, 458–486 CrossRef CAS.
- D. Patra, Appl. Spectrosc. Rev., 2003, 38, 155–185 CrossRef CAS.
- O. Devos, M. Ghaffari, R. Vitale, A. Juan, M. Sliwa and C. Ruckebusch, Anal. Chem., 2021, 93, 12504–12513 CrossRef CAS PubMed.
- H. Gu, M. Q. Feng, J. Li, J. Lu, H. Y. Gu, J. H. Chen, S. H. He, X. P. Qi, W. J. Chen and T. Chen, Anal. Lett., 2022, 55, 1217–1234 CrossRef.
- Y. Nakaya, S. Nakashima, M. Moriizumi, M. Oguchi, S. Kashiwagi and N. Naka, Spectrochim. Acta, Part A, 2020, 233, 118188 CrossRef CAS PubMed.
- J. Xu, X. F. Liu and Y. T. Wang, Food Chem., 2016, 212, 72–77 CrossRef CAS PubMed.
- R. J. Yang, G. M. Dong, X. S. Sun, Y. R. Yang, Y. P. Yu, H. X. Liu and W. Y. Zhang, Spectrochim. Acta, Part A, 2018, 190, 342–346 CrossRef CAS PubMed.
- R. Huang, N. J. Zhao, D. S. Meng, Z. L. Zuo, Z. Li, Y. N. Chen and X. W. Xiao, Chin. J. Lasers, 2020, 47, 1011002 CrossRef.
- A. M. Li, Z. Y. Lian, R. J. Yang and G. M. Dong, Environ. Chem., 2018, 37, 910–912 CAS.
- S. f. Ye, D. Wang and S. G. Min, Chemom. Intell. Lab. Syst., 2008, 91, 194–199 CrossRef CAS.
- X. H. Bian, S. J. Li, L. Lin, X. Y. Tan, Q. J. Fan and M. Li, Anal. Chim. Acta, 2016, 925, 16–22 CrossRef CAS PubMed.
- H. D. Li, Y. Z. Liang and Q. S. Xu, Anal. Chim. Acta, 2009, 648, 77–84 CrossRef CAS PubMed.
- Q. Q. Li, Y. Huang, X. Z. Song, J. X. Zhang and S. G. Min, Spectrochim. Acta, Part A, 2019, 214, 129–138 CrossRef CAS PubMed.
- Y. Yang, Y. Jin, Y. J. Wu and Y. Chen, J. Near Infrared Spectrosc., 2016, 24, 171–178 CrossRef CAS.
- R. G. Brereton, J. Jansen, J. Lopes, F. Marini, A. Pomerantsev, O. Rodionova, J. M. Roger, B. Walczak and R. Tauler, Anal. Bioanal. Chem., 2018, 410, 6691–6704 CrossRef CAS PubMed.
- E. Szymańska, Anal. Chim. Acta, 2018, 1028, 1–10 CrossRef PubMed.
- S. Wold, M. Sjöström and L. Eriksson, Chemom. Intell. Lab. Syst., 2001, 58, 109–130 CrossRef CAS.
- M. S. Dhanoa, S. J. Lister, R. Sanderson and R. J. Barnes, J. Near Infrared Spectrosc., 1995, 2, 43–47 CrossRef.
- P. D. Geladi, D. B. Macdougall and H. Martens, Appl. Spectrosc., 1985, 39, 491–500 CrossRef.
- B. Jiang, W. Li and Y. D. J. Huang, Appl. Polym. Sci., 2012, 124, 1529–1533 CrossRef CAS.
- Y. Xi, Y. Li, Z. Z. Duan and Y. Lu, Appl. Spectrosc., 2018, 72, 1752–1763 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra08279a |
| This journal is © The Royal Society of Chemistry 2023 |