Open Access Article
Open Access ArticleCytotoxic cytochalasans from cultures of the fungus Metarhizium brunneum TBRC-BCC 79240†
Jittra Kornsakulkarn,
Patchanee Auncharoen,
Artit Khonsanit,
Nattawut Boonyuen and
Chawanee Thongpanchang *
*
National Center for Genetic Engineering and Biotechnology (BIOTEC), National Science and Technology Development Agency (NSTDA), 111 Thailand Science Park, Phahonyothin Road, Khlong Nueng, Khlong Luang, Pathum Thani 12120, Thailand. E-mail: chawanee@biotec.or.th
First published on 4th April 2023
Abstract
Fourteen new cytochalasans, brunnesins A–N (1–14), along with eleven known compounds, were isolated from the culture extracts of the insect pathogenic fungus Metarhizium brunneum strain TBRC-BCC 79240. The compound structures were established by spectroscopy, X-ray diffraction analysis, and electronic circular dichroism. Compound 4 exhibited antiproliferative activity against all cell lines tested (mammalian), with 50% inhibition concentration (IC50) values ranging from 2.09 to 16.8 μg mL−1. Compounds 6 and 16 were shown to be bioactive only against non-cancerous Vero cells (IC50 4.03 and 0.637 μg mL−1, respectively) whereas compounds 9 and 12 were bioactive only against NCI-H187 small-cell lung cancer cells (IC50 18.59 and 18.54 μg mL−1, respectively). Compounds 7, 13, and 14 showed cytotoxicity against NCI-H187 and Vero cell lines with IC50 values ranging from 3.98–44.81 μg mL−1.
Introduction
Cytochalasans constitute a large family of fungal polyketide–amino acid hybrid metabolites. They share a tricyclic core structure, in which a diverse macrocyclic ring is fused to a perhydroisoindole moiety. These compounds have attracted much attention owing to their intriguing structures and a broad spectrum of biological activities, including inhibition of monosaccharide transport1–3 and thyroid secretion systems,4 and growth inhibition of microbial,5,6 human,7–9 and plant cells.10,11 Since the discovery of cytochalasins A and B from Phoma strain S298![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 and Helminthosporium dematioideum13 in 1966, more than 300 related compounds have been reported from various fungal sources14,15 among the genera Aspergillus, Metarhizium, Xylaria, Diaporthe, Phomopsis, and Hypoxylon.16–21 As part of our ongoing research on the discovery of chemically diverse and bioactive compounds from entomopathogenic fungi, we investigated crude extracts of the fungus Metarhizium brunneum TBRC-BCC 79240. The bioactivity of the crude extracts of this fungus was assessed by growth inhibition assay against human cells, including small-cell lung cancer (NCI-H187) and human breast cancer (MCF-7), with activities measured as 50% inhibition concentration (IC50) values of 2.80 μg mL−1 and 17.84 μg mL−1, respectively. The antibacterial (Staphylococcus aureus) and anti-plant pathogen (Curvularia lunata) activities were tested, with minimum inhibitory concentration (MIC) values of 6.25 μg mL−1 and 50 μg mL−1, respectively. The bioactivity of the crude extracts suggested the presence of constituent bioactive metabolites, which were isolated and structurally characterized.
12 and Helminthosporium dematioideum13 in 1966, more than 300 related compounds have been reported from various fungal sources14,15 among the genera Aspergillus, Metarhizium, Xylaria, Diaporthe, Phomopsis, and Hypoxylon.16–21 As part of our ongoing research on the discovery of chemically diverse and bioactive compounds from entomopathogenic fungi, we investigated crude extracts of the fungus Metarhizium brunneum TBRC-BCC 79240. The bioactivity of the crude extracts of this fungus was assessed by growth inhibition assay against human cells, including small-cell lung cancer (NCI-H187) and human breast cancer (MCF-7), with activities measured as 50% inhibition concentration (IC50) values of 2.80 μg mL−1 and 17.84 μg mL−1, respectively. The antibacterial (Staphylococcus aureus) and anti-plant pathogen (Curvularia lunata) activities were tested, with minimum inhibitory concentration (MIC) values of 6.25 μg mL−1 and 50 μg mL−1, respectively. The bioactivity of the crude extracts suggested the presence of constituent bioactive metabolites, which were isolated and structurally characterized.
Results and discussion
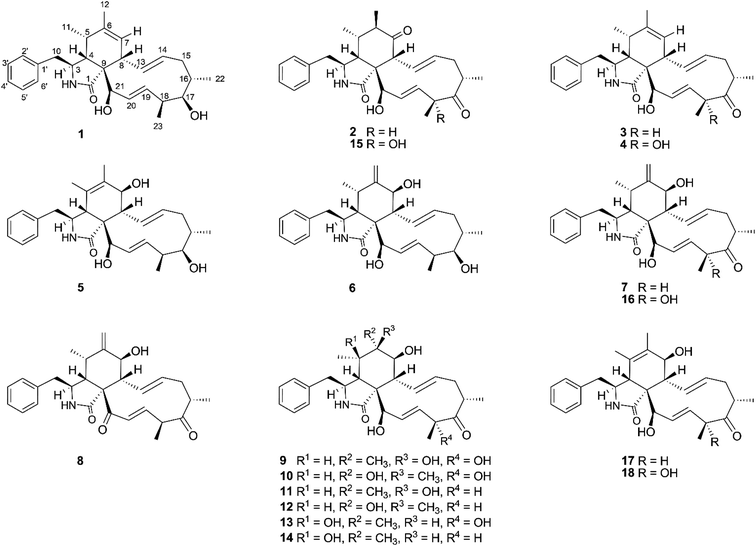
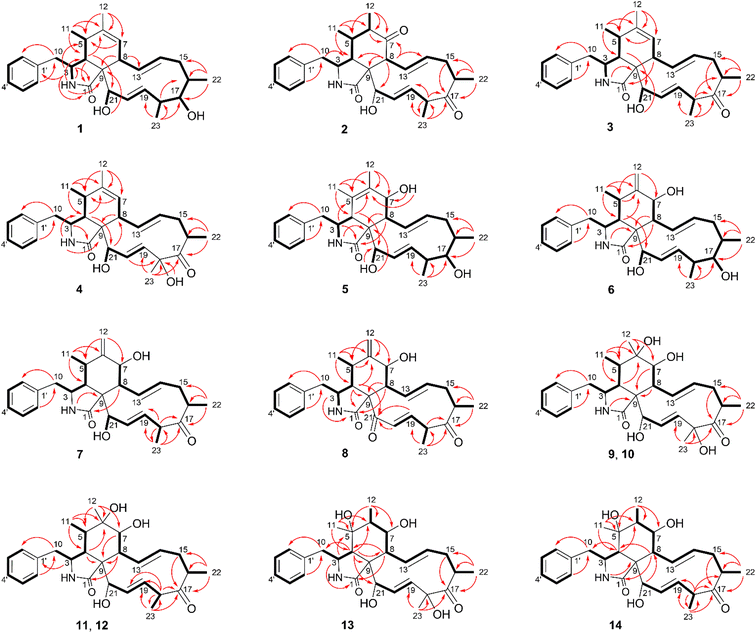
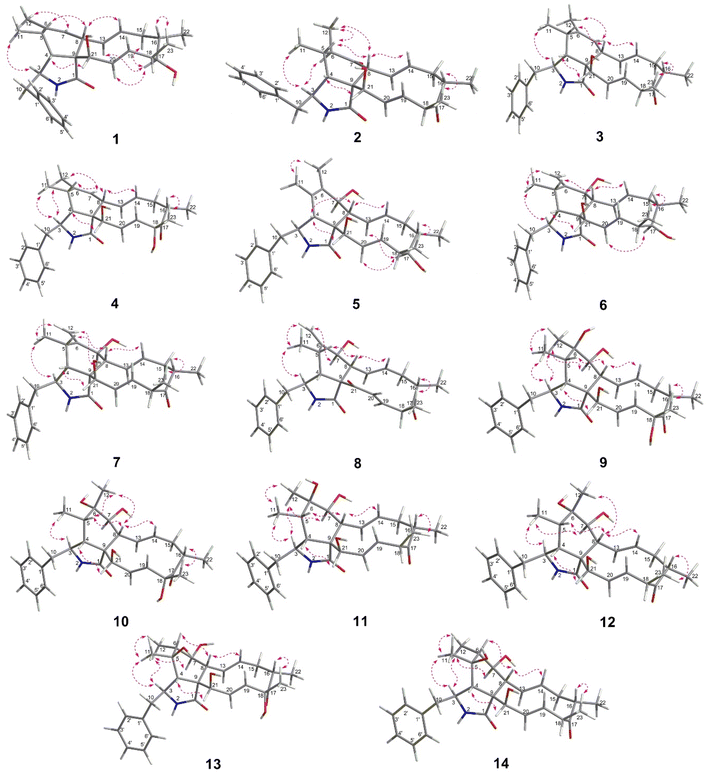
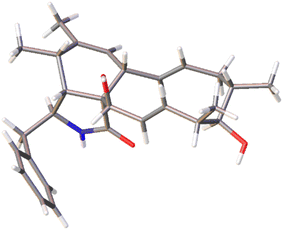
The chemical constituents of Metarhizium brunneum TBRC-BCC 79240 crude extract were separated, which led to the isolation of fourteen new cytochalasans (brunnesins A–N (1–14); Fig. 1), together with eleven known compounds.Brunnesin A (1) was obtained as an off-white solid. Its molecular formula was determined as C28H37NO3 from HRESIMS data. The 1H and 13C NMR data (Table 1) as well as the HSQC spectrum showed the presence of a mono-substituted phenyl group, thirteen methines including five olefinic methines, two oxymethines, one nitrogen-bearing methine, two methylenes, four methyls, three D2O-exchangeable protons, and one carbonyl carbon. The cross peaks in the 1H–1H COSY spectrum indicated three independent spin–spin coupling systems, as shown in Fig. 2. These data suggested that 1 is a 10-phenyl cytochalasin. The key HMBC correlations from H-3 to C-1, H-4 to C-1/C-6, H-7 to C-5, H-8 to C-9, H3-11 to C-4/C-5/C-6, H3-12 to C-5/C-6/C-7, and N–H to C-3/C-4/C-9 revealed the presence of a hydrogenated isoindolone moiety. Further correlations from H-8 to C-13, H-13 to C-7, H-4 to C-21, H3-22 to C-15/C-16/C-17, H3-23 to C-17/C-18/C-19, OH-21 to C-9, H2-10 to C-2′/C-6′, and H-2′/H-6′ to C-10 in the HMBC spectrum indicated the planar structure of 10-phenyl cytochalasin. The relative configuration of 1 was determined from the coupling constants and NOESY data. The large coupling constants between H-13/H-14 (J = 15.5 Hz) and H-19/H-20 (J = 15.9 Hz) indicated the E configuration of C-13/C-14 and C-19/C-20 double bonds. The presence of NOESY correlations between H-3/H3-11, H-4/H-21, H-5/H-8, H-8/H-14, H-16/H3-23, and H-17/H-13/H-20 (Fig. 3) suggested that H-4, H-5, H-8, H-16, and H3-23 are all β-oriented, whereas H-3, H-17, and H-21 are α-oriented. X-ray diffraction analysis (Fig. 4) established the absolute configuration of 1 (based on the use of the Flack parameter), as depicted in Fig. 1.
| Position | 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|---|
| δC, type | δH mult. (J in Hz) | δC, type | δH mult. (J in Hz) | δC, type | δH mult. (J in Hz) | δC, type | δH mult. (J in Hz) | |
| 1 | 177.2 | — | 175.2 | — | 176.8 | — | 176.7 | — |
| 3 | 55.6 | 3.35 dd (10.2, 5.1) | 53.1 | 3.74 dd (11.0, 5.8) | 55.4 | 3.34 dd (9.9, 5.0) | 55.5 | 3.35 dd (9.8, 4.7) |
| 4 | 53.5 | 2.65 t (4.4) | 50.0 | 2.72 t (4.2) | 52.9 | 2.67 t (4.6) | 53.1 | 2.65 t (4.4) |
| 5 | 36.1 | 2.40–2.50 m | 36.3 | 2.23–2.27 m | 36.1 | 2.43–2.48 m | 36.2 | 2.45 m |
| 6 | 138.2 | — | 46.1 | 1.96 dd (11.8, 7.1) | 138.5 | — | 138.3 | — |
| 7 | 129.0 | 5.25 s | 214.4 | — | 128.6 | 5.22 s | 128.5 | 5.21 s |
| 8 | 42.6 | 3.18 d (10.2) | 51.5 | 3.82 d (9.5) | 42.3 | 3.15 d (10.2) | 42.5 | 3.17 d (9.7) |
| 9 | 59.4 | — | 56.5 | — | 59.6 | — | 59.9 | — |
| 10 | 45.3 | (a) 2.71 dd (13.5, 6.4) | 45.1 | (a) 2.77 dd (13.6, 6.0) | 45.1 | (a) 2.70 dd (13.6, 6.3) | 45.2 | (a) 2.71 dd (13.5, 6.3) |
| (b) 2.91 dd (13.5, 5.1) | (b) 2.97 dd (13.6, 5.2) | (b) 2.91 dd (13.6, 5.0) | (b) 2.89 dd (13.5, 5.1) | |||||
| 11 | 13.9 | 1.08 d (7.4) | 15.7 | 0.95 d (6.9) | 14.0 | 1.09 d (7.4) | 13.9 | 1.07 d (7.4) |
| 12 | 19.9 | 1.70 s | 15.8 | 1.03 d (7.15) | 19.9 | 1.70 s | 19.9 | 1.69 s |
| 13 | 131.7 | 5.82 ddd (15.5, 10.1, 0.9) | 128.4 | 5.65 ddd (15.6, 9.5, 1.2) | 133.5 | 5.79 ddd (15.3, 10.1, 1.3) | 133.6 | 5.76 ddd (15.3, 10.2, 0.8) |
| 14 | 133.2 | 5.01 ddd (15.5, 10.8, 4.8) | 133.5 | 4.97 ddd (15.6, 11.0, 4.6) | 131.8 | 5.04 ddd (15.3, 10.9, 4.4) | 131.0 | 5.10 ddd (15.3, 10.9, 4.6) |
| 15 | 41.9 | (a) 1.73 obsc. | 38.6 | (a) 1.85 m | 38.5 | (a) 1.86 m | 38.8 | (a) 1.91 ddd (12.6, 4.6, 1.6) |
| (b) 1.81 ddd (12.4, 4.9, 1.4) | (b) 2.20 dt (12.4, 10.9) | (b) 2.17 dt (12.3, 11.0) | (b) 2.26 dt (12.6, 11.6) | |||||
| 16 | 33.3 | 1.48–1.53 m | 43.2 | 2.62 ddd (10.6, 6.9, 1.9) | 43.4 | 2.62 ddd (10.2, 6.9, 2.0) | 42.7 | 2.80 m |
| 17 | 78.3 | 3.67 br s | 210.5 | — | 210.8 | — | 211.9 | — |
| 18 | 43.8 | 2.36–2.39 m | 51.5 | 3.02 m | 51.4 | 3.04 ddd (9.1, 7.1, 1.3) | 78.4 | — |
| 19 | 130.4 | 5.12 ddd (15.9, 7.3, 2.4) | 124.3 | 5.11 ddd (15.9, 7.3, 2.4) | 123.6 | 5.09 ddd (15.8, 7.2, 2.4) | 126.7 | 5.45 dd (15.8, 2.3) |
| 20 | 135.9 | 6.02 d (16.0) | 138.5 | 6.01 d (15.9) | 139.2 | 6.06 dd (15.8, 1.4) | 139.0 | 6.12 dd (15.8, 2.6) |
| 21 | 75.9 | 3.85 br s | 76.2 | 3.76–3.78 m | 75.5 | 3.77 br s | 75.6 | 3.85 t (2.0) |
| 22 | 17.8 | 0.94 d (6.9) | 19.7 | 1.05 d (6.9) | 19.7 | 1.05 d (6.9) | 19.6 | 1.10 d (6.8) |
| 23 | 12.7 | 0.93 d (7.2) | 16.4 | 1.21 d (7.1) | 16.4 | 1.21 d (7.1) | 24.8 | 1.42 s |
| 1′ | 138.7 | — | 138.2 | — | 138.4 | — | 138.6 | — |
| 2′, 6′ | 130.9 | 7.25 d (7.3) | 131.0 | 7.27 d (7.4) | 131.0 | 7.23–7.26 m | 130.9 | 7.25 d (6.6) |
| 3′, 5′ | 129.0 | 7.28 t (7.3) | 129.1 | 7.29 t (7.7) | 129.0 | 7.26–7.29 m | 129.0 | 7.28 t (7.6) |
| 4′ | 127.2 | 7.20 ddd (7.0, 6.3, 1.8) | 127.4 | 7.19–7.23 m | 127.2 | 7.20 t (7.0) | 127.2 | 7.20 dt (6.9, 1.8) |
| 17-OH | — | 3.48 d (4.1) | — | — | — | — | — | — |
| 18-OH | — | — | — | — | — | — | 4.46 br s | |
| 21-OH | — | 4.00 d (5.9) | — | 4.65 d (5.8) | — | 4.24 d (5.9) | — | 4.28 br s |
| NH | — | 6.69 br s | — | 6.90 br s | — | 6.69 br s | — | 6.70 br s |
Brunnesin B (2) was obtained as an off-white solid. Its molecular formula was deduced as C28H35NO4 from HRESIMS data. The 1H and 13C NMR spectra (Table 1) matched those of the known co-metabolite 5,6-dihydroxy-7-oxo-deacetyl cytochalasin C (15).17 Analysis of the 2D NMR spectroscopic data of 2 revealed the same structural features as those of 15, except for the absence of the hydroxyl group at C-18 (Fig. 1). The structural features of 2 were supported by the HMBC correlations from H-18 to C-17/C-19/CH3-23 (Fig. 2). The absolute configuration of 2 was determined by X-ray diffraction analysis (Fig. 5).
Brunnesin C (3) was determined to have a molecular formula of C28H35NO3, 16 amu lower than that of 2, based on HRESIMS data. A comparison of the 1H NMR spectra of 3 and 2 revealed the presence of an additional olefinic methine in the 1H NMR spectrum of 3 instead of the sp3 methine in 2. Analysis of the 13C NMR spectra revealed the absence of one carbonyl carbon and the presence of two more olefinic carbons in 3 than in 2. The correlations from an additional methine (δH 5.22) to C-5 and H3-11/H3-12 to the olefinic carbons (δC 128.6, 138.5) in the HMBC spectrum of 3 established a double bond at C-6/C-7 in 3 instead of a carbonyl group at C-7 in 2. The NOESY correlations between H-3/H3-11, H-4/H-21, H-5/H-8, and H-16/H3-23 (Fig. 3) indicated that 3 and 2 share the same relative configuration. Moreover, the CD spectra of 3 [Δε (nm) +13.10 (200), 0 (209), −5.14 (228)] correspond to those of 2 [Δε (nm) +9.7 (199), 0 (210), −2.45 (230)], which suggested the same absolute configuration. In addition, since these compounds have a common biosynthetic origin, they were expected to have the same absolute configuration (for equivalent chiral centers among different compounds).
The 1H NMR spectrum of brunnesin D (4) was similar to that of 3, except for the presence of one more hydroxyl proton in 4 instead of one methine proton in 3. The molecular formula of 4 was determined as C28H35NO4 from HRESIMS, which is 16 amu greater than that of 3. This difference indicates that 4 has one additional hydroxyl group compared with 3. This hydroxyl group was deduced at C-18 from the HMBC correlation of the hydroxyl proton (δH 4.46) to C-18/C-17. The structure of 4 was elucidated from 1H–1H COSY, HMBC and NOESY spectroscopic data (Fig. 2 and 3).
The molecular formula of brunnesin E (5) was determined as C28H37NO4 by HRESIMS. Signals for a mono-substituted phenyl group, two double bonds, four methyls, two methylenes, one amide, and one ketone group were present in the 1H and 13C NMR spectra (Table 2). The 1H–1H COSY and HMBC correlations revealed the spin systems and the connection between the fragments (Fig. 2), which suggested the same structural features as the known co-metabolite 18,22 except for the presence of a hydroxyl group at C-17 in 5 instead of a carbonyl group in 18. The NOESY correlations between H-4/H-8, H-16/H3-23, H-17/H-13/H-20, and H-21/H-4 indicated that H-4, H-8, H-16, H-17, H3-23, and H-21 exhibited β, β, β, α, β, and α orientations, respectively. The α orientation of H-7 was determined from the coupling constant between H-7/H-8 (J = 10.0 Hz). The structure of 5 was established using these data (Fig. 1).
| Position | 5a | 6b | 7b | 8b | ||||
|---|---|---|---|---|---|---|---|---|
| δC, type | δH mult. (J in Hz) | δC, type | δH mult. (J in Hz) | δC, type | δH mult. (J in Hz) | δC, type | δH mult. (J in Hz) | |
| a In DMSO-d6.b In acetone-d6.c The signals may be exchanged. | ||||||||
| 1 | 176.4 | — | 175.8 | — | 176.3 | — | 173.6 | — |
| 3 | 59.6 | 3.12 dd (9.7, 5.0) | 52.5 | 3.10 t (7.5) | 53.8 | 3.32–3.35 m | 53.4 | 3.35–3.39 m |
| 4 | 48.5 | 2.84 s | 47.7 | 2.45 dd (5.1, 3.3) | 49.4 | 2.66 t (4.3) | 44.4 | 3.31 dd (6.0, 2.0) |
| 5 | 126.3 | — | 32.1 | 2.54 m | 33.7 | 2.77 m | 32.1 | 2.59–2.64 m |
| 6 | 132.4 | — | 151.8 | — | 151.9 | — | 151.7 | — |
| 7 | 68.3 | 3.56 m | 70.3 | 3.55 dd (10.8, 4.7) | 71.1 | 3.73 dd (10.9, 3.9) | 72.5 | 3.99 dd (9.7, 4.8) |
| 8 | 48.0 | 2.30 t (10.0) | 45.1 | 2.64–2.68 m | 46.5 | 2.84 obsc. | 52.1 | 2.33–2.36 m |
| 9 | 53.7 | — | 54.0 | — | 55.4 | — | 65.1 | — |
| 10 | 44.5 | (a) 2.71 dd (13.0, 9.5) | 43.6 | 2.63–2.69 m | 45.0 | (a) 2.74 m | 44.2 | (a) 2.47 dd (13.1, 7.9) |
| (b) 2.90 dd (13.0, 5.4) | (b) 2.86–2.88 m | (b) 2.69 dd (13.1, 5.8) | ||||||
| 11 | 16.5 | 0.97 s | 13.2 | 0.59 d (6.7) | 14.1 | 0.87 d (6.7) | 13.4 | 0.72 d (6.8) |
| 12 | 14.3 | 1.50 s | 110.6 | (a) 4.80 d (0.8) | 112.0 | (a) 4.94 d (3.3) | 112.5 | (a) 4.93 d (2.2) |
| (b) 5.03 d (1.2) | (b) 5.17 d (3.0) | (b) 5.12 d (2.5) | ||||||
| 13 | 129.8 | 5.71 dd (15.6, 10.0) | 129.4 | 5.49 dd (15.5, 9.5) | 131.9 | 5.36 dd (15.7, 9.7) | 131.4 | 5.79 ddd (15.4, 9.7, 1.3) |
| 14 | 132.6 | 4.91 ddd (15.6, 10.5, 5.5) | 132.8 | 4.90 ddd (15.5, 10.5, 5.2) | 133.7 | 5.12 ddd (15.7, 11.1, 4.8) | 134.7 | 5.02 ddd (15.4, 11.0, 4.1) |
| 15 | 41.3 | (a) 1.69 m | 41.1 | (a) 1.65 dt (12.1, 10.9) | 38.6 | (a) 1.84 m | 38.9 | (a) 1.91–1.96 m |
| (b) 1.74–1.77 m | (b) 1.73 dd (12.2, 5.2) | (b) 2.22 dt (12.6, 10.9) | (b) 2.33–2.36 m | |||||
| 16 | 31.7 | 1.42 m | 31.9 | 1.38 m | 43.2 | 2.62 m | 44.1 | 2.64–2.67 m |
| 17 | 77.0 | 3.53 br s | 76.9 | 3.49 br s | 210.3 | — | 209.6 | — |
| 18 | 41.8 | 2.34–2.38 m | 41.9 | 2.27–2.31 m | 51.6 | 3.03 m | 51.9 | 3.23–3.28 m |
| 19 | 130.4 | 5.13 ddd (15.8, 7.3, 2.3) | 129.7 | 5.03 m | 124.0 | 5.13 ddd (15.5, 6.8, 2.1) | 140.3 | 6.09 dd (16.1, 6.3) |
| 20 | 134.8 | 5.84 d (16.1) | 134.8 | 3.85 dd (16.3, 1.5) | 139.1 | 6.02 d (15.9) | 135.3 | 6.85 dd (16.1, 1.7) |
| 21 | 73.2 | 4.33 d (3.4) | 74.8 | 3.65 br s | 76.2 | 3.81 s | 196.3 | — |
| 22 | 17.8 | 0.86 d (7.2) | 17.6 | 0.84 d (7.2) | 19.9 | 1.05 d (6.9) | 20.4 | 1.10 d (6.9) |
| 23 | 12.5 | 0.91 d (6.8) | 12.4 | 0.88 d (6.9) | 16.4 | 1.23 d (7.1) | 15.7 | 1.44 d (7.2) |
| 1′ | 138.0 | — | 137.4 | — | 138.7 | — | 138.5 | — |
| 2′, 6′ | 129.6 | 7.27 d (6.8) | 129.9 | 7.20 d (7.6) | 130.8 | 7.24 d (6.8) | 130.6 | 7.22 d (7.6) |
| 3′, 5′ | 128.3 | 7.32 t (7.4) | 128.1 | 7.29 t (7.5) | 129.1 | 7.29 t (7.3) | 129.2 | 7.30 t (7.2) |
| 4′ | 126.4 | 7.24 t (7.2) | 126.3 | 7.21 t (6.6) | 127.3 | 7.21 t (7.1) | 127.3 | 7.22 t (7.6) |
| 7-OH | — | 3.88 d (6.7) | — | 4.28 d (5.9)c | — | 4.38 d (6.1)c | — | 3.71 d (5.2) |
| 17-OH | 4.40 d (4.4) | — | 4.35 d (3.9)c | — | — | — | — | |
| 21-OH | — | 5.18 d (6.4) | — | 4.97 d (6.0)c | — | 4.73 sc | — | — |
| NH | — | 7.87 br s | — | 7.75 br s | — | 6.80 br s | — | 7.05 br s |
The molecular formula of brunnesin F (6) was determined from HRESIMS as C28H37NO4, which is the same as that of 5. The 1H and 13C NMR spectra of these compounds were also similar. 2D NMR spectroscopic data analysis revealed a shift of double bond from C-5/C-6 in 5 to C-6/C-12 in 6. The 1H–1H COSY correlation between H-4 and H-5 and the HMBC correlations from H-4/H-5 to C-6, H3-11 to C-4/C-5/C-6, and H-12 to C-5/C-7 (Fig. 2) confirmed the assignment of the double bond at C-6/C-12 in 6. The NOESY correlations (Fig. 3) and the CD spectrum of 6 [Δε (nm) +33.3 (190), 0 (202), −15.68 (213)] corresponded to those of 5 [Δε (nm) +6.85 (196), 0 (211), −3.89 (220)], which suggested the configurations of 6 (Fig. 1).
The molecular formula of brunnesin G (7) was determined as C28H35NO4 by HRESIMS. The 1H, 13C and 2D NMR spectroscopic data analyses (Table 2, Fig. 2) suggested a structure similar to that of the known co-metabolite zygosporin D (16). The only difference was the absence of a hydroxyl group at C-18 in 7, which was confirmed by the correlations from H-18 to C-17/C-19/CH3-23 in the HMBC spectrum. The NOESY correlations (Fig. 3) and the CD spectrum of 7 [Δε (nm) +57.61 (198), 0 (207), −16.81 (219)] were in close agreement with those of 6 [Δε (nm) +33.3 (190), 0 (202), −15.68 (213)]. The structure of 7 was determined using these data (Fig. 1).
Brunnesin H (8) was obtained as an off-white solid. The molecular formula was determined as C28H33NO4 by HRESIMS. Compared with 7, the 1H NMR spectrum of 8 revealed the absence of one oxymethine and one hydroxyl proton, and the 13C NMR spectrum showed the presence of one more carbonyl group in 8. The correlations from H-4 and H-19 to this additional carbonyl in the HMBC spectrum (Fig. 2) indicated a carbonyl group at C-21. The coupling constants between H-7/H-8 (J = 9.7 Hz), H-13/H-14 (J = 15.5 Hz), H-19/H-20 (J = 16 Hz), and the NOESY correlations (Fig. 3), established the planar structure of 8. The CD spectrum of 8 [Δε (nm) +181.9 (192), 0 (212), −20.1 (221)] was highly similar to that of 7 [Δε (nm) +57.61 (198), 0 (207), −16.81 (219)], which suggested the configurations of 8 (Fig. 1).
Brunnesin I (9) was obtained as an off-white solid. Its molecular formula was determined as C28H37NO6 from HRESIMS. The 1H, 13C, and 2D NMR spectroscopic data analyses suggested a structure similar to that of the known compound cytochalasin P. The 1H NMR spectrum of 9 revealed an additional hydroxyl proton, the absence of acetyl protons, and an upfield shift of the H-21 chemical shift. These observations, together with the absence of one carbonyl carbon in the 13C NMR spectrum, indicated that the only difference between 9 and cytochalasin P was the presence of a hydroxyl group at C-21 in 9, instead of an acetyl group in cytochalasin P. The NOESY correlations (Fig. 3) suggested that the relative configuration of 9 was the same as that of cytochalasin P. Therefore, 9 was determined to be deacetylcytochalasin P.
The molecular formula of brunnesin J (10) was determined from HRESIMS as C28H37NO6, which is the same as that of 9. Analysis of the NMR spectroscopic data, including 1H, 13C, 1H–1H COSY, and HMBC (Table 3, Fig. 2), indicated that 10 has the same planar structure as 9, which suggests that it could be a stereo isomer. The additional correlations between H-5/H3-12 and H3-12/H-8 observed in the NOESY spectrum of 10 (Fig. 3) indicate different configurations at C-6 between these compounds. Thus, 10 was determined to be 6-epi 9. The CD spectrum of 10 [Δε (nm) +20.85 (198), 0 (209), −8.20 (219)] was also in agreement with that of 9 [Δε (nm) +26.26 (198), 0 (210), −6.99 (219)].
| Position | 9 | 10 | 11 | 12 | ||||
|---|---|---|---|---|---|---|---|---|
| δC, type | δH mult. (J in Hz) | δC, type | δH mult. (J in Hz) | δC, type | δH mult. (J in Hz) | δC, type | δH mult. (J in Hz) | |
| a The signals may be exchanged. | ||||||||
| 1 | 176.7 | — | 176.6 | — | 176.4 | — | 176.3 | — |
| 3 | 53.8 | 3.72 m | 54.1 | 4.18 dd (9.7, 6.0) | 53.6 | 3.73 dd (10.9, 4.9) | 54.0 | 4.20–4.25 m |
| 4 | 49.3 | 2.48 t (5.1) | 50.8 | 2.31 t (5.4) | 49.0 | 2.48 t (5.2) | 50.7 | 2.32 t (5.1) |
| 5 | 39.8 | 2.24 m | 39.4 | 1.97–2.03 m | 39.8 | 2.26 dd (8.3, 5.5) | 39.4 | 1.99–2.03 m |
| 6 | 72.6 | — | 76.8 | — | 72.5 | — | 76.6 | — |
| 7 | 73.3 | 3.07 d (11.7) | 76.3 | 3.42 d (11.7) | 73.2 | 3.06 d (11.4) | 76.5 | 3.45 d (11.9) |
| 8 | 43.0 | 2.94 m | 45.0 | 2.68 dd (11.7, 10.3) | 42.9 | 2.90 dd (11.4, 10.2) | 45.0 | 2.65–2.67 m |
| 9 | 56.8 | — | 57.0 | — | 56.5 | — | 56.7 | — |
| 10 | 45.5 | (a) 2.72 dd (13.5, 6.2) | 44.9 | (a) 2.59 dd (13.7, 6.9) | 45.5 | (a) 2.69 dd (13.5, 6.2) | 45.0 | (a) 2.58 dd (13.7, 6.8) |
| (b) 2.93 m | (b) 3.02 dd (13.7, 3.6) | (b) 2.94 dd (13.5, 4.8) | (b) 3.04 dd (13.7, 3.8) | |||||
| 11 | 13.5 | 0.96 d (7.3) | 13.1 | 1.12 d (6.5) | 13.7 | 0.99 d (7.3) | 13.3 | 1.12 d (6.9) |
| 12 | 25.3 | 1.13 s | 22.7 | 1.13 s | 25.3 | 1.18 s | 22.7 | 1.13 s |
| 13 | 130.8 | 5.51 dd (15.6, 10.1) | 130.6 | 5.46 dd (15.2, 10.4) | 130.8 | 5.55 ddd (15.5, 10.0, 1.0) | 130.6 | 5.51 dd (15.4, 10.0) |
| 14 | 133.8 | 5.13 ddd (15.6, 10.9, 4.7) | 134.2 | 5.09 ddd (15.2, 11.0, 4.5) | 134.7 | 5.06 ddd (15.5, 11.1, 4.5) | 135.3 | 5.00–5.04 m |
| 15 | 38.9 | (a) 1.93 ddd (12.8, 3.0, 1.6) | 39.0 | (a) 1.93 dd (12.5, 4.4) | 38.6 | (a) 1.85–1.89 m | 38.6 | (a) 1.88–1.93 m |
| (b) 2.30 m | (b) 2.28 t (12.5) | (b) 2.22 m | (b) 2.19 m | |||||
| 16 | 42.6 | 2.79 m | 42.7 | 2.78 m | 43.5 | 2.62 ddd (10.4, 6.9, 2.0) | 43.8 | 2.61–2.64 m |
| 17 | 211.9 | — | 212.0 | — | 210.6 | — | 210.8 | — |
| 18 | 78.3 | — | 78.2 | — | 51.2 | 3.05 m | 50.9 | 3.06 m |
| 19 | 126.7 | 5.45 dd (15.9, 2.4) | 126.3 | 5.40 dd (15.9, 2.3) | 123.6 | 5.09 ddd (15.9, 6.9, 2.6) | 123.4 | 5.00–5.06 m |
| 20 | 139.3 | 6.15 dd (15.9, 2.6) | 139.8 | 6.19 dd (15.9, 2.7) | 139.3 | 6.10 dd (15.9, 1.6) | 139.6 | 6.18 d (16.2) |
| 21 | 76.8 | 3.72 m | 77.0 | 3.64 t (2.4) | 76.6 | 3.64 br t (1.9) | 76.9 | 3.61 br s |
| 22 | 19.7 | 1.10 d (6.9) | 19.7 | 1.09 d (7.1) | 19.7 | 1.05 d (6.9) | 19.6 | 1.06 d (6.9) |
| 23 | 24.7 | 1.44 s | 24.7 | 1.42 s | 16.4 | 1.22 d (7.1) | 16.4 | 1.21 d (7.1) |
| 1′ | 138.3 | — | 138.7 | — | 138.3 | — | 138.8 | — |
| 2′, 6′ | 130.9 | 7.25 m | 130.9 | 7.22 d (7.4) | 131.0 | 7.24 d (8.0) | 130.9 | 7.22 d (7.0) |
| 3′, 5′ | 129.1 | 7.28 m | 129.0 | 7.29 t (7.7) | 129.1 | 7.27 t (7.5) | 129.0 | 7.27 t (7.4) |
| 4′ | 127.3 | 7.21 dt (6.8, 1.7) | 127.2 | 7.20 t (7.2) | 127.3 | 7.21 t (7.0) | 127.2 | 7.19 t (7.2) |
| 6-OH | — | 3.25 br sa | — | — | — | — | — | — |
| 7-OH | 3.51 br sa | — | — | — | — | — | — | |
| 18-OH | 4.42 br sa | — | — | — | — | — | — | |
| 21-OH | — | 4.51 br sa | — | 4.49 br s | — | — | — | — |
| NH | — | 7.03 br s | — | 6.93 br s | — | 6.89 br s | — | 6.75 br s |
Brunnesins K (11) and L (12) possess the same molecular formula of C28H37NO5 as deduced by HRESIMS. Their 1H and 13C NMR spectra were comparable, which were similar to those of 9 and 10 (Table 3). The key difference between 11/12 and 9/10 was the presence of one more methine proton in the 1H NMR spectra of 11/12. The HMBC correlations from this additional methine proton (δH 3.05) to C-17/C-19/C-20/C-23 in 11/12 (Fig. 2), in combination with the molecular formula which indicated the absence of one oxygen atom compared with 9/10, suggested the absence of the hydroxyl group at C-18 in 11/12. The relative configurations of 11 and 12 were deduced from NOESY correlations (Fig. 3). Therefore, 11 and 12 are epimers (Fig. 1). Similarities of CD spectra among 9–12 were also observed.
The molecular formula of brunnesin M (13) was determined by HRESIMS as C28H37NO6, which is the same as 9. Their 1H and 13C NMR spectra are also highly similar (Table 4). 2D NMR spectroscopic data analysis revealed that these compounds shared the same structural features, except for the position of one hydroxyl group. The 1H–1H COSY correlations from H-6 to H-7 and H3-12 together with the HMBC correlations from H-3 to C-5, H-4 to C-5/C-6/C-8/CH3-11, H3-11 to C-4/C-5/C-6, H3-12 to C-5/C-6/C-7, and H-8 to C-6 (Fig. 2) suggested the position of the hydroxyl group at C-5 in 13 instead of C-6 in 9. The NOESY correlations (Fig. 3) established the relative configuration (Fig. 1). The CD spectrum of 13 [Δε (nm) +26.29 (193), 0 (209), −7.16 (219)] resembled that of 9 [Δε (nm) +26.26 (198), 0 (210), −6.99 (219)].
| Position | 13 | 14 | ||
|---|---|---|---|---|
| δC, type | δH mult. (J in Hz) | δC, type | δH mult. (J in Hz) | |
| a The signals may be exchanged. | ||||
| 1 | 175.2 | — | 175.3 | — |
| 3 | 54.1 | 3.78 dd (9.8, 5.3) | 54.0 | 3.78 ddd (9.6, 5.5, 3.8) |
| 4 | 57.6 | 2.33 d (4.6) | 57.0 | 2.35 dd (5.5, 1.2) |
| 5 | 73.8 | — | 73.7 | — |
| 6 | 50.4 | 1.75 m | 50.4 | 1.75 m |
| 7 | 73.3 | 3.03 dd (11.5, 8.5) | 73.3 | 3.02 m |
| 8 | 46.2 | 3.29 t (10.7) | 46.1 | 3.24 dd (11.4, 10.1) |
| 9 | 57.3 | — | 57.1 | — |
| 10 | 43.9 | (a) 2.74 dd (13.8, 6.2) | 43.8 | (a) 2.72 dd (13.9, 6.1) |
| (b) 3.09 dd (13.8, 3.7) | (b) 3.10 dd (13.9, 3.7) | |||
| 11 | 26.6 | 1.37 s | 26.7 | 1.38 s |
| 12 | 18.0 | 1.17 d (7.5) | 18.0 | 1.18 d (7.4) |
| 13 | 130.5 | 5.51 dd (15.6, 10.5) | 130.5 | 5.53 dd (15.1, 10.0) |
| 14 | 135.1 | 5.12 ddd (15.6, 11.0, 4.4) | 135.8 | 5.05 m |
| 15 | 39.1 | (a) 1.94 dd (12.3, 4.5) | 38.7 | (a) 1.90 m |
| (b) 2.27 dt (12.3, 11.2) | (b) 2.16 ddd (12.5, 11.6, 8.4) | |||
| 16 | 42.7 | 2.81 m | 43.6 | 2.60–2.67 m |
| 17 | 212.1 | — | 210.9 | — |
| 18 | 78.5 | — | 51.4 | 3.05 m |
| 19 | 127.1 | 5.44 dd (15.8, 3.6) | 124.2 | 5.10 ddd (15.8, 7.3, 2.4) |
| 20 | 137.8 | 6.06 dd (15.8, 2.8) | 138.0 | 6.01ddd (15.8, 2.4, 1.3) |
| 21 | 76.1 | 3.48 s | 75.7 | 3.40 br s |
| 22 | 19.6 | 1.09 d (6.8) | 19.5 | 1.06 d (6.9) |
| 23 | 24.6 | 1.42 s | 16.3 | 1.20 d (7.1) |
| 1′ | 139.3 | — | 137.8 | — |
| 2′, 6′ | 131.2 | 7.28 d (7.0) | 129.1 | 7.27 dd (7.0, 1.7) |
| 3′, 5′ | 129.1 | 7.30 t (7.5) | 131.2 | 7.26 t (8.1) |
| 4′ | 127.5 | 7.22 dd (7.1, 1.9) | 127.4 | 7.22 dd (7.9, 1.5) |
| 5-OH | — | 4.96 br sa | — | — |
| 7-OH | — | 4.98 br sa | ||
| 18-OH | 4.46 br s | — | ||
| 21-OH | — | — | — | 5.02 br sa |
| NH | — | 7.02 br s | — | 7.00 br s |
Brunnesin N (14) was obtained as an off-white solid. The NMR spectroscopic data suggested that 14 was closely related to 13, except for the absence of one hydroxyl group in 14. This was supported by the molecular formula of 14, determined from HRESIMS as C28H37NO5, which was 16 amu lower than that of 13. The upfield shift of the C-18 chemical shift in the 13C NMR spectrum of 14 and the HMBC correlations from H-18 to C-16/C-17/C-19/CH3-23 indicated the absence of a hydroxyl group at C-18 in 13. The NOESY correlations (Fig. 3), as well as the similarity of CD spectra between 14 [Δε (nm) +16.10 (199), 0 (209), −8.08 (219)] and 13 [Δε (nm) +26.29 (193), 0 (209), −7.16 (219)], established the structure of 14 (Fig. 1).
The structures of the known compounds were dereplicated from HRMS and NMR (1H and 13C) data. The known compounds were identified as 6,7-dihydro-7-oxo-deacetylcytochalasin C (15),17 zygosporin D (16),17 deacetylcytochalasin C (17),17 18-deshydroxyl-deacetylcytochalasin C (18),22 destruxins A23 and B,24 helvolic acid,25,26 12-dihydrohelvolic acid,25 meromuside I,27 ustilaginoidins D28 and K.29
All isolated compounds (apart from 8 and ustilaginoidin K owing to the limited amount of the sample) were evaluated for bioactivity against bacteria (Mycobacterium tuberculosis, Staphylococcus aureus and Acinetobacter baumannii), phytopathogenic fungi (Alternaria brassicicola, Colletotrichum acutatum, and Curvularia lunata), and mammalian cells (NCI-H187, MCF-7 and Vero) (Table 5). None of the tested compounds was active against A. baumannii or C. lunata (MIC > 50 μg mL−1). Only helvolic acid showed significant activity against M. tuberculosis (anti-TB). Destruxins A and B were cytotoxic to NCI-H187 and Vero cells. Ustilaginoidin D displayed cytotoxicity against all cells, except A. baumannii and C. lunata. Cytochalasans are known to exhibit cytotoxicity; however, among the isolated cytochalasins, compounds 1, 2, 3, 5, 10, 11, 17, and 18 were inactive against all tested mammalian cell lines (IC50 > 50 μg mL−1). Only compound 4 was active against all mammalian cell lines, with IC50 values ranging from 2.09 to 16.8 μg mL−1. Compounds 6 and 16 were shown to be selectively active against non-cancerous Vero cells, whereas compounds 9 and 12 were active only against NCI-H187 cells. Compounds 7, 13, and 14 exhibited cytotoxicity against both NCI-H187 and Vero cell lines with IC50 values in the range 3.98–44.81 μg mL−1.
| Compounds | Cytotoxicity (IC50, μg mL−1) | Anti-TB MIC, μg mL−1 | Anti-bacteria (MIC, μg mL−1) | Anti-phytopathogenic fungal (MIC, μg mL−1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| NCI-H187 | MCF-7 | Vero | S. aureus | A. baumannii | C. acutatum | A. brassicicola | C. lunata | ||
| a Positive control for cytotoxicity assay.b Positive control for anti-TB assay.c Positive control for antibacterial assay.d Positive control for antifungal assay. | |||||||||
| 1 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| 2 | >50 | >50 | >50 | 25.0 | >50 | >50 | >50 | >50 | >50 |
| 3 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| 4 | 3.71 | 16.8 | 2.09 | 50.0 | >50 | >50 | >50 | >50 | >50 |
| 5 | >50 | >50 | >50 | 50.0 | >50 | >50 | >50 | >50 | >50 |
| 6 | >50 | >50 | 4.03 | 50.0 | >50 | >50 | >50 | >50 | >50 |
| 7 | 29.94 | >50 | 3.98 | 50.0 | >50 | >50 | >50 | >50 | >50 |
| 9 | 18.59 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| 10 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| 11 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| 12 | 18.54 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| 13 | 5.58 | >50 | 44.81 | >50 | >50 | >50 | >50 | >50 | >50 |
| 14 | 4.89 | >50 | 27.99 | >50 | >50 | >50 | >50 | >50 | >50 |
| 15 | >50 | >50 | >50 | 25.0 | >50 | >50 | >50 | >50 | >50 |
| 16 | >50 | >50 | 0.637 | 50.0 | >50 | >50 | >50 | >50 | >50 |
| 17 | >50 | >50 | >50 | 50.0 | >50 | >50 | >50 | >50 | >50 |
| 18 | >50 | >50 | >50 | 25.0 | >50 | >50 | >50 | >50 | >50 |
| Helvolic acid | >50 | >50 | >50 | 6.25 | 12.5 | >50 | >50 | >50 | >50 |
| Dihydrohelvolic acid | >50 | >50 | >50 | 25.0 | 50.0 | >50 | >50 | >50 | >50 |
| Destruxin A | 5.57 | >50 | 22.67 | 50.0 | >50 | >50 | >50 | >50 | >50 |
| Destruxin B | 4.75 | >50 | 9.37 | 50.0 | >50 | >50 | >50 | >50 | >50 |
| Ustilaginoidin D | 2.10 | 35.91 | 3.94 | 50.0 | 12.5 | >50 | 25.0 | 50.0 | >50 |
| Doxorubicina | 0.077 | 9.14 | — | — | — | — | — | — | — |
| Ellipticinea | 3.51 | — | 0.70 | — | — | — | — | — | — |
| Tamoxifena | — | 8.73 | — | — | — | — | — | — | — |
| Isoniazidb | — | — | — | 0.0937 | — | — | — | — | — |
| Rifampicinb,c | — | — | — | 0.0063 | 0.0391 | 3.13 | — | — | — |
| Vancomycinc | — | — | — | — | 1.00 | — | — | — | — |
| Erythromycinc | — | — | — | — | — | 25.0 | — | — | — |
| Amphotericin Bd | — | — | — | — | — | — | 0.781–1.56 | 0.781 | 0.781–1.56 |
Correlations between structures of cytochalasans and their biological activities have been reported.14,30–32 Similarly, the structure–activity relationship (SAR) of the new cytochalasans can be observed in this study. The C-6/C-7 double bond, the carbonyl group at C-17, and 18-OH are important for cytotoxicity against MCF-7 cells. The shift of the C-6/C-7 double bond to C-6/C-12, and the hydroxylation of C-6 and C-7 reduce cytotoxicity against NCI-H187 cells. However, the hydroxyl group at C-5 is important for this activity. The presence of the C-5/C-6 double bond and the carbonyl group at C-7 result in the loss of cytotoxicity.
Conclusions
This study demonstrated that Metarhizium brunneum TBRC-BCC 79240 is a rich source of bioactive cytochalasans. The SAR of cytochalasans for cytotoxicity against NCI-H187 and MCF-7 cell lines was determined. The knowledge generated from this study together with the SAR observations for the various set of cytochalasans and biological assays from the previous studies in the literature can be used to guide the synthesis of derivatives with improved biological activity.Experimental
General experimental procedures
Melting points were measured using a Mettler MP90 melting point apparatus, and reported as uncorrected. Optical rotation measurements were conducted by using a JASCO P-2000 digital polarimeter. UV and FT-IR spectra were recorded on an JASCO V-730 spectrophotometer and a Bruker Alpha spectrometer, respectively. Circular dichroism (CD) spectra were recorded on a JASCO J-810 spectropolarimeter. NMR spectra were recorded on a Bruker AV500D spectrometer. ESITOF MS data were obtained using a Bruker micrOTOF mass spectrometer. Column chromatography was performed using silica gel 60 (70–230 mesh ASTM, Merck). HPLC experiments were performed using a Dionex-Ultimate 3000 series instrument equipped with a binary pump, an autosampler, and a diode array detector.Fungal material
Metarhizium sp. was found on a dead insect (Lepidoptera) sample collected on October 2, 2015, in Khok Pa Si Community Forest, Phu Si Than Wildlife Sanctuary, Kuchinarai District, Kalasin Province, Thailand, by Ms. Kanoksri Tasanathai. The axenic fungal culture of the isolate was deposited at the Thailand Bioresource Research Center as TBRC-BCC 79240, and the Fungarium BIOTEC Bangkok Herbarium (BBH) as BBH40291. The DNA sequences of the internal transcribed spacer (ITS), RNA polymerase II second largest subunit (RPB2), beta-tubulin gene (β-tubulin), and translation elongation factor 1 alpha (TEF) genes of this isolate were obtained using standard methods. The sequences are available from GenBank with ITS, RPB2, β-tubulin, and TEF sequence accession numbers OQ080035, OQ116606, OQ116604, and OQ116605, respectively. The DNA sequences were used in Basic Local Alignment Search Tool (BLAST) search for related sequences using the National Center for Biotechnology Information (NCBI) web tool (available at https://blast.ncbi.nlm.nih.gov/Blast.cgi). The ITS sequence of the isolate matched Metarhizium brunneum (98.20% identity) with the accession numbers MH856876, NR132023, and MT078888. The RPB2, β-tubulin, and TEF sequences of the isolate show 98.15–99.17% identity with M. brunneum sequences in NCBI. From the DNA sequence analysis, Metarhizium sp. TBRC-BCC 79240 was therefore identified as M. brunneum TBRC-BCC 79240 (Clavicipitaceae, Hypocreales, Hypocreomycetidae, Sordariomycetes, Pezizomycotina, Ascomycota).Fermentation and isolation
The fungus, TBRC-BCC 79240, was maintained on potato dextrose agar at 25 °C. The agar was cut into pieces (1 × 1 cm) and inoculated into 8 × 250 mL Erlenmeyer flasks containing 25 mL of potato dextrose broth (PDB, potato starch 4.0 g L−1, dextrose 20.0 g L−1). After incubation at 25 °C for 7 days on a rotary shaker (200 rpm), each primary culture was transferred into 1 L Erlenmeyer flask containing 250 mL of the same liquid medium (PDB) and incubated under the same conditions for 7 days. Each 25 mL portion of the secondary culture was transferred into 80 × 1 L Erlenmeyer flasks containing 250 mL of M102 medium [80 × 250 mL, composition: sucrose (30.0 g L−1), malt extract (20.0 g L−1), bacto-peptone (2.0 g L−1), yeast extract (1.0 g L−1), KCl (0.5 g L−1), MgSO4·7H2O (0.5 g L−1), and KH2PO4 (0.5 g L−1)] and the fermentation was carried out under shaking conditions at 200 rpm, 25 °C for 10 days.After filtration of the culture, the mycelia were macerated in MeOH (5 L) for three days, and then in CH2Cl2 (5 L) for three days. MeOH and CH2Cl2 extracts were combined and evaporated under reduced pressure. The residue was diluted with H2O (800 mL) and the mixture was repeatedly extracted with hexane (3 × 800 mL), followed by EtOAc (3 × 800 mL). The combined EtOAc extract was concentrated under reduced pressure to obtain a brown gum (extract A, 6.02 g). The filtrate of the cultures (broth) was extracted with EtOAc (3 × 20 L) and evaporated to dryness, leaving a dark brown gum (extract B, 2.96 g).
Extract A was fractionated using Sephadex LH-20 and eluted with MeOH to give nine fractions (A1–A9). Further purification of fraction A3 by preparative HPLC using a reverse-phase column (gradient elution with MeCN–H2O, 20–70%) yielded compounds 7 (6.1 mg), 16 (9.3 mg), 17 (12.4 mg), 18 (7.3 mg), helvolic acid (41.7 mg), 1,2-dihydrohelvolic acid (11.8 mg), destruxin A (11.8 mg), and destruxin B (4.3 mg). Trituration of fraction A4 (1.56 g) with MeOH, followed by filtration, afforded a brown solid (0.78 g), which was subjected to preparative HPLC (gradient elution with MeCN–H2O, 30–65%) to yield compounds 7 (3.3 mg), 16 (8.7 mg), 17 (116.7 mg), 18 (96.1 mg), helvolic acid (55.0 mg), and 1,2-dihydro helvolic acid (1.5 mg). Consecutive purification of the filtrate (0.79 g) using preparative HPLC yielded compounds 1 (4.6 mg), 2 (2.4 mg), 3 (2.5 mg), 4 (43.5 mg), 5 (2.9 mg), 7 (14.8 mg), 9 (2.7 mg), 11 (2.0 mg), 12 (1.0 mg), 13 (6.5 mg), 15 (3.4 mg), 16 (38.2 mg), 17 (67.1 mg), 18 (55.0 mg), helvolic acid (39.3 mg), and 1,2-dihydrohelvolic acid (8.6 mg). Ustilaginoidin D (151.5 mg) and K (1.4 mg) were obtained from fractions A7 and A9, respectively, after purification by preparative HPLC (gradient elution with MeCN–H2O, 45–100%).
Extract B (broth extract, 2.96 g) was triturated with MeOH and filtered to obtain a brown solid (0.48 g), which was further purified by preparative HPLC (gradient elution with MeCN–H2O, 25–60%) to afford compounds 7 (9.8 mg), 16 (14.6 mg), 17 (101.2 mg), and 18 (91.1 mg). The filtrate (2.47 g) was fractionated using a Sephadex LH-20 column and eluted with MeOH to obtain five fractions (B1–B5). Fraction B2 was purified by preparative HPLC, which yielded compounds 2 (6.2 mg), 4 (8.2 mg) 5 (7.6 mg), 6 (3.4 mg), 7 (61.7 mg), 8 (2.9 mg), 9 (33.3 mg), 10 (12.0 mg), 11 (6.6 mg), 12 (7.2 mg), 13 (9.4 mg), 14 (5.1 mg), 15 (13.6 mg), 16 (73.4 mg), 17 (118.7 mg), 18 (47.7 mg), and helvolic acid (20.1 mg). Meromuside I (3.2 mg) was obtained from fraction B4 after purification by preparative HPLC (gradient elution with MeCN–H2O, 18–60%).
Brunnesin A (1): white solid; [α]25D –13.5 (c 0.13, EtOH); UV (CH3CN) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 210 (3.90), 220 (3.53) nm; CD (CH3CN) Δε (nm) −9.90 (209), 0 (230), +1.62 (239); IR (ATR) νmax 3402, 2962, 2926, 1680, 1455, 1437, 1381, 1304, 1126, 1026 cm−1; 1H and 13C NMR data, see Table 1; HRMS (ESITOF) m/z 458.2664 [M + Na]+ (calcd for: C28H37NO3 + Na, 458.2666).
ε) 210 (3.90), 220 (3.53) nm; CD (CH3CN) Δε (nm) −9.90 (209), 0 (230), +1.62 (239); IR (ATR) νmax 3402, 2962, 2926, 1680, 1455, 1437, 1381, 1304, 1126, 1026 cm−1; 1H and 13C NMR data, see Table 1; HRMS (ESITOF) m/z 458.2664 [M + Na]+ (calcd for: C28H37NO3 + Na, 458.2666).
Brunnesin B (2): off-white solid; [α]25D –25.9 (c 0.24, EtOH); UV (CH3CN) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 210 (3.49), 218 (3.26), 231 (2.74) nm; CD (CH3CN) Δε (nm) +9.7 (199), 0 (210), −2.45 (230); IR (ATR) νmax 3287, 2963, 2928, 1703, 1686, 1455, 1374, 1295, 1139, 1057 cm−1; 1H and 13C NMR data, see Table 1; HRMS (ESITOF) m/z 472.2454 [M + Na]+ (calcd for: C28H35NO4 + Na, 472.2458).
ε) 210 (3.49), 218 (3.26), 231 (2.74) nm; CD (CH3CN) Δε (nm) +9.7 (199), 0 (210), −2.45 (230); IR (ATR) νmax 3287, 2963, 2928, 1703, 1686, 1455, 1374, 1295, 1139, 1057 cm−1; 1H and 13C NMR data, see Table 1; HRMS (ESITOF) m/z 472.2454 [M + Na]+ (calcd for: C28H35NO4 + Na, 472.2458).
Brunnesin C (3): off-white solid; [α]25D –17.4 (c 0.19, EtOH); UV (CH3CN) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 210 (3.89), 220 (3.57), 231 (3.16) nm; CD (CH3CN) Δε (nm) +13.10 (200), 0 (209), −5.14 (228); IR (ATR) νmax 3368, 2930, 2874, 1701, 1687, 1455, 1377, 1302, 1135, 1061 cm−1; 1H and 13C NMR data, see Table 1; HRMS (ESITOF) m/z 456.2508 [M + Na]+ (calcd for: C28H35NO3 + Na, 456.2509).
ε) 210 (3.89), 220 (3.57), 231 (3.16) nm; CD (CH3CN) Δε (nm) +13.10 (200), 0 (209), −5.14 (228); IR (ATR) νmax 3368, 2930, 2874, 1701, 1687, 1455, 1377, 1302, 1135, 1061 cm−1; 1H and 13C NMR data, see Table 1; HRMS (ESITOF) m/z 456.2508 [M + Na]+ (calcd for: C28H35NO3 + Na, 456.2509).
Brunnesin D (4): white solid; [α]25D –26.8 (c 0.21, EtOH); UV (CH3CN) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 210 (3.88), 219 (3.59), 231 (3.19) nm; CD (CH3CN) Δε (nm) +10.23 (200), 0 (210), −2.24 (230); IR (ATR) νmax 3368, 2966, 2932, 1699, 1685, 1454, 1438, 1376,1267, 1124, 1089, 1031 cm−1; 1H and 13C NMR data, see Table 1; HRMS (ESITOF) m/z 472.2462 [M + Na]+ (calcd for: C28H35NO4 + Na, 472.2458).
ε) 210 (3.88), 219 (3.59), 231 (3.19) nm; CD (CH3CN) Δε (nm) +10.23 (200), 0 (210), −2.24 (230); IR (ATR) νmax 3368, 2966, 2932, 1699, 1685, 1454, 1438, 1376,1267, 1124, 1089, 1031 cm−1; 1H and 13C NMR data, see Table 1; HRMS (ESITOF) m/z 472.2462 [M + Na]+ (calcd for: C28H35NO4 + Na, 472.2458).
Brunnesin E (5): white solid; [α]25D +34.5 (c 0.10, EtOH); UV (CH3CN) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 210 (3.56), 220 (3.30), 231 (2.76) nm; CD (CH3CN) Δε (nm) +6.85 (196), 0 (211), −3.89 (220); IR (ATR) νmax 3360, 2924, 2853, 1686, 1596, 1454, 1354, 1262, 1128, 1036 cm−1; 1H and 13C NMR data, see Table 2; HRMS (ESITOF) m/z 474.2613 [M + Na]+ (calcd for: C28H37NO4 + Na, 474.2615).
ε) 210 (3.56), 220 (3.30), 231 (2.76) nm; CD (CH3CN) Δε (nm) +6.85 (196), 0 (211), −3.89 (220); IR (ATR) νmax 3360, 2924, 2853, 1686, 1596, 1454, 1354, 1262, 1128, 1036 cm−1; 1H and 13C NMR data, see Table 2; HRMS (ESITOF) m/z 474.2613 [M + Na]+ (calcd for: C28H37NO4 + Na, 474.2615).
Brunnesin F (6): off-white solid; [α]25D +43.6 (c 0.10, EtOH); UV (CH3CN) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 210 (4.35), 219 (4.07), 231 (3.41) nm; CD (CH3CN) Δε (nm) +33.3 (190), 0 (202), −15.68 (213); IR (ATR) νmax 3379, 2962, 2926, 1681, 1454, 1274, 1124, 1021 cm−1; 1H and 13C NMR data, see Table 2; HRMS (ESITOF) m/z 474.2618 [M + Na]+ (calcd for: C28H37NO4 + Na, 474.2615).
ε) 210 (4.35), 219 (4.07), 231 (3.41) nm; CD (CH3CN) Δε (nm) +33.3 (190), 0 (202), −15.68 (213); IR (ATR) νmax 3379, 2962, 2926, 1681, 1454, 1274, 1124, 1021 cm−1; 1H and 13C NMR data, see Table 2; HRMS (ESITOF) m/z 474.2618 [M + Na]+ (calcd for: C28H37NO4 + Na, 474.2615).
Brunnesin G (7): white solid; [α]25D +43.2 (c 0.13, EtOH); UV (CH3CN) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 210 (4.36), 220 (4.00), 231 (3.49) nm; CD (CH3CN) Δε (nm) +57.61 (198), 0 (207), −16.81 (219); IR (ATR) νmax 3380, 2925, 2854, 1693, 1684, 1603, 1455, 1377, 1137, 1028 cm−1; 1H and 13C NMR data, see Table 2; HRMS (ESITOF) m/z 472.2456 [M + Na]+ (calcd for: C28H35NO4 + Na, 472.2458).
ε) 210 (4.36), 220 (4.00), 231 (3.49) nm; CD (CH3CN) Δε (nm) +57.61 (198), 0 (207), −16.81 (219); IR (ATR) νmax 3380, 2925, 2854, 1693, 1684, 1603, 1455, 1377, 1137, 1028 cm−1; 1H and 13C NMR data, see Table 2; HRMS (ESITOF) m/z 472.2456 [M + Na]+ (calcd for: C28H35NO4 + Na, 472.2458).
Brunnesin H (8): off-white solid; [α]25D +27.2 (c 0.06, EtOH); UV (CH3CN) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 210 (4.83), 250 (4.27) nm; CD (CH3CN) Δε (nm) +181.9 (192), 0 (212), −20.1 (221); IR (ATR) νmax 3298, 2966, 2930, 1703, 1685, 1613, 1454, 1363, 1262, 1221, 1028 cm−1; 1H and 13C NMR data, see Table 2; HRMS (ESITOF) m/z 470.2311 [M + Na]+ (calcd for: C28H33NO4 + Na, 470.2302).
ε) 210 (4.83), 250 (4.27) nm; CD (CH3CN) Δε (nm) +181.9 (192), 0 (212), −20.1 (221); IR (ATR) νmax 3298, 2966, 2930, 1703, 1685, 1613, 1454, 1363, 1262, 1221, 1028 cm−1; 1H and 13C NMR data, see Table 2; HRMS (ESITOF) m/z 470.2311 [M + Na]+ (calcd for: C28H33NO4 + Na, 470.2302).
Brunnesin I (9): off-white solid; [α]25D –77.5 (c 0.19, EtOH); UV (CH3CN) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 210 (4.40), 232 (3.53) nm; CD (CH3CN) Δε (nm) +26.26 (198), 0 (210), −6.99 (219); IR (ATR) νmax 3389, 2970, 2933, 1697, 1681, 1454, 1375, 1346, 1122, 1083, 1038 cm−1; 1H and 13C NMR data, see Table 3; HRMS (ESITOF) m/z 506.2519 [M + Na]+ (calcd for: C28H37NO6 + Na, 506.2513).
ε) 210 (4.40), 232 (3.53) nm; CD (CH3CN) Δε (nm) +26.26 (198), 0 (210), −6.99 (219); IR (ATR) νmax 3389, 2970, 2933, 1697, 1681, 1454, 1375, 1346, 1122, 1083, 1038 cm−1; 1H and 13C NMR data, see Table 3; HRMS (ESITOF) m/z 506.2519 [M + Na]+ (calcd for: C28H37NO6 + Na, 506.2513).
Brunnesin J (10): white solid; [α]25D –68.2 (c 0.16, EtOH); UV (CH3CN) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 210 (3.94), 218 (3.65), 231 (3.10) nm; CD (CH3CN) Δε (nm) +20.85 (198), 0 (209), −8.20 (219); IR (ATR) νmax 3416, 2971, 2923, 1697, 1679, 1454, 1374, 1348, 1126, 1089, 1040 cm−1; 1H and 13C NMR data, see Table 3; HRMS (ESITOF) m/z 506.2512 [M + Na]+ (calcd for: C28H37NO6 + Na, 506.2513).
ε) 210 (3.94), 218 (3.65), 231 (3.10) nm; CD (CH3CN) Δε (nm) +20.85 (198), 0 (209), −8.20 (219); IR (ATR) νmax 3416, 2971, 2923, 1697, 1679, 1454, 1374, 1348, 1126, 1089, 1040 cm−1; 1H and 13C NMR data, see Table 3; HRMS (ESITOF) m/z 506.2512 [M + Na]+ (calcd for: C28H37NO6 + Na, 506.2513).
Brunnesin K (11): white solid; [α]25D –52.6 (c 0.13, EtOH); UV (CH3CN) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 210 (4.21), 218 (3.94), 231 (3.31) nm; CD (CH3CN) Δε (nm) +29.7 (198), 0 (210), −11.32 (219); IR (ATR) νmax 3368, 2965, 2929, 1697, 1680, 1454, 1380, 1350, 1116, 1089, 1038 cm−1; 1H and 13C NMR data, see Table 3; HRMS (ESITOF) m/z 490.2569 [M + Na]+ (calcd for: C28H37NO5 + Na, 490.2564).
ε) 210 (4.21), 218 (3.94), 231 (3.31) nm; CD (CH3CN) Δε (nm) +29.7 (198), 0 (210), −11.32 (219); IR (ATR) νmax 3368, 2965, 2929, 1697, 1680, 1454, 1380, 1350, 1116, 1089, 1038 cm−1; 1H and 13C NMR data, see Table 3; HRMS (ESITOF) m/z 490.2569 [M + Na]+ (calcd for: C28H37NO5 + Na, 490.2564).
Brunnesin L (12): white solid; [α]25D –51.0 (c 0.11, EtOH); UV (CH3CN) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 210 (4.26), 218 (3.99), 231 (3.32) nm; CD (CH3CN) Δε (nm) +24.25 (199), 0 (209), −12.54 (219); IR (ATR) νmax 3380, 2966, 2930, 1694, 1680, 1454, 1378, 1352, 1117, 1089, 1033 cm−1; 1H and 13C NMR data, see Table 3; HRMS (ESITOF) m/z 490.2565 [M + Na]+ (calcd for: C28H37NO5 + Na, 490.2564).
ε) 210 (4.26), 218 (3.99), 231 (3.32) nm; CD (CH3CN) Δε (nm) +24.25 (199), 0 (209), −12.54 (219); IR (ATR) νmax 3380, 2966, 2930, 1694, 1680, 1454, 1378, 1352, 1117, 1089, 1033 cm−1; 1H and 13C NMR data, see Table 3; HRMS (ESITOF) m/z 490.2565 [M + Na]+ (calcd for: C28H37NO5 + Na, 490.2564).
Brunnesin M (13): white solid; [α]25D –50.9 (c 0.16, EtOH); UV (CH3CN) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 210 (4.29), 231 (3.46) nm; CD (CH3CN) Δε (nm) +26.29 (193), 0 (209), −7.16 (219); IR (ATR) νmax 3348, 2972, 2933, 1697, 1686, 1454, 1375, 1348, 1124, 1060, 1036 cm−1; 1H and 13C NMR data, see Table 4; HRMS (ESITOF) m/z 506.2515 [M + Na]+ (calcd for: C28H37NO6 + Na, 506.2513).
ε) 210 (4.29), 231 (3.46) nm; CD (CH3CN) Δε (nm) +26.29 (193), 0 (209), −7.16 (219); IR (ATR) νmax 3348, 2972, 2933, 1697, 1686, 1454, 1375, 1348, 1124, 1060, 1036 cm−1; 1H and 13C NMR data, see Table 4; HRMS (ESITOF) m/z 506.2515 [M + Na]+ (calcd for: C28H37NO6 + Na, 506.2513).
Brunnesin N (14): off-white solid; [α]25D –34.8 (c 0.10, EtOH); UV (CH3CN) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 210 (4.20), 220 (3.85), 231 (3.39) nm; CD (CH3CN) Δε (nm) +16.10 (199), 0 (209), −8.08 (219); IR (ATR) νmax 3350, 2967, 2931, 1688, 1454, 1380, 1349, 1128, 1059, 1015 cm−1; 1H and 13C NMR data, see Table 4; HRMS (ESITOF) m/z 490.2562 [M + Na]+ (calcd for: C28H37NO5 + Na, 490.2564).
ε) 210 (4.20), 220 (3.85), 231 (3.39) nm; CD (CH3CN) Δε (nm) +16.10 (199), 0 (209), −8.08 (219); IR (ATR) νmax 3350, 2967, 2931, 1688, 1454, 1380, 1349, 1128, 1059, 1015 cm−1; 1H and 13C NMR data, see Table 4; HRMS (ESITOF) m/z 490.2562 [M + Na]+ (calcd for: C28H37NO5 + Na, 490.2564).
X-ray crystallographic analysis of 1 and 2
X-ray diffraction data were measured using a Bruker D8 Venture diffractometer equipped with a graphite monochromated CuKα radiation source (λ = 1.54178 Å) at 273 K during data collection. Using Olex2,33 the structure was solved with the olex2.solve34 structure solution program using charge flipping and refined with the XL35 refinement package using least squares minimization.Crystal data for compound 1: C28H37NO3, MW = 435.58, tetragonal, 0.10 × 0.05 × 0.05 mm3, D = 1.099 g cm−3, space group P43212, Z = 8, a = 14.0756(16) Å, c = 26.584(4) Å, α = β = γ = 90°, V = 5266.8(15) Å3, reflections collected/unique: 73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 517/5006, number of observations [I > 2σ(I)] 4687, R1 = 0.0620, wR2 = 0.1702. Flack parameter = −0.02(4). Crystallographic data were deposited at the Cambridge Crystallographic Data Centre under the reference number CCDC 2231938.
517/5006, number of observations [I > 2σ(I)] 4687, R1 = 0.0620, wR2 = 0.1702. Flack parameter = −0.02(4). Crystallographic data were deposited at the Cambridge Crystallographic Data Centre under the reference number CCDC 2231938.
Crystal data for compound 2: C28H35NO4, MW = 449.57, orthorhombic, 0.08 × 0.05 × 0.05 mm3, D = 1.188 g cm−3, space group P212121, Z = 4, a = 7.3344(2) Å, b = 14.2208(4) Å, c = 24.1002(6) Å, α = β = γ = 90°, V = 2513.68(12) Å3, reflections collected/unique: 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 405/4765, number of observations [I > 2σ(I)] 4508, R1 = 0.0408, wR2 = 0.1158. Flack parameter = 0.02(5). Crystallographic data were deposited at the Cambridge Crystallographic Data Centre under the reference number CCDC 2231937.†
405/4765, number of observations [I > 2σ(I)] 4508, R1 = 0.0408, wR2 = 0.1158. Flack parameter = 0.02(5). Crystallographic data were deposited at the Cambridge Crystallographic Data Centre under the reference number CCDC 2231937.†
Biological assays
Growth inhibitory activity against M. tuberculosis H37Ra and non-cancerous Vero cells (African green monkey kidney fibroblast, ATCC CCL-81) was evaluated using the green fluorescent protein microplate assay (GFPMA),36,37 The resazurin microplate assay (REMA)38 was used to evaluate cytotoxicity against cancerous cells, including MCF-7 (human breast cancer, ATCC HTC-22) and NCI-H187 (human small-cell lung cancer, ATCC CRL-5804). Antibacterial activity against S. aureus (ATCC 29213) and A. baumannii (ATCC 19606) was evaluated by using standard protocols published by the Clinical and Laboratory Standard Institute.39,40 5(6)-Carboxyfluorescein diacetate (CFDA)41–43 fluorometric assay was used to evaluate anti-phytopathogenic fungal activity against C. acutatum (BCC 58146), A. brassicicola (BCC 42724) and C. lunata DOAC 1479 (BCC 15558).Conflicts of interest
All authors declare that they have no conflicts of interest.Acknowledgements
This work was financially supported by National Center for Genetic Engineering and Biotechnology (BIOTEC), National Science and Technology Development Agency (NSTDA) (Grant No. P2050093). We thank Dr Philip J. Shaw for manuscript editing.References
- J. F. Griffin, A. L. Rampal and C. Y. Jung, Proc. Natl. Acad. Sci. U. S. A., 1982, 79, 3759–3763 CrossRef CAS PubMed.
- R. F. Kletzien, J. F. Perdue and A. Springer, J. Biol. Chem., 1972, 247, 2964–2966 CrossRef CAS PubMed.
- A. L. Rampal, H. B. Pinkofsky and C. Y. Jung, Biochemistry, 1980, 19, 679–683 CrossRef CAS PubMed.
- J. A. Williams and J. Wolff, Biochem. Biophys. Res. Commun., 1971, 44, 422–425 CrossRef CAS PubMed.
- A. Makioka, M. Kumagai, S. Kobayashi and T. Takeuchi, Parasitol. Res., 2004, 93, 68–71 CrossRef PubMed.
- W. Pongcharoen, V. Rukachaisirikul, S. Phongpaichit, N. Rungjindamai and J. Sakayaroj, J. Nat. Prod., 2006, 69, 856–858 CrossRef CAS PubMed.
- E. L. Kim, J. L. Li, H. T. Dang, J. Hong, C.-O. Lee, D.-K. Kim, W. D. Yoon, E. Kim, Y. Liu and J. H. Jung, Bioorg. Med. Chem. Lett., 2012, 22, 3126–3129 CrossRef CAS PubMed.
- J. Wang, Z. Wang, Z. Ju, J. Wan, S. Liao, X. Lin, T. Zhang, X. Zhou, H. Chen, Z. Tu and Y. Liu, Planta Med., 2015, 81, 160–166 CrossRef CAS PubMed.
- D. Zhang, H. Ge, D. Xie, R. Chen, J.-h. Zou, X. Tao and J. Dai, Org. Lett., 2013, 15, 1674–1677 CrossRef CAS PubMed.
- A. Berestetskiy, A. Dmitriev, G. Mitina, I. Lisker, A. Andolfi and A. Evidente, Phytochemistry, 2008, 69, 953–960 CrossRef CAS PubMed.
- A. Cimmino, A. Andolfi, A. Berestetskiy and A. Evidente, J. Agric. Food Chem., 2008, 56, 6304–6309 CrossRef CAS PubMed.
- W. Rothweiler and C. Tamm, Experientia, 1966, 22, 750–752 CrossRef CAS.
- D. C. Aldridge, J. J. Armstrong, R. N. Speake and W. B. Turner, Chem. Commun., 1967, 26–27, 10.1039/C19670000026.
- K. Scherlach, D. Boettger, N. Remme and C. Hertweck, Nat. Prod. Rep., 2010, 27, 869–886 RSC.
- E. Skellam, Nat. Prod. Rep., 2017, 34, 1252–1263 RSC.
- C. R. d. Carvalho, A. Ferreira-D'Silva, D. E. Wedge, C. L. Cantrell and L. H. Rosa, Can. J. Microbiol., 2018, 64, 835–843 CrossRef PubMed.
- Y. Fujii, H. Tani, M. Ichinoe and H. Nakajima, J. Nat. Prod., 2000, 63, 132–135 CrossRef CAS PubMed.
- S. A. Patwardhan, R. C. Pandey, S. Dev and G. S. Pendse, Phytochemistry, 1974, 13, 1985–1988 CrossRef CAS.
- B.-C. Yan, W.-G. Wang, D.-B. Hu, X. Sun, L.-M. Kong, X.-N. Li, X. Du, S.-H. Luo, Y. Liu, Y. Li, H.-D. Sun and J.-X. Pu, Org. Lett., 2016, 18, 1108–1111 CrossRef CAS PubMed.
- Q. Zhang, J. Xiao, Q.-Q. Sun, J.-C. Qin, G. Pescitelli and J.-M. Gao, J. Agric. Food Chem., 2014, 62, 10962–10969 CrossRef CAS PubMed.
- H. Zhu, C. Chen, Y. Xue, Q. Tong, X.-N. Li, X. Chen, J. Wang, G. Yao, Z. Luo and Y. Zhang, Angew. Chem., Int. Ed. Engl., 2015, 54, 13374–13378 CrossRef CAS PubMed.
- X.-G. Li, W.-D. Pan, H.-Y. Lou, R.-M. Liu, J.-H. Xiao and J.-J. Zhong, Bioorg. Med. Chem. Lett., 2015, 25, 1823–1826 CrossRef CAS PubMed.
- S. Gupta, D. W. Roberts and J. A. A. Renwick, J. Chem. Soc., Perkin Trans. 1, 1989, 2347–2357, 10.1039/P19890002347.
- M. Païs, B. C. Das and P. Ferron, Phytochemistry, 1981, 20, 715–723 CrossRef.
- H. Fujimoto, E. Negishi, K. Yamaguchi, N. Nishi and M. Yamazaki, Chem. Pharm. Bull., 1996, 44, 1843–1848 CrossRef CAS.
- S.-Y. Lee, H. Kinoshita, F. Ihara, Y. Igarashi and T. Nihira, J. Biosci. Bioeng., 2008, 105, 476–480 CrossRef CAS PubMed.
- A. Fan, W. Mi, Z. Liu, G. Zeng, P. Zhang, Y. Hu, W. Fang and W.-B. Yin, Org. Lett., 2017, 19, 1686–1689 CrossRef CAS PubMed.
- K. Koyama and S. Natori, Chem. Pharm. Bull., 1988, 36, 146–152 CrossRef CAS.
- S. Lu, W. Sun, J. Meng, A. Wang, X. Wang, J. Tian, X. Fu, J. Dai, Y. Liu, D. Lai and L. Zhou, J. Agric. Food Chem., 2015, 63, 3501–3508 CrossRef CAS PubMed.
- R. Kretz, L. Wendt, S. Wongkanoun, J. J. Luangsa-ard, F. Surup, S. E. Helaly, S. R. Noumeur, M. Stadler and T. E. B. Stradal, Biomolecules, 2019, 9, 73 CrossRef CAS PubMed.
- W.-X. Wang, Z.-H. Li, J. He, T. Feng, J. Li and J.-K. Liu, Fitoterapia, 2019, 137, 104278 CrossRef CAS PubMed.
- X. Yang, P. Wu, J. Xue, H. Li and X. Wei, Fitoterapia, 2020, 145, 104611 CrossRef CAS PubMed.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, J. Appl. Crystallogr., 2009, 42, 339–341 CrossRef CAS.
- L. J. Bourhis, O. V. Dolomanov, R. J. Gildea, J. A. K. Howard and H. Puschmann, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71, 59–75 CrossRef CAS PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112–122 CrossRef CAS PubMed.
- C. Changsen, S. G. Franzblau and P. Palittapongarnpim, Antimicrob. Agents Chemother., 2003, 47, 3682–3687 CrossRef CAS PubMed.
- L. Hunt, M. Jordan, M. De Jesus and F. M. Wurm, Biotechnol. Bioeng., 1999, 65, 201–205 CrossRef CAS PubMed.
- J. O'Brien, I. Wilson, T. Orton and F. Pongnan, Eur. J. Biochem., 2000, 267, 5421–5426 CrossRef PubMed.
- P. A. Wayne, Methods for dilution antimicrobial susceptibility test for bacteria that growth aerobically; approve standard, Clinical and Laboratory Standards Institute, CLSI document M7-A7, 7th edn, 2006 Search PubMed.
- P. A. Wayne, Performance standards for antimicrobial susceptibility testing; 16th informational supplement, Clinical and Laboratory Standards Institute, CLSI document M100-S16, 2006 Search PubMed.
- E. A. Aremu, T. Furumai, Y. Igarashi, Y. Sato, H. Akamatsu, M. Kodama and H. Otani, J. Gen. Plant Pathol., 2003, 69, 211–217 CrossRef CAS.
- J. Guarro, I. Pujol, C. Aguilar, C. Llop and J. Fernández-Ballart, J. Antimicrob. Chemother., 1998, 42, 385–387 CrossRef CAS PubMed.
- R. P. Haugland, in Handbook of fluorescent probes and research products, ed. J. Gregory, Molecular Probes, Inc., Oregon, USA, 2022, p. 966 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2231937 and 2231938. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d3ra00042g |
| This journal is © The Royal Society of Chemistry 2023 |