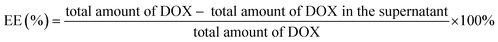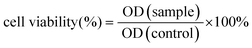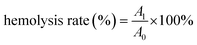Open Access Article
Open Access ArticleAptamer modified Zr-based porphyrinic nanoscale metal–organic frameworks for active-targeted chemo-photodynamic therapy of tumors†
Haidi Fenga,
Lu Zhaoa,
Zhiqiang Baiab,
Zhihui Xinab,
Chaoyu Wanga,
Lizhen Liua,
Jinping Songa,
Haifei Zhang d,
Yunfeng Bai
d,
Yunfeng Bai *a and
Feng Feng
*a and
Feng Feng *abc
*abc
aSchool of Chemistry and Chemical Engineering, Shanxi Provincial Key Laboratory of Chemical Biosensing, Shanxi Datong University, Datong 037009, P. R. China. E-mail: baiyunfeng1130@126.com; feng-feng64@263.net
bSchool of Chemistry and Material Science, Shanxi Normal University, Linfen 041004, P. R. China
cSchool Department of Energy Chemistry and Materials Engineering, Shanxi Institute, P. R. China
dDepartment of Chemistry, University of Liverpool, Crown Street, Liverpool, L69 7ZD, UK
First published on 11th April 2023
Abstract
Active-targeted nanoplatforms could specifically target tumors compared to normal cells, making them a promising therapeutic agent. The aptamer is a kind of short DNA or RNA sequence that can specifically bind to target molecules, and could be widely used as the active targeting agents of nanoplatforms to achieve active-targeted therapy of tumors. Herein, an aptamer modified nanoplatform DOX@PCN@Apt-M was designed for active-targeted chemo-photodynamic therapy of tumors. Zr-based porphyrinic nanoscale metal organic framework PCN-224 was synthesized through a one-pot reaction, which could produce cytotoxic 1O2 for efficient treatment of tumor cells. To improve the therapeutic effect of the tumor, the anticancer drug doxorubicin (DOX) was loaded into PCN-224 to form DOX@PCN-224 for tumor combination therapy. Active-targeted combination therapy achieved by modifying the MUC1 aptamer (Apt-M) onto DOX@PCN-224 surface can not only further reduce the dosage of therapeutic agents, but also reduce their toxic and side effects on normal tissues. In vitro, experimental results indicated that DOX@PCN@Apt-M exhibited enhanced combined therapeutic effect and active targeting efficiency under 808 nm laser irradiation for MCF-7 tumor cells. Based on PCN-224 nanocarriers and aptamer MUC1, this work provides a novel strategy for precisely targeting MCF-7 tumor cells.
Introduction
Metal organic frameworks (MOFs) are a promising category of hybrid porous structural materials that have attracted significant attention in recent years.1–3 MOFs, which have been used extensively in the fields of catalysis, energy storage, sensing and drug delivery, have the advantages of ultra-high porosity and enormous surface area.4–6 Nanoscale metal organic frameworks (NMOFs), with particle sizes down to nanometres, have been widely applied in biomedical studies such as imaging and therapeutics.7–9 Some advantages are as follows: (1) the structural diversity and physicochemical properties of MOFs are retained in NMOFs;10 (2) low biomedical toxicity, high drug-loading performance and superior biodegradability;11,12 (3) the enhanced permeability and retention (EPR) effect allows NMOFs to accumulate at the tumor site;13 (4) the surface is easily modified with targeting agents to improve the therapeutic effect.14,15Porphyrin-based NMOFs are excellent photosensitizers, which have received increasing attention in photodynamic therapy (PDT).16–18 As an emerging treatment strategy for a malignant tumor, PDT can produce reactive oxygen species (ROS) to destroy cell function and ultimately lead to cancer cell apoptosis.19,20 PDT has the characteristics of minimally invasive, precise ablation of the local tumor and avoiding damage to healthy tissues.21,22 However, PDT is easily limited by the depth of light penetration and causes photoallergic reactions at the skin of the tumor site, which limits its tumor therapeutic effects.23,24 Therefore, PDT is often combined with other therapies like chemotherapy (CT), photothermal (PTT), and so on, to further improve the efficacy of tumor treatment.25–28 Due to highly conjugated π-electron systems of porphyrin, porous coordination network-224 (PCN-224) has shown tremendous advantages in the effective treatment of tumors.18,25,29–33 At present, porphyrin-based NMOFs PCN-224 photosensitizer has attracted a lot of attention for PDT with high efficiency.34 To realize chemo-photodynamic combination therapy, CT drugs were encapsulated in PCN-224, which not only improved treatment efficiency, but also greatly minimized the side effects of high doses.35
Through passive and active targeting, NMOFs used in cancer therapy can enter tumor cells. In contrast to passive targeting, active targeting strategies take advantage of the specific recognition ability to target ligands, which realize the precise delivery and promote drug uptake by cancer cells.36,37 Up to now, various targeting ligands were used to fabricate the targeting delivery system, such as folic acid,38–40 hyaluronic acid,36,41,42 cRGD polypeptides,37,43 cancer cell membrane,25,44 aptamer45–47 and so on. Among them, the aptamer is a kind of short DNA or RNA sequence that binds specifically to target molecules, which could be widely used as active targeting agents of nanoplatform to achieve active-targeted therapy of tumors.48,49 Due to the merits of aptamer, Pei et al.45 designed a novel MOFs@RBCs@AS1411 nanorice containing tetrakis(4-carboxyphenyl)porphyrin (TCPP) molecules. With the aid of aptamer AS1411, MOFs@RBCs@AS1411 could enter tumor cells after binding to them specifically. In vitro and vivo results confirmed that MOFs@RBCs@AS1411 nanorice could show effective improvement for the therapeutic effect of KB tumor cells.
In this work, an aptamer modified nanoplatform DOX@PCN@Apt-M was designed and fabricated for active-targeted therapy of tumor. Various tumors overexpressed MUC1 which is an important tumor biomarker. So, DOX@PCN@Apt-M could enter MCF-7 cells after binding to them specifically by using Apt-M. When exposed to 808 nm laser irradiation, DOX@PCN@Apt-M produced cytotoxic 1O2 to kill tumor cells. The in vitro experimental results indicated that DOX@PCN@Apt-M exhibited enhanced combined therapeutic effects and active targeting efficiency (Scheme 1). Therefore, the proposed tumor active-targeted nanoplatform DOX@PCN@Apt-M could achieve a high tumor therapeutic effect.
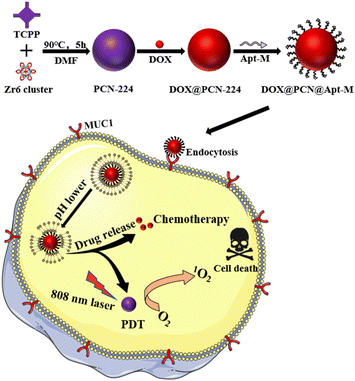 | ||
| Scheme 1 The preparation and tumor active targeted chemotherapy and photodynamic combination therapy of nanoplatform DOX@PCN@Apt-M. | ||
Results and discussion
Synthesis and characterization
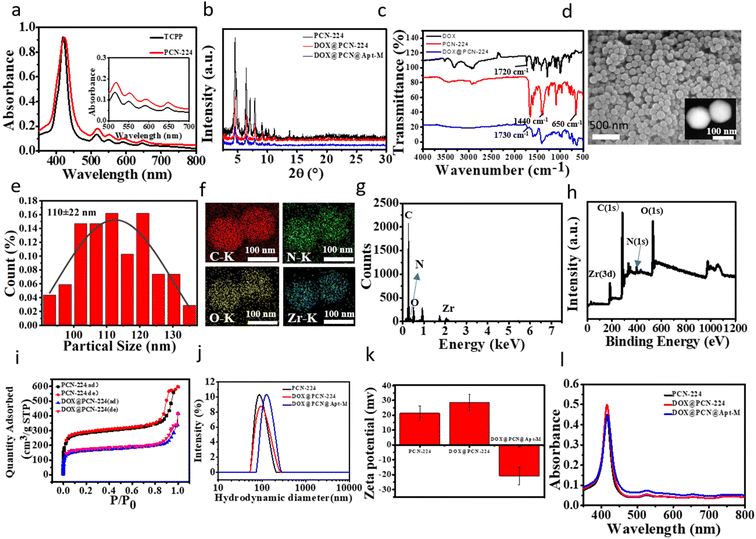
Firstly, a low toxicity Zr-based NMOFs PCN-224 was synthesized through a one-pot reaction by mixing the Zr6 clusters and TCPP. In order to improve its therapeutic effect, the CT drug doxorubicin (DOX) was loaded on PCN-224 through electrostatic adsorption. Finally, to achieve active targeting ability, the carboxy-modified MUC1 aptamer (Apt-M) was successfully coupled to the surface of DOX@PCN-224 through the strong coordination with Zr6 clusters of PCN-224 to form DOX@PCN@Apt-M.In this study, the UV-vis spectra showed that PCN-224 had a Soret peak and four Q-band peaks similar to TCPP (Fig. 1a), indicating that Zr6 clusters did not coordinate with the nitrogen atom in the center of TCPP, but only with the four carboxyl groups of TCPP.32,50 Powder X-ray diffraction (PXRD) results suggested that PCN-224 had good crystallinity (Fig. 1b and S1†). The PXRD analysis results showed that the characteristic peaks of PCN-224 were consistent with those of the simulation curve,51 which indicated that PCN-224 was successfully synthesized. In addition, the successful synthesis of PCN-224 was confirmed by the Fourier Transform Infrared (FT-IR) spectrum, which showed O–H bonds at 1440 cm−1 and Zr–OH bonds at 650 cm−1 (Fig. 1c).52 Dynamic Light Scattering (DLS), consistent with results of scanning electron microscopy (SEM) and transmission electron microscopy (TEM) (Fig. 1d), indicated that the hydrodynamic diameter of PCN-224 was approximately 100 nm. It can also be seen from SEM that PCN-224 was spherical with uniform particle size (Fig. 1e). Moreover, the successful coordination between Zr6 and TCPP was confirmed by elemental mapping (Fig. 1f), indicating Zr6 was uniformly distributed on PCN-224 framework. The energy dispersive X-ray spectroscopy (EDS) (Fig. 1g) and X-ray photoelectron spectroscopy (XPS) (Fig. 1h and S2†) patterns further showed the successful synthesis of PCN-224. Taken together, these results proved that PCN-224 was successfully prepared.
In Fig. 1i, the N2 sorption specific surface area of PCN-224 was 1070 m2 g−1 and the pore volume was 0.3056 cm3 g−1. This demonstrated that PCN-224 can be used as a carrier for delivering drugs due to its high surface area and large pore size. When the concentration ratio of PCN-224 and DOX was 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the encapsulation efficiency of DOX was determined to be about 99% according to the corresponding calibration curve (Fig. S3a and b†), suggesting that DOX were loaded onto PCN-224 with high capacity. After loading of DOX, the BET surface area (613 m2 g−1) and pore volume (0.1701 cm3 g−1) of DOX@PCN-224 were smaller than that those of PCN-224, indicating successful loading of DOX into PCN-224 (Fig. 1i and S4a†). The typical peak at 1730 cm−1 in DOX@PCN-224 was attributed to the stretching vibration of aromatic rings in the DOX, which indicated that DOX was successfully loaded on the PCN-224. The hydrodynamic diameter of DOX@PCN@Apt-M increased dramatically to about 150 nm after modification of Apt-M (Fig. 1j), which indicates that DOX@PCN@Apt-M was successfully synthesized. Furthermore, compared with PCN-224 (+21.3 mV), DOX@PCN@Apt-M had a negative zeta potential (Fig. 1k), which can stably exist in the tumor microenvironment, ensuring DOX@PCN@Apt-M to successfully reach the tumor sites.46 In addition, the UV-vis absorption of DNA at 260 nm decreased significantly, suggesting the successful modification of Apt-M (Fig. S4b†). It can be seen from UV-vis absorption spectra that DOX@PCN@Apt-M and PCN-224 have similar structures, indicating that DOX loading and Apt modification will not affect the structure of PCN-224 (Fig. 1l). Moreover, PXRD data revealed that the crystallinity of DOX@PCN@Apt-M had no obvious difference with that of PCN-224 (Fig. 1b). These results indicate that DOX loading and Apt modification do not destroy the crystallinity of PCN-224. Besides, the DOX@PCN@Apt-M showed no obvious aggregation for 48 h in PBS, H2O, saline, DMEM, 1640 medium (Fig. S4c†), indicating that DOX@PCN@Apt-M could exist stably in various solutions.
1, the encapsulation efficiency of DOX was determined to be about 99% according to the corresponding calibration curve (Fig. S3a and b†), suggesting that DOX were loaded onto PCN-224 with high capacity. After loading of DOX, the BET surface area (613 m2 g−1) and pore volume (0.1701 cm3 g−1) of DOX@PCN-224 were smaller than that those of PCN-224, indicating successful loading of DOX into PCN-224 (Fig. 1i and S4a†). The typical peak at 1730 cm−1 in DOX@PCN-224 was attributed to the stretching vibration of aromatic rings in the DOX, which indicated that DOX was successfully loaded on the PCN-224. The hydrodynamic diameter of DOX@PCN@Apt-M increased dramatically to about 150 nm after modification of Apt-M (Fig. 1j), which indicates that DOX@PCN@Apt-M was successfully synthesized. Furthermore, compared with PCN-224 (+21.3 mV), DOX@PCN@Apt-M had a negative zeta potential (Fig. 1k), which can stably exist in the tumor microenvironment, ensuring DOX@PCN@Apt-M to successfully reach the tumor sites.46 In addition, the UV-vis absorption of DNA at 260 nm decreased significantly, suggesting the successful modification of Apt-M (Fig. S4b†). It can be seen from UV-vis absorption spectra that DOX@PCN@Apt-M and PCN-224 have similar structures, indicating that DOX loading and Apt modification will not affect the structure of PCN-224 (Fig. 1l). Moreover, PXRD data revealed that the crystallinity of DOX@PCN@Apt-M had no obvious difference with that of PCN-224 (Fig. 1b). These results indicate that DOX loading and Apt modification do not destroy the crystallinity of PCN-224. Besides, the DOX@PCN@Apt-M showed no obvious aggregation for 48 h in PBS, H2O, saline, DMEM, 1640 medium (Fig. S4c†), indicating that DOX@PCN@Apt-M could exist stably in various solutions.
pH responsive release of DOX and photodynamic performance of DOX@PCN@Apt-M
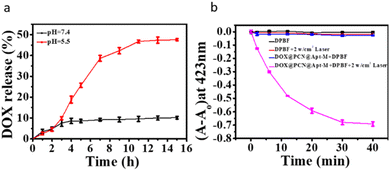
DOX@PCN@Apt-M was evaluated under different pH and time conditions for its DOX release performance. As shown in Fig. 2a, the release of DOX depends on pH and time, and the release ratio was 47.56% at pH = 5.5, while the release ratio was 10.16% at pH = 7.4. These results indicated that the DOX can be efficiently release from DOX@PCN@Apt-M at low pH. This might be advantageous for DOX release in an acidic tumor environment.To evaluate the photodynamic performance of DOX@PCN@Apt-M, 1,3-diphenylisobenzofuran (DPBF) was used as an indicator for the determination of the 1O2.33 The DPBF absorption of PCN-224 decreased significantly under 808 nm laser irradiation, suggesting PCN-224 exhibits a remarkable photodynamic performance for generation 1O2 (Fig. S5a–d†). Furthermore, the absorption of DPBF for DOX@PCN@Apt-M also decayed significantly with 808 nm laser irradiation (Fig. 2b and S5e and f†), which confirmed that DOX@PCN@Apt-M does not affect the photodynamic performance of PCN-224.
Internalization of PCN-224 and release of DOX in vitro
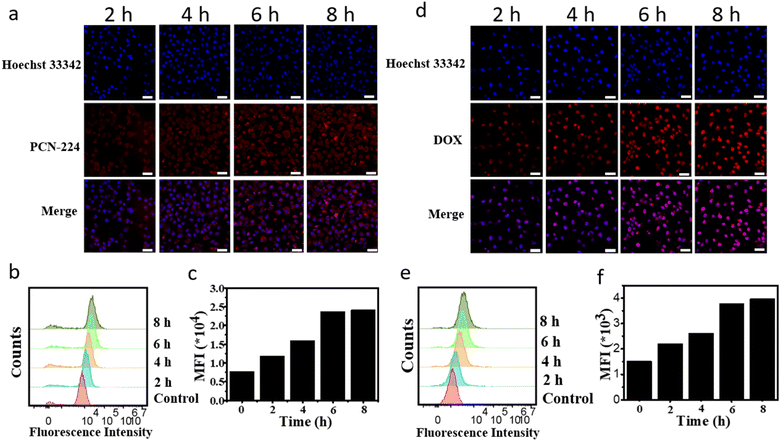
Nanoparticles were effectively internalized into cells, which play a key role in therapeutic effect. PCN-224 can emit red fluorescence when excited at 543 nm. To investigate the uptake capacity of PCN-224 by MCF-7 cells, confocal laser scanning microscopy (CLSM) and flow cytometry were used. As shown in Fig. 3a, PCN-224 was effectively taken up by MCF-7 cells and was able to enter the cell nucleus. With extensive time, more and more PCN-224 entered the cells, and red fluorescence showed hardly change at 8 h, indicating that PCN-224 reached saturation at 6 h. Moreover, flow cytometric uptake results were consistent with that of CLSM (Fig. 3b and c). The above results confirmed that PCN-224 was well taken up by the cells. To prove the advantage of aptamer, MCF-7 cells were treated with PCN-224, DOX@PCN-224 and DOX@PCN@Apt-M for 4 hours. MCF-7 cells treated with DOX@PCN@Apt-M showed stronger fluorescence of PCN-224 than those treated with PCN-224 and DOX@PCN-224 (Fig. S6†).To evaluate the ability of DOX@PCN@Apt-M to DOX release in the MCF-7 cells. DOX@PCN@Apt-M was incubated separately with MCF-7 cells for 2 h, 4 h, 6 h and 8 h. CLSM results showed the red fluorescence signals of DOX increased with incubation time increased, and cellular uptake reached saturation at 6 h (Fig. 3d). Furthermore, it could be inferred that DOX@PCN@Apt-M had a pH-responsive release capacity in MCF-7 cells. Meanwhile, the results have also been confirmed by flow cytometry (Fig. 3e and f). In summary, DOX could release from DOX@PCN-224 and enter the nuclei successfully.
Intracellular target evaluation mediated by Apt-M
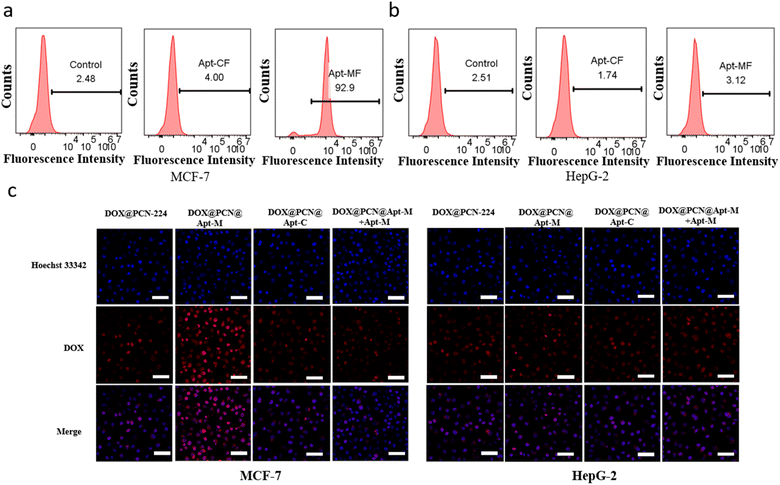
To evaluate the specific targeting ability of Apt-M to MCF-7 cells, Apt-M was modified with FAM (Apt-MF) and incubated with the cells for a period of 2.0 h. As shown in flow cytometry, compared with the control sequence (Apt-CF), the fluorescence intensity of Apt-MF was significantly higher (Fig. 4a). However, after incubation with HepG-2 cells, the fluorescence intensity of Apt-MF and Apt-CF was almost the same (Fig. 4b). These results indicated a high specificity and affinity of Apt-M for MCF-7 cells. Therefore, this study used MCF-7 as positive cells and HepG-2 cells as a negative control.As shown in Fig. 4c, MCF-7 cells incubated with DOX@PCN@Apt-M exhibited stronger fluorescence of DOX than those incubated with DOX@PCN-224, indicating that Apt-M′ recognition ability facilitates DOX@PCN@Apt-M entry into MCF-7 cells. To verify the active targeting ability of Apt-M on MCF-7 cells, an excess of free Apt-M was added to cells in competition with DOX@PCN@Apt-M. We found a reduction in the red fluorescence of DOX due to the addition of free Apt-M, suggesting a reduced uptake of DOX@PCN@Apt-M by MCF-7 cells.
This experimental result indicates that Apt-M can indeed bind to the MUC1 protein on the surface of MCF-7 cells for specific active target recognition of MCF-7 cells. To further demonstrate the performance of DOX@PCN@Apt-M active-targeted, similar treatments were applied to HepG-2 cells. We found that in the DOX@PCN@Apt-M group, HepG-2 cells exhibited weaker DOX red fluorescence compared to MCF-7 cells, and the addition of free Apt-M did not affect the fluorescence intensity of HepG-2 cells. These results indicate that DOX@PCN@Apt-M could enter MCF-7 cells after binding to them specifically, which was mediated by the active targeting ability of Apt-M.
DCFH-DA was a green fluorescent probe for detecting intracellular ROS. DOX@PCN@Apt-M active-targeted performance is also verified by evaluating the photodynamic of its. As shown in Fig. S7a,† the green fluorescence observed in the PCN-224 and DOX@PCN@Apt-M groups of MCF-7 cells were very faint and almost negligible. However, when exposed to laser irradiation at 808 nm, DOX@PCN@Apt-M showed significant green fluorescence compared to PCN-224, demonstrating that DOX@PCN@Apt-M can indeed active-targeted MCF-7 cells to increase their accumulation in the cell. As shown in Fig. S7b,† in HepG-2 cells, the same green fluorescence was observed when the PCN-224 and DOX@PCN@Apt-M groups were exposed to 808 nm laser, which indirectly proves that Apt-M does have a specific recognition ability on MCF-7 cells.
Biocompatibility evaluation
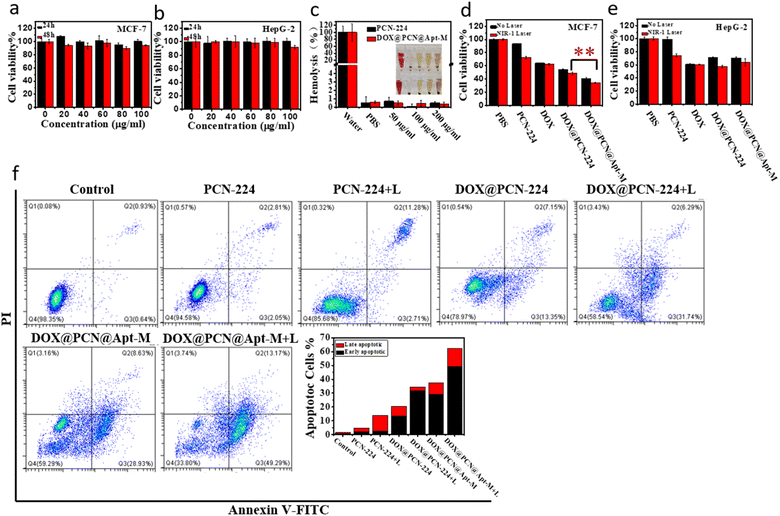
To evaluate the cytotoxicity of PCN-224, the MTT test was performed by incubating PCN-224 with MCF-7, HepG-2 and 7721 cells for 24 h and 48 h. As shown in Fig. 5a and b and S8,† after 48 h incubation with 100 μg mL−1 PCN-224, there was no evident cytotoxicity, revealing the good biocompatibility of PCN-224. In addition, hemolysis rates were found to remain low at concentrations as high as 200 μg mL−1 for PCN-224 and DOX@PCN@Apt-M (Fig. 5c), indicating that they had no negative effects on the integrity of erythrocyte. These results both demonstrated the biosafety and biocompatibility of PCN-224 and DOX@PCN@Apt-M.Cellular cytotoxicity evaluation
The active-targeted chemo-photodynamic therapeutic effect of DOX@PCN@Apt-M was investigated using the MTT assay. As revealed in Fig. 5d and e, the viability of cells was as high as 60% when treated with monotherapy, but decreased to about 50% when treated with chemo-photodynamic therapy. These results show that combination therapy is superior to monotherapy. When incubated with HepG-2 cells, the cell viability in the DOX@PCN@Apt-M group was almost the same as that in the DOX@PCN-224 group under 808 nm laser irradiation. However, after incubation of DOX@PCN@Apt-M with cells, cell viability was only 34% when exposed to the 808 nm laser. Therefore, the active-targeted chemo-photodynamic therapy indeed had a very good therapeutic effect with the aid of Apt-M.To gain deeper insight into the mechanism by which cell proliferation was inhibited, apoptosis induced by samples was studied using flow cytometry. As shown in Fig. 5f, the apoptosis rate of MCF-7 cells treated with the DOX@PCN-224 under laser irradiation was 37.56%, which was higher than that of PDT and CT groups. Besides, the proportion of apoptosis cells treated with DOX@PCN@Apt-M under laser irradiation further increased to 62.46%, indicating that DOX@PCN@Apt-M showed active-targeted combination activity in promoting the apoptosis of MCF-7 cells. As shown in Fig. S9,† when incubated with HepG-2 cells, the apoptosis rate in the DOX@PCN@Apt-M group was almost the same as that in the DOX@PCN-224 group under laser irradiation. These results of the apoptosis analysis by flow cytometry were in agreement with those of the MTT assay.
Moreover, a live/dead cell staining assay further confirmed the active-targeted chemo-photodynamic therapeutic effect of DOX@PCN@Apt-M on MCF-7 cells. As shown in Fig. S10,† compared with the other groups, the majority of dead cells appeared in the DOX@PCN@Apt-M+L group, demonstrating the superiority of active-targeted combination therapy compared to the monotherapy approach. These results demonstrated the great potential of DOX@PCN@Apt-M for MCF-7 tumor cells inhibition by active-targeted chemo-photodynamic therapy.
Conclusion
In conclusion, aptamer MUC1-modified porphyrinic NMOFs was a versatile therapeutic delivery platform with enhanced active-targeted combination therapeutic effect for MCF-7 cells. When DOX@PCN@Apt-M was incubated with MCF-7 cells, with the help of Apt-M, DOX@PCN@Apt-M was able to active-targeted MCF-7 cells to extensively improve the aggregation at the tumour sites. And DOX@PCN@Apt-M could continuously and effectively release DOX under acidic conditions. In addition, DOX@PCN@Apt-M also could produce a large number of 1O2 under laser irradiation. Therefore, active-targeted chemo-photodynamic therapy had a good therapeutic effect on MCF-7 cells, thus reducing the side effects on normal tissues. Based on PCN-224 nanocarriers and aptamer MUC1, this work provides a novel strategy for precisely targeting MCF-7 tumor cells.Experimental
Reagents and materials
N,N-Dimethylformamide (DMF), dimethyl sulfoxide (DMSO) and doxorubicin (DOX) were provided from Shanghai Macklin Biochemical Co., Ltd. Benzoic acid (BA) and zirconium chloride octahydrate (ZrOCl2·8H2O, 99.99%) were provided by Aladdin Biochemical Technology Co., Ltd. Tetrakis(4-carboxyphenyl)porphyrin (TCPP) were obtained from Beijing Innochem Technology Co., Ltd. 1,3-Diphenylisobenzofuran (DPBF) were acquired from Adamas Reagent Co., Ltd. Penicillin/streptomycin, Dulbecco's phosphate buffered saline (PBS), [4′5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium-bromide (MTT), and trypsin were provided by Shanghai Sigma Aldrich Trading Co., Ltd. 2′,7′-Dichlorodihydrofluorescein diacetate (DCFH-DA), Hoechst 33342, Annexin V-FITC/PI cell apoptosis kit and Calcein-AM/PI double staining kit were provided by Shanghai Beyotime Biotechnology Co., Ltd. Fetal bovine serum (FBS) was obtained from Gibco reagent Co., Ltd. Roswell Park Memorial Institute 1640 (RPMI-1640) medium and Dulbecco's modified Eagle's medium (DMEM) were purchased from Biosharp reagent Co., Ltd. All chemicals were of analytical grade and were not purified before use. Nucleic acid sequences were synthesized and HPLC purified by Shanghai Sangon Biotech Co., Ltd as Table S1.†Characterization
A JEM-2100F transmission electron microscope (JEOL, Japan) was used for TEM images, elemental mapping and EDS of nanomaterials. SEM images were taken of the samples covered with Au, using a MAIA3 microscope (TESCAN, Czech Republic). PXRD patterns of samples were acquired from a SmartLab SE diffractometer (Rigaku, Japan). The detailed chemical structure of nanomaterials was analysed by XPS that was detected using a K-Alpha XPS energy spectrometer (Thermo Scientific, USA). The Zetasizer Nano ZS90 Nano Particle Size and Zeta Potential Analyser (Malvern Panalytical, UK) was used to determine the zeta potential and hydrodynamic size of the samples. A Lambda35 spectrometer (PerkinElmer, USA) was used to record the UV-vis absorption spectra of the samples. FT-IR spectroscopy was carried out on NICOLET iS20 Spectrometer (Thermo Scientific, USA). N2 sorption isotherms at 77 K of the nanomaterials were carried out on ASAP 2020 PLUS HD88 Surface Area and Porosity Analyzer (Micromeritics, USA). MTT assay was carried out using an Infinite M200PRO microplate reader (TECAN, Switzerland). An FV1200V CLSM (Olympus, Japan) was used for CLSM images. A CytoFLEX flow cytometer (Beckman Coulter, USA) was used for flow cytometry experiments.Cell cultures
The MCF-7 cells were incubated with RPMI-1640 medium with 10% FBS and 1% penicillin/streptomycin and grown in an incubator containing 37 °C and 5% CO2. The HepG-2 and SMMC-7721 cells were incubated with DMEM medium with 10% FBS and 1% penicillin/streptomycin and grown in an incubator containing 37 °C and 5% CO2.Preparation of PCN-224
PCN-224 was prepared on the basis of the method in the literature.18 In short, 300 mg of ZrOCl2·8H2O, 100 mg of TCPP and 2.8 g of BA were placed in a round bottom flask and dispersed in DMF. And the mixture was stirred (300 rpm) at 90 °C for 5 h. After the solution had cooled, the product PCN-224 was collected by centrifugation and washed several times with DMF to remove unreacted reactants.Preparation of DOX@PCN-224
Briefly, 5 mg of PCN-224 was placed in an EP tube and dispersed in DOX solution of 250 μL, 1 mg mL−1. The reaction was sonicated for 15 min and was placed in a dark shaker at 25 °C for 24 h.53 The DOX@PCN-224 product was then collected by centrifugation. It was washed several times with water to remove the unreacted DOX. Finally, DOX@PCN-224 were dispersed in the neutral PBS solution and used for the following experiments. To calculate encapsulation efficiency (EE) of DOX, the UV absorption of the supernatant at 480 nm was measured and the EE of DOX was calculated using the following formula:Preparation of DOX@PCN@Apt-M
The DOX@PCN-224 (1 mg) was dispersed in Apt-M aqueous solution (2 OD) and ultrasonically treated for 1 min. The solution was gently shaken for 24 h in the dark at 25 °C.47 Then, the DOX@PCN@Apt-M was collected by centrifugation and washed several times with water to remove the unloaded Apt-M. Finally, DOX@PCN@Apt-M was dispersed in the neutral PBS solution.Release characteristics of DOX
To obtain the DOX’ release performance, DOX@PCN@Apt-M (1 mg) was dispersed in PBS solution with pH = 5.5 or 7.4 and gently shaken in the dark at 37 °C. At different time points, the supernatant was collected by centrifugation.54 The concentration of DOX was calculated by measuring UV-vis absorbance of the supernatant at 480 nm. The DOX release rate was calculated by the equation.Photodynamic characteristics of DOX@PCN@Apt-M
DPBF was used as the indicator to evaluate the photodynamic performance of the DOX@PCN@Apt-M in solution according to the previous method.33 60 μL of 500 μg mL−1 DOX@PCN@Apt-M was added to 3 mL of 14 μg mL−1 DPBF in DMSO solution. After exposure to laser irradiation at 808 nm (2 W cm−2), the UV-Vis absorption spectra of the mixture were recorded.The photodynamic performance of DOX@PCN@Apt-M in cells was evaluated using DCFH-DA as a capture agent.55 MCF-7 or HepG-2 cells were seeded into a confocal dish at a density of 1 × 105 cells per mL and cultured overnight. Incubate for 6 hours with 100 μg mL−1 of fresh medium containing PCN-224 or DOX@PCN@Apt-M in a confocal dish. Cells were washed with PBS, serum-free medium containing DCFH-DA (1 μM) was added and after 30 minutes incubation, the cells were treated with a laser at 808 nm (2 W cm−2) for 15 minutes. Eventually, the cells were washed several times with PBS and observed with CLSM.
Internalization of PCN-224 and release of DOX in vitro
MCF-7 cells were seeded into a confocal dish at a density of 1 × 105 cells per mL and cultured overnight. Incubate for 2, 4, 6, 8 hours with 100 μg mL−1 of fresh medium containing PCN-224 in a confocal dish.56 The medium was discarded, the cells were washed with PBS and the dye Hoechst 33342 was added. After 15 minutes of incubation, the cells were washed several times with PBS and observed with CLSM. Moreover, to further demonstrate that PCN-224 can indeed enter the cells, we performed further flow cytometric analysis experiments. MCF-7 cells were first seeded into 6-well plates and grown overnight. Fresh medium containing PCN-224 was added to the 6-well plates. After incubation for 2, 4, 6 and 8 hours, the medium was discarded and the cells were digested for flow cytometry.MCF-7 cells were seeded into a confocal dish and grown overnight. Afterward, fresh medium containing DOX@PCN@Apt-M of 100 μg mL−1 was added to the confocal dishes. After 2, 4, 6 and 8 hours of incubation, the medium was discarded and Hoechst 33342 was added.57 After 15 minutes of incubation, the cells were washed several times with PBS and observed with CLSM. Moreover, flow cytometric analysis was performed to detect the release of DOX. DOX@PCN@Apt-M was added to the cells and analysed by flow cytometry after incubation for various times.
Aptamer affinity analysis
MCF-7 and HepG-2 cells were selected as cell models to study the active targeted ability of Apt-M. The cells were harvested and incubated with Apt-MF or Apt-CF for 2 hours respectively.58 The fluorescence intensity of the cells was determined by flow cytometry after five washes with PBS.Intracellular target evaluation mediated by Apt-M
MCF-7 and HepG-2 cells were seeded into a confocal dish at a density of 1 × 105 cells per mL and cultured overnight. Afterward, cells were incubated with fresh medium containing DOX@PCN-224 (100 μg mL−1), DOX@PCN@Apt-M (100 μg mL−1) and DOX@PCN@Apt-C (100 μg mL−1). For competitive binding assays, prior to incubation with DOX@PCN@Apt-M, MCF-7 and HepG-2 cells were treated with a tenfold excess of Apt-M solution.59 Fluorescence of the cells was detected by CLSM after incubation for 4 hours.Biocompatibility of PCN-224
The MTT method was used to determine the cell biocompatibility of PCN-224. Briefly, the cells were seeded into 96-well plates at a density of 5 × 104 cells per mL and grown overnight. This was followed by incubation with fresh medium containing various concentrations of PCN-224 for 24 h or 48 h.60 After that, the cells were then incubated for 4 hours with MTT solution (5 mg mL−1, 10 μL). Next, the medium was discarded and 150 μL of DMSO solution was added and shaken for about 10 minutes on a shaker. Finally, a microplate reader was then used to read the optical density (OD) at 570 nm. For the calculation of cell viability, untreated cells were used as controls. The viability of cells was calculated as follows:where OD (control) and OD (sample) represent the absorbance in the absence of the samples and with the addition of the samples at different concentrations.
Hemolysis assay
To assess the hemolysis rate of PCN-224 and DOX@PCN@Apt-M, different concentrations of the material were incubated with blood cells (obtained from BALB/c mice) at 37 °C.58 After 30 minutes, the supernatant was collected by centrifugation and the hemolysis rate was calculated by measuring the absorption peak at 540 nm using UV-Vis absorption spectroscopy according to the following formula:where A0 and A1 are the absorbance of the supernatant when incubation with H2O and other solutions, respectively.
Cytotoxicity of DOX@PCN@Apt-M
The cytotoxicity of DOX@PCN@Apt-M was assessed using the MTT method. Briefly, the MCF-7 and HepG-2 cells were seeded into 96-well plates. And the cells were grown overnight. This was followed by incubation with samples of different concentrations. Laser irradiation (808 nm, 2 W cm−2) was performed for 15 minutes after 6 hours of incubation. After a further 24 hours of incubation, the viability of cells was determined using the MTT method.A live/dead cell staining assay was used to further assess the cytotoxicity of DOX@PCN@Apt-M. In general, MCF-7 cells were seeded into confocal dishes. And the cells were grown overnight. The cells were then cultured with the samples for 4 hours, after which laser involved groups were irradiated with a laser of 808 nm for 15 min. The cells treated as described above were then stained with a Calcium AM/PI solution for 20 minutes and then imaged using a CLSM.
In addition, the cytotoxicity was also assessed by means of flow cytometry. A 6-well plate was seeded with MCF-7 and HepG-2 cells. After incubation with samples for 6 h, laser-involved groups were irradiated for 15 min followed by 24 hours incubation. The cells were then collected. And they were stained for 20 min with Annexin V-FITC and PI. Finally, for apoptosis analysis, the cells were measured by flow cytometry.
Statistical analysis
Data were expressed as the mean ± standard error of the mean. Statistical analysis was performed using Student's t-test. In addition, the statistical significance of the difference was indicated as *p < 0.05, **p < 0.01, and ***p < 0.001.Author contributions
Haidi Feng: conceptualization, synthesis and characterization, cell experiments, data curation, writing-original draft. Lu Zhao: data curation, writing-original draft. Zhiqiang Bai: cell experiments. Zhihui Xin: cell experiments. Chaoyu Wang: characterization. Lizhen Liu: cell experiments. Jinping Song: characterization and cell experiments. Haifei Zhang: writing-original draft. Yunfeng Bai and Feng Feng: conceptualization, validation, supervision, writing-reviewing and editing, project administration, funding acquisition.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could affect the work described in this article.Acknowledgements
This work was supported by the Cultivate Scientific Research Excellence Programs of Higher Education Institutions in Shanxi (Grant No. 2020KJ023), Shanxi Scholarship Council of China (Grant No. 2020-133), Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi (Grant No. 2021L368, 2022L424), Fundamental Research Program of Shanxi Province (Grant No. 202303021211324), Postgraduate science and technology innovation project of Shanxi Datong University (Grant No. 21CX19).Notes and references
- A. J. Howarth, Y. Liu, P. Li, Z. Li, T. C. Wang, J. T. Hupp and O. K. Farha, Nat. Rev. Mater., 2016, 1, 1–15 CrossRef.
- A. C. McKinlay, R. E. Morris, P. Horcajada, G. Ferey, R. Gref, P. Couvreur and C. Serre, Angew. Chem., Int. Ed., 2010, 49, 6260–6266 CrossRef CAS PubMed.
- T. Simon-Yarza, A. Mielcarek, P. Couvreur and C. Serre, Adv. Mater., 2018, 30, e1707365 CrossRef PubMed.
- P. Horcajada, C. Serre, M. Vallet-Regi, M. Sebban, F. Taulelle and G. Ferey, Angew. Chem., Int. Ed., 2006, 45, 5974–5978 CrossRef CAS PubMed.
- H. Zheng, Y. Zhang, L. Liu, W. Wan, P. Guo, A. M. Nystrom and X. Zou, J. Am. Chem. Soc., 2016, 138, 962–968 CrossRef CAS PubMed.
- Y. Wang, J. Yan, N. Wen, H. Xiong, S. Cai, Q. He, Y. Hu, D. Peng, Z. Liu and Y. Liu, Biomaterials, 2020, 230, 119619 CrossRef CAS PubMed.
- P. Geng, N. Yu, D. K. Macharia, R. Meng, P. Qiu, C. Tao, M. Li, H. Zhang, Z. Chen and W. Lian, Chem. Eng. J., 2022, 441, 135964 CrossRef CAS.
- X. Ren, Y. Han, Y. Xu, T. Liu, M. Cui, L. Xia, H. Li, Y. Gu and P. Wang, Coord. Chem. Rev., 2021, 431, 213676 CrossRef CAS.
- X. Huang, X. Sun, W. Wang, Q. Shen, Q. Shen, X. Tang and J. Shao, J. Mater. Chem. B, 2021, 9, 3756–3777 RSC.
- I. Imaz, M. Rubio-Martinez, L. Garcia-Fernandez, F. Garcia, D. Ruiz-Molina, J. Hernando, V. Puntes and D. Maspoch, Chem. Commun., 2010, 46, 4737–4739 RSC.
- Y. Yang, Y. Deng, J. Huang, X. Fan, C. Cheng, C. Nie, L. Ma, W. Zhao and C. Zhao, Adv. Funct. Mater., 2019, 29, 1900143 CrossRef.
- D. Zhang, M. Wu, Z. Cai, N. Liao, K. Ke, H. Liu, M. Li, G. Liu, H. Yang, X. Liu and J. Liu, Adv. Sci., 2018, 5, 1700648 CrossRef PubMed.
- G. Lan, K. Ni and W. Lin, Coord. Chem. Rev., 2019, 379, 65–81 CrossRef CAS PubMed.
- C. He, D. Liu and W. Lin, Chem. Rev., 2015, 115, 11079–11108 CrossRef CAS PubMed.
- F. Chen, H. Hong, Y. Zhang, H. F. Valdovinos, S. Shi, G. S. Kwon, C. P. Theuer, T. E. Barnhart and a. W. Cai, ACS Nano, 2013, 7, 9027–9039 CrossRef CAS PubMed.
- L. Zhang, J. Lei, F. Ma, P. Ling, J. Liu and H. Ju, Chem. Commun., 2015, 51, 10831–10834 RSC.
- K. Lu, C. He, N. Guo, C. Chan, K. Ni, R. R. Weichselbaum and W. Lin, J. Am. Chem. Soc., 2016, 138, 12502–12510 CrossRef CAS PubMed.
- J. Park, Q. Jiang, D. Feng, L. Mao and H. C. Zhou, J. Am. Chem. Soc., 2016, 138, 3518–3525 CrossRef CAS PubMed.
- A. P. Castano, P. Mroz and M. R. Hamblin, Nat. Rev. Cancer, 2006, 6, 535–545 CrossRef CAS PubMed.
- M. Ethirajan, Y. Chen, P. Joshi and R. K. Pandey, Chem. Soc. Rev., 2011, 40, 340–362 RSC.
- D. K. Chatterjee, L. S. Fong and Y. Zhang, Adv. Drug Delivery Rev., 2008, 60, 1627–1637 CrossRef CAS PubMed.
- D. W. Felsher, Nat. Rev. Cancer, 2003, 3, 380–387 CrossRef PubMed.
- Q. Jiang, M. Zhang, Q. Sun, D. Yin, Z. Xuan and Y. Yang, Mol. Pharmaceutics, 2021, 18, 3026–3036 CrossRef CAS.
- Z. Wang, B. Liu, Q. Sun, S. Dong, Y. Kuang, Y. Dong, F. He, S. Gai and P. Yang, ACS Appl. Mater. Interfaces, 2020, 12, 17254–17267 CrossRef CAS PubMed.
- S. Y. Li, H. Cheng, B. R. Xie, W. X. Qiu, J. Y. Zeng, C. X. Li, S. S. Wan, L. Zhang, W. L. Liu and X. Z. Zhang, ACS Nano, 2017, 11, 7006–7018 CrossRef CAS PubMed.
- J. Lu, X. Zhu, M. Li, C. Fu, Y. Li, J. Zhang, J. Liu and Y. Zhang, ACS Appl. Bio Mater., 2021, 4, 6316–6325 CrossRef CAS PubMed.
- P. Geng, N. Yu, X. Liu, M. Wen, Q. Ren, P. Qiu, D. K. Macharia, H. Zhang, M. Li and Z. Chen, ACS Appl. Mater. Interfaces, 2021, 13, 31440–31451 CrossRef CAS PubMed.
- Z. H. Zhou, J. Zhao, Z. Di, B. Liu, Z. Li, X. Wu and L. Li, Nanoscale, 2021, 13, 131–137 RSC.
- K. Lu, C. He and W. Lin, J. Am. Chem. Soc., 2014, 136, 16712–16715 CrossRef CAS PubMed.
- K. Lu, C. He and W. Lin, J. Am. Chem. Soc., 2015, 137, 7600–7603 CrossRef CAS PubMed.
- W. Yu, W. Zhen, Q. Zhang, Y. Li, H. Luo, J. He and Y. Liu, ChemMedChem, 2020, 15, 1766–1775 CrossRef CAS PubMed.
- Y. Zhang, F. Wang, C. Liu, Z. Wang, L. Kang, Y. Huang, K. Dong, J. Ren and X. Qu, ACS Nano, 2018, 12, 651–661 CrossRef CAS PubMed.
- J. Hu, W. Wu, Y. Qin, C. Liu, P. Wei, J. Hu, P. H. Seeberger and J. Yin, Adv. Funct. Mater., 2020, 30, 1910084 CrossRef CAS.
- P. Yuan, G. L. Fan, L. P. Zhao, L. S. Liu, F. A. Deng, X. Y. Jiang, A. H. Hu, X. Y. Yu, A. L. Chen, H. Cheng and S. Y. Li, Acta Biomater., 2020, 117, 349–360 CrossRef CAS PubMed.
- X. Zeng, B. Chen, Y. Song, X. Lin, S.-F. Zhou and G. Zhan, ACS Appl. Bio Mater., 2021, 4, 6417–6429 CrossRef CAS PubMed.
- Q. Sun, H. Bi, Z. Wang, C. Li, X. Wang, J. Xu, H. Zhu, R. Zhao, F. He, S. Gai and P. Yang, Biomaterials, 2019, 223, 119473 CrossRef CAS PubMed.
- J. Liu, M. Wu, Y. Pan, Y. Duan, Z. Dong, Y. Chao, Z. Liu and B. Liu, Adv. Funct. Mater., 2020, 30, 1908865 CrossRef CAS.
- R. Abazari, F. Ataei, A. Morsali, A. M. Z. Slawin and C. L. Carpenter-Warren, ACS Appl. Mater. Interfaces, 2019, 11, 45442–45454 CrossRef CAS PubMed.
- V. Nejadshafiee, H. Naeimi, B. Goliaei, B. Bigdeli, A. Sadighi, S. Dehghani, A. Lotfabadi, M. Hosseini, M. S. Nezamtaheri, M. Amanlou, M. Sharifzadeh and M. Khoobi, Mater. Sci. Eng., C, 2019, 99, 805–815 CrossRef CAS PubMed.
- A. Hashemzadeh, F. Amerizadeh, F. Asgharzadeh, M. Darroudi, A. Avan, S. M. Hassanian, M. Landarani and M. Khazaei, Toxicol. Appl. Pharmacol., 2021, 423, 115573 CrossRef CAS PubMed.
- H. Zhang, Q. Li, R. Liu, X. Zhang, Z. Li and Y. Luan, Adv. Funct. Mater., 2018, 28, 1802830 CrossRef.
- W. Cai, H. Gao, C. Chu, X. Wang, J. Wang, P. Zhang, G. Lin, W. Li, G. Liu and X. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 2040–2051 CrossRef CAS PubMed.
- D. Wang, H. Wu, J. Zhou, P. Xu, C. Wang, R. Shi, H. Wang, H. Wang, Z. Guo and Q. Chen, Adv. Sci., 2018, 5, 1800287 CrossRef PubMed.
- S. Y. Li, H. Cheng, W. X. Qiu, L. Zhang, S. S. Wan, J. Y. Zeng and X. Z. Zhang, Biomaterials, 2017, 142, 149–161 CrossRef CAS PubMed.
- Y. Zhao, J. Wang, X. Cai, P. Ding, H. Lv and R. Pei, ACS Appl. Mater. Interfaces, 2020, 12, 23697–23706 CrossRef CAS PubMed.
- Q. G. Zhao, J. Wang, Y. P. Zhang, J. Zhang, A. N. Tang and D. M. Kong, J. Mater. Chem. B, 2018, 6, 7898–7907 RSC.
- Y. Zhang, Q. Wang, G. Chen and P. Shi, Inorg. Chem., 2019, 58, 6593–6596 CrossRef CAS PubMed.
- H. Alijani, A. Noori, N. Faridi, S. Z. Bathaie and M. F. Mousavi, J. Solid State Chem., 2020, 292, 121680 CrossRef CAS.
- B. Liu, M. Jiang, D. Zhu, J. Zhang and G. Wei, Chem. Eng. J., 2022, 428, 131118 CrossRef CAS.
- S. S. Wan, Q. Cheng, X. Zeng and X. Z. Zhang, ACS Nano, 2019, 13, 6561–6571 CrossRef CAS PubMed.
- Y. Tao, Y. Sun, K. Shi, P. Pei, F. Ge, K. Yang and T. Liu, Biomater. Sci., 2021, 9, 2947–2954 RSC.
- J. Wang, Y. Fan, Y. Tan, X. Zhao, Y. Zhang, C. Cheng and M. Yang, ACS Appl. Mater. Interfaces, 2018, 10, 36615–36621 CrossRef CAS PubMed.
- R. Zeng, T. He, L. Lu, K. Li, Z. Luo and K. Cai, J. Mater. Chem. B, 2021, 9, 4143–4153 RSC.
- L. Yao, Y. Tang, W. Cao, Y. Cui and G. Qian, ACS Biomater. Sci. Eng., 2021, 7, 4999–5006 CrossRef CAS PubMed.
- T. Luo, G. T. Nash, Z. Xu, X. Jiang, J. Liu and W. Lin, J. Am. Chem. Soc., 2021, 143, 13519–13524 CrossRef CAS PubMed.
- Y. Zhu, Y. Yang, N. Li, C. Luo and X. Hou, Chem. Eng. J., 2022, 446, 137333 CrossRef CAS.
- R. Xie, P. Yang, S. Peng, Y. Cao, X. Yao, S. Guo and W. Yang, J. Mater. Chem. B, 2020, 8, 6128–6138 RSC.
- Z. Bai, L. Zhao, H. Feng, H. Xu, N. Zhang, Y. Li, J. Song, Y. Bai, R. Yang and F. Feng, Mater. Des., 2023, 226, 111656 CrossRef CAS.
- B. Wang, Y. Dai, Y. Kong, W. Du, H. Ni, H. Zhao, Z. Sun, Q. Shen, M. Li and Q. Fan, ACS Appl. Mater. Interfaces, 2020, 12, 53634–53645 CrossRef CAS PubMed.
- Q. Ren, N. Yu, L. Wang, M. Wen, P. Geng, Q. Jiang, M. Li and Z. Chen, J. Colloid Interface Sci., 2022, 614, 147–159 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra00753g |
| This journal is © The Royal Society of Chemistry 2023 |