Open Access Article
Open Access ArticleEvaluation of dual potentiality of 2,4,5-trisubstituted oxazole derivatives as aquaporin-4 inhibitors and anti-inflammatory agents in lung cells†
Maniarasu Meenakshia,
Arun Kannanb,
Muralidharan Jothimanic,
Thangavel Selvia,
Muthusamy Karthikeyan c,
Chidambaram Prahalathan*b and
Kannupal Srinivasan
c,
Chidambaram Prahalathan*b and
Kannupal Srinivasan *a
*a
aSchool of Chemistry, Bharathidasan University, Tiruchirappalli-620024, Tamil Nadu, India. E-mail: srinivasank@bdu.ac.in
bDepartment of Biochemistry, Bharathidasan University, Tiruchirappalli-620024, Tamil Nadu, India
cDepartment of Bioinformatics, Alagappa University, Karaikudi-630003, Tamil Nadu, India
First published on 1st September 2023
Abstract
Inflammation is a multifaceted “second-line” adaptive defense mechanism triggered by exo/endogenous threating stimuli and inter-communicated by various inflammatory key players. Unresolved or dysregulated inflammation in lungs results in manifestation of diseases and leads to irreparable damage. Aquaporins (AQPs) are a ubiquitously expressed superfamily of intrinsic transmembrane water channel proteins that modulate the fluid homeostasis. In addition to their conventional functions, AQPs have clinical relevance to inflammation prevailing under the infectious conditions of various lung diseases and this proclaims them as appropriate biomarkers to be targeted. Hence an endeavor was undertaken to identify potential ligands to target AQP4 for the treatment of lung diseases. Oxazole being a versatile bio-potent core, a series of 2,4,5-trisubstituted oxazoles 3a–j were synthesized by a Lewis acid mediated reaction of aroylmethylidene malonates with nitriles. In silico studies conducted using the protein data bank (PDB) structure 3gd8 for AQP4 revealed that compound 3a would serve as a suitable candidate to inhibit AQP4 in human lung cells (NCI-H460). Further, in vitro studies demonstrated that compound 3a could effectively inhibit AQP4 and inflammatory cytokines in lung cells and hence it may be considered as a viable drug candidate for the treatment of various lung diseases.
Introduction
Inflammation is a complex and dynamic surveillance mechanism which responds to various noxious stimuli. This crucial mechanism involves multiple inflammatory players including pro-inflammatory cytokines (like TNFα, IFNγ, IL-1β, IL-6 and IL-2), chemokines, adhesion molecules and anti-inflammatory factors (such as TGF-β, IL-4, IL-10 and IL-13), which have a profound impact both at cellular and molecular levels.1 Conversely to its protective role, any failures in feedback inhibition mechanisms are responsible for the attenuation of the process leading to excessive inflammatory cell recruitment. The induction of a “cytokine storm” is known to be a hallmark of various lung diseases including influenza A virus (IAV) infection and Covid-19.2 The exaggerated inflammatory state provoked by hypercytokinemia engenders alveolar damage, acute respiratory distress (ARDS), and pulmonary edema leading to severe comorbidity and mortality.3 Alleviation of the hyper-inflammatory state and fluid clearance remain significant protocols to improve the prognosis of lung diseases.Aquaporins (AQPs) – “the plumbing system for cells” constitute a large family of membrane water channel proteins (WCPs), which play a vital role in regulation of water fluid homeostasis in response to osmotic gradients. There are 13 prevalent isoforms of AQPs, AQP0–AQP12 and they are quite ubiquitous in diverse mammalian cells. On the basis of function and phylogenesis, this super family of WCPs are subcategorized into (i) “orthodox” which are confined only to transportation of water (AQP0, AQP1, AQP2, AQP4, AQP5), (ii) “aquaglyceroporins” which are permeable to water and other small non-polar molecules like glycerol (AQP3, AQP7, AQP9, AQP10) (iii) “superaquaporins” which can transport certain charged and neutral solutes (AQP6, AQP8, AQP11, AQP12).4 In addition to their secretive and absorptive functions, AQPs also mediate other physiological processes such as cell migration, cell proliferation, adipocyte metabolism, neuro excitation etc.5 Extensive data indicate that AQPs play a prominent role in several pathological conditions like intestinal bowel disease (IDB),6 cerebral edema,7 neuroinflammation8 and hypoxia.9 Amongst various isoforms, AQP1, AQP3, AQP4 and AQP5 are expressed in lungs and regulate cell function via transcellular pathway of water across the lung microvascular endothelium and epithelia.10 Though the underlying signaling pathways and molecular mechanisms driving osmotic stress-induced inflammatory response still remain untangled, the pertinence of AQPs to inflammation during the onset of various lung diseases like asthma, pneumonia, pulmonary edema, bronchitis and Covid-19 are well documented.11 Thus it comes to light that AQPs participate in pathophysiological processes apart from their conventional activity of fluid trafficking between inter cellular spaces and this makes them suitable biomarkers to be targeted for the diagnosis and/or treatment of various inflammatory diseases.12 Studies on various models of lung inflammation show the differential regulation of all the four isoforms of AQPs that are expressed in lungs. Recent factual evidence shows that AQP4 is found to be upregulated in numerous inflammatory conditions of lung diseases.13 Drugs which target AQPs are anticipated to possess a wide spectrum of pharmacological applications because of the ubiquitous nature and pathophysiological implication of AQP in inflammation. Recent advancement in the discovery of drugs for AQP and future orientation towards the identification of appropriate AQP inhibitors were well documented.14–16
AQP inhibitors like tetraethylammonium (TEA) interacts with tyrosine residues at pore-lining thereby inhibits water permeability reversibly and selectively through AQP1, AQP2 and AQP4.17,18 Acetazolamide, a first generation carbonic anhydrase inhibitor which is primarily used for the treatment of glaucoma is found to inhibit AQP1 and AQP4 in Xenopus oocytes.19 Other drugs like TGN-020, phloretin, bumetanide have drawn considerable attraction as potential AQP inhibitors.14,20 Although several other putative small molecule aquaporin inhibitors were reported and patented for therapeutic usage, their lack of selectivity and toxic side effects impede their application in clinical trials.14
Non-steroidal anti-inflammatory drugs (NSAIDs) are extensively used for symptomatic relief by controlling pain, fever and inflammation in infected patients. Commercially available drugs like ibuprofen and naproxen continue to be the choice of drugs for treating lung inflammation.21 Also, heterocyclic compounds such as oxazoles, thiazoles and imidazoles have been accredited for their anti-inflammatory potential. Especially, oxazole is a significant structural motif found in many natural products22 and several oxazole derivatives exhibit a wide range of biological activities including anti-inflammatory, antibacterial, antifungal, antiviral, antitubercular, anticancer, antidepressant and neuroprotective activities.23 Despite their wide spectrum of biological activities, the therapeutic efficacy of oxazoles as anti-inflammatory drugs through inhibition of AQP4 has not yet been reported to the best of our knowledge. Further, lack of appropriate drugs to downregulate the AQP4 expression opens up opportunities for the research community to develop new pharmacological therapies through modulation of AQPs. In the present study, we have synthesized a series of 2,4,5-trisubstituted oxazole derivatives and investigated their ability to modulate the expressions of AQP4 and inflammatory cytokines.
Results and discussion
Chemistry
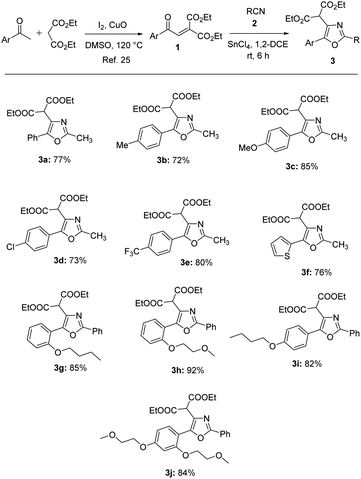
Aroylmethylidene malonates are versatile building blocks in organic synthesis for the access of various heterocyclic compounds.24 They could be easily prepared by the oxidative condensation of commercially available aryl methyl ketones with diethyl malonate in the presence of iodine and cupric oxide in DMSO.25 Aroylmethylidene malonates when treated with nitriles in the presence of tin(IV) chloride undergo conjugate addition followed by ring closure to give oxazoles in 72–92% yields (Table 1).24b The synthesized compounds 3a–j were characterized by IR, NMR and HRMS techniques. The IR spectrum of 3a displayed absorbances at 2982, 1735, 1581, 1446, 1372, 1296, 1238, 1148, 1028, 942, 780, 688 and 595 cm−1. The 1H NMR spectrum of 3a showed a six-proton triplet at δ 1.26 for the two ester methyl protons, a three-proton singlet at δ 2.54 for methyl protons attached to the oxazole ring, a four-proton multiplet at δ 4.22–4.28 for two ester methylene protons and a one-proton singlet at δ 4.88 for malonate methine proton in addition to five aromatic protons in the region of δ 7.39–7.57. In the 13C NMR spectrum of 3a, the two ester methyl carbons appeared at δ 13.9, the methyl carbon attached to the oxazole ring at δ 14.1, malonate methine carbon δ 50.7, the two ester methylene carbons at δ 62.1 and the two ester carbonyl carbons at δ 166.9 in addition to seven aromatic carbons in the region of δ 126.4–160.3. Similar NMR patterns were observed for other oxazole derivatives 3b–j. The HRMS spectra of the tri-substituted oxazoles displayed peaks corresponding to the [M + H]+ ion.| a Isolated yield. |
|---|
 |
In silico analysis on aquaporin-4 inhibition
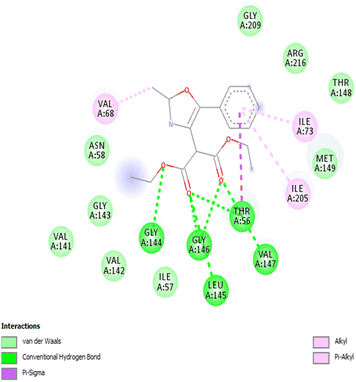
We employed a combination of computational methods to examine the intracellular inhibition of AQP4 by 2,4,5-trisubstituted oxazoles 3a–j. Using AutodockVinasoftware, putative binding sites on the putative binding sites on the AQP4 subunit for the compounds were proposed based on molecular modeling and docking (Fig. 1 and Table 2). All the studied compounds showed good binding scores and they have a binding energy in the range of −6.1 to −7.3 kcal mol−1, with compound 3a having the highest binding energy of −7.3 kcal mol−1. The binding energy was calculated based on the hydrophobic side chain interaction and specific energy that contributes to the molecule binding. Out of curiosity, we performed structure activity relationship studies on hypothetical oxazoles 3k–l by replacing the R group of the oxazole ring by –CF3/–Et group and also the diester group by mono/di carboxylic acid/amide group (see Table 1 in ESI†). To our surprise, the in silico analysis revealed that compound 3a had a better binding affinity towards AQP4 protein when compared to all the other synthetic and hypothetical oxazole derivatives.| S. no. | Compound | Binding energy (kcal mol−1) | Interacting residues |
|---|---|---|---|
| 1 | 3a | −7.3 | GLY144, GLY146, ILE73, ILE205, LEU145, THR56, VAL68, VAL147 |
| 2 | 3b | −6.1 | HIS201, ILE57, ILE73, ILE205, THR56 |
| 3 | 3c | −6.5 | ARG216, GLY209, HIS201, ILE57, ILE73, ILE205, MET149, PHE77, THR56 |
| 4 | 3d | −6.3 | ARG216, GLY209, HIS201, ILE57, ILE73, ILE205, MET149, THR56 |
| 5 | 3e | −6.9 | ARG216, ASN206, HIS201, GLY209, ILE57, ILE73, ILE205, MET149, PHE77, THR56, VAL150 |
| 6 | 3f | −6.2 | ALA176, ALA192, ARG182, ILE189, PHE172, THR173, VAL185 |
| 7 | 3g | −6.6 | GLY143, GLY144, GLY146, ILE73, ILE205, MET149, THR56, VAL147, VAL68 |
| 8 | 3h | −6.8 | GLY146, ILE73, ILE205, THR56, VAL147, VAL68 |
| 9 | 3i | −6.5 | ALA176, ALA192, ARG182, LEU191, PHE172, PHE195 |
| 10 | 3j | −7.1 | ASN58, ASP69, ASN206, ARG216, GLY144, GLY209, VAL68 |
ADME prediction
In silico prediction of ADME (absorption, distribution, metabolism and excretion) properties is essential to predict the biological effect of the molecules, before taking them into the experimental testing. So, the ADME properties of oxazoles 3a–j were predicted using the QikProp module of the Schrodinger suite. The drug-likeness descriptors such as molecular weight, Qplogpo/w, QplogBB, QPPMDCK, QPPCaCo and percentage human oral absorption based on Lipinski's rule of five are given in Table 3. The QplogPo/w is the partial coefficient factor which examines crucial intake of absorption and estimation inside the body. The values of QplogPo/w range from 3.1 to 6.0 for the screened oxazole derivatives 3a–j. The cell permeability factor QPPCaco determines the metabolism reaction of the drug inside the body condition and the values range from 1401 to 2002 for 3a–j. Except for 3j, the human oral absorption for all other oxazoles was estimated to be 100%. The compounds also satisfy the Lipinski's rule of 5. Thus, the ADME results show that the oxazoles 3a–j have pharmacokinetic features within the acceptable range and potentially possess a drug-likeness nature for human use.| Compound | Molecular weight (g mol−1) | QPlogS | QPlogHERG | QPPCaco | QPPMDCK | QPlogKp | QPlogPo/w | Human oral absorption | Rule of five |
|---|---|---|---|---|---|---|---|---|---|
| 3a | 317.341 | −4.244 | −5.504 | 1583.218 | 812.886 | −1.901 | 3.156 | 100 | 0 |
| 3b | 331.368 | −4.965 | −5.527 | 1401.242 | 712.383 | −2.199 | 3.453 | 100 | 0 |
| 3c | 347.367 | −4.367 | −5.385 | 1606.292 | 825.7 | −1.989 | 3.215 | 100 | 0 |
| 3d | 351.786 | −5.06 | −5.474 | 1579.846 | 1999.186 | −2.061 | 3.671 | 100 | 0 |
| 3e | 385.339 | −5.754 | −5.498 | 1593.481 | 3607.933 | −2.123 | 4.165 | 100 | 0 |
| 3f | 323.363 | −3.892 | −5.107 | 1759.474 | 1241.048 | −1.951 | 2.992 | 100 | 0 |
| 3g | 451.518 | −7.156 | −6.945 | 1972.184 | 1030.75 | −0.897 | 5.768 | 100 | 1 |
| 3h | 453.491 | −6.028 | −6.91 | 1793.371 | 930.117 | −0.965 | 4.753 | 100 | 0 |
| 3i | 451.518 | −8.102 | −7.531 | 2002.569 | 1047.926 | −0.836 | 6.004 | 100 | 1 |
| 3j | 527.57 | −6.335 | −7.094 | 1845.361 | 959.296 | −0.761 | 5.002 | 88.772 | 2 |
In vitro analysis on AQP4 inhibition
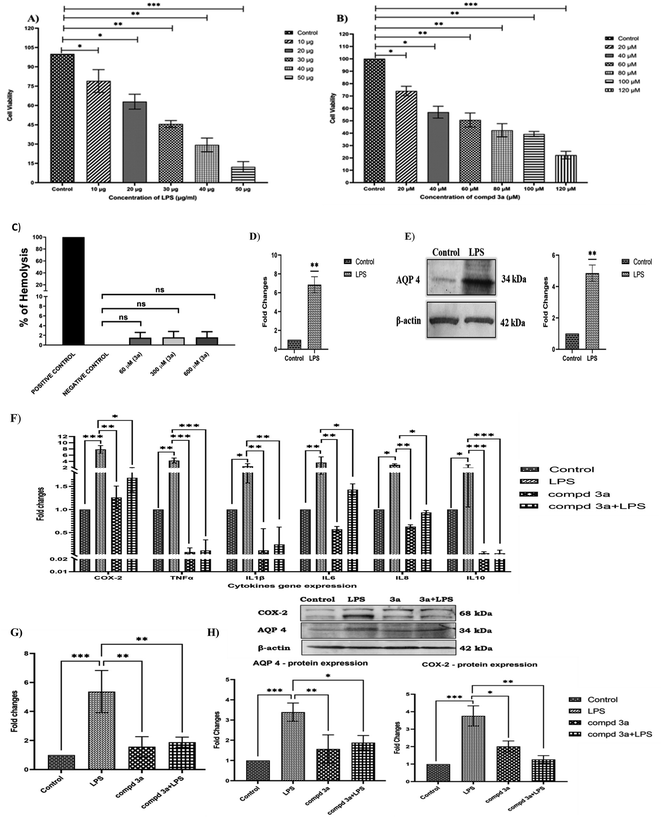
For in vitro studies, we selected oxazole 3a as a representative compound as it had the highest binding energy with AQP4. The dose-dependent effect of lipopolysaccharide (LPS) and compound 3a on cell viability in NCI-H460 cells were analyzed using MTT assay. In the present study, NCI-H460 cells were treated with different concentrations of LPS for 48 h and IC50 value at 30 μg showed 50% decrease in cell viability (Fig. 2A). Further, NCI-H460 cells treated with oxazole 3a at different concentrations for 24 h significantly decreased the cell viability at IC50 concentration of 60 μM (Fig. 2B). We therefore treated NCI-H460 cells with LPS at 30 μg for 48 h and compound 3a at 60 μM for 24 h for all the subsequent experiments.The free hemoglobin in plasma may cause damage to vital organs, such as heart, kidney, and liver. Hence, it is essential to investigate the hemolytic activity of biologically active compounds. So, in vitro hemolysis assay was carried out as a preclusive test for assessing the toxicity of compound 3a. The hemoglobin produced as a result of membrane disruption upon treatment with 3a was assessed. Accordingly, the hemocompatibility of 3a was evaluated against human RBC. As shown in Fig. 2C, the compound 3a was tested at different concentrations (60 μM, 300 μM and 600 μM). The hemolytic activity was found to be 1.5% for 60 μM (IC50 concentration), 1.6% for 300 μM (5 times greater than IC50) and 1.6% for 600 μM (10 times greater than IC50). The value is near to the one obtained by using phosphate-buffered saline (PBS) (negative control). On the other hand 0.1% Triton X-100 (positive control) treated group displays 100% hemolysis. According to ISO/TR 7406, samples with a hemolytic rate of less than 5% (the critical safe hemolytic ratio for biomaterials) are considered non-hemolytic. Hence, compound 3a possesses a good blood compatibility without the cell membrane-disrupting activity.
AQP4 is one of the key players in edema formation and resolution. AQP4 is expressed in the ciliated columnar cells of various epithelial regions (nasopharyngeal, tracheal, bronchial) in the upper airways. Several processes including airway hydration, alveolar transport and submucosal gland secretion are mediated by water transport across epithelia and endothelia in the peripheral lung. In the present study, inflammation was induced in NCI-H460 cells by treating with 30 μg of LPS and the effect of LPS on AQP4 gene and protein expressions were studied. Our results indicated that AQP4 m-RNA expression was significantly upregulated (7-fold) on LPS treatment as shown in the Fig. 2D. Furthermore, we investigated the protein expression of AQP4 under LPS treatment and was found to be significantly increased compared to controls as shown in Fig. 2E. Thus, the results confirmed that AQP4 expression was increased under inflammation in NCI-H460 cells. It has been shown earlier that increased expression of AQP4 upregulates cytokine expression under inflammation. Thus, we examined the effect of compound 3a on cytokine expressions in NCI-H460 cells. Accordingly, NCI-H460 cells were treated with LPS, compound 3a and also LPS pretreated with compound 3a and the cytokine expressions of cyclooxygenase-2 (COX-2), TNF-α, IL-1β, IL-6, IL-8 and IL-10 were studied using real time PCR (qRT-PCR). The m-RNA expression of the cytokines COX-2, TNF-α, IL-1β, IL-6, IL-8, IL-10 were significantly increased on LPS treatment. NCI-H460 cells treated with compound 3a and LPS pre-treated with compound 3a significantly downregulated the gene expressions for the cytokines COX-2, TNF-α, IL-1β, IL-6, IL-8, IL-10 as compared to LPS treated groups as shown in the Fig. 2F. Overall results indicated that increased expression of AQP4 increased cytokine expressions and pre-treatment or treatment with compound 3a in turn decreased these cytokines under inflammation.
To envisage the effect of compound 3a on AQP4 expression, we studied gene expression of AQP4 on NCI-H460 cells treated with LPS, compound 3a and also LPS pretreated with compound 3a by using qRT-PCR. Our results demonstrated that LPS significantly increased AQP4 expression and surprisingly, on compound 3a treatment and pre-treatment significantly downregulated AQP4 m-RNA expression as shown in the Fig. 2G. Further, the same was confirmed analyzing the protein expressions of COX-2 and AQP4 on NCI-H460 cells treated with compound 3a and pre-treatment. As shown in Fig. 2H, LPS drastically increased the protein expressions of AQP4 and COX-2, but compound 3a treatment and pre-treatment significantly decreased the expressions of the same. Thus, our data suggest that compound 3a inhibits the expression of AQP4 and thus decreases the expressions of cytokines under inflammation in NCI-H460 cells.
Inflammation is a complex protective mechanism which provoke against any offending agent. Imbalanced inflammatory response leads to amplification of cytokines production, which in turn leads to organ damage. AQPs which are primarily known for their standard functionality of mediating transcellular fluid across the cell. Although several studies have demonstrated the differential regulation of AQP1, AQP3, AQP4 and AQP5 in various inflammatory models,26 AQP4 is specifically found to be constantly upregulated during inflammatory condition. Liu et al. have demonstrated that AQP4 elevates brain edema by increasing the water transfer into astrocyte during hypoxia ischemia (HI) and the downregulation of the AQP4 induces the anti-inflammatory response thereby helps in restoration of normal cerebral condition. Moreover, by silencing the AQP4, the expression of pro-inflammatory cytokines TNF-α, IL-1β and IL-10 were found to decrease.27 Studies by Li et al. disclosed the pro-inflammatory role of AQP4 in experimental autoimmune encephalomyelitis (EAE). Intracerebral injection of LPS induced greater neuroinflammation in wild-type than in AQP4-knockout mice. Astrocyte culture from AQP4-knock out mice showed reduced expression of cytokines. The result suggest that the reduction in AQP4 expression plays a protective role in neuroinflammatory CNS diseases by inhibiting water transport.28 Further, Dai and his co-workers elucidated that AQP4 knockout led to the inhibition of mitogen-activated protein kinase (MAPK)/protein kinase B (AKT) pathway and alleviates the fluctuation of sphingosine kinase 1 (SPHK1) which eventually leads to decrease in cytokine secretion. The results of the experiments carried out in two genotypes, AQP4+/+ and AQP4−/− exhibit increased production of TNF-α, IL-6 in LPS treated astrocytes. But significant low level of expressions of the same could be observed in AQP4−/−. It is evident from the results that AQP4 deficiency alleviates the release of proinflammatory cytokines from astrocytes in association with SPHK1/MAPK/AKT pathway and underlines the role of AQP4 in neuroinflammation.29 AQP4 deletion under insulin-induced hypoglycemic condition in mice was shown to exhibit positive neurological effect by attenuation of inflammatory response release (TNF-α, IL-6 and IL-1β) through the upregulation of PPAR-γ (peroxisome proliferator-activated receptor γ) and reduced brain edema thereby establishing blood–brain barrier (BBB) integrity.30 The participation of AQP4 in modulation of seizure activity was clear from the outcome of studies by Binder et al. The seizure activity was monitored after the administration of chemoconvulsant pentylenetetrazol (PTZ) and the AQP4−/− mice did not exhibit seizure activity. AQP4 deficient mice showed decreased cerebral edema and improved neurological outcome in comparison with wild-type control.31
Among various heterocyclic compounds, oxazole scaffolds were reported to have valuable therapeutic applications because of their profound anti-inflammatory activity. Dunder et al., have synthesized a series of 4,5-diphenyloxazolone derivatives and evaluated their COX-2 inhibitory potential. Among them, 4-(4-phenyl-3-methyl-2-oxo-3H-1,3-oxazol-5-yl)benzensulfonamide was found to possess the highest selective COX-2 inhibition.32 Chobanian et al., have identified a tri-substituted oxazole derivative, MK-4409 as potential selective fatty acid amide hydrolase (FAAH) inhibitor. In vivo studies showed that MK-4409 had excellent efficacy in various rodent inflammatory and neuropathic pain assays.33 Seth et al. have reported 2-(2-arylphenyl)benzoxzole motifs as selective inhibitors of COX-2 with in vivo anti-inflammatory potential better than the standard drugs like diclofenac and celecoxib.34 The compound 3a used in the present study was also found to possess significant anti-inflammatory activity.
Conclusion
A series of highly substituted oxazole derivatives were prepared from aroylmethylidene malonates and nitriles through a conjugate addition followed by cyclization. The in silico analysis revealed that the synthesized compounds possess good AQP4 inhibitory activity and acceptable ADME properties. The in vitro studies performed taking oxazole 3a as a representative compound demonstrated that it effectively inhibits the expression of AQP4 and cytokines and thus would serve as an appropriate drug candidate to reduce edema and suppress cytokines. The study opens up new scenarios in pharmacological therapies for treating various lung diseases including Covid-19.Experimental section
General remarks
Melting points were determined by the open capillary tube method and were uncorrected. 1H and 13C NMR spectra were recorded on a 400 MHz NMR spectrometer. High-resolution mass spectra (ESI) were recorded on Q-TOF mass spectrometer. Thin layer chromatography (TLC) was performed on precoated alumina sheets and detection was done under UV light. Silica gel (100–200 mesh) was used for column chromatography.Synthesis of oxazoles 3a–j
To a mixture of aroylmethylidene malonate 1 (ref. 25) (1 mmol) and nitriles 2 (1 mmol) in 1,2-dichloroethane (5 mL) was added SnCl4 (1 mmol). The reaction mixture was stirred at room temperature for 6 h. After the reaction was complete, the reaction mixture was quenched with ice-water and extracted with dichloromethane. The organic layer was washed with water, dried (anhydrous Na2SO4) and the solvent was removed under reduced pressure. The crude product was purified by column chromatography using ethyl acetate/hexane to give pure oxazole 3.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 v/v). IR (ATR): ν = 2982, 1735, 1581, 1446, 1372, 1296, 1238, 1148, 1028, 942, 780, 688, 595 cm−1. 1H NMR (400 MHz, CDCl3): δ 7.57 (d, J = 7.2 Hz, 2H), 7.45 (t, J = 7.6 Hz, 2H), 7.39 (t, J = 7.2 Hz, 1H), 4.88 (s, 1H), 4.28–4.22 (m, 4H), 2.54 (s, 3H), 1.26 (t, J = 7.0 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 166.9, 160.3, 148.4, 128.9, 128.7, 127.9, 127.7, 126.4, 62.1, 50.7, 14.1, 13.9 ppm.
7 v/v). IR (ATR): ν = 2982, 1735, 1581, 1446, 1372, 1296, 1238, 1148, 1028, 942, 780, 688, 595 cm−1. 1H NMR (400 MHz, CDCl3): δ 7.57 (d, J = 7.2 Hz, 2H), 7.45 (t, J = 7.6 Hz, 2H), 7.39 (t, J = 7.2 Hz, 1H), 4.88 (s, 1H), 4.28–4.22 (m, 4H), 2.54 (s, 3H), 1.26 (t, J = 7.0 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 166.9, 160.3, 148.4, 128.9, 128.7, 127.9, 127.7, 126.4, 62.1, 50.7, 14.1, 13.9 ppm.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 v/v). IR (ATR): ν = 2995, 2699, 2307, 1736, 1584, 1508, 1373, 1267, 1151, 1094, 1032, 822, 755, 599 cm−1. 1H NMR (400 MHz, CDCl3): δ 7.47 (d, J = 8.0 Hz, 2H), 7.27 (d, J = 8.0 Hz, 2H), 4.87 (s, 1H), 4.29–4.24 (m, 4H), 2.54 (s, 3H), 2.41 (s, 3H), 1.28 (t, J = 7.2 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 167.0, 160.1, 148.5, 138.8, 129.6, 127.2, 126.4, 125.1, 62.1, 50.7, 21.3, 14.1, 14.0 ppm. HRMS (ESI-QTOF) m/z: [M + H]+ calcd for C18H21NO5, 332.1492; found, 332.1506.
7 v/v). IR (ATR): ν = 2995, 2699, 2307, 1736, 1584, 1508, 1373, 1267, 1151, 1094, 1032, 822, 755, 599 cm−1. 1H NMR (400 MHz, CDCl3): δ 7.47 (d, J = 8.0 Hz, 2H), 7.27 (d, J = 8.0 Hz, 2H), 4.87 (s, 1H), 4.29–4.24 (m, 4H), 2.54 (s, 3H), 2.41 (s, 3H), 1.28 (t, J = 7.2 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 167.0, 160.1, 148.5, 138.8, 129.6, 127.2, 126.4, 125.1, 62.1, 50.7, 21.3, 14.1, 14.0 ppm. HRMS (ESI-QTOF) m/z: [M + H]+ calcd for C18H21NO5, 332.1492; found, 332.1506.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 v/v). IR (ATR): ν = 1975, 1728, 1588, 1509, 1297, 1240, 1154, 1024, 839, 778, 723, 607 cm−1. 1H NMR (400 MHz, CDCl3): δ 7.51 (d, J = 8.4 Hz, 2H), 6.99 (d, J = 8.8 Hz, 2H), 4.84 (s, 1H), 4.29–4.23 (m, 4H), 3.87 (s, 3H), 2.54 (s, 3H), 1.28 (t, J = 7.2 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 167.1, 159.9, 159.8, 148.4, 128.1, 126.6, 120.5, 114.3 62.1, 55.4, 50.7, 14.1, 14.0 ppm.
3 v/v). IR (ATR): ν = 1975, 1728, 1588, 1509, 1297, 1240, 1154, 1024, 839, 778, 723, 607 cm−1. 1H NMR (400 MHz, CDCl3): δ 7.51 (d, J = 8.4 Hz, 2H), 6.99 (d, J = 8.8 Hz, 2H), 4.84 (s, 1H), 4.29–4.23 (m, 4H), 3.87 (s, 3H), 2.54 (s, 3H), 1.28 (t, J = 7.2 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 167.1, 159.9, 159.8, 148.4, 128.1, 126.6, 120.5, 114.3 62.1, 55.4, 50.7, 14.1, 14.0 ppm.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 v/v). IR (ATR): ν = 2985, 2355, 1737, 1580, 1318, 1241, 1161, 1065, 1026, 947, 844, 679, 597 cm−1. 1H NMR (400 MHz, CDCl3): δ 7.51 (d, J = 8.8 Hz, 2H), 7.40 (d, J = 8.4 Hz, 2H), 4.83 (s, 1H), 4.27–4.21 (m, 4H), 2.52 (s, 3H), 1.25 (t, J = 7.2 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 166.8, 160.5, 147.3, 134.7, 129.1, 128.2, 127.6, 126.4, 62.2, 50.8, 14.0, 13.9 ppm.
7 v/v). IR (ATR): ν = 2985, 2355, 1737, 1580, 1318, 1241, 1161, 1065, 1026, 947, 844, 679, 597 cm−1. 1H NMR (400 MHz, CDCl3): δ 7.51 (d, J = 8.8 Hz, 2H), 7.40 (d, J = 8.4 Hz, 2H), 4.83 (s, 1H), 4.27–4.21 (m, 4H), 2.52 (s, 3H), 1.25 (t, J = 7.2 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 166.8, 160.5, 147.3, 134.7, 129.1, 128.2, 127.6, 126.4, 62.2, 50.8, 14.0, 13.9 ppm.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 v/v). IR (ATR): ν = 2995, 2699, 1737, 1585, 1483, 1373, 1267, 1152, 1095, 1031, 942, 834, 755, 598 cm−1. 1H NMR (400 MHz, CDCl3): δ 7.66 (s, 4H), 4.85 (s, 1H), 4.24–4.18 (m, 4H), 2.50 (s, 3H), 1.21 (t, J = 7.2 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 166.6, 161.0, 146.8, 131.2, 129.5, 126.4, 125.8, 125.7, 62.2, 50.9, 13.9, 13.8 ppm. HRMS (ESI-QTOF) m/z: [M + H]+ calcd for C18H18F3NO5, 386.1210; found, 386.1216.
7 v/v). IR (ATR): ν = 2995, 2699, 1737, 1585, 1483, 1373, 1267, 1152, 1095, 1031, 942, 834, 755, 598 cm−1. 1H NMR (400 MHz, CDCl3): δ 7.66 (s, 4H), 4.85 (s, 1H), 4.24–4.18 (m, 4H), 2.50 (s, 3H), 1.21 (t, J = 7.2 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 166.6, 161.0, 146.8, 131.2, 129.5, 126.4, 125.8, 125.7, 62.2, 50.9, 13.9, 13.8 ppm. HRMS (ESI-QTOF) m/z: [M + H]+ calcd for C18H18F3NO5, 386.1210; found, 386.1216.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 v/v). IR (ATR): ν = 3100, 2980, 2360, 1736, 1657, 1580, 1444, 1368, 1235, 1151, 1028, 846, 708, 592 cm−1. 1H NMR (400 MHz, CDCl3): δ 7.40 (d, J = 4.8 Hz, 1H), 7.29 (d, J = 4.0 Hz, 1H), 7.11 (d, J = 4.4 Hz, 1H), 4.89 (s, 1H), 4.30–4.24 (m, 4H), 2.53 (s, 3H), 1.29 (t, J = 7.2 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 166.7, 160.2, 143.8, 128.9, 127.8, 127.6, 126.6, 125.9, 62.2, 50.6, 14.1, 14.0 ppm. HRMS (ESI-QTOF) m/z: [M + H]+ calcd for C15H17NO5S, 324.0900; found, 324.0911.
7 v/v). IR (ATR): ν = 3100, 2980, 2360, 1736, 1657, 1580, 1444, 1368, 1235, 1151, 1028, 846, 708, 592 cm−1. 1H NMR (400 MHz, CDCl3): δ 7.40 (d, J = 4.8 Hz, 1H), 7.29 (d, J = 4.0 Hz, 1H), 7.11 (d, J = 4.4 Hz, 1H), 4.89 (s, 1H), 4.30–4.24 (m, 4H), 2.53 (s, 3H), 1.29 (t, J = 7.2 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 166.7, 160.2, 143.8, 128.9, 127.8, 127.6, 126.6, 125.9, 62.2, 50.6, 14.1, 14.0 ppm. HRMS (ESI-QTOF) m/z: [M + H]+ calcd for C15H17NO5S, 324.0900; found, 324.0911.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 v/v). IR (ATR): ν = 3058, 2967, 2353, 1739, 1587, 1486, 1452, 1371, 1258, 1154, 1031, 1258, 1154, 1031,894, 734, 609 cm−1. 1H NMR (400 MHz, CDCl3): δ 8.45–8.12 (m, 2H), 7.62–7.60 (m, 1H), 7.49–7.39 (m, 4H), 7.10–7.06 (m, 1H), 7.03 (d, J = 8.4 Hz, 1H), 5.05 (s, 1H), 4.29–4.20 (m, 4H), 4.07 (t, J = 7.0 Hz, 2H), 1.83–1.76 (m, 2H), 1.51–1.41 (m, 2H), 1.29 (t, J = 7.2 Hz, 6H), 0.95 (t, J = 7.4 Hz, 3H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 167.3, 160.5, 156.2, 146.0, 131.2, 130.8, 130.6, 130.2, 128.6, 127.5, 126.6, 120.7, 117.4, 112.4, 68.6, 61.8, 51.4, 30.9, 19.2, 14.0, 13.8 ppm. HRMS (ESI-QTOF) m/z: [M + H]+ calcd for C26H29NO6, 452.2068; found, 452.2078.
7 v/v). IR (ATR): ν = 3058, 2967, 2353, 1739, 1587, 1486, 1452, 1371, 1258, 1154, 1031, 1258, 1154, 1031,894, 734, 609 cm−1. 1H NMR (400 MHz, CDCl3): δ 8.45–8.12 (m, 2H), 7.62–7.60 (m, 1H), 7.49–7.39 (m, 4H), 7.10–7.06 (m, 1H), 7.03 (d, J = 8.4 Hz, 1H), 5.05 (s, 1H), 4.29–4.20 (m, 4H), 4.07 (t, J = 7.0 Hz, 2H), 1.83–1.76 (m, 2H), 1.51–1.41 (m, 2H), 1.29 (t, J = 7.2 Hz, 6H), 0.95 (t, J = 7.4 Hz, 3H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 167.3, 160.5, 156.2, 146.0, 131.2, 130.8, 130.6, 130.2, 128.6, 127.5, 126.6, 120.7, 117.4, 112.4, 68.6, 61.8, 51.4, 30.9, 19.2, 14.0, 13.8 ppm. HRMS (ESI-QTOF) m/z: [M + H]+ calcd for C26H29NO6, 452.2068; found, 452.2078.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 v/v). IR (ATR): ν = 2993, 2299, 1737, 1583, 1486, 1450, 1366, 1268, 1150, 1029, 927, 856, 756, 608 cm−1. 1H NMR (400 MHz, CDCl3): δ 8.13–8.12 (m, 2H), 7.64–7.62 (m, 1H), 7.47–7.45 (m, 3H), 7.43–7.38 (m, 1H), 7.10 (t, J = 7.6 Hz, 1H), 7.05 (d, J = 8.4 Hz, 1H), 5.22 (s, 1H), 4.29–4.21 (m, 6H), 3.76 (t, J = 5.0 Hz, 2H), 3.40 (s, 3H), 1.29 (t, J = 7.0 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 167.4, 160.5, 155.7, 145.7, 131.5, 130.7, 130.5, 130.2, 128.6, 127.6, 126.5, 121.2, 117.7, 112.7, 70.6, 68.2, 61.7, 59.1, 51.3, 14.0 ppm. HRMS (ESI-QTOF) m/z: [M + H]+ calcd for C25H27NO7, 454.1860; found, 454.1870.
3 v/v). IR (ATR): ν = 2993, 2299, 1737, 1583, 1486, 1450, 1366, 1268, 1150, 1029, 927, 856, 756, 608 cm−1. 1H NMR (400 MHz, CDCl3): δ 8.13–8.12 (m, 2H), 7.64–7.62 (m, 1H), 7.47–7.45 (m, 3H), 7.43–7.38 (m, 1H), 7.10 (t, J = 7.6 Hz, 1H), 7.05 (d, J = 8.4 Hz, 1H), 5.22 (s, 1H), 4.29–4.21 (m, 6H), 3.76 (t, J = 5.0 Hz, 2H), 3.40 (s, 3H), 1.29 (t, J = 7.0 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 167.4, 160.5, 155.7, 145.7, 131.5, 130.7, 130.5, 130.2, 128.6, 127.6, 126.5, 121.2, 117.7, 112.7, 70.6, 68.2, 61.7, 59.1, 51.3, 14.0 ppm. HRMS (ESI-QTOF) m/z: [M + H]+ calcd for C25H27NO7, 454.1860; found, 454.1870.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 v/v). IR (ATR): ν = 2931, 1732, 1505, 1303, 1235, 1175, 1031, 789, 701 cm−1. 1H NMR (400 MHz, CDCl3): δ 8.15–8.12 (m, 2H), 7.62 (d, J = 8.8 Hz, 2H), 7.49–7.48 (m, 3H), 7.02 (d, J = 8.8 Hz, 2H), 4.98 (s, 1H), 4.31–7.24 (m, 4H), 4.05 (t, J = 6.4 Hz, 2H), 1.87–1.80 (m, 2H), 1.59–1.51 (m, 2H), 1.29 (t, J = 7.2 Hz, 6H), 1.03 (t, J = 7.4 Hz, 3H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 167.1, 159.8, 159.6, 148.6, 130.3, 128.7, 128.2, 128.16, 127.3, 126.5, 120.2, 114.8, 67.9, 62.1, 51.3, 31.2, 19.3, 14.0, 13.9 ppm. HRMS (ESI-QTOF) m/z: [M + H]+ calcd for C26H29NO6, 452.2068; found, 452.2077.
7 v/v). IR (ATR): ν = 2931, 1732, 1505, 1303, 1235, 1175, 1031, 789, 701 cm−1. 1H NMR (400 MHz, CDCl3): δ 8.15–8.12 (m, 2H), 7.62 (d, J = 8.8 Hz, 2H), 7.49–7.48 (m, 3H), 7.02 (d, J = 8.8 Hz, 2H), 4.98 (s, 1H), 4.31–7.24 (m, 4H), 4.05 (t, J = 6.4 Hz, 2H), 1.87–1.80 (m, 2H), 1.59–1.51 (m, 2H), 1.29 (t, J = 7.2 Hz, 6H), 1.03 (t, J = 7.4 Hz, 3H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 167.1, 159.8, 159.6, 148.6, 130.3, 128.7, 128.2, 128.16, 127.3, 126.5, 120.2, 114.8, 67.9, 62.1, 51.3, 31.2, 19.3, 14.0, 13.9 ppm. HRMS (ESI-QTOF) m/z: [M + H]+ calcd for C26H29NO6, 452.2068; found, 452.2077.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 v/v). IR (ATR): ν = 2994, 2886, 2699, 1737, 1598, 1455, 1369, 1268, 1185, 1122, 1031, 916, 858, 755 cm−1. 1H NMR (400 MHz, CDCl3): δ 8.11–8.09 (m, 2H), 7.51 (d, J = 8.4 Hz, 1H), 7.45–7.44 (m, 3H), 6.66–6.62 (m, 2H), 5.14 (s, 1H), 4.26–7.18 (m, 8H), 3.81–3.79 (m, 2H), 3.75–3.73 (m, 2H), 3.49 (s, 3H), 3.39 (s, 3H), 1.28 (t, J = 7.2 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 167.5, 161.0, 160.2, 157.0, 145.9, 131.3, 130.5, 130.0, 128.6, 127.6, 126.5, 110.6, 105.9, 70.9, 70.5, 68.2, 67.5, 61.7, 59.3, 59.1, 51.2, 14.0 ppm. HRMS (ESI-QTOF) m/z: [M + H]+ calcd for C28H33NO9, 528.2228; found, 528.2241.
3 v/v). IR (ATR): ν = 2994, 2886, 2699, 1737, 1598, 1455, 1369, 1268, 1185, 1122, 1031, 916, 858, 755 cm−1. 1H NMR (400 MHz, CDCl3): δ 8.11–8.09 (m, 2H), 7.51 (d, J = 8.4 Hz, 1H), 7.45–7.44 (m, 3H), 6.66–6.62 (m, 2H), 5.14 (s, 1H), 4.26–7.18 (m, 8H), 3.81–3.79 (m, 2H), 3.75–3.73 (m, 2H), 3.49 (s, 3H), 3.39 (s, 3H), 1.28 (t, J = 7.2 Hz, 6H) ppm. 13C{1H}NMR (100 MHz, CDCl3) δ 167.5, 161.0, 160.2, 157.0, 145.9, 131.3, 130.5, 130.0, 128.6, 127.6, 126.5, 110.6, 105.9, 70.9, 70.5, 68.2, 67.5, 61.7, 59.3, 59.1, 51.2, 14.0 ppm. HRMS (ESI-QTOF) m/z: [M + H]+ calcd for C28H33NO9, 528.2228; found, 528.2241.Materials and methods for in silico studies
Materials and methods for in vitro studies
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 3 min. The supernatant was collected and the optical density (OD) value was observed at 595 nm. The percentage of hemolysis was calculated as follows: % of hemolysis = (OD(test) − OD(negative control))/(OD(positive control) − OD(negative control)) × 100%.
000 rpm for 3 min. The supernatant was collected and the optical density (OD) value was observed at 595 nm. The percentage of hemolysis was calculated as follows: % of hemolysis = (OD(test) − OD(negative control))/(OD(positive control) − OD(negative control)) × 100%.| Gene | Forward primer | Reverse primer |
|---|---|---|
| COX-2 | CCGGGTACAATCGCACTTAT | GGCGCTCAGCCATACAG |
| TNFα | CCCAGGGACCTCTCTCTAATCA | AGCTGCCCCTCAGCTTGAG |
| IL-1β | ACAGATGAAGTGCTCCTTCCA | GTCGGAGATTCGTAGCTGGAT |
| IL-6 | GGTACATCCTCGACGGCATCT | GTGCCTCTTTGCTGCTTTCAC |
| IL-8 | GGCACAAACTTTCAGAGACAGCAG | GTTTCTTCCTGGCTCTT GTCCTAG |
| IL-10 | GGTTGCCAAGCCTTGTCTGA | AGGGAGTTCACATGCGCCT |
| β-Actin | CGGGAAATCGTGCGTGAC | CGGGAAATCGTGCGTGAC |
Data analysis
The values are expressed as mean ± standard deviation (SD). Differences between the groups were assessed by one-way ANOVA using GraphPad Prism 9.0 software. Post hoc testing was performed for inter-group comparisons using Tukey's multiple comparisons test. Values were considered significant at P < 0.05. **P < 0.01 and ***P < 0.001.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Tamil Nadu State Council for Higher Education (TANSCHE) for financial support and DST-FIST for infrastructure/instrumentation facilities at Bharathidasan University. K. S. also thanks Council of Scientific and Industrial Research (CSIR), India for financial support. T. S. thanks Department of Science and Technology (DST) for a WOS-A fellowship.Notes and references
- (a) B. Moldoveanu, P. Otmishi, P. Jani, J. Walker, X. Sarmiento, J. Guardiola, M. Saad and J. Yu, J. Inflammation Res., 2009, 2, 1–11 CAS; (b) M. Antonelli and I. Kushner, FASEF J., 2017, 31, 1787–1797 CrossRef CAS PubMed; (c) M.-G. Netea, F. Balkwill, M. Chonchol, F. Cominelli, M.-Y. Donath, G. Bourboulis, D. Golenbock, M.-S. Gresnigt, M.-T. Heneka, H.-M. Hoffman, R. Hotchkiss, A.-B. Joosten, D.-L. Kastner, M. Korte, E. Latz, P. Libby, T.-M. Poulsen, A. Mantovani, H.-G. Mills, K.-L. Nowak, L.-A. O'Neill, P. Pickkers, T.-V. Poll, P.-M. Ridker, J. Schalkwijk, D.-A. Schwartz, B. Siegmund, C.-J. Steer, H. Tilg, W.-M. Meer, F.-L. Veerdonk and C.-A. Dinarello, Nat. Immunol., 2017, 18, 826–832 CrossRef CAS PubMed; (d) D.-R. Germolec, K.-A. Shipkowski, R.-P. Frawley and E. Evans, Methods Mol. Biol., 2018, 1803, 57–79 CrossRef CAS PubMed; (e) J.-M. Zhang and J. An, Int. Anesthesiol. Clin., 2007, 45, 27–37 CrossRef PubMed.
- B. Hu, S. Huang and L. Yin, J. Med. Virol., 2021, 93, 250–256 CrossRef CAS PubMed.
- (a) C. Pelaia, C. Tinello, A. Vatrella, G.-D. Sarro and G. Pelaia, Ther. Adv. Respir. Dis., 2020, 14, 1–19 Search PubMed; (b) C. Peteranderl, J.-I. Sznajder, S. Herold and E. Lecuona, Front. Immunol., 2017, 8, 446–456 Search PubMed.
- G. Benga, Mol. Aspects Med., 2012, 33, 514–517 CrossRef CAS PubMed.
- (a) M.-C. Papadapoulos, S. Saadoun and A. S. Verkman, Pflügers Archiv: European Journal of Physiology, 2008, 456, 693–700 CrossRef PubMed; (b) M.-C. Papadopoulos and S. Saadoun, Biochimica et Biophysica, 2015, 1848, 2576–2583 CrossRef CAS PubMed; (c) S. Edamana, F.-H. Login, S. Yamada, T.-H. Kwon and L.-N. Nejsum, Am. J. Physiol.: Cell Physiol., 2021, 320, 771–777 CrossRef PubMed.
- H. Lv, Y. Li, C. Xue, N. Dang, C. Bi and A. Shan, J. Anim. Physiol. Anim. Nutr., 2022, 106, 167–180 CrossRef PubMed.
- (a) G. Tang and G.-Y. Yang, Int. J. Mol. Sci., 2016, 17, 1413–1424 CrossRef PubMed; (b) A.-M. Fukuda and J. Badaut, J. Neuroinflammation, 2012, 9, 279–288 CAS.
- (a) A.-S. Verkman, J. Ratelade, A. Rossi, H. Zhang and L. Tradtrantip, Acta Pharmacol. Sin., 2011, 32, 702–710 CrossRef CAS PubMed; (b) Y.-L. Lan, D.-Y. Fang, J. Zhao, T.-H. Ma and S. Li, Acta Neurol. Belg., 2016, 116, 127–134 CrossRef PubMed; (c) H.-I. Kataoka, Int. J. Mol. Sci., 2016, 17, 1306–1322 CrossRef PubMed.
- S. Liu, J. Mao, T. Wang and X. Fu, Mol. Neurobiol., 2017, 54, 6426–6435 CrossRef CAS PubMed.
- (a) A.-S. Verkman, Respir. Physiol. Neurobiol., 2007, 159, 324–330 CrossRef CAS PubMed; (b) A. Zannetti, G. Benga, A. Brunetti, F. Napolitano, L. Avallone and A. Pelagalli, Cells, 2020, 9, 2678–2695 CrossRef CAS PubMed.
- (a) Z. Borok and A.-S. Verkman, J. Appl. Physiol., 2002, 93, 2199–2206 CrossRef CAS PubMed; (b) R. Meli, C. Pirozzi and A. Pelagalli, Front. Physiol., 2018, 9, 101–112 CrossRef PubMed; (c) D. Vecchio, C. Solaro, E. Virgilio, P. Naldi, R. Bottero, F. Masuccio, M. Capabianco and R. Cantello, Sclerosis, 2023, 1, 1–4 CrossRef.
- (a) E. Yadav, N. Yadav, A. Hus and J.-S. Yadav, Respir. Med., 2020, 174, 106193–106210 CrossRef PubMed; (b) I.-V. da Silva and G. Soveral, Int. J. Mol. Sci., 2021, 22, 1845–1860 CrossRef CAS PubMed.
- C. Guo, T. Wu, H. Zhu and L. Gao, Inflammation, 2019, 42, 1401–1412 CrossRef CAS PubMed.
- M.-M. Salman, P. Kitchen, A.-J. Yool and R.-M. Bill, Trends Pharmacol. Sci., 2022, 43, 30–42 CrossRef CAS PubMed.
- L. Tradtrantip, B.-J. Jin, X. Yao, M.-O. Anderson and A.-S. Verkman, Adv. Exp. Med. Biol., 2017, 967, 239–250 CrossRef PubMed.
- A.-S. Verkman, A.-J. Smith, P.-W. Phuan, L. Tradtrantip and M.-O. Anderson, Expert Opin. Ther. Targets, 2017, 21, 1161–1170 CrossRef CAS PubMed.
- F.-J.-M. Detmers, B.-L. de-Groot, E.-M. Muller, A. Hinton, I.-B.-M. Konings, M. Sze, S.-L. Filtsch, H. Grubmuller and P.-M.-T. Deen, J. Biol. Chem., 2006, 281, 14207–14214 CrossRef CAS PubMed.
- M.-A. Awan, P. Kitchen, M.-M. Salman, M.-T. Conner, A.-C. Conner and R.-M. Bill, Int. J. Mol. Sci., 2009, 20, 1589–1608 CrossRef PubMed.
- V.-J. Huber, M. Tsujita, M. Yamazaki, K. Sakimura and T. Nakada, Bioorg. Med. Chem. Lett., 2007, 17, 1270–1273 CrossRef CAS PubMed.
- I. Pirici, T.-A. Balsanu, C. Bogdan, C. Margaritescu, T. Divan, V. Vitalie, L. Magoanta, D. Pirici, R.-O. Carare and D.-F. Muresanu, Int. J. Mol. Sci., 2018, 19, 46–64 CrossRef PubMed.
- N. Azh, F. Barzkar, N.-M. Gorji, P.-P. Talatappeh, Y. Moradi, R.-V. Azad, M. Ranjbar and H.-R. Baradaran, Pharmacol. Res. Perspect., 2022, 10, 925–956 Search PubMed.
- (a) J.-T. Mhlongo, E. Brasil, B.-G. Torre and F. Albericio, Mar. Drugs, 2020, 18, 203–228 CrossRef CAS PubMed; (b) Z. Jin, Nat. Prod. Rep., 2011, 28, 1143–1191 RSC.
- (a) H.-Z. Zhang, Z.-L. Zhao and C.-H. Zhou, Eur. J. Med. Chem., 2018, 144, 444–492 CrossRef CAS PubMed; (b) R. Kuand, H.-J. Shue, D.-J. Blythin, N.-Y. Shih, D. Gu, X. Chen, J. Schwerdt, L. Lin, P.-C. Ting, X. Zhu, R. Aslanian, J.-J. Piwinski, L. Xiao, D. Prelusky, P. Wu, J. Zhang, X. Zhang, C.-S. Celly, M. Minnicozzi, M. Billah and P. Wang, Bioorg. Med. Chem. Lett., 2007, 17, 5150–5154 CrossRef PubMed; (c) H.-R. Chobanian, Y. Guo, P. Liu, M.-D. Chioda, S. Fung, T.-J. Lanza, L. Chang, R.-K. Bakshi, J.-P. Dellureficio, Q. Hong, M. McLaughlin, K.-M. Belyk, S.-W. Krska, A.-K. Makarewicz, E.-J. Martel, J.-F. Leone, L. Frey, B. Karanam, M. Maderia, R. Alvaro, J. Shuman, G. Salituro, J.-L. Terebetski, N. Jochnowitz, S. Mistry, E. McGowan, R. Hajdu, M. Rosenbach, C. Abbadie, J.-P. Alexander, L.-L. Shiao, K.-M. Sullivan, R.-P. Nargund, M.-J. Wyvratt, L.-S. Lin and R.-J. DeVita, ACS Med. Chem. Lett., 2014, 5, 717–721 CrossRef CAS PubMed; (d) D. Zhang, J. Guo, M. Zhang, X. Liu, M. Ba, X. Tao, L. Yu, Y. Guo and J. Dai, J. Nat. Prod., 2017, 80, 3241–3246 CrossRef CAS PubMed; (e) N.-C. Desai, D.-V. Vaja, S.-B. Joshi and V.-M. Khedkar, Res. Chem. Intermed., 2021, 47, 573–587 CrossRef CAS; (f) N.-Y. Pepinosa, M.-G. Trujillo, S.-C. Castano, L.-A. Veloza and J.-C. Arias, Biomed. Pharmacother., 2021, 138, 111495–111510 CrossRef PubMed; (g) H. Ahmad, S. Ullah, F. Rahman, A. Saeed, J. Pelletier, J. Sevigny, A. Hassan and J. Iqbal, Eur. J. Med. Chem., 2020, 208, 112759–112768 CrossRef CAS PubMed; (h) C. Maria, L. Gluseppe, C. Ugo, G. Salvatore, R. Roberto, I. Daniela and L. Laura, Curr. Med. Chem., 2019, 26, 7337–7371 Search PubMed.
- (a) T. Selvi and K. Srinivasan, J. Org. Chem., 2014, 79, 3653–3658 CrossRef CAS PubMed; (b) T. Selvi and K. Srinivasan, Chem. Commun., 2014, 50, 10845–10848 RSC; (c) M. Meenakshi and K. Srinivasan, Org. Biomol. Chem., 2022, 20, 8741–8746 RSC.
- M. Meenakshi, P. Antojenifer, M. Karthikeyan, C. Prahalathan and K. Srinivasan, J. Heterocycl. Chem., 2022, 59, 351–358 CrossRef CAS.
- (a) L.-F. M. Antony, A. Kannan, A. Panneerselvam, C. Loganathan, E.-M. Shankar, K. Anbarasu and C. Prahalathan, Inflammation, 2020, 43, 1599–1609 CrossRef PubMed; (b) L.-F. M. Antony, A. Kannan, A. Panneerselvam, C. Loganathan, K. Anbarasu and C. Prahalathan, Med. Hypotheses, 2020, 143, 110201–110206 CrossRef PubMed.
- S. Liu, J. Mao, T. Wang and X. Fu, Mol. Neurobiol., 2017, 54, 6426–6435 CrossRef CAS PubMed.
- L. Li, H. Zhang, M.-V. Doyer, S.-S. Zamvil and A.-S. Verkman, FASEB J., 2011, 25, 1556–1566 CrossRef CAS PubMed.
- W. Dai, J. Yan, G. Chen, G. Hu, X. Zhou and X. Zeng, Int. J. Mol. Med., 2018, 42, 1716–1722 CAS.
- F. Zhao, J. Deng, X. Xu, F. Cao, K. Lu, D. Li, X. Cheng, X. Wang and Y. Zhao, J. Neuroinflammation, 2018, 15, 157–170 CrossRef PubMed.
- D.-K. Binder, K. Oshio, T. Ma, A.-S. Verkman and G. Manley, NeuroReport, 2004, 15, 259–262 CrossRef CAS PubMed.
- Y. Dunder, S. Unlu, E. Banoglu, A. Entrena, G. Costantino, M.-T. Nunez, F. Ledo, M.-F. Sahin and N. Noyanalpan, Eur. J. Med. Chem., 2009, 44, 1830–1837 CrossRef PubMed.
- H.-R. Chobanian, Y. Guo, P. Liu, M.-D. Chioda, S. Fung, T.-J. Lanza, L. Chang, R.-K. Bakshi, J.-P. Dellureficio, Q. Hong, M. McLaughlin, K.-M. Belyk, S.-W. Krska, A.-K. Makarewicz, E.-J. Martel, J.-F. Leone, L. Frey, B. Karanam, M. Madeira, R. Alvaro, J. Shuman, G. Salituro, J.-L. Terebetski, N. Jochnowitz, S. Mistry, E. McGowan, R. Hajdu, M. Rosenbach, C. Abbadie, J.-P. Alexander, L.-L. Shiao, K.-M. Sullivian, R.-P. Nargund, M.-J. Wyvratt, L.-S. Lin and R.-J. DeVita, ACS Med. Chem. Lett., 2014, 5, 717–721 CrossRef CAS PubMed.
- K. Seth, S.-K. Garg, R. Kumar, P. Purohit, V.-S. Meena, R. Goyal, U.-C. Banerjee and A.-K. Chakraborti, ACS Med. Chem. Lett., 2014, 5, 512–516 CrossRef CAS PubMed.
- L. Loganathan, K. Muthusamy, J.-M. Jayaraj, A. Kajamaideen and J.-J. Balthasar, J. Biomol. Struct. Dyn., 2018, 37, 3637–3648 CrossRef.
- Q. Xue, X. Liu, P. Russell, J. Li, W. Pan, J. Fu and A. Zhang, Ecotoxicol. Environ. Saf., 2022, 233, 113323 CrossRef CAS PubMed.
- A. Markou, L. Unger, M. Abir-Awan, A. Saadallah, A. Halsey, Z. Balklava, M. Conner, S. T Horsefield, S.-D. Greenhill, A. Conner and R.-M. Bill, Biochim. Biophys. Acta, Biomembr., 2022, 1864, 183853 CrossRef CAS.
- Y. Abe and M. Yasui, Biomolecules, 2022, 12, 591 CrossRef CAS PubMed.
- Schrödinger Release 2020-1, qikprop Schrödinger, LLC, New York, NY, 2020 Search PubMed.
- J.-M. Jayaraj, M. Jothimani, C.-P. Palanisamy, O.-T. Pentikäinen, M. Pannipara, A.-G. Al-Sehemi, K. Muthusamy and K. Gopinath, Bioinorg. Chem. Appl., 2022, 8635054 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Copies of 1H and 13C NMR spectra of all products. See DOI: https://doi.org/10.1039/d3ra03989g |
| This journal is © The Royal Society of Chemistry 2023 |