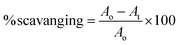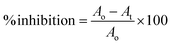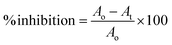Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Metabolomics and molecular networking approach for exploring the anti-diabetic activity of medicinal plants†
Arjun Prasad Timilsina‡
a,
Bimal Kumar Raut‡a,
Chen Huo‡b,
Karan Khadayata,
Prakriti Budhathokia,
Mandira Ghimirea,
Rabin Budhathokia,
Niraj Aryalc,
Ki Hyun Kim *b and
Niranjan Parajuli
*b and
Niranjan Parajuli *a
*a
aBiological Chemistry Lab, Central Department of Chemistry, Tribhuvan University, Kirtipur, Kathmandu, 44618, Nepal. E-mail: niranjan.parajuli@cdc.tu.edu.np; Tel: +977-1-4332034
bSchool of Pharmacy, Sungkyunkwan University, Suwon 16419, Republic of Korea. E-mail: khkim83@skku.edu; Tel: +82-31-290-7700
cDepartment of Biology, University of Florida, Gainesville, FL 32611, USA
First published on 19th October 2023
Abstract
Metabolomics and molecular networking approaches have expanded rapidly in the field of biological sciences and involve the systematic identification, visualization, and high-throughput characterization of bioactive metabolites in natural products using sophisticated mass spectrometry-based techniques. The popularity of natural products in pharmaceutical therapies has been influenced by medicinal plants with a long history of ethnobotany and a vast collection of bioactive compounds. Here, we selected four medicinal plants Cleistocalyx operculatus, Terminalia chebula, Ficus lacor, and Ficus semicordata, the biochemical characteristics of which remain unclear owing to the inherent complexity of their plant metabolites. In this study, we aimed to evaluate the potential of these aforementioned plant extracts in inhibiting the enzymatic activity of α-amylase and α-glucosidase, respectively, followed by the annotation of secondary metabolites. The methanol extract of Ficus semicordata exhibited the highest α-amylase inhibition with an IC50 of 46.8 ± 1.8 μg mL−1, whereas the water fraction of Terminalia chebula fruits demonstrated the most significant α-glucosidase inhibition with an IC50 value of 1.07 ± 0.01 μg mL−1. The metabolic profiling of plant extracts was analyzed through Liquid Chromatography-Mass Spectrometry (LC-HRMS) of the active fractions, resulting in the annotation of 32 secondary metabolites. Furthermore, we applied the Global Natural Product Social Molecular Networking (GNPS) platform to evaluate the MS/MS data of Terminalia chebula (bark), revealing that there were 205 and 160 individual ion species observed as nodes in the methanol and ethyl acetate fractions, respectively. Twenty-two metabolites were tentatively identified from the network map, of which 11 compounds were unidentified during manual annotation.
Introduction
Despite considerable advancements in synthetic chemistry, the search for new and innovative pharmaceuticals is perpetual, and nature continues to be a crucial source of potential new medicines. Recent instances of natural product drug development have highlighted the use of genomic and metabolomics methodologies to supplement conventional methods of natural product research. Medicinal plants contain a range of bioactive metabolites with therapeutic properties. Therefore, robust techniques and technological advancements have been used to explore phytochemistry.The field of metabolomics has been rapidly growing over the past two decades owing to its ability to provide information on dozens to hundreds of metabolites in a single experiment. It includes the identification and quantification of endogenous and exogenous metabolites present in biological samples using highly sensitive analytical tools and bioinformatics.1 Significant advancements in metabolomics have enhanced its applicability to biomarker investigations, toxicological analysis, drug discovery, and natural product chemistry.2 The metabolic data acquired in plant metabolomics using single analytical tools are often insufficient and incomplete, which has led the scientific community to utilize MS-based techniques such as GC-MS and LC-MS.3 Complex MS/MS data acquired in metabolomics experiments can be visualized and analyzed using a computer-based approach, such as molecular networking. The Global Natural Product Social Molecular Networking (GNPS) is an online bioinformatics platform currently being utilized in research to perform molecular networking. It tends to detect possible similarities among all MS/MS spectra in the dataset and extend annotation to unknown but closely related metabolites.4,5
Diabetes is an unassailable burden in low and high-income countries.6 Managing diabetes remains a significant challenge for the scientific community since individuals with this condition are vulnerable to various health complications resulting in the annual mortality of 1.5 million people.7 Consequently, promoting research on medicinal plants for the identification of new alternative therapeutics for diabetes is currently relevant due to favorable outcomes associated with plant-based therapeutics in terms of long-term safety, mode of action, and metabolic activity.
Based on their ethnobotanical pharmaceutical properties, four medicinal plants, namely C. operculatus, T. chebula, F. lacor, and F. semicordata, were selected in this study. Although these plants have a wide range of pharmacological applications, this study intended to investigate the applicability of these species and their related metabolites as anti-diabetic drugs. The efficacy and toxicity of herbal formulations must be thoroughly explored even though they are prepared and consumed locally. Finding viable drug candidates from crude extracts is more challenging due to the complexity of biological samples.8
Various classes of drugs like insulin, α-glucosidase inhibitors (acarbose, miglitol, voglibose), biguanides (metformin, phenformin), thiazolidinediones, sulfonylureas, dipeptidyl-peptidase-4 inhibitors, meglitinides, dopamine agonists have currently being used to treat diabetes. However, these are reported to have several side effects such as allergic reactions, increased risk of bladder cancer, hypoglycemia, bloating, flatulence, etc.9 As a result, the issue demands the development of new potent drugs with no or minimal human side effects.
The purpose of this research was to explore secondary metabolites using a mass spectrometry-based metabolomics approach with the GNPS platform and to evaluate the possible enzyme inhibitory activity of C. operculatus, T. chebula, F. semicordata, and F. lacor in targeting diabetes and its complications.
Materials and methods
Chemicals
Methanol, ethanol, ethyl acetate, dichloromethane, and hexane were purchased from Thermo Fisher Scientific (India). Gallic acid and quercetin were procured from HiMedia, an Indian supplier. α-Glucosidase (Saccharomyces cerevisiae), 4-nitrophenyl-α-D-glucopyranoside (pNPG), pancreatic porcine α-amylase (PPA), 2-chloro-4-nitrophenyl-α-D-maltotrioside (CNPG3), and acarbose were ordered from Sigma-Aldrich (Germany).Medicinal plants
Medicinal plants were collected from four distinct locations in Nepal, and the herbarium of each plant was submitted to the Central Herbarium of the Central Botany Department at Tribhuvan University, where voucher specimens were subsequently registered (Table 1). The entire collection of plant samples was shade-dried and pulverized into powder.| Voucher specimen | Medicinal plant | Family | Ethnobotanical uses | Chemical constituents |
|---|---|---|---|---|
| TUCH 210018 | Terminalia chebula | Combretaceae | It is used for antidiabetic, anticancer, cardioprotective, neuroprotective, anti-inflammatory, and antiarthritic activities.10 It is also used to treat dyspepsia, piles, hepatosplenomegaly, irritable bowel syndrome, and heart failure.11 | Gallic acid, digallic acid, ellagic acid, caffeic acid, pyragallol, rutin, quercetin, isoquercetin, chebulagic acid, chebulinic acid, chebulic acid, eugenol, terflavin A, terchebulin10 |
| TUCH 210017 | Cleistocalyx operculatus | Myrtaceae | It is used for its cytotoxic,12 antitumor,13 antihyperglycemic,14 cardiotonic,15 and anti-inflammatory16 activities | 2′,4′-Dihydroxy-6′-methoxy-3′,5′-dimethylchalcone and ursolic acid, 5,7,8,4′-tetrahydroxy-3′-5′-dimethoxyflavone-3-O-β-D-galactopyranoside, gossypetin-8, 3′-dimethyl ether-3-O-β-D-galactoside, myricetin-3′-methyl ether-3-O-β-D-galactopyranoside, myricetin-3′-methyl ether, quercetin, kaempferol16 |
| TUCH 201022 | Ficus lacor | Moraceae | The powdered ripe fruit is utilized as an anti-diabetic.17 It is also used for wound healing18 ulcers,19 hay fever, dysentery, and stomach disorder.18–21 | β-Sitosterol, lupeol, α-amyrin, β-amyrin, stigmasterol, campesterol, scutellarein glucoside, scutellarein, infection, sorbifolin, bergaptol, and bergapten22,23 |
| TUCH 201020 | Ficus semicordata | Moraceae | For headaches and diarrhea, raw fruit juice and fruits are employed. Fruits are also used for visceral ulcers, jaundice, colic pain, and hepatitis. Externally, the juice of the leaves is useful to treat scabies. It has been shown to have antioxidant, antibacterial, and anticancer properties.24 | Catechin, rutin, quercetin, dodecane, indole, linalool, gallic acid and germacrene25 |
Methanolic extracts and fractionation
Methanolic extracts of four plants (Cleistocalyx operculatus, Terminalia chebula, Ficus lacor, and Ficus semicordata) were prepared using the cold percolation method, which involves the absorption of the powder for a day in methanol before percolation. The same procedure was repeated continuously for 3 days, and the collected solvent was dried at temperatures below 50 °C in the rotary evaporator. After 50 g of methanolic extracts were dissolved in distilled water for the fractionation process, the extracts were separated using the following solvents: hexane, DCM, and ethyl acetate in order of polarity to obtain four fractions from each plant. This procedure was performed three times for each solvent.26Determination of polyphenolic and flavonoid contents
Total polyphenol and flavonoid contents (TPC and TFC) were quantified using standard Folin–Ciocalteu and aluminum trichloride methods.27–29 Both reactions were performed in 200 μL final volume, and absorbance was measured using a microplate reader (SynergyLX, BioTek, Instruments, Inc., USA). Calibration curves were generated by using various concentrations of gallic acid and quercetin. The extract concentrations were calculated and expressed as gallic acid and quercetin equivalents (mg GAE per g and mg QE per g, respectively).Antioxidant assay
The antioxidant assay was performed as described previously in literature.30 The reaction was performed using an equal volume of plant extracts with 0.1 mM DPPH to maintain a final volume of 200 μL. After 15 min of incubation at room temperature in the dark, the absorbance was recorded at 517 nm. The percentage scavenging activity was determined using the following formula:where Ao = optical density of the negative control (50% DMSO), and At = optical density of the test or reference sample.
Assay of α-glucosidase inhibition
The α-glucosidase inhibition was performed by dissolving the enzyme and substrate in a 50 mM phosphate buffer (pH 6.8). The process was initiated by mixing 20 μL of plant extracts of varying concentrations with 20 μL enzyme (0.2 units) along with 120 μL phosphate buffer followed by 10 min incubation at 37 °C. After incubation, pNPG substrate was added (0.7 mM) and incubated for 15 min at the same temperature.31 The absorbance of the released p-nitrophenyl was measured at 405 nm using Synergy LX (BioTek Instruments, Inc., USA). Each test was performed in triplicate. The following formula was used to calculate the % α-glucosidase inhibitory activitywhere Ao is the optical density of the negative control with 30% DMSO and At is the optical density of the test/positive control.
Assay of α-amylase inhibition
The reaction was performed by dissolving the enzyme and substrate in 50 mM phosphate-buffered saline (pH; 0.9% NaCl, pH 7.0). The assay was inducted by mixing 20 μL plant extracts at different concentrations and adding 80 μL PPA (1.5 units per mL) followed by 10 min incubation at 37 °C. After incubation, CNPG3 (0.5 mM) was added and the cells were incubated for 15 min.26 The absorbance was measured at 405 nm using a microplate reader (Synergy LX, BioTek Instruments, Inc., USA). The percentage of inhibition was determined using the following formula:where Ao is the optical density of the negative control with 30% DMSO and At is the optical density of the test/positive control.
Liquid chromatography-mass spectrometry analysis
LC-HRMS of the ethyl acetate and water fractions of T. chebula fruits, C. operculatus, and F. lacor, F. semicordata were analyzed at the Sophisticated Analytical Instrument Facility (SAIF), CSIR-Central Drug Research Institute, Lucknow (Lot No. 1607642300 and 1606747677). Compounds were identified using an Agilent 1200 series HPLC system (Agilent Technologies, Santa Clara, CA, USA) equipped with an Agilent 6520 I Accurate-Mass Q-TOF LC/MS system (Agilent Technologies, Santa Clara, CA, USA). An Agilent 6520 Accurate-Mass Q-TOF Mass Spectrometer equipped with a G1311A quaternary pump, G1329A autosampler, and G1315D diode array detector (DAD) was used for further analysis. The elution solvent comprised a mixture of acetonitrile, 5 mM acetate buffer, and water flowing at 1.5 mL min−1, following a gradient from 5% to 30% acetonitrile over 10 minutes, reaching 80% acetonitrile over 32 minutes, and returning to the initial conditions, with the column temperature consistently held at 30 °C throughout the process. For positive mode analysis of electrospray ionization, the source gas temperature was maintained at 30 °C, with a gas flow rate of 11.01 L min−1, and a nebulizer pressure of 40 psi. VCap voltage was set to 3500 V, fragmentor at 175 V, skimmer at 65.0 V, and Octapole RF peak at 750 V. The MS data acquisition was done in the range of 100–2000 Da at a scan rate of 1.03 for T. chebula (fruit), C. operculatus, F. lacor, and F. semichordata.32 Moreover, the molecular annotation of the methanolic extract (AT1) and ethyl acetate fraction (AT2) of T. chebula bark was completed in SIRIUS 5.6.3, using the CSI: FingerID interface, where the efficiency of the predicted molecular formula, functional group, and fragmentation pattern was evaluated in terms of the Sirius score.33HRMS and GNPS-based metabolomic analysis
Samples of AT1 and AT2 were prepared for MS/MS analysis by dissolving them in HPLC-grade (1 mg mL−1), and then 150 μL volume of each sample was transferred to an HPLC autosampler vial. Metabolomic profiling of the two samples was performed using an Agilent G6545B quadrupole time-of-flight (Q-TOF) mass spectrometer (Agilent Technologies, Santa Clara, CA, USA) equipped with a heated electrospray ion source (HESI). For chromatographic separation, an Acquity® UPLC BEH reverse-phase column C18 (150 mm × 2.1 mm, 1.7 μM) was used. The mobile phase was acidified with 0.1% formic acid and comprised of H2O (A) and acetonitrile (B). The composition of organic solvent was 5% at 0.00–2.00 min, 20% at 5.00 min, 100% at 20.00 min, and back again to 5% at 23.00–25.00 min. The injection volume of two samples was kept at 3 μL and a flow rate of 0.5 mL min−1 was maintained. MS/MS analysis was performed using electrospray ionization (ESI) in positive ion mode. Spectral hits were performed using a modified version of the method reported by Bashir et al. (2021)34 with an m/z range of 50–1200, collision energies of 15 eV and 40 eV, and a full width at half maximum (FWHM) of 3000. The RAW files were converted to open-source ‘.mzXML’ file format using the MSConvert tool of ProteoWizard MSConvert Version 3 software. The files were uploaded to the GNPS platform using WinSCP, the recommended FTP client. The acquired MS2 data were visualized using a GNPS-based visualization following an established procedure (accessed on January 17, 2023). The molecular networks generated were further exported from GNPS to Cytoscape in ‘.graphml’ format to enable customized visualization.Data analysis
The collected data were processed using Gen5 Microplate Software, followed by MS Excel. GraphPad Prism Software version 8 was used for IC50 calculation and data were represented as a mean ± standard error of the mean of triplicate. The Mnova software ver. 12.0 (Mestrelab Research, Santiago de Compostela, Spain) was used to process the raw data files obtained from the HPLC-QTOF-MS.Results
Estimation of polyphenol contents and antioxidant potential
Phytochemical and antioxidant assays of the medicinal plants were performed in triplicate using methanolic extracts. The total phenol, flavonoid, and antioxidant activities of the plants are shown in Table 2. T. chebula (Bark and Fruit) showed the highest TPC (164.534 ± 3.70 mg GAE per g) and TFC values (158.67 ± 14.68 mg QE per g), respectively. Similarly, T. chebula (bark) showed the lowest IC50 value of 5.144 ± 0.06 μg mL−1 for DPPH radical scavenging activity as compared with the control candidate, quercetin (6.3 ± 1.0 μg mL−1).| Sample | TPC (mg GAE per g) | TFC (mg QE per g) | Antioxidant IC50 values (μg mL−1) |
|---|---|---|---|
| Terminalia chebula (fruit) | 161.56 ± 0.39 | 158.67 ± 14.68 | 34.82 ± 1.12 |
| Terminalia chebula (bark) | 164.53 ± 3.70 | 115.34 ± 4.07 | 5.144 ± 0.06 |
| Cleistocalyx operculatus | 154.26 ± 1.01 | 69.17 ± 0.73 | 30.34 ± 1.12 |
| Ficus lacor | 108.67 ± 6.37 | 131.16 ± 7.75 | 130 ± 1.02 |
| Ficus semicordata | 107.76 ± 3.21 | 77.18 ± 0.73 | 22.76 ± 0.19 |
| Quercetin (control) | — | — | 6.3 ± 1.00 |
Inhibition of diabetic targets α-amylase and α-glucosidase
Preliminary screening was performed using 500 μg mL−1 of the extracts to determine the percentage inhibition of α-glucosidase and α-amylase, respectively. Only fractions with more than 50% inhibition were used to determine the IC50 values. As compared to the positive control, acarbose (6.1 ± 0.10 μg mL−1), and the lowest IC50 value towards α-amylase inhibition was found for F. semicordata in methanolic extract (46.8 ± 1.8 μg mL−1). Similarly, the lowest IC50 value for α-glucosidase inhibition was observed for T. chebula (fruits) in aqueous fraction (1.07 ± 0.01 μg mL−1) and for C. operculatus in methanolic extract (1.26 ± 0.03 μg mL−1). The results of α-glucosidase and α-amylase inhibition by different plant fractions are shown in Table 3.| Medicinal plants | Minimum inhibitory concentration, IC50 (μg mL−1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Methanol ext. | Hexane fr. | Dichloromethane fr. | Ethyl acetate fr. | Water fr. | ||||||
| A | G | A | G | A | G | A | G | A | G | |
| a Extract: ext. Fraction: fr. | ||||||||||
| Terminalia chebula (fruit) | 153.2 ± 1.91 | 95.37 ± 0.30 | <50% | 236.06 ± 4.78 | <50% | <50% | 95.5 ± 1.5 | 12.72 ± 0.40 | 309.1 ± 0.9 | 1.07 ± 0.01 |
| Terminalia chebula (bark) | 180.4 ± 10.22 | 8.251 ± 1.08 | 167.5 ± 9.47 | <50% | 141.2 ± 1.43 | <50% | 140.6 ± 2.61 | 6.633 ± 1.82 | 219.2 ± 3.26 | 31.96 ± 0.74 |
| Cleistocalyx operculatus | 62.49 ± 1.89 | 1.26 ± 0.03 | 414.9 ± 7.8 | 80.31 ± 1.22 | 429.4 ± 5.2 | 95.19 ± 0.75 | 100.8 ± 1.9 | 8.67 ± 0.55 | 53.2 ± 1.0 | 4.91 ± 0.02 |
| Ficus lacor | 485.6 ± 1.8 | 33.78 ± 3.6 | <50% | 76.57 ± 0.5 | 355.2 ± 11.2 | <50% | 240.1 ± 3.4 | 94.2 ± 8.1 | <50% | 76.6 ± 9.7 |
| Ficus semicordata | 46.8 ± 1.8 | 46.3 ± 3.07 | <50% | <50% | <50% | <50% | 241.7 ± 4.3 | 34.7 ± 1.1 | <50% | 5.0 ± 0.2 |
| Acarbose | 6.1 ± 0.10 (α-amylase) | 344.23 ± 1.03 (α-glucosidase) | ||||||||
Liquid chromatography and molecular annotation
Compound identification and characterization were based on the comparison of molecular masses, retention times, and MS spectra via authentic databases (PubChem, and Dictionary of Natural Products), as well as relevant literature. Because of the potent anti-diabetic activity exhibited by the ethyl acetate and water fractions of the four plants, the ethyl acetate and water fractions were selected for further molecular annotation analysis. Thirty-two compounds were identified from the ethyl acetate and water fraction of plant species, and the mass spectra of these compounds showed peaks at m/z 357.0455 (ESI Fig. S1†), 371.0619 (Fig. S2), 303.0140 (Fig. S3), 619.0970 (Fig. S4), 385.0788 (Fig. S5), 667.4054 (Fig. S6), 657.3627 (Fig. S7), 471.0197 (Fig. S8), 323.0762 (Fig. S9), 489.3570 (Fig. S10), 519.3313 (Fig. S11), 505.3529 (Fig. S12), 443.0978 (Fig. S13), 563.1549 (Fig. S14), 503.3363 (Fig. S15), 273.0759 (Fig. S16), 307.0815 (Fig. S17), 331.0463 (Fig. S18), 465.1023 (Fig. S19), 291.0859 (Fig. S20), 579.1504 (Fig. S21), 435.0578 (Fig. S22), 193.0705 (Fig. S23), 487.22 (Fig. S24), 467.0872 (Fig. S25), 649.1082 (Fig. S26), 393.0435 (Fig. S27), 279.1601 (Fig. S28), 387.1817 (Fig. S29), 293.1736 (Fig. S30), 331.2842 (Fig. S31), and 463.3794 (Fig. S32†) as protonated molecular ions [M + H]+ as shown in Table 4. In ESI figures, ESI (+) indicates data collected in the positive mode of electrospray ionization mass spectrometry, centroid TIC and centroid MS indicate the data are presented in centroid mode only where in centroid mode each ion's mass and intensity are presented as discrete values, while profile mode indicates the entire distribution of intensity of ions recorded at each m/z value. Therefore, centroid MS data offers simpler spectra and helps in the interpretation of spectra. The annotated compounds and their fragment's peak were compared with previous findings as listed in Table 4. The structures of annotated molecules were drawn by using http://www.chemspider.com/. The structures of the annotated secondary metabolites are shown in Fig. 1.| Purposed compound | Calculated mass | Observed mass | Formula | RDBE | Absolute error (ppm) | (Rt) Min | Fragment peak | Solvent fraction and plant species | CSI: Finger ID (score%) |
|---|---|---|---|---|---|---|---|---|---|
| a EA: ethyl acetate, RDBE: ring double bond equivalents. | |||||||||
| Chebulic acid | 356.0374 | 357.0455 | C14H12O11 | 9 | 0.77 | 2.84 | 339, 321, 293, 203 | EA T. chebula (fruits) | — |
| 2-O-Caffeoyl hydroxy citric acid | 370.0530 | 371.0619 | C15H14O11 | 9 | 2.88 | 2.93 | 191, 163, 145 | EA T. chebula (fruits) | — |
| Ellagic acid | 302.0057 | 303.0140 | C14H6O8 | 12 | 1.52 | 8.98 | 193 | EA T. chebula (fruits) | — |
| 302.0057 | 303.0143 | C14H6O8 | 12 | 2.60 | 8.70 | — | Water C. operculatus | — | |
| 302.0057 | 303.0134 | C14H6O8 | 12 | 0.31 | 8.62 | 201.01, 229.01, 247.02, 201.01, 173.02, 145.02 | T. chebula (bark) | 98.05 | |
| Trigalloyllevoglucosan IX | 618.0910 | 619.0970 | C20H26O22 | 8 | 2.83 | 10.62 | 153 | EA T. chebula (fruits) | — |
| 2-O-Feruloylhydroxycitric acid | 384.0690 | 385.0788 | C16H16O11 | 9 | 4.39 | 4.58 | 385, 209, 195 | EA T. chebula (fruits) | — |
| Arjunglucoside I | 666.3973 | 667.4054 | C36H58O11 | 8 | 0.46 | 10.06 | 527.33, 483.34 | T. chebula (bark) | 99.41 |
| 23-Galloylterminolic acid | 656.3555 | 657.3627 | C37H52O10 | 12 | 0.94 | 10.33 | 639.35, 469.32, 451.31, 405.31, 187.14 | T. chebula (bark) | 55.46 |
| Flavogallonic acid | 470.0115 | 471.0197 | C21H10O13 | 17 | 0.73 | 5.68 | 453.09, 379.00, 363.01 | T. chebula (bark) | 72.99 |
| Leucodelphidin | 322.0683 | 323.0762 | C15H14O8 | 9 | 0.24 | 6.85 | 305.06, 139.03, 217.04, 263.05, 163.03 | T. chebula (bark) | 54.41 |
| Arjunolic acid | 488.3496 | 489.3570 | C30H48O5 | 7 | 0.92 | 11.50 | 471.34, 453.33, 205.15, 135.04 | T. chebula (bark) | 92.54 |
| Bartogenic acid | 518.3238 | 519.3313 | C30H46O7 | 8 | 0.47 | 11.18 | 501.32, 215.14, 119.08, 187.14, 437.30 | T. chebula (bark) | 90.33 |
| Arjungenin | 504.3445 | 505.3529 | C30H48O6 | 7 | 1.10 | 10.06 | 469.33, 451.32, 297.17, 187.14, 173.13, 405.31, 423.32 | T. chebula (bark) | 95.0 |
| (−)-Epicatechin-3-O-gallate | 442.0894 | 443.0978 | C22H18O10 | 14 | 1.41 | 8.95 | 139.03, 123.04, 153.01, 273.07, 11.04 | T. chebula (bark) | 99.31 |
| Gambiriin B1 | 562.1469 | 563.1549 | C30H26O11 | 18 | 0.29 | 9.02 | 437.12, 411.10, 139.03, 123.04, 273.07 | T. chebula (bark) | 93.01 |
| Rotundioic acid | 502.3288 | 503.3363 | C30H46O6 | 8 | 0.77 | 13.40 | 449.30, 421.31, 485.32, 173.13, 119.08, 95.08, 201.16 | T. chebula (bark) | 82.97 |
| Butin | 272.0679 | 273.0759 | C12H22O4 | 10 | 0.77 | 8.95 | 123.04, 77.03, 51.02 | T. chebula (bark) | 83.65 |
| Gallocatechin or epigallocatechin | 306.0734 | 307.0815 | C15H14O7 | 9 | 0.89 | 7.22 | 289, 307, 139, 151 | EA C. operculatus | — |
| 306.0734 | 307.0814 | C15H14O7 | 9 | 0.87 | 6.66 | 289.07, 139.03, 163.03, 181.04, 65.03 | T. chebula (bark) | 100 | |
| 4,4′-Di-O-methylellagic acid | 330.0370 | 331.0463 | C16H10O8 | 12 | 4.61 | 17.48 | 225, 245, 270, 299, 300.9, 316 | EA C. operculatus | — |
| Isoquercitrin | 464.0949 | 465.1023 | C21H20O12 | 12 | 3.31 | 8.80 | 303.04, 137.05 | EA F. lacor | — |
| Catechin | 290.0784 | 291.0859 | C15H14O6 | 9 | 4.30 | 11.56 | 123, 139, 165, 207 | Water F. semicordata | — |
| 290.0784 | 291.0866 | C15H14O6 | 9 | 0.29 | 7.77 | 139.03, 123.04, 207.06, 91.05, 95.05 | T. chebula (bark) | 100 | |
| Procyanidin B2 | 578.1418 | 579.15 | C30H26O12 | 18 | 3.84 | 10.79 | 427.09 | Water F. semicordata | 93.73 |
| 578.1418 | 579.1504 | C30H26O12 | 1.24 | 7.44 | 427.10, 409.09, 139.03, 127.03, 123.04, 163.03, 247.06 | T. chebula (bark) | |||
| Ellagic acid-O-pentoside | 434.0479 | 435.0578 | C19H14O12 | 14 | 4.67 | 7.27 | 303.01 | Water C. operculatus | — |
| Quinic acid | 192.0628 | 193.07 | C7H12O6 | 2 | 3.95 | 3.02 | 175.06, 157.05, 113.02 | EA F. semicordata | — |
| 192.0628 | 193.0705 | C7H12O6 | — | 0.53 | 1.05 | — | Water F. lacor | — | |
| Hydroxyl-6-gingerol-O-β-D-glucuronide | 486.20 | 487.22 | C23H34O11 | 7 | 4.90 | 12.60 | 311.15, 295.11 | Water F. semicordata | — |
| 3-O-Galloylnorbergenin | 466.0700 | 467.0872 | C20H18O13 | 1.37 | 11.18 | 171.03, 53.01 | EA C. operculatus | — | |
| 3,4,5-Tri-O-galloylquinic acid | 648.0915 | 649.1082 | C28H24O18 | 1.76 | 10.17 | 495, 343 | EA C. operculatus | — | |
| Tri-O-methoxyellagic acid | 392.0374 | 393.0435 | C17H12O11 | 4.29 | 3.21 | — | EA C. operculatus | — | |
| Butyl isobutyl phthalate | 278.1512 | 279.1601 | C16H22O4 | 6 | 3.66 | 30.01 | — | Water C. operculatus | — |
| 278.1512 | 279.1603 | C16H22O4 | 6 | 4.72 | 29.94 | — | Water T. chebula (fruits) | — | |
| 278.1512 | 279.1590 | C16H22O4 | 6 | 0.30 | 22.65 | — | Water, EA F. lacor | — | |
| 278.1512 | 279.1592 | C16H22O4 | 6 | 0.40 | 18.64 | — | T. chebula (bark) | 84.29 | |
| Eudesmin | 386.1723 | 387.1817 | C22H26O6 | 10 | 3.98 | 17.45 | — | Water F. lacor | — |
| Lasiodiplodin | 292.1669 | 293.1736 | C17H24O4 | 6 | 3.00, 3.22 | 24.28, 24.01 | — | Water, EA F. lacor | — |
| 1-Monopalmitin | 330.2764 | 331.2842 | C19H38O4 | 1 | 3.54, 0.72 | 27.76, 27.78 | 125.09 | Water, EA F. lacor | — |
| α-Tocospiro B | 462.3703 | 463.3794 | C29H50O4 | 5 | 2.75 | 36.62 | — | Water F. lacor | — |
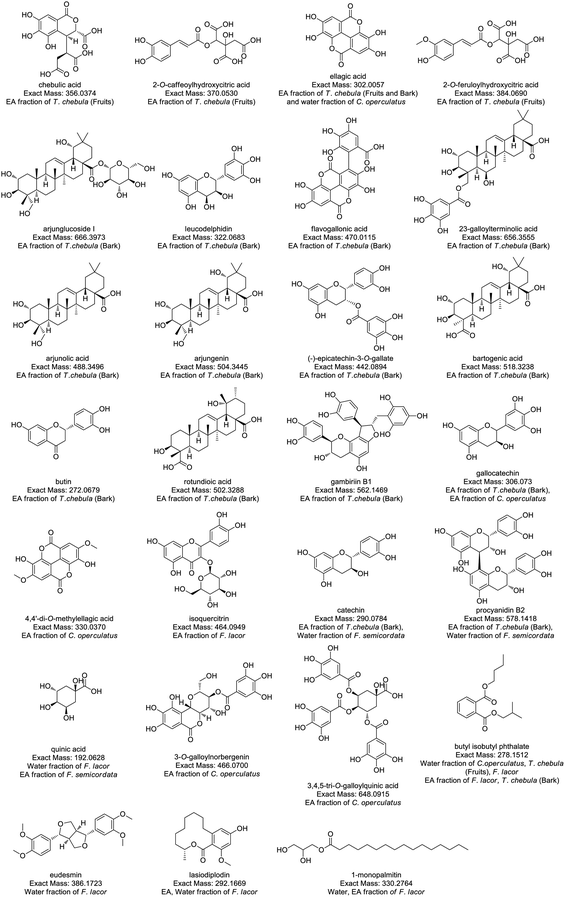 | ||
| Fig. 1 Structure of annotated compounds from T. chebula, F. lacor, C. operculatus, and F. semicordata based on LC-HRMS. | ||
GNPS-based metabolomic profiling
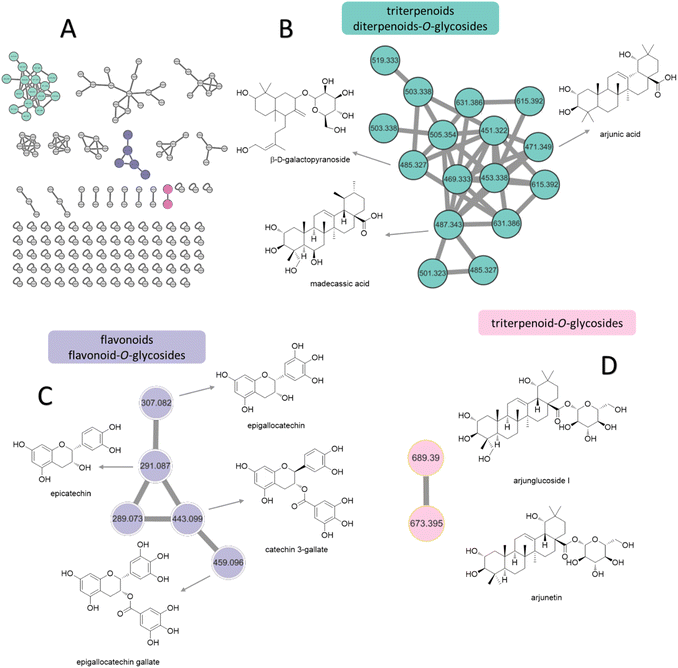
Based on the anti-diabetic activity of the methanol extract and ethyl acetate fraction of the bark of T. chebula, a comprehensive and detailed phytochemical profile was investigated using MS/MS and a GNPS-based metabolomics platform. A total of 205 and 160 individual ion species (MS/MS spectra) were present as nodes in the network map for the methanol extract and the EA fraction, respectively. Moreover, 22 compounds were tentatively identified from T. chebula bark, belonging to various groups of phytochemicals including phenolic compounds, diterpenoids-O-glycosides, triterpenoids, triterpenoid-O-glycosides, flavonoids, flavonoid-O-glycosides, fatty acids, and ceramides (Fig. 2, Table 5).| Methanol (AT1) and EA (AT2) extract of the bark of T. chebula | Fractions | |||||
|---|---|---|---|---|---|---|
| Comp. name | RT | m/z | MS2 fragmentation pattern | Molecular formula | Exact mass | |
| Epigallocatechin | 4.87 | 307.07 | 181.04, 139.03 | C15H14O7 | 306.07 | AT2 |
| Epicatechin | 7.42 | 291.09 | 165.05, 139.04, 123.04 | C15H14O6 | 290.08 | AT1, 2 |
| Catechin | 7.44 | 291.09 | 162.05, 139.04, 123.04 | C15H14O6 | 290.08 | AT1, 2 |
| Procyanidin B2 | 7.50 | 579.15 | 409.09, 287.05, 247.06, 163.04, 127.04 | C30H26O12 | 578.14 | AT1, 2 |
| Arjunglucoside I | 10.47 | 689.39 | 527.33, 185.04, 89.06 | C36H58O11 | 666.40 | AT1, 2 |
| Arjunetin | 11.66 | 673.39 | 511.34, 185.04 | C36H58O10 | 650.40 | AT1, 2 |
| Sesamin | 15.60 | 337.11 | 319.09, 289.08, 267.06, 203.08, 185.06, 135.04 | C20H18O6 | 354.11 | AT1 |
| Epigallocatechin gallate | 7.93 | 459.09 | 139.04 | C22H18O11 | 458.08 | AT2 |
| Quercitrin | 8.34 | 449.09 | 303.04, 85.02 | C21H20O11 | 448.10 | AT1, 2 |
| Ellagic acid | 8.53 | 303.01 | 291.09, 273.07, 153.02 | C14H6O8 | 302.01 | AT1 |
| (−)-Catechin 3-gallate | 8.93 | 443.09 | 273.08, 153.02, 123.04 | C22H18O10 | 442.09 | AT1, 2 |
| Epicatechin gallate | 8.99 | 443.10 | 273.07, 139.04, 123.04 | C22H18O10 | 442.09 | AT1, 2 |
| Arjunic acid | 11.66 | 471.35 | 407.33, 201.16, 107.08 | C30H48O5 | 488.35 | AT2 |
| Madecassic acid | 12.19 | 487.342 | 451.32, 187.15, 145.10, 119.08, 95.08 | C30H48O6 | 504.35 | AT1, 2 |
| β-D-Galactopyranoside | 12.25 | 485.31 | 199.15, 171.12, 145.10, 105.07, 81.07 | C26H44O8 | 484.30 | AT2 |
| Phytosphingosine | 15.38 | 318.30 | 282.28, 60.04 | C18H39NO3 | 317.29 | AT1 |
| Palmitic acid | 17.38 | 402.36 | 283.26 | C22H42O6 | 402.30 | AT1 |
| Dibutyl phthalate | 18.64 | 279.16 | 149.02, 121.03 | C16H22O4 | 278.15 | AT1, 2 |
| Linolenic acid | 20.53 | 279.23 | 109.10, 95.05, 81.07, 67.05 | C18H30O2 | 278.22 | AT1, 2 |
| Oleanolic acid | 20.74 | 439.36 | 249.18, 217.19, 191.18, 95.08 | C30H48O3 | 456.36 | AT1 |
| Dioctyl phthalate | 22.52 | 391.28 | 167.03, 149.02, 71.08, 57.07 | C24H38O4 | 390.28 | AT1 |
| Erucamide | 23.80 | 675.67 | 338.34, 321.31 | C22H43NO | 337.33 | AT1, 2 |
Molecular networking analysis involves representing each metabolite as a node labeled with its m/z value. The metabolites were then grouped into several clusters based on the similarity of their fragmentation patterns, indicating similarities in their core chemical structures.35 From molecular networking and chart of categorized compounds (Fig. 2A), the major clusters included diterpenoid-O-glycosides, triterpenoids (Fig. 2B), flavonoid derivatives (Fig. 2C), and triterpenoid-O-glycosides (Fig. 2D).
The terpenoid clusters, characterized by precursor ions at m/z 471.3449, 485.3109, and 487.3403, were identified as arjunic acid, β-D-galactopyranoside, and madecassic acid, respectively. The flavonoids were annotated for m/z 291.0869 and 307.0736 clusters of epicatechin and epigallocatechin, respectively. Moreover, the neutral loss of 152 Da from both precursor ions at m/z 443.0947 and 459.0964 indicated the presence of gallate substitution, suggesting the presence of catechin gallate and epigallocatechin gallate. Finally, the networking of glycosylated triterpenoids, namely arjunglucoside I and arjunetin, was attributed to the presence of intense product ion signals at m/z 527.3334 and 511.3390, respectively, originating from the aglycone moiety. Additionally, the fragmentation patterns exhibited prominent neutral losses of 162 Da owing to the elimination of glucose. The various constituents observed in the two fractions from T. chebula bark are listed in Table 5 along with their m/z values in positive ion mode, retention times, MS/MS fragmentation patterns, and molecular formulas.
Discussion
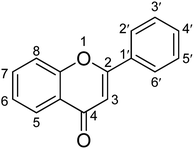
This study aimed to explore the secondary metabolites, TPC, TFC, and antioxidant activities of extracts of C. operculatus, T. chebula, F. semicordata, and F. lacor. In addition, the enzyme inhibitory activities of the different fractions against α-glucosidase and α-amylase were determined and are presented in Tables 2 and 3. The results suggest that methanolic extracts, ethyl acetate, and water fractions showed potent activity against diabetic enzymes, which might be due to the synergistic effects of different chemical constituents. The ethyl acetate fractions exhibit a high flavonoid content, thereby attributing to their role in the inhibition of enzymes.36,37 Previous studies have also suggested that the methanolic, ethyl acetate, and water fractions significantly inhibit the activities of both α-glucosidase and α-amylase, respectively.32High blood glucose levels lead to the formation of free radicals that damage macromolecules within the cell and increase the risk of complications related to hyperglycemia. Thus, compounds with antioxidant properties can aid in the mitigation of diabetes-related problems.38 Phenolic compounds are an important class of secondary metabolites with antioxidants as well as antidiabetic activity. Polyphenols with several hydroxyl groups can act as a source of hydrogen and electron donor to radicals to stabilize them and hence reduce oxidative stress which plays an important role in the mitigation of diabetes.39 The introduction of hydroxyl and methoxyl groups to different positions of flavonoid structures is the most common form in natural products. The patterns of hydroxyl and methoxyl groups on aryl rings A and B of flavonoids were reported to be closely associated with their biological activity towards enzymes. The hydroxylation of flavonoids at positions C3, C6, C3′, and C4′ all exhibited better inhibitory activity on α-glucosidase than the unhydroxylated forms at the corresponding positions.39 The antidiabetic effect of flavonoids also can be increased by replacing the hydroxyl groups at the C3 position with various functional group moieties, such as methoxy, sugar-like rhamnose, galloyl group, and chlorine atom. However, the glycosyl and geranyl moieties at C7 can suppress the antidiabetic effect. The presence of a C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3 double bond, along with the C4-oxo group and aryl ring A, is essential for maintaining the planar structure of flavonoids. Hydrogenation of this double bond weakens the enzyme's inhibitory activity likely because the planar molecular structure significantly influences the binding conformation within the enzyme's active site (Fig. 3).40,41
C3 double bond, along with the C4-oxo group and aryl ring A, is essential for maintaining the planar structure of flavonoids. Hydrogenation of this double bond weakens the enzyme's inhibitory activity likely because the planar molecular structure significantly influences the binding conformation within the enzyme's active site (Fig. 3).40,41
Among four selected medicinal plants, T. chebula (bark) has shown excellent free radical scavenging activity with IC50 values of 5.144 ± 0.06 μg mL−1 corresponding to control quercetin (6.3 ± 1.00 μg mL−1). This may be due to the presence of high TPC and TFC values in T. chebula. Such promising antioxidant potential could be ascribed to ellagic acid (IC50 = 2.012 ± 0.10 μg mL−1), catechin (IC50 = 6.7 μM), epicatechin (IC50 = 6.8 μM),42 arjunglucoside I, quercitrin, flavogallonic acid,43 bartogenic acid and arjunic acid identified in the T. chebula bark extracts.
Since α-glucosidase and α-amylase enzymes are thought to be promising targets for the treatment of diabetes mellitus,44 we assessed the antidiabetic potential of four medicinal plants. The promising inhibitory activity towards α-glucosidase was observed in the water fraction of T. chebula (fruits) with an IC50 value of 1.07 ± 0.01 μg mL−1 followed by methanolic and water fraction of C. operculatus (IC50 = 1.26 ± 0.03 μg mL−1 and IC50 = 4.91 ± 0.02 μg mL−1 respectively). Moreover, the water fraction of F. semicordata (IC50 = 5.0 ± 0.2 μg mL−1) and ethyl acetate fraction of the T. chebula bark (IC50 = 6.633 ± 1.82 μg mL−1) displayed excellent activity as compared to acarbose (positive control, IC50 = 344.23 ± 1.03 μg mL−1). The high polyphenol and flavonoid contents in these plant extracts may be responsible for their promising α-glucosidase-inhibitory activity. Previously, α-glucosidase inhibitory activity was observed with an IC50 value of 38.2 μg mL−1 (methanolic extract), and 9.0 μg mL−1 (water extract) for T. chebula, 68.2 ± 3.4 μg mL−1 for C. operculatus,14 4.226 ± 0.43 μg mL−1 (ethyl acetate) for F. semicordata.45
Similarly, the significant α-amylase inhibitory activity was shown by the methanolic extract of F. semicordata (IC50 = 46.8 ± 1.8 μg mL−1) followed by water and methanolic extract of C. operculatus (53.2 ± 1.0 μg mL−1 and 62.49 ± 1.89 μg mL−1 respectively) corresponding positive control acarbose (6.1 ± 0.10 μg mL−1). In previous work, α-amylase inhibitory activity was observed in ethyl acetate fraction of F. semicordata and T. chebula extract with IC50 values of 4.861 ± 0.41 and 15.1 ± 1.4 μg mL−1 respectively.45,46 The variation between our results and the literature data might be attributed to various factors, including the degree of ripeness at the time of harvesting, environmental factors, processing, and storage.47
The MS-based metabolomics approach was employed in our study to putatively identify all the secondary metabolites present in the plant extracts responsible for the anti-diabetic activity. Most of the compounds annotated in our study were similar to those in the literature; the ethyl acetate extract of T. chebula showed a base peak at 357.0455 [M + H]+ and fragment peaks at 339, 321, 293, and 203 corresponding to chebulic acid.48 It has been reported as a potent compound for preventing the vascular complications associated with diabetes.49 The base peak at 371.0619, with fragment peaks at 191, 163, and 145, was predicted to be 2-O-caffeoyl hydroxy citric acid, which correlates with the fragmentation pattern reported in the literature.50 The molecular formula C14H6O8, with a base peak of 303.0140, has been proposed to be ellagic acid based on a comparative analysis of the spectral findings of Wu et al.51 As per the existing literature, ellagic acid demonstrates anti-diabetic effects by targeting the β-cells in the pancreas.52 Similarly, [M + H]+ at m/z 619.0970 along with a fragment peak at 153 was annotated as trigalloevaloglucosan IX based on the results of Abu-Reidah et al.53 Likewise, molecular ion [M + H]+ at m/z 385.07 was annotated as 2-O-feruloyl hydroxy citric acid with fragment peaks at 385, 209, and 195. The spectral data were consistent with the literature.50
A molecular ion at m/z 667.4054 [M + H]+ detected at retention time 10.33 min was identified as arjunglucoside I in T. chebula bark, isolated previously from T. arjuna.54 Previous study indicates that arjunglucoside I exhibits potential antidiabetic effects against the α-glucosidase enzyme (IC50 = 1074 ± 32 μM).55 Similarly, a protonated ion detected at m/z 657.3627 [M + H]+ was tentatively identified as galloylterminolic acid which was reported already in T. albida.56 Another molecular ion at m/z [M + H]+ was identified as flavogallonic acid, and was consistent with the literature found in the negative ESI-mode of T. chebula.57 The results of HPLC-MS analysis of T. chebula (fruits) showed a protonated ion with m/z 323.0762 [M + H]+ and sodiated ion as adduct at m/z 345.0580 [M + Na]+. Based on the literature, this compound was putatively identified as leucodelphidin.58 Arjunolic acid, previously identified in the T. arjuna,59 was also identified in (+)-ESI-MS/MS analysis at m/z 489.3570 [M + H]+. Against α-glucosidase isolated from S. cerevisiae, arjunolic acid demonstrated anti-diabetic activity (IC50 = 18.63 ± 0.32 g mL−1).60 In addition, bartogenic acid was annotated for a molecular ion at m/z 519.3313 [M + H]+. This compound has demonstrated anti-diabetic properties against intestinal-glucosidase (IC50 = 168.09 μg mL−1).61 Another molecular ion at m/z 505.3529 [M + H]+ was identified as arjungenin, previously reported by Honda and coworkers.54 It has been reported to show moderate antibacterial activity and beta-glucuronidase inhibitor activity.62,63 From T. chebula bark extract, a molecular ion with m/z 443.0978 [M + H]+ was tentatively annotated as (−)-epicatechin-3-O-gallate which was already reported by Singh et al.64 Based on a literature survey, gambiriin B1 was identified as a molecular ion with m/z 563.1549 [M + H]+ in the T. chebula bark.65 Similarly, rotundioic acid was isolated and identified.66,67 These data are consistent with our results, molecular ion peaks at 503.3363 [M + H]+ for rotundioic acid and 273.0759 [M + H]+ for butin. From the data obtained from the extract of C. operculatus and T. chebula bark, a protonated ion was detected at m/z 307.0815 and putatively identified as epigallocatechin or gallocatechin, which is similar to the study carried out by Lee and colleagues.68 Its efficacy for the treatment of prostate cancer is being currently studied and completed in phase 2 trial.69 It has been reported that epigallocatechin modifies glucose and lipid metabolism in rat hepatoma cell line H4IIE and notably enhances glucose tolerance in diabetic rodents.70
The compound exhibiting a base peak of 465.1023 and fragment peaks at 303 and 137 was annotated as isoquercitrin sourced from F. lacor.71 Isoquercitrin is currently being studied in a phase 2 clinical trial for its efficacy on Coronavirus Disease 2019.72 Moreover, it has been reported to demonstrate a prophylactic impact on diabetes mellitus.73 Similarly, from F. semicordata and T. chebula (bark) molecular ions at m/z 579.1504 and at m/z 291.0866 were annotated as procyanidin B2 and catechin based on the result analysis.74,75 In vitro-glucosidase activity showed that procyanidin B2 could be used to reduce blood glucose levels.76 Whereas, in the α-glucosidase inhibitory assay, it has been found that catechin has possessed significant antidiabetic activity (IC50 = 87.55 ± 2.23 μg mL−1).77 We annotated quinic acid from F. semicordata and F. lacor with a protonated ion at m/z 193.0705 which was similar to those reported in the literature.78 A previous study showed that quinic acid potentially stimulates insulin secretion and enhances glucose tolerance.79 Hydroxyl-6-gingerol-O-β-D-glucuronide was annotated for a protonated ion with m/z 487.22 from the extract of F. semicordata with comparison to the spectral results from Zeng et al.80 The compound with molecular ion at m/z 435.0578 [M + H]+ was tentatively identified as ellagic acid-O-pentoside from C. operculatus based on the results obtained by Di Stefano and co-workers.81 Similarly, a compound with protonated ion at m/z 467.0872 [M + H]+ was identified as 3-O-galloylnorbergenin, which was previously reported.53 Moreover, molecular ions at m/z 649.1082 and 393.0435 were annotated as 3,4,5-tri-O-galloylquinic acid and tri-O-methoxyellagic acid, respectively from C. operculatus, as reported by Bindra et al. and Maldini et al.82,83 A molecular ion at m/z 279.1601 [M + H]+ and sodiated ion as adduct at m/z 301.1 [M + Na]+ was tentatively annotated as butyl isobutyl phthalate from C. operculatus, T. chebula, and F. lacor. This was previously reported by Bu et al. through LC/MS analysis, which was further supported by ESI-MS and NMR analysis.84 It was observed that butyl isobutyl phthalate exhibits a hypoglycemic effect when tested in living organisms, and therefore, acts as a non-competitive inhibitor of α-glucosidase (IC50 = 10.6 ± 1.1 μg mL−1).84 Likewise, molecular ion at m/z 387.1817 with retention time at 17.45 min was tentatively annotated as eudesmin from the water fraction of F. lacor and was already reported.85 Furthermore, based on literature data, a molecular ion at m/z of 293.1736 [M + H]+ was annotated as lasiodiplodin,86 while the compound analyzed in the ESI-MS (+) mode molecular ion at m/z 331.2842 was tentatively identified as 1-monopalmitin.87 Similarly, molecular ion at m/z 463.3794 was putatively identified as α-tocospiro B from the water fraction of F. lacor, as per the analysis performed by Chiang et al.88
GNPS-based molecular networking can be performed to validate manually annotated compounds and aid in the exploration of secondary metabolite derivatives according to similarities in the core chemical structure.89 Hence, this approach improves the visualization of the chemical constituents present in biological samples, forming clusters in molecular networking. In this study, the methanolic extract and EA fraction of the bark of T. chebula was subjected to comprehensive phytochemical profiling using MS/MS and a GNPS-based metabolomics platform, resulting in the identification of 22 compounds based on GNPS analysis and previous reports. Among these, 11 compounds, including linolenic acid, phytosphingosine, sesamin, dioctyl phthalate, palmitic acid, oleanolic acid, quercitrin, epigallocatechin gallate, arjunic acid, β-D-galactopyranoside, madecassic acid, and arjunetin, were annotated based on GNPS that were not identified by manual annotation. These compounds were categorized into three distinct molecular network clusters: diterpenoid-O-glycosides, triterpenoids, flavonoid derivatives, and triterpenoid-O-glycosides clusters (Fig. 2B–D). All compounds were previously reported to be isolated from T. chebula, indicating that further investigation is necessary for the unknown nodes and edges of T. chebula.
Conclusion
Since ancient times, many traditional plants have been used by various ethnic groups to manage diabetes. Consequently, their scientific exploration is necessary. Four ethnically selected plants C. operculatus, T. chebula, F. lacor, and F. semicordata, exhibited significant enzyme inhibitory activity. In addition, LC/MS-based metabolomics and GNPS-based molecular networking were employed to explore and correlate secondary metabolites with the observed activities in these medicinal plants. Metabolic profiling revealed the presence of different classes of bioactive metabolites, including phenolic compounds, diterpenoids-O-glycosides, triterpenoids, triterpenoid-O-glycosides, flavonoids, flavonoid-O-glycosides, fatty acids, and ceramides. Additional investigation is recommended to isolate and identify potential inhibitors from the active fractions of plant extracts to develop a therapeutic candidate for diabetes and to explore the underlying mechanisms responsible for the observed anti-diabetic action.Author contributions
Niranjan Parajuli and Ki Hyun Kim designed and supervised the research; Arjun Prasad Timilsina, Bimal Kumar Raut, Chen Huo, and Rabin Budhathoki conducted computational metabolomics work; Bimal Kumar Raut, Arjun Prasad Timilsina, Karan Khadayat, and Niranjan Parajuli wrote the manuscript; Arjun Prasad Timilsina Prakriti Budhathoki, Mandira Ghimire and Karan Khadayat conducted in vitro assays; Niraj Aryal, Ki Hyun Kim, and Niranjan Parajuli edited the manuscript.Conflicts of interest
There are no conflicts of interest among authors.Acknowledgements
This project was supported by the University Grants Commission, Nepal (Award No. CRIG-78/79-S&T-01) to Niranjan Parajuli. We are thankful to the Sophisticated Analytical Instrument Facility (SAIF), CSIR-Lucknow, India, for the LC-HRMS data acquisition. We are also thankful to Kusum Basnet for her research assistance.References
- S. C. Booth, A. M. Weljie and R. J. Turner, Computational Tools For The Secondary Analysis of Metabolomics Experiments, Comput. Struct. Biotechnol. J., 2013, 4, e201301003 CrossRef PubMed.
- Q. Yang, et al., Metabolomics biotechnology, applications, and future trends: a systematic review, RSC Adv., 2019, 9, 37245–37257 RSC.
- M. K. Patel, et al., Plants Metabolome Study: Emerging Tools and Techniques, Plants, 2021, 10, 2409 CrossRef PubMed.
- L.-F. Nothias, et al., Feature-based molecular networking in the GNPS analysis environment, Nat. Methods, 2020, 17, 905–908 CrossRef CAS PubMed.
- F. Vincenti, et al., Molecular Networking: A Useful Tool for the Identification of New Psychoactive Substances in Seizures by LC–HRMS, Front. Chem., 2020, 8, 572952 CrossRef CAS PubMed.
- N. Shrestha, S. R. Mishra, S. Ghimire, B. Gyawali and S. Mehata, Burden of Diabetes and Prediabetes in Nepal: A Systematic Review and Meta-Analysis, Diabetes Ther., 2020, 11, 1935–1946 CrossRef PubMed.
- WHO, Diabetes, 2023, https://www.who.int/news-room/fact-sheets/detail/diabetes Search PubMed.
- N. E. Thomford, et al., Natural Products for Drug Discovery in the 21st Century: Innovations for Novel Drug Discovery, Int. J. Mol. Sci., 2018, 19, 1578 CrossRef PubMed.
- Md. A. Hossain and R. Pervin, Chapter 34 – Current Antidiabetic Drugs: Review of Their Efficacy and Safety, in Nutritional and Therapeutic Interventions for Diabetes and Metabolic Syndrome, ed. D. Bagchi and S. Nair, Academic Press, 2nd edn, 2018, pp. 455–473, DOI:10.1016/B978-0-12-812019-4.00034-9.
- M. Nigam, et al., Fruits of Terminalia chebula Retz.: a review on traditional uses, bioactive chemical constituents and pharmacological activities, Phytother. Res., 2020, 34, 2518–2533 CrossRef CAS PubMed.
- N. N. Rege, U. M. Thatte and S. A. Dahanukar, Adaptogenic properties of six rasayana herbs used in Ayurvedic medicine, Phytother. Res., 1999, 13, 275–291 CrossRef CAS PubMed.
- C.-L. Ye, Y.-H. Lu, X.-D. Li, D.-Z. Wei and C. S. Whitehead, HPLC analysis of a bioactive chalcone and triterpene in the buds of Cleistocalyx operculatus, S. Afr. J. Bot., 2005, 71, 312–315 CrossRef CAS.
- C.-L. Ye, J.-W. Liu, D.-Z. Wei, Y.-H. Lu and F. Qian, In vivo antitumor activity by 2′,4′-dihydroxy-6′-methoxy-3′,5′-dimethylchalcone in a solid human carcinoma xenograft model, Cancer Chemother. Pharmacol., 2005, 56, 70–74 CrossRef CAS PubMed.
- T. T. Mai and N. V. Chuyen, Anti-Hyperglycemic Activity of an Aqueous Extract from Flower Buds of Cleistocalyx operculatus (Roxb.) Merr and Perry, Biosci., Biotechnol., Biochem., 2007, 71, 69–76 CrossRef CAS PubMed.
- A. Y. H. Woo, M. M. Y. Waye, H. S. Kwan, M. C. Y. Chan, C. F. Chau and C. H. K. Cheng, Inhibition of ATPases by Cleistocalyx operculatus. A possible mechanism for the cardiotonic actions of the herb, Vasc. Pharmacol., 2002, 38, 163–168 CrossRef CAS PubMed.
- B.-S. Min, T. D. Cuong, J.-S. Lee, M.-H. Woo and T. M. Hung, Flavonoids from Cleistocalyx operculatus buds and their cytotoxic activity, Bull. Korean Chem. Soc., 2010, 31, 2392–2394 CrossRef CAS.
- K. Y. Khan, et al., Ethno-medicinal Species of genus Ficus L. Used to Treat Diabetes in Pakistan, J. Appl. Pharm. Sci., 2011, 3, 209–211 Search PubMed.
- P. K. Mukherjee, K. Mukherjee, M. Rajesh Kumar, M. Pal and B. P. Saha, Evaluation of wound healing activity of some herbal formulations, Phytother. Res., 2003, 17, 265–268 CrossRef PubMed.
- S. K. Rai, S. Subedi and S. Mishra, Utilization pattern of medicinal plants in Thumpakhar, Village Development Committee, Sindhupalchok, Nepal, Botanica Orientalis: Journal of Plant Science, 2004, 4, 75–78 Search PubMed.
- D. Bajracharya, S. J. B. Rana and A. K. Shrestha, A general survey and biochemical analysis of fodder plants found in Nagarjun hill forest of Kathmandu Valley [Nepal], J. Nat. Hist. Mus., 1978, 2, 105–116 Search PubMed.
- Y. Uprety, et al., Diversity of use and local knowledge of wild edible plant resources in Nepal, J. Ethnobiol. Ethnomedicine, 2012, 8, 16 CrossRef PubMed.
- P. Ghimire, S. Pandey and A. Shrestha, A comprehensive review on phytochemical, pharmacognostical properties and pharmacological activities of Ficus lacor L. (Moraceae), Int. J. Tradit. Herb. Med., 2020, 8, 96–102 Search PubMed.
- R. K. Sindhu and S. Arora, Phytochemical and Pharmacognostical Investigations on Aerial roots of Ficus lacor Buch. Ham, Int. J. Phytomed., 2013, 5, 267 Search PubMed.
- S. Gupta, Ethnomedicinal claims of Ficus semicordata Buch.-Ham. ex Sm.: a review, Int. J. Green Pharm., 2018, 12, 206–213 Search PubMed.
- P. Deepa, K. Sowndhararajan, S. Kim and S. J. Park, A role of Ficus species in the management of diabetes mellitus: A review, J. Ethnopharmacol., 2018, 215, 210–232 CrossRef CAS PubMed.
- K. Khadayat, B. P. Marasini, H. Gautam, S. Ghaju and N. Parajuli, Evaluation of the alpha-amylase inhibitory activity of Nepalese medicinal plants used in the treatment of diabetes mellitus, Clin. Phytosci., 2020, 6, 34 CrossRef.
- E. A. Ainsworth and K. M. Gillespie, Estimation of total phenolic content and other oxidation substrates in plant tissues using Folin–Ciocalteu reagent, Nat. Protoc., 2007, 2, 875–877 CrossRef CAS PubMed.
- C.-C. Chang, M.-H. Yang, H.-M. Wen and J.-C. Chern, Estimation of total flavonoid content in propolis by two complementary colometric methods, J. Food Drug Anal., 2020, 10, 178–182 Search PubMed.
- X. Lu, et al., Determination of total phenolic content and antioxidant capacity of onion (Allium cepa) and shallot (Allium oschaninii) using infrared spectroscopy, Food Chem., 2011, 129, 637–644 CrossRef CAS PubMed.
- T. Sabudak, O. Demirkiran, M. Ozturk and G. Topcu, Phenolic compounds from Trifolium echinatum Bieb. and investigation of their tyrosinase inhibitory and antioxidant activities, Phytochemistry, 2013, 96, 305–311 CrossRef CAS PubMed.
- H. Fouotsa, et al., Xanthones inhibitors of α-glucosidase and glycation from Garcinia nobilis, Phytochem. Lett., 2012, 5, 236–239 CrossRef CAS.
- B. Aryal, et al., LC-HRMS Profiling and Antidiabetic, Antioxidant, and Antibacterial Activities of Acacia catechu (L.f.) Willd, BioMed Res. Int., 2021, 2021, e7588711 Search PubMed.
- K. Dührkop, K. Scheubert and S. Böcker, Molecular Formula Identification with SIRIUS, Metabolites, 2013, 3, 506–516 CrossRef PubMed.
- K. Bashir, et al., Assessing the ethnobotanical potential of Carissa opaca berries by merging outcomes from metabolomics profiling, enzyme assays, and in silico docking studies, Food Chem., 2021, 363, 130259 CrossRef CAS PubMed.
- R. A. Quinn, et al., Molecular Networking As a Drug Discovery, Drug Metabolism, and Precision Medicine Strategy, Trends Pharmacol. Sci., 2017, 38, 143–154 CrossRef CAS PubMed.
- G. Pistia-Brueggeman and R. I. Hollingsworth, A preparation and screening strategy for glycosidase inhibitors, Tetrahedron, 2001, 57, 8773–8778 CrossRef CAS.
- F. Sobreira, et al., Gastroprotective activity of the hydroethanolic extract and ethyl acetate fraction from Kalanchoe pinnata (Lam.) Pers, Braz. J. Pharm. Sci., 2017, 53, e16027 Search PubMed.
- Y.-Z. Cai, M. Sun, J. Xing, Q. Luo and H. Corke, Structure–radical scavenging activity relationships of phenolic compounds from traditional Chinese medicinal plants, Life Sci., 2006, 78, 2872–2888 CrossRef CAS PubMed.
- K. E. Heim, A. R. Tagliaferro and D. J. Bobilya, Flavonoid antioxidants: chemistry, metabolism and structure-activity relationships, J. Nutr. Biochem., 2002, 13, 572–584 CrossRef CAS PubMed.
- J. Xiao, G. Kai, K. Yamamoto and X. Chen, Advance in Dietary Polyphenols as α-Glucosidases Inhibitors: A Review on Structure-Activity Relationship Aspect, Crit. Rev. Food Sci. Nutr., 2013, 53, 818–836 CrossRef CAS PubMed.
- N. F. Shamsudin, et al., Flavonoids as Antidiabetic and Anti-Inflammatory Agents: A Review on Structural Activity Relationship-Based Studies and Meta-Analysis, Int. J. Mol. Sci., 2022, 23, 12605 CrossRef CAS PubMed.
- J. E. Hayes, P. Allen, N. Brunton, M. N. O'Grady and J. P. Kerry, Phenolic composition and in vitro antioxidant capacity of four commercial phytochemical products: Olive leaf extract (Olea europaea L.), lutein, sesamol and ellagic acid, Food Chem., 2011, 126, 948–955 CrossRef CAS.
- J. Chen, et al., Structural elucidation and antioxidant activity evaluation of key phenolic compounds isolated from longan (Dimocarpus longan Lour.) seeds, J. Funct. Foods, 2015, 17, 872–880 CrossRef CAS.
- A. Mocan, et al., Biological effects and chemical characterization of Iris schachtii Markgr. extracts: A new source of bioactive constituents, Food Chem. Toxicol., 2018, 112, 448–457 CrossRef CAS PubMed.
- V. Kaur, K. Upadhyaya and M. Pande, Bioassay-Guided Evaluation of Ficus Semicordata For Antidiabetic Activity, Int. J. Pharm. Pharm. Sci., 2017, 71–77, DOI:10.22159/ijpps.2017v9i3.16441.
- D. Y. Lee, H. Yang, H. W. Kim and S. H. Sung, New polyhydroxytriterpenoid derivatives from fruits of Terminalia chebula Retz. and their α-glucosidase and α-amylase inhibitory activity, Bioorg. Med. Chem. Lett., 2017, 27, 34–39 CrossRef CAS PubMed.
- C. Manach, A. Scalbert, C. Morand, C. Rémésy and L. Jiménez, Polyphenols: food sources and bioavailability, Am. J. Clin. Nutr., 2004, 79, 727–747 CrossRef CAS PubMed.
- J. Yin, et al., Characterization of chemical composition and prebiotic effect of a dietary medicinal plant Penthorum chinense Pursh, Food Chem., 2020, 319, 126568 CrossRef CAS PubMed.
- H.-S. Lee, Y.-C. Koo, H. J. Suh, K.-Y. Kim and K.-W. Lee, Preventive effects of chebulic acid isolated from Terminalia chebula on advanced glycation endproduct-induced endothelial cell dysfunction, J. Ethnopharmacol., 2010, 131, 567–574 CrossRef CAS PubMed.
- I. Parveen, A. Winters, M. D. Threadgill, B. Hauck and P. Morris, Extraction, structural characterisation and evaluation of hydroxycinnamate esters of orchard grass (Dactylis glomerata) as substrates for polyphenol oxidase, Phytochemistry, 2008, 69, 2799–2806 CrossRef CAS PubMed.
- L. Wu, Y. Liu, Y. Qin, L. Wang and Z. Wu, HPLC-ESI-qTOF-MS/MS Characterization, Antioxidant Activities and Inhibitory Ability of Digestive Enzymes with Molecular Docking Analysis of Various Parts of Raspberry (Rubus ideaus L.), Antioxidants, 2019, 8, 274 CrossRef CAS PubMed.
- N. Fatima, et al., Ellagic acid in Emblica officinalis exerts anti-diabetic activity through the action on β-cells of pancreas, Eur. J. Nutr., 2017, 56, 591–601 CrossRef PubMed.
- I. M. Abu-Reidah, M. S. Ali-Shtayeh, R. M. Jamous, D. Arráez-Román and A. Segura-Carretero, HPLC–DAD–ESI-MS/MS screening of bioactive components from Rhus coriaria L. (Sumac) fruits, Food Chem., 2015, 166, 179–191 CrossRef CAS PubMed.
- T. Honda, T. Murae, T. Tsuyuki, T. Takahashi and M. Sawai, Arjungenin, Arjunglucoside I, and Arjunglucoside II. A New Triterpene and New Triterpene Glucosides from Terminalia arjuna, Bull. Chem. Soc. Jpn., 1976, 49, 3213–3218 CrossRef CAS.
- H. Yuca, et al., α-Glucosidase and α-amylase inhibitory potential of main compounds and drug candidates from Elaeagnus rhamnoides (L.) A. Nelson, Chem. Pap., 2022, 76, 913–922 CrossRef CAS.
- A. Camara, et al., Terminalia albida treatment improves survival in experimental cerebral malaria through reactive oxygen species scavenging and anti-inflammatory properties, Malar. J., 2019, 18, 431 CrossRef CAS PubMed.
- Y. Wang, et al., Hydrolyzable tannins from Balanophora polyandra, Acta Pharm. Sin. B, 2013, 3, 46–50 CrossRef.
- M. B. T. Tali, et al., In vitro antiplasmodial activity-directed investigation and UPLC–MS fingerprint of promising extracts and fractions from Terminalia ivorensis and Terminalia brownii Fresen, J. Ethnopharmacol., 2022, 296, 115512 CrossRef CAS PubMed.
- T. M. Manu, T. Anand, M. D. Pandareesh, P. B. Kumar and F. Khanum, Terminalia arjuna extract and arjunic acid mitigate cobalt chloride–induced hypoxia stress–mediated apoptosis in H9c2 cells, Naunyn-Schmiedeberg's Arch. Pharmacol., 2019, 392, 1107–1119 CrossRef CAS PubMed.
- W. Hou, et al., Triterpene acids isolated from Lagerstroemia speciosa leaves as α-glucosidase inhibitors, Phytother. Res., 2009, 23, 614–618 CrossRef CAS PubMed.
- P. M. Gowri, A. K. Tiwari, A. Z. Ali and J. M. Rao, Inhibition of α-glucosidase and amylase by bartogenic acid isolated from Barringtonia racemosa Roxb. seeds, Phytother. Res., 2007, 21, 796–799 CrossRef CAS PubMed.
- D. P. A. Gossan, et al., Antibacterial and cytotoxic triterpenoids from the roots of Combretum racemosum, Fitoterapia, 2016, 110, 89–95 CrossRef CAS PubMed.
- A. Rahman, et al., Some Chemical Constituents Of Terminalia Glaucescens And Their Enzymes Inhibition Activity, Z. Naturforsch., B: J. Chem. Sci., 2005, 60, 347–350 CrossRef.
- A. Singh, S. Kumar and B. Kumar, LC-MS Identification of Proanthocyanidins in Bark and Fruit of six Terminalia species, Nat. Prod. Commun., 2018, 13, 1934578X1801300 Search PubMed.
- S. Taniguchi, et al., Revised Structures of Gambiriins A1, A2, B1, and B2, Chalcane-Flavan Dimers from Gambir (Uncaria gambir Extract), Chem. Pharm. Bull., 2007, 55, 268–272 CrossRef CAS PubMed.
- B. Yang, et al., A facile and selective approach to the qualitative and quantitative analysis of triterpenoids and phenylpropanoids by UPLC/Q-TOF-MS/MS for the quality control of Ilex rotunda, J. Pharm. Biomed. Anal., 2018, 157, 44–58 CrossRef CAS PubMed.
- R. Chokchaisiri, et al., Bioactive Flavonoids of the Flowers of Butea monosperma, Chem. Pharm. Bull., 2009, 57, 428–432 CrossRef CAS PubMed.
- J.-H. Lee, J. V. Johnson and S. T. Talcott, Identification of Ellagic Acid Conjugates and Other Polyphenolics in Muscadine Grapes by HPLC-ESI-MS, J. Agric. Food Chem., 2005, 53, 6003–6010 CrossRef CAS PubMed.
- University of Southern California, A Phase II Trial of a Combination Herbal Therapy for Men With Biochemical Recurrence of Prostate Cancer After Initial Local Therapy, 2017, https://clinicaltrials.gov/study/NCT00669656 Search PubMed.
- S. Wolfram, et al., Epigallocatechin Gallate Supplementation Alleviates Diabetes in Rodents1, J. Nutr., 2006, 136, 2512–2518 CrossRef CAS PubMed.
- Y. Cai, L. Wu, X. Lin, X. Hu and L. Wang, Phenolic profiles and screening of potential α-glucosidase inhibitors from Polygonum aviculare L. leaves using ultra-filtration combined with HPLC-ESI-qTOF-MS/MS and molecular docking analysis, Ind. Crops Prod., 2020, 154, 112673 CrossRef CAS.
- AB Science, Randomized, Phase 2 Clinical Trial to Evaluate the Safety and Efficacy of Masitinib Combined With Isoquercetin, and Best Supportive Care in Hospitalized Patients With Moderate and Severe COVID-19, 2023, https://clinicaltrials.gov/study/NCT04622865 Search PubMed.
- S. Panda and A. Kar, Antidiabetic and antioxidative effects of Annona squamosa leaves are possibly mediated through quercetin-3-O-glucoside, BioFactors, 2007, 31, 201–210 CrossRef CAS PubMed.
- A. Gahlaut, A. Sharma, A. Shirolkar and R. Dabur, Non-targeted identification of compounds from regenerated bark of Terminalia tomentosa by HPLC-(+) ESI-QTOFMS, J. Pharm. Res., 2013, 6, 415–418 CAS.
- T. Li, et al., Analysis of Chemical Constituents of Guizhi Shaoyao Zhimu Granules by UPLC-Q-Orbitrap HRMS, 2020, https://www.researchsquare.com/article/rs-73167/v1 Search PubMed.
- C. H. J. Yu, Z. Migicovsky, J. Song and H. P. V. Rupasinghe, (Poly)phenols of apples contribute to in vitro antidiabetic properties: Assessment of Canada's Apple Biodiversity Collection, Plants People Planet, 2023, 5, 225–240 CrossRef.
- H. N. Mrabti, et al., Separation, Identification, and Antidiabetic Activity of Catechin Isolated from Arbutus unedo L. Plant Roots, Plants, 2018, 7, 31 CrossRef PubMed.
- K. Masike, et al., Highlighting mass spectrometric fragmentation differences and similarities between hydroxycinnamoyl-quinic acids and hydroxycinnamoyl-isocitric acids, Chem. Cent. J., 2017, 11, 29 CrossRef PubMed.
- E. Heikkilä, et al., The plant product quinic acid activates Ca2+-dependent mitochondrial function and promotes insulin secretion from pancreatic beta cells, Br. J. Pharmacol., 2019, 176, 3250–3263 CrossRef PubMed.
- S.-L. Zeng, P. Li and E.-H. Liu, Metabolic profile of Guge Fengtong tablet in rat urine and bile after oral administration, using high-performance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry, Chin. J. Nat. Med., 2015, 13, 954–960 CAS.
- V. Di Stefano, S. Scandurra, A. Pagliaro, V. Di Martino and M. G. Melilli, Effect of Sunlight Exposure on Anthocyanin and Non-Anthocyanin Phenolic Levels in Pomegranate Juices by High Resolution Mass Spectrometry Approach, Foods, 2020, 9, 1161 CrossRef CAS PubMed.
- R. S. Bindra, N. K. Satti and O. P. Suri, Isolation and structures of ellagic acid derivatives from Euphorbia acaulis, Phytochemistry, 1988, 27, 2313–2315 CrossRef CAS.
- M. Maldini, P. Montoro and C. Pizza, Phenolic compounds from Byrsonima crassifolia L. bark: Phytochemical investigation and quantitative analysis by LC-ESI MS/MS, J. Pharm. Biomed. Anal., 2011, 56, 1–6 CrossRef CAS PubMed.
- T. Bu, M. Liu, L. Zheng, Y. Guo and X. Lin, α-glucosidase inhibition and the in vivo hypoglycemic effect of butyl-isobutyl-phthalate derived from the Laminaria japonica rhizoid, Phytother. Res., 2010, 24, 1588–1591 CrossRef PubMed.
- S.-M. Seo, et al., Furofuran lignans from the bark of Magnolia kobus, Chem. Nat. Compd., 2008, 44, 419–423 CrossRef CAS.
- K. Z. W. Lae, S. S. Su, N. N. Win, N. N. Than and H. Ngwe, Isolation of Lasiodiplodin and Evaluation of some Biological Activities of the Stem Barks of Phyllanthus Albizzioides (Kurz) Hook.f., SciMed. J., 2019, 1, 199–216 CrossRef.
- M. E. Hergueta-Castillo, R. López-Ruiz, A. Garrido Frenich and R. Romero-González, Characterization of the composition of plant protection products in different formulation types employing suspect screening and unknown approaches, J. Sci. Food Agric., 2022, 102, 5995–6004 CrossRef CAS PubMed.
- Y.-M. Chiang and Y.-H. Kuo, Two Novel α-Tocopheroids from the Aerial Roots of Ficus microcarpa, J. Cheminf., 2003, 34, 5125–5128 Search PubMed.
- R. Schmid, et al., Ion identity molecular networking for mass spectrometry-based metabolomics in the GNPS environment, Nat. Commun., 2021, 12, 3832 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1: mass spectrum of chebulic acid from Terminalia chebula; Fig. S2: mass spectrum of 2-O-caffeoyl hydroxycitric acid from Terminalia chebula; Fig. S3: mass spectrum of ellagic acid from Terminalia chebula; Fig. S4: mass spectrum of Trigalloyllevoglucosan IX from Terminalia chebula; Fig. S5: mass spectrum of 2-O-feruloylhydroxycitric acid from Terminalia chebula; Fig. S6: mass spectrum of arjunglucoside I from Terminalia chebula; Fig. S7: mass spectrum of 23-galloylterminolic acid from Terminalia chebula; Fig. S8: mass spectrum of flavogallonic acid from Terminalia chebula; Fig. S9: mass spectrum of Leucodelphidin from Terminalia chebula; Fig. S10: mass spectrum of arjunolic acid from Terminalia chebula; Fig. S11: mass spectrum of bartogenic acid from Terminalia chebula; Fig. S12: mass spectrum of arjungenin from Terminalia chebula; Fig. S13: mass spectrum of (−)-epicatechin-3-O-gallate from Terminalia chebula; Fig. S14: mass spectrum of gambiriin B1 from Terminalia chebula; Fig. S15; mass spectrum of rotundioic acid from Terminalia chebula; Fig. S16; mass spectrum of butin from Terminalia chebula; Fig. S17: mass spectrum of gallocatechin or epigallocatechin from Cleistocalyx operculatus; Fig. S18: mass spectrum of 4,4′-di-O-methylellagic acid from Cleistocalyx operculatus; Fig. S19: mass spectrum of isoquercitrin from Ficus lacor; Fig. S20: mass spectrum of catechin from Ficus semicordata; Fig. S21: mass spectrum of procyanidin B2 from Ficus semicordata; Fig. S22: mass spectrum of ellagic acid-O-pentoside from Cleistocalyx operculatus; Fig. S23: mass spectrum of quinic acid from Ficus lacor; Fig. S24: mass spectrum of hydroxyl-6-gingerol-O-β-D-glucuronide from Ficus semicordata; Fig. S25: mass spectrum of 3-O-galloylnorbergenin from Cleistocalyx operculatus; Fig. S26: mass spectrum of 3,4,5-tri-O-galloylquinic acid from Cleistocalyx operculatus; Fig. S27: mass spectrum of tri-O-methoxyellagic acid from Cleistocalyx operculatus; Fig. S28: mass spectrum of butyl isobutyl phthalate from Cleistocalyx operculatus; Fig. S29: mass spectrum of eudesmin from Ficus lacor; Fig. S30 mass spectrum of lasiodiplodin from Ficus lacor; Fig. S31 mass spectrum of 1-monopalmitin from Ficus lacor; Fig. S32 mass spectrum of α-tocospiro B from Ficus lacor. See DOI: https://doi.org/10.1039/d3ra04037b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |