Open Access Article
Open Access ArticleInvestigating the hepatoprotective potentiality of marine-derived steroids as promising inhibitors of liver fibrosis†
Mohamed A. Tammam‡
 a,
Florbela Pereira‡
a,
Florbela Pereira‡ b,
Omnia Aly‡c,
Mohamed Sebak
b,
Omnia Aly‡c,
Mohamed Sebak d,
Yasser M. Diaba,
Aldoushy Mahdye and
Amr El-Demerdash
d,
Yasser M. Diaba,
Aldoushy Mahdye and
Amr El-Demerdash *fg
*fg
aDepartment of Biochemistry, Faculty of Agriculture, Fayoum University, Fayoum 63514, Egypt
bLAQV REQUIMTE, Department of Chemistry, NOVA School of Science and Technology, Universidade Nova de Lisboa, 2829516 Caparica, Portugal
cDepartment of Medical Biochemistry, National Research Centre, Cairo 12622, Egypt
dMicrobiology and Immunology Department, Faculty of Pharmacy, Beni-Suef University, Egypt
eDepartment of Zoology, Faculty of Science, Al-Azhar University (Assiut Branch), Assiut 71524, Egypt
fDivision of Organic Chemistry, Department of Chemistry, Faculty of Sciences, Mansoura University, Mansoura 35516, Egypt. E-mail: a_eldemerdash83@mans.edu.eg; Amr.El-Demerdash@jic.ac.uk
gDepartment of Biochemistry and Metabolism, the John Innes Centre, Norwich Research Park, Norwich NR4 7UH, UK
First published on 13th September 2023
Abstract
It has been reported that organic extracts derived from soft corals belonging to the genus Sarcophyton have exhibited a wide range of therapeutic characteristics. Based on biochemical and histological techniques, we aimed to assess the hepatoprotective role of the organic extract and its principal steroidal contents derived from the Red Sea soft coral Sarcophyton glaucum on acetaminophen-induced liver fibrosis in rats. Serum liver function parameters (ALT, AST, ALP and total bilirubin) were quantified using a spectrophotometer, and both alpha-fetoprotein (AFP) and carcinoembryonic antigen (CEA) levels were determined by using enzyme-linked immunosorbent assay (ELISA) kits while transformed growth factor beta (TGF-β) and tumor necrosis factor α (TNF-α) in liver tissue homogenate were determined using ELISA, and TGF-β and TNF-α gene expression in liver tissue was determined using real-time PCR following extraction and purification. Histopathological alterations in hepatic tissue were also examined under a microscope. In order to prioritize the isolation and characterization of the most promising marine steroids from the organic extract of the Red Sea soft coral Sarcophyton glaucum as hepatoprotective agents, a computational approach was employed. This approach involved molecular docking (MDock) and analysis of the structure–activity relationship (SAR) against glutathione-S-transferase (GST) and Cu–Zn human superoxide dismutase (Cu–ZnSOD) enzymes. Although the major role in the detoxification of foreign chemicals and toxic metabolites of GST and SOD enzymes is known, there is a lack of knowledge about the mode of action of the hepatoprotective process and those of the targets involved. The present study investigated the multiple interactions of a series of marine steroids with the GST and SOD enzymes, in order to reveal insights into the process of hepatoprotection.
1. Introduction
The liver is essential for maintaining homeostasis and a number of other biochemical and physiological processes, including the breakdown and elimination of substances produced inside as well as those introduced from the outside (such as medicines and other xenobiotics).1 Hepatotoxic substances, such as medications, alcohol, and viral infections, can induce liver damage.2 Acetaminophen, also known as N-acetyl-para-aminophenol, paracetamol, and acetaminophen (APAP), is a popular over-the-counter pain reliever and fever reducer. While acetaminophen is often well tolerated at therapeutic doses, it has been shown to cause serious liver damage in both experimental animals and humans.3Liver disease is worldwide problem that affects people all over the world. Traditional liver disease treatments are sometimes ineffective and can have serious repercussions. As a result, new treatments to treat liver illnesses must be identified in order to obtain an alternative pharmaceutical with questionable safety and efficacy.4,5 Hepatotoxicity is thought to be related to decreased activity of antioxidant enzymes6,7 e.g., glutathione-S-transferase (GST),6,8 human Cu–Zn superoxide dismutase (Cu–ZnSOD).9 This decrease is believed to come as a result of the harmful effects of free radicals produced after exposure to foreign chemicals and toxic metabolites such as ethanol. Kim et al. also reported a study in which GST activity in the liver was significantly inhibited by acetaminophen, but its inhibition was reversed by pretreatment with silymarin in mice.10
In terms of liver inflammation and fibrosis, biomarkers are used to assess the severity of liver disease. Liver enzymes such as ALT, AST, ALP and total bilirubin are important, as well as transforming growth factor beta (TGF-β) and tumor necrosis factor alpha (TNF-α) are two cytokines that are crucial to the development of liver inflammation and fibrosis. TNF-α is a pro-inflammatory cytokine which produced as response to infection or injury by the immune cells. TNF-α may stimulate hepatic stellate cells (HSCs) in the liver, which are responsible for creating excess collagen and other extracellular matrix proteins that cause fibrosis. TNF-α may also cause liver cell death, exacerbating liver damage and inflammation. TGF-β, on the other hand, is a multifunctional cytokine with anti-inflammatory and pro-fibrotic properties. TGF-β may stimulate tissue repair in the early stages of liver injury by promoting the formation of extracellular matrix proteins and activating HSCs. However, in chronic liver injury, persistent TGF-β activation can result in excessive collagen deposition and fibrosis.11 Also, AFP (alpha-fetoprotein) and CEA (carcinoembryonic antigen) are glycoproteins that are often used as tumor markers for different types of liver cancer, including hepatocellular carcinoma (HCC).12 Additionally, liver biopsies can be performed to directly assess the extent of liver fibrosis.
Marine organisms are distinguished by their ability to produce unique and novel metabolites with higher structural diversity in comparison with the terrestrial organisms, due to the extreme marine environment, which resulted development of unique physiological and metabolic capabilities.13–15 The higher structural diversity of the marine natural products (MNPs) allowed them to be unique supplier of several metabolites with several biological activities either in the pharmaceutical and or the cosmetology industrial applications.16 The global marine pharmaceutical clinical pipeline already comprises 17 small molecules and peptides, which are drugs approved by the most representative approval agencies such as the US FDA, the European Medicines Agency (EMEA), the Japanese Ministry of Health and the Administration of Therapeutic Products of Australia.17 For examples, within these approved drugs, there are two nucleoside derivatives (cytarabine (Ara-C)-anticancer 1969 and vidarabine (Ara-A)-antiviral 1976), three omega-3 fatty acid derivatives (all of them hypertriglyceridemia agents; omega-3-acid ethyl esters – 2004, eicosapentaenoic acid ethyl ester – 2012, and omega-3-carboxylic acid – 2014), two peptide derivatives (ziconotide – chronic pain 2004) and plitidepsin – anticancer 2018 (Australia), one macrocyclic ketone derivative (eribulin mesylate – anticancer 2010), and two alkaloid derivatives (the two anticancer agents; trabectedin – 2015 and lurbinectedin – 2020).17,18 MNPs are also used in cosmetics, namely – Resilience – a skin cream made by Estée Lauder, contains an anti-inflammatory natural extract of the Caribbean soft coral Pseudopterogorgia elisabethae, that was discovery by Prof. William Fenical.19 The anti-inflammatory activity was attributed to the presence of methopterosin, a simple synthetic derivative of the natural sesquiterpenes pseudopterosins, which have a variety of important pharmacological properties e.g., arthritis, psoriasis, inflammatory bowel disease.19
Among the soft corals, the genus of Sarcophyton (order: Alcyonacea, family: Alcyoniidae), including approximately 36 accepted species is considered one of the richest sources of new and unique natural products from the marine environment.20–22 Chemical examination of the several species of the genus Sarcophyton resulted in the identification of approximately 828 divers' marine natural products,22 including terpenes,23,24 steroids,25,26 quinones,27 and other classes of secondary metabolites.20 The species of glaucum, trocheliophorum and ehrenbergi with 143, 119 and 75 different isolated metabolites represented the most chemically investigated among the different identified species of the genus Sarcophyton.21,22 The high diversity of the secondary metabolites produced by the genus of Sarcophyton resulted in displaying wide spectrum of intriguing pharmacological activities including anti-inflammatory, cytotoxicity, antimicrobial, anti-angiogenic, neuroprotective, immunomodulatory, ichthyotoxic, antitumor and antifouling.13,20,24
As a part of our continuing research program to identify pharmacologically active natural products,4,15,28–31 here, we detail the isolation and structural elucidation of three steroid-containing metabolites from the Red Sea soft coral organic extract Sarcophyton glaucum, which was collected from the reefs of Hurghada, Egypt. Additionally, to provide valuable insights into finding the most promising marine steroids obtained from the Red Sea soft coral S. glaucum organic extract as hepatoprotective agents for liver disorder, a computational approach was employed, including molecular docking (MDock) and analysis of structure–activity relationship (SAR) against glutathione-S-tranferase (PDB ID 18GS) and Cu–Zn human superoxide dismutase (PDB ID 2C9V). This computational approach was employed to a focused chemical library of 26 steroidal derivatives, which were previously reported from soft corals and plants belonging to the genera Lobophytum, Euphorbia, Sinularia, Sarcophyton, Plexaurella and Klyxum.
2. Materials and methods
2.1. General experimental procedures
Bruker DRX 400 (Bruker BioSpin GmbH, Rheinstetten, Germany) spectrometers were used for NMR spectra recording. Chemical shifts are given on the δ (ppm) scale with reference to the solvent signals. Column chromatography separations were performed with Kieselgel 60 (Merck, Darmstadt, Germany). Kieselgel 60 F254 aluminium plates (Merck, Darmstadt, Germany) have been used for TLC performance, and spots were detected after spraying with 25% H2SO4 in MeOH reagent and heating at 100 °C for 1 min. HPLC separations were conducted on a Waters 515 liquid chromatography pump equipped with a RI-102 Shodex refractive index detector (ECOM spol. s r.o., Prague, Czech Republic), using a Kromasil 10-TBB-Chiral (10 × 250 mm) column (Eka Chemicals AB, Bohus, Sweden).2.2. Biological material
Specimens of S. glaucum were collected by SCUBA diving at a depth of 8 m from the reefs of Hurghada, (GPS coordinates 33°46′24′′E, 27°17′06′′N), Egypt in July 2015 where they were transported to the laboratory in ice bags, afterword they were stored at −20 °C until analysed. A voucher specimen has been deposited at the animal collection of NOIF in Hurghada.2.3. In vivo evaluation of the hepatoprotective ability of S. glaucum extract
Both alpha-fetoprotein (AFP) and carcinoembryonic antigen (CEA) levels were determined in serum using ELISA kits (Avi-Bion ELISA Kit; Orgenium Laboratories, Finland) for rats according to Chandler et al.36 and Abeyounis et al.,37 respectively.
Liver glutathione S-transferases (GSTs) and superoxide dismutase (SOD) were determined colorimetrically according Habig et al.38 and Nishikimi et al.,39 respectively using commercial kits from Biodiagnostics, Cairo-Egypt.
Each rat's liver tissue was used to extract RNA using a RNeasy Mini kit (Qiagen). Reverse transcription was performed using an RT kit (Promega). The transcription of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was studied by real-time polymerase chain reaction (PCR) using the specific primers shown in Table 1. The levels of gene expression were measured using a SYBR Green kit (Qiagen, Hilden, Germany).40 The data was analysed using MxPro qPCR (Agilent Technologies). Avi-Bion ELISA Kit (Orgenium Laboratories, Finland) was used to detect TNF- and TGF- in rat liver homogenate using ELISA kits developed in accordance with the methods described by Corti et al.41 and Kim et al.42
| Target | Sequence |
|---|---|
| TNF-α | F, 5′-AACTCGAGTGACAAGCCCGTAG-3′R, 5′-GTACCACCAGTTGGTTGTCTTTGA-3′ |
| TGF-β | F, 5′-TGCGCCTGCAGAGATTCAAG-3′R, 5′-AGGTAACGCCAGGAATTGTTGCTA-3′ |
| GAPDH | F, 5′-ACCACAGTCCATGCCATCAC-3′R, 5′-TCCACCACCCTGTTGCTGTA-3′ |
2.4. Extraction and purification
250 g of the freeze-dried coral were exhaustively extracted with mixtures of (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) CHCl3/MeOH, to afford 11 g green oily residue after solvent evaporation under vacuum. The obtained residue was submitted to normal phase vacuum column chromatography using mixtures of cHex/EtOAc and EtOAc/MeOH of increasing polarity as eluent system to afford 14 fractions (A–N). Fraction G (25–30% EtOAc in cHex, 600 mg) was further fractionated by normal phase gravity column chromatography using cHex/Ac, of increasing polarity as eluent, to yield 11 fractions (G1–G11). Fraction G3 (12% Me2CO in cHex, 68 mg) was subjected to normal phase SEP-PAC using cHex/EtoAc of increasing polarity as eluent, to afford 3 fractions (G3A–G3C). Fraction G3A (20% EtOAc in cHex, 57.4 mg) was further fractionated by normal phase SEP-PAC using mixtures of cHex/CHCl3 with increasing polarity to afford six fractions (G3A1–G3A6). Fractions G3A5 and G3A6 (80 and 100 CHCl3 in cHex, 27.9 and 20.0 mg, respectively) were pooled together and submitted to gravity column chromatography on silica gel using cHex/CHCl3 30
1) CHCl3/MeOH, to afford 11 g green oily residue after solvent evaporation under vacuum. The obtained residue was submitted to normal phase vacuum column chromatography using mixtures of cHex/EtOAc and EtOAc/MeOH of increasing polarity as eluent system to afford 14 fractions (A–N). Fraction G (25–30% EtOAc in cHex, 600 mg) was further fractionated by normal phase gravity column chromatography using cHex/Ac, of increasing polarity as eluent, to yield 11 fractions (G1–G11). Fraction G3 (12% Me2CO in cHex, 68 mg) was subjected to normal phase SEP-PAC using cHex/EtoAc of increasing polarity as eluent, to afford 3 fractions (G3A–G3C). Fraction G3A (20% EtOAc in cHex, 57.4 mg) was further fractionated by normal phase SEP-PAC using mixtures of cHex/CHCl3 with increasing polarity to afford six fractions (G3A1–G3A6). Fractions G3A5 and G3A6 (80 and 100 CHCl3 in cHex, 27.9 and 20.0 mg, respectively) were pooled together and submitted to gravity column chromatography on silica gel using cHex/CHCl3 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 as the mobile phase, to yield 9 fractions among which compound 3 was obtained in a pure form (2.5 mg). Fraction G3A5H was fractionated by chiral HPLC using nHex/i-PrOH (97
70 as the mobile phase, to yield 9 fractions among which compound 3 was obtained in a pure form (2.5 mg). Fraction G3A5H was fractionated by chiral HPLC using nHex/i-PrOH (97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) as eluent, to afford five fractions (G3A5H1–G3A5H5). Fractions G3A5H-(2, 4 and 5), 1.0, 1.1 and 1.2 mg, respectively) was further purified by chiral HPLC using nHex/i-PrOH (97
3) as eluent, to afford five fractions (G3A5H1–G3A5H5). Fractions G3A5H-(2, 4 and 5), 1.0, 1.1 and 1.2 mg, respectively) was further purified by chiral HPLC using nHex/i-PrOH (97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) as the mobile phase, to afford compounds 1 (1.1 mg) and 2 (0.9 mg).
3) as the mobile phase, to afford compounds 1 (1.1 mg) and 2 (0.9 mg).
2.5. Identification of steroid library
A focused library of 26 steroid derivatives (1–26) were previously reported from soft corals and plant, belonging to the genera Lobophytum, Euphorbia, Sinularia, Sarcophyton, Plexaurella and Klyxum (Fig. 2 which summarized in Scheme S1, ESI†).2.6. Preparation of the screening library
Steroids compounds have been drawn based on the references that discussed their isolation, which can be found in the ESI data Scheme S1;† the silymarin chemical structure was extracted from the PubChem database in the MDL SDF format. The 26 steroids and positive control compound, silymarin (a known amino-transferase inhibitor), had their 3D structures optimized with the Gaussian 09 program,44 utilizing the hybrid approach B3LYP and the base set 6-31G(d,p).45,46 We utilized OpenBabel (2.3.1)47 to transform the mol2 files into PDBQT format. All steroids and the positive control (silymarin) were treated fully flexible in terms of torsional degrees of freedom.2.7. Preparation of the protein structures and molecular docking (MDock)
Docking was performed with AutoDock Vina (version 1.1) using PDBQT files for the glutathione-S-transferase (GST) enzyme (PDB ID: 18GS) and the Cu–Zn human superoxide dismutase (Cu–ZnSOD) enzyme (PDB ID: 2C9V).48 In the AutoDock Vina program, the protein molecules were kept rigid, and the ligands were treated as flexible molecules with the number of active rotatable bonds ranging from 0 to 32. AutoDock Vina uses a gradient optimization method in its local optimization procedure using the Broyden–Fletcher–Goldfarb–Shanno (BFGS) method, which is an efficient quasi-Newton method. AutoDock Vina has implemented a hybrid scoring function (empirical plus knowledge-based function) inspired by the X-Score function, mainly differing in the terms and in the parametrization method. Water molecules, ions and ligands were removed from 18GS and 2C9V prior to docking using the AutoDockTools (http://mgltools.scripps.edu/). The search boxes were set to cover the electrophilic residues (Tyr7-Trp38-Gly205) and the A and F interface side chain residues (Val7-Asn53-Val148) for GST and SOD enzymes, respectively, with size 40 × 40 × 40 Å3 centred at (7.861, 6.722, 26.194) Å and (16.570, −15.508, 16.942) Å, respectively. The enzymes GST and SOD have their ligands tethered to them by adjusting the parameters of a genetic algorithm (GA) over the course of 10 iterations. PyMOL v2.0 Schrödinger, LLC, UCSF Chimera,49 and LigPlot+ v2.2.5 (ref. 50) were used to depict the docking binding postures.2.8. In silico prediction of physicochemical properties, pharmacokinetic and toxicity profiles
The pharmacokinetic properties of the most promising marine steroids (12, 14, 17 and 24) and the positive control (silymarin) were calculated using the SWISS-absorption, distribution, metabolism and excretion (ADME) platform (https://www.swissadme.ch/, accessed on 14 August 2023).513. Results and discussion
3.1. In vivo evaluation of the hepatoprotective ability of S. glaucum extract
Liver fibrosis is regarded as a major health condition due to the liver's prominent involvement as an organ in several physiological processes such as metabolism and detoxification of a variety of medications and xenobiotics. Fibrosis is caused by an insufficient tissue repair process that occurs after chronic liver injuries such as alcohol-induced damage, chronic viral hepatitis B or C, parasite and metabolic illnesses. If fibrosis is not medicated, it can develop to cirrhosis, which can lead to hepatic failure and potentially liver cancer. Despite the severity of the condition, there are currently few therapy options for liver fibrosis.52Table 2 demonstrated that as compared to the control group, both the acetaminophen group and the treatment group (acetaminophen + silymarin) had a substantial rise in liver enzymes (ALT, AST, and ALP) and total bilirubin, but no difference in soft coral extract group and treated group (acetaminophen + soft coral) when compared to control group. However, as compared to the acetaminophen group, the treated groups (acetaminophen + soft coral) and (acetaminophen + silymarin) exhibited considerable improvement after therapy. Our data demonstrated that the group treated with soft coral extract restored liver enzymes to normal levels better than the group treated with silymarin. The increased activity of liver enzymes (ALT and AST) and bilirubin was caused by hepatocyte membrane damage and leakage of cytosol enzymes such as ALT, AST, and total bilirubin into the bloodstream.
| Groups | Control | Acetaminophen | Soft coral | Acetaminophen + soft coral | Acetaminophen + silymarin |
|---|---|---|---|---|---|
| a The mean ± standard error (SE) is used to express the values. The number n, denotes the number of rats (10 rats), in each group. Value of p < 0.05 was considered statistically significant. a, b, c significant at p < 0.05 in comparison to the control group, acetaminophen group; acetaminophen + soft coral group, respectively. | |||||
| ALT (IU L−1) | 24.14 ± 0.56 | 75.05 ± 1.08a | 24.95 ± 0.84b | 30.66 ± 0.60a,b | 46.77 ± 0.88a,b,c |
| AST (IU L−1) | 37.30 ± 1.34 | 95.39 ± 0.91a | 34.25 ± 0.81b | 39.31 ± 0.89b | 60.42 ± 1.03a,b,c |
| ALP (IU L−1) | 116.49 ± 1.31 | 219.48 ± 0.68a | 112.89 ± 0.98b | 117.94 ± 1.44b | 153.92 ± 1.18a,b,c |
| Total bilirubin (mg dl−1) | 0.71 ± 0.01 | 1.16 ± 0.01a | 0.70 ± 0.01b | 0.75 ± 0.01b | 0.86 ± 0.01a,b,c |
The levels of these enzymes are indicators of liver damage and are used to evaluate acetaminophen-induced liver damage.53 Consistent with our findings, an increased blood level of ALP is indicative of hepatobiliary and hepatocellular damage. Faulty liver discharges or enhanced ALP synthesis in hepatic parenchymal or duct cells may be the cause of the elevated ALP levels.54 Our findings are corroborated those of Zidan et al.55 and Kim et al.10 discovered that silymarin groups resulted in a significant improvement when compared to acetaminophen groups. This is due to silymarin's ability to scavenge the active free radical, allowing it to limit lipid peroxide generation and protect hepatocytes from injury. In comparison with the acetaminophen group, the treated group with soft coral extract demonstrated significant drop of the liver enzyme (ALT, AST, ALP) and total bilirubin and a return to normal levels of liver enzymes and total bilirubin, as shown in Table 2. This might be explained that the soft coral extract helps to maintain the structural integrity of the hepatocellular membrane making it less permeable to enzymes and so preventing their release into the bloodstream. Our findings support the widely held belief that transaminase levels revert to normal with hepatic parenchymal repair and hepatocyte regeneration.1 In addition, Abdel-Wahhab et al.32 agree that soft coral extract had a better effect because of the significant reduction in serum ALP levels that it induced in hepatotoxic rats.
Table 3 showed significant increase in tumor markers (AFP and CEA) in both acetaminophen and treated groups (acetaminophen + silymarin) in contrast to the control group while there was non-significant change in the soft coral extract group and treated group (acetaminophen + soft coral) when compared to control group. However, as compared to the acetaminophen group, the treated groups (acetaminophen + soft coral) and (acetaminophen + silymarin) exhibited considerable improvement after therapy. These results indicated that the soft coral extract has a powerful anti-cancer effect more potent than silymarin as positive control.
| Groups | Control | Acetaminophen | Soft coral | Acetaminophen + soft coral | Acetaminophen + silymarin |
|---|---|---|---|---|---|
| a The mean ± standard error (SE) is used to express the values. The number n denotes the number of rats (10 rats) in each group. Value of p < 0.05 was considered statistically significant. a, b, c significant at p < 0.05 in comparison to the control group, acetaminophen group; acetaminophen + soft coral group, respectively. | |||||
| AFP (IU ml−1) | 0.36 ± 0.01 | 1.56 ± 0.01a | 0.33 ± 0.01b | 0.39 ± 0.01b | 0.70 ± 0.01a,b,c |
| CEA (mg ml−1) | 0.28 ± 0.01 | 1.13 ± 0.01a | 0.27 ± 0.01b | 0.33 ± 0.01a,b | 0.66 ± 0.01a,b,c |
Liver fibrosis is a chronic liver disease characterized by the accumulation of excess extracellular matrix in the liver. Both AFP and CEA are glycoproteins that are often used as tumor markers for different types of liver cancer, including hepatocellular carcinoma (HCC). While AFP levels can be elevated in cases of HCC, which is a potential complication of liver fibrosis, AFP levels are not typically elevated in cases of liver fibrosis itself. Similarly, while CEA levels may be increased in some cases of liver fibrosis in rats, it is not a reliable marker for the disease and cannot be used as a diagnostic tool.12 Several prior research have reported the anti-cancer activity of soft corals against several forms of cancer, including breast, lung, colon, and liver cancer.20
Because of its different modes of action in liver cancer, the group treated with silymarin following development of liver fibrosis demonstrated improvement in AFP and CEA when compared to the acetaminophen group. It has been shown to slow the growth of liver cancer cells by causing cell cycle arrest and death. Indeed, silymarin has been demonstrated to have anti-inflammatory effects, which may help lessen the chance of developing liver cancer. Furthermore, silymarin possesses antioxidant capabilities that may help protect liver cells from damage caused by free radicals and other hazardous substances. Also, silymarin has showed promise as a supplementary treatment for liver cancer.56 However, in this study, the group treated with the soft coral after induction of liver fibrosis showed amazing results, coming back to near-normal levels. This may be because of the bioactive substances found in soft corals, such as terpenoids, steroids, alkaloids, and polyketides, which have been linked to the corals' anti-cancer action. These naturally occurring organic chemicals have been found to have anti-cancer cytotoxic, apoptotic, and anti-angiogenic properties. While these studies show promising results, further research is needed to fully understand the potential anti-cancer activity of the soft corals extract.
Liver fibrosis in rats is associated with oxidative stress and the detoxification pathways involving glutathione S-transferases (GSTs) and superoxide dismutase (SOD) enzymes. GSTs are a group of enzymes that help eliminate toxins from the body, including reactive oxygen species (ROS) and various xenobiotics. They catalyze the conjugation of glutathione, a potent antioxidant, to electrophilic compounds, facilitating their elimination from the body by increasing their solubility in water. In liver fibrosis, GSTs can be upregulated as a protective response to oxidative stress and the increased production of ROS. The enhanced activity of GSTs helps in the removal of toxic metabolites, thereby reducing the burden of oxidative damage and preventing further injury to liver cells.57
Superoxide dismutase enzymes are a family of antioxidant enzymes that catalyze the conversion of superoxide radicals (O2−) into less harmful hydrogen peroxide (H2O2). They are essential for cellular redox homeostasis and defense against oxidative damage. In liver fibrosis, oxidative stress is a hallmark feature resulting from too much reactive oxygen species (ROS) production and not enough antioxidant defense systems, SOD enzymes, including copper–zinc SOD (CuZnSOD) and manganese SOD (MnSOD), are involved in neutralizing superoxide radicals and preventing their harmful effects. However, the activity of SOD enzymes can be influenced by the severity and progression of liver fibrosis. Initially, SOD activity may increase as a compensatory response to counteract the elevated ROS production. However, in advanced stages of fibrosis, the antioxidant defense system may become overwhelmed, leading to decreased SOD activity and further oxidative damage.58
Table 4 showed significant decline in antioxidant enzyme (SOD and GST) in both acetaminophen and treated groups (acetaminophen + silymarin) when compared to control group while there was non-significant change in soft coral extract group and treated group (acetaminophen + soft coral) in comparison to the control group. However, in comparison to acetaminophen group, the treated groups (acetaminophen + soft coral) and (acetaminophen + silymarin) exhibited considerable improvement after therapy. These results indicated that soft coral extract has a powerful antioxidant effect more potent than silymarin drug.
| Groups | Control | Acetaminophen | Soft coral | Acetaminophen + soft coral | Acetaminophen + silymarin |
|---|---|---|---|---|---|
| a The mean ± standard error (SE) is used to express the values. The number n denotes the number of rats in each group (10 rats). Value of p < 0.05 was considered statistically significant. a, b, c significant at p < 0.05 in comparison to the control group, acetaminophen group; acetaminophen + Soft coral group, respectively. | |||||
| GSTs (U per g tissue) | 514.54 ± 11.56 | 275.05 ± 8.05a | 511.75 ± 9.93b | 488.16 ± 15.60b | 410.67 ± 13.48a,b,c |
| SOD (U per g tissue) | 439.25 ± 9.34 | 244.32 ± 11.91a | 437.25 ± 8.81b | 405.88 ± 9.89b | 320.59 ± 11.73a,b,c |
From our results we found that the expression of GSTs and SOD in liver fibrosis can have protective effect. Because they can promote the detoxification of pro-fibrogenic agents, such as oxidative stress-inducing compounds, and thereby protect liver cells from injury.
As in Table 5, result of this research found a growth significantly in TGF-β and TNF-α in both acetaminophen and the treatment group (acetaminophen + silymarin) as compared to the control group, indicating that it may cause liver inflammation in rats. Through the comparison with the control group, there was no observable difference in the soft coral extract group or the treated group (acetaminophen + soft coral). However, when compared to the acetaminophen group, the treated groups (acetaminophen + soft coral) and (acetaminophen + silymarin) demonstrated significant improvement after therapy due to the hepatoprotective effect of the soft coral extract and silymarin against liver inflammation caused a decrease in TGF-β and TNF-α liver content.
| Groups | Control | Acetaminophen | Soft coral | Acetaminophen + soft coral | Acetaminophen + silymarin |
|---|---|---|---|---|---|
| a The mean ± standard error (SE) is used to express the values. The number n denotes the number of rats (10 rats), in each group. Value of p < 0.05 was considered statistically significant. a, b, c significant at p < 0.05 in comparison to the control group, acetaminophen group; acetaminophen + soft coral group, respectively. | |||||
| TNF-α ng g−1 | 131.60 ± 1.18 | 535.03 ± 3.08a | 129.91 ± 0.91b | 137.64 ± 1.63b | 250.60 ± 0.84a,b,c |
| TGF-β ng g−1 | 228.66 ± 0.97 | 950.23 ± 2.24a | 220.19 ± 1.72a,b | 230.35 ± 0.96b | 317.41 ± 1.36a,b,c |
| TNF-α relative | 1.08 ± 0.07 | 9.01 ± 0.13a | 0.88 ± 0.06b | 1.33 ± 0.05b | 2.80 ± 0.13a,b,c |
| TGF-β relative | 1.08 ± 0.07 | 6.83 ± 0.12a | 0.92 ± 0.06b | 0.94 ± 0.06b | 1.91 ± 0.08a,b,c |
Transformed growth factor beta and TNF-α both play vital roles in the development and progression of liver inflammation and fibrosis. TNF-α is a pro-inflammatory cytokine which is produced due to Kupffer cells activation, hepatic stellate cells, and infiltrating immune cells. TNF-α promotes liver fibrosis by inducing the activation of hepatic stellate cells, which are the main effector cells in liver fibrosis. TGF-β is a multifunctional cytokine that also plays a role in the development and progression of liver inflammation and fibrosis. TGF-β is produced by many cell types, including hepatic stellate cells, Kupffer cells, and infiltrating immune cells. TGF-β promotes liver fibrosis by inducing the activation of hepatic stellate cells and stimulating extracellular matrix production. TGF-β can also promote the differentiation of fibroblasts into myofibroblasts, which are cells that are involved in extracellular matrix production.59
As demonstrated in Table 5, TGF-β and TNF-α gene expression was considerably higher in the acetaminophen and treatment groups (acetaminophen + silymarin) than in the control group. No significant difference was observed in the soft coral extract group or the treated group (acetaminophen + soft coral), in comparison with the control group; However, as compared to the acetaminophen group, the treated groups (acetaminophen + soft coral extract) and (acetaminophen + silymarin) improved significantly after therapy. These findings showed that acetaminophen induction could cause liver injury and that soft coral extract and silymarin treatment reduced TGF-β and TNF-α expression. Silymarin has been reported to inhibit the production of TNF-α in a variety of cells, including liver and immune cells, by blocking the activation of the NF-κB signaling pathway. Also, silymarin has been shown to inhibit the activity of TGF-β in various types of cancer cells by blocking its signaling pathway. By modulating the activity of these cytokines, Silymarin may help reduce inflammation and inhibit cancer progression.56
The soft coral organic extract has been found to have anti-inflammatory and anti-fibrotic effects. The therapeutic effect of soft corals was due to contain bioactive compounds that can modulate the activity of various signaling pathways involved in inflammation and fibrosis, including the TGF-β and TNF-α pathways. The anti-TNF-α activity of soft coral compounds is thought to be mediated by the inhibition of NF-κB, a transcription factor that plays a key role in the production of pro-inflammatory cytokines such as TNF-α. The anti-TGF-β activity of soft coral compounds is thought to be mediated by the inhibition of Smad signaling, a downstream pathway activated by TGF-β that promotes fibrosis and deposition of extracellular matrix.60
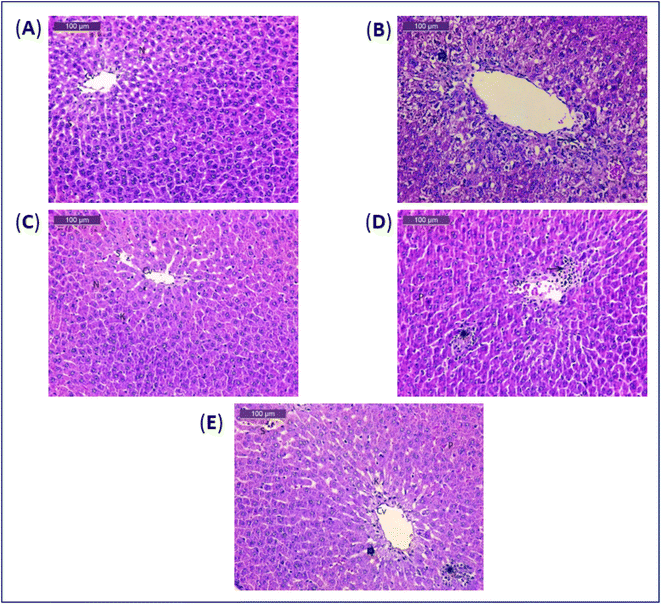
The histopathological findings from this research (Fig. 1 and Table 6), revealed that the control group's liver slices revealed typical hepatic architecture, with hepatocytes distributed in cords radiating from the central veins and spherical vesicular nuclei with blood sinusoids (Fig. 1A). Histological examination of acetaminophen-treated liver tissues revealed severe degenerative alterations, necrosis, mononuclear cell infiltration, interstitial haemorrhage with pyknotic nuclei, light dilated blood sinusoids, and minor activation of Kupffer cells (Fig. 1B). Liver tissues of the group treated with the soft corals only, had almost normal structure and modest activation of Kupffer cells (Fig. 1C). The examination sections in the acetaminophen-treated and soft coral groups were basically normal, with moderate degenerative changes with pyknotic nuclei and mild Kupffer cells (Fig. 1D). Liver tissues of acetaminophen and silymarin group showed more or less usual degenerative alterations with pyknotic nuclei and moderate Kupffer cells (Fig. 1E). Histopathological findings corroborated the biochemical conclusions.
| Groups | Control | Acetaminophen | Soft coral | Acetaminophen + soft coral | Acetaminophen + silymarin |
|---|---|---|---|---|---|
| a Histological grading was made according to four severity grades: − (none); + (mild); ++ (moderate) and +++ (severe). | |||||
| Hepatic necrosis | — | +++ | — | + | + |
| Inflammatory infiltration | — | +++ | — | — | — |
| Congestion and dilatation of sinusoids | — | +++ | — | — | — |
| Pyknotic nuclei | — | +++ | — | + | + |
| Proliferation of Kupffer cells | — | +++ | — | + | + |
3.2. Identification of the isolated metabolites
S. glaucum organic extract, collected from the reef of Hurghada in Egypt was submitted to a series of chromatographic purifications to yield 3 known metabolites (1–3) (Fig. 2). Compounds 1–3 were identified by comparison of their spectroscopic and physical characteristics i.e., 1H-NMR data and their ESI-MS spectra (Fig. S1–S6†), with those reported in the literature as 24(S)-methyl-cholest-5-en-3β-ol (1),61 gorgostan-5,25-dien-3β-ol (2)62 and gorgosterol (3),61 previously isolated from the soft corals Lobophytum crassum and Lobophytum lobophytum, respectively.3.3. Molecular docking (MDock) and binding energies studies
The binding mechanism of a focused library of 26 steroid derivatives (1–26), (Fig. 2) along with a positive control, silymarin, against GST and SOD for hepatoprotective activity was determine through the performance of a molecular docking experiment. The steroid dataset comprises: 21 derivatives with core I (1–9, 11–18, 22, 24–26), 4 derivatives with core II (10, 19–21), and only one derivative with core III (23) (Fig. 1 and Scheme S1†). The outcomes of molecular docking with the AutoDock Vina program on GST and SOD targets were displayed in Table S1 of the ESI data.† Fig. 3 displays the optimal docked poses for silymarin (the positive control), were shown on GST and SOD enzymes for hepatoprotective activity.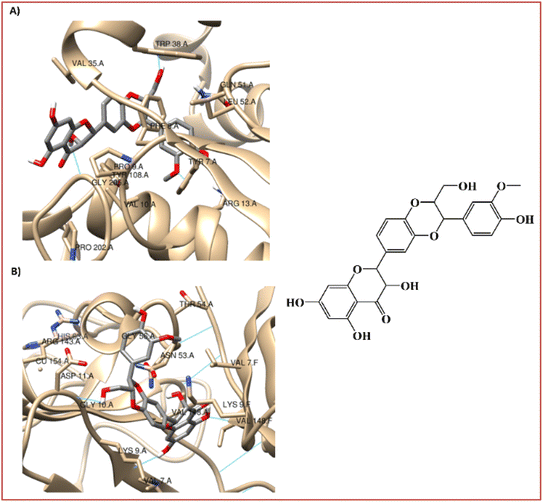 | ||
| Fig. 3 Interaction profiles of the best-docked poses for the silymarin on (A) GST and (B) SOD enzymes. | ||
It is known that GST play a pivotal role in the detoxication of foreign chemicals and toxic metabolites.6,8 Fig. 3A shows a clear interaction between the electrophilic residues, Tyr7-Pro9-Val35-Trp38-Gly205, of the GST enzyme with silymarin in a very similar position to that found for the sulfasalazine, a known inhibitor of GST.6,8 The 3-(4′-hydroxyphenyl), 3-(3′-methoxyphenyl), and 2-hydroxymethyl substitutes in the 1,4-benzodioxine moiety of silymarin apparently participate in the interaction of the hydrogen-bond with the carboxylate oxygen atom of Leu52, with the 4-hydoxylphenyl side chain of Tyr7 and with the indole side chain of Trp38, respectively (Fig. 3A).
On the other hand, the 3-hydroxyl substitute in the 2,3-dihydro-4H-chromon-4-one moiety of silymarin is within hydrogen bonding distance of Gly205, Fig. 3A. Also, the hydrophobic interactions of the benzene ring in the 1,4-benzodioxine and 2,3-dihydro-4H-chromon-4-one fused systems of silymarin with the Phe8-Val35 and Pro9 residues of the GST enzyme, respectively are very relevant (Fig. 3A).
SOD is a homodimer that converts toxic oxygen radicals to less harmful species.63,64 The catalytic pocket is defined by Cu ion and residues of His63 and Arg143,63 Fig. 3B. The dimerization of SOD1 is essential to the stability of the protein.63 Fig. 3B shows a clear interaction between the residues, Val7-Asn53-Val148, of the A and F interface sidechain in SOD with silymarin. The 3,5,7-trihydroxyl substitutes in the 2,3-dihydro-4H-chromon-4-one moiety of silymarin apparently interact in hydrogen-bonding interactions with the carboxylate oxygen atom of Asn53 (A), with the amine nitrogen atom of Val148 (F) and with the carboxylate oxygen atom of Val148(A) – with amide nitrogen atom of Val7(A), respectively (Fig. 3B).
In addition to these interface side chain interactions, the 2-hydroxymethyl substituent in the 1,4-benzodioxin moiety of silymarin participates in hydrogen bonding interactions with both the carboxylate oxygen atom of Gly10 (A) and the carboxylate side chain of Asp11 (A), Fig. 3B. The hydrophobic interactions of the 4′-hydroxy-3′-methoxyphenyl in the 1,4-benzodioxin moiety of silymarin with residues Lis9 (F), Thr54 (A), Gly56 (A), and Ala55 (A) of the SOD enzyme appear to be very relevant as well (Fig. 3B).
To detect the most favourable binding interactions, a selection of three steroid derivatives that were recovered experimentally in the current work, and six steroid derivatives were done for each target as well as the positive control (silymarin) through a virtual screening using a flexible molecular docking. The calculated free binding energies by the set of search space coordinates are reported in Table 7.
| Steroid derivatives | ΔGB, in kcal mol−1 | H-Bond residues | Hydrophobic interaction residues | |||
|---|---|---|---|---|---|---|
| GST | SOD | GST | SOD | GST | SOD | |
| a The steroid derivatives recovered experimentally in current work.b The steroid derivatives selected (highlighted in bold) have a calculated ΔGB ≤ −8.9 kcal mol−1 and −9.0 kcal mol−1 for GST and SOD, respectively.c Positive control.d Chain B of the GST.e Chains A and F of the SOD.f Chain F of the SOD. | ||||||
| 1a | −7.7 | −7.8 | — | — | Tyr7, Phe8, Val10, Arg13, Val35, Gln51, Leu52, Ile104, Tyr108, Gly205, Asp98d | Leu106, Ser107, Gly108e, Asp109e, Cys111e, Ile113e, Arg115, Ile151f |
| 2a | −8.4 | −8.2 | — | — | Tyr7, Phe8, Pro9, Val10, Arg13, Val35, Gln51, Leu52, Ile104, Tyr108, Gly205 | Gly49, Gly108, Asp109, Cys111f, Ile113f, Arg115, Ile151 |
| 3a | −8.4 | −8.8 | — | — | Tyr7, Phe8, Val10, Arg13, Val35, Gln51, Leu52, Ile104, Tyr108, Gly205, Asp98d | Leu106f, Ser107, Gly108f, Asp109e, Cys111e, Ile113e, Arg115, Ile151f |
| 12b | −8.9 | −9.1 | Arg13 | — | Tyr7, Phe8, Pro9, Val10, Val35, Gln51, Ile104, Tyr108, Gly205 | Ser107f, Gly108f, Asp109, Ile113f, Arg115, Ile151 |
| 13b | −9.1 | −9.0 | Try108 | — | Tyr7, Phe8, Pro9, Val10, Arg13, Val35, Gln51, Ile104, Gly205 | Asp109, Ile113e, Arg115, Ile151e |
| 14b | −9.3 | −9.4 | Arg13 | Gly108f | Tyr7, Phe8, Pro9, Val10, Val35, Gln51, Ile104, Tyr108, Gly205 | Ser107f, Asp109, Ile113e, Arg115, Ile151f |
| 16b | −8.4 | −9.0 | — | Gly108f | Tyr7, Phe8, Pro9, Val10, Arg13, Val3, Val35, Thr34, Ile104, Tyr108, Gly205 | Ser107, Gly108, Asp109, Ile113f, Arg115, Ile151 |
| 17b | −9.3 | −8.7 | Arg13, Try108 | Cys111f | Tyr7, Phe8, Pro9, Val10, Val35, Gln51, Ile104, Gly205 | Ser107, Gly108, Asp109, Arg115, Ile151e |
| 24b | −9.1 | −9.1 | — | — | Tyr7, Phe8, Pro9, Val10, Arg13, Val35, Ile104, Tyr108, Gly205 | Ser107f, Asp109, Ile113f, Arg115, Ile151 |
| Silymarinc | −8.5 | −8.6 | Tyr7, Trp38, Leu52, Gly205 | Val7, Gly10, Asp11, Asn53, Val14e | Phe8, Pro9, Arg13, Val36, Tyr108 | Lys9e, Thr54, Gly56, Ala55 |
As shown in Table 7 and Fig. 2, all the steroid derivatives selected to be the most promising GST and SOD modulators are from core I (12–14, 16, 17, 24). Double bond presence in the B ring of the tetracyclic system of the steroidal nucleus seems to potentiate the activity against the GST and SOD enzymes. Likewise, all the seven steroid derivatives selected against the GST and SOD enzymes present a cyclopropane ring in the side chain, Scheme S1† and Table 7.
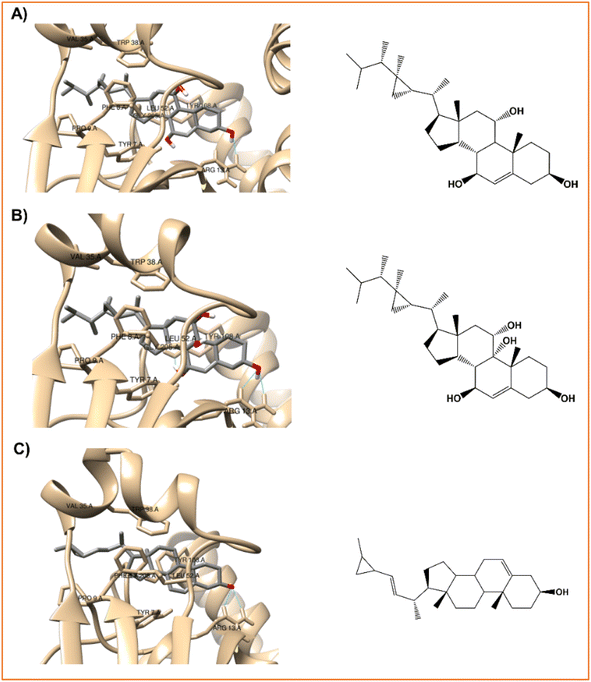 | ||
| Fig. 4 Interaction profiles of the best-docked poses for the (A) klyflaccisteroids G (14), (B) klyflaccisteroids I (17), and (C) glaucasterol (24). | ||
As verified with the positive control, silymarin (Fig. 3A), in the case of the three steroid derivatives selected as the most promising hepatoprotective agents (14, 17 and 24) there is a clear interaction between these and the electrophilic residues, Tyr7-Pro9-Val35-Gly205, from the GST enzyme as shown in Fig. 4 and Table 7.
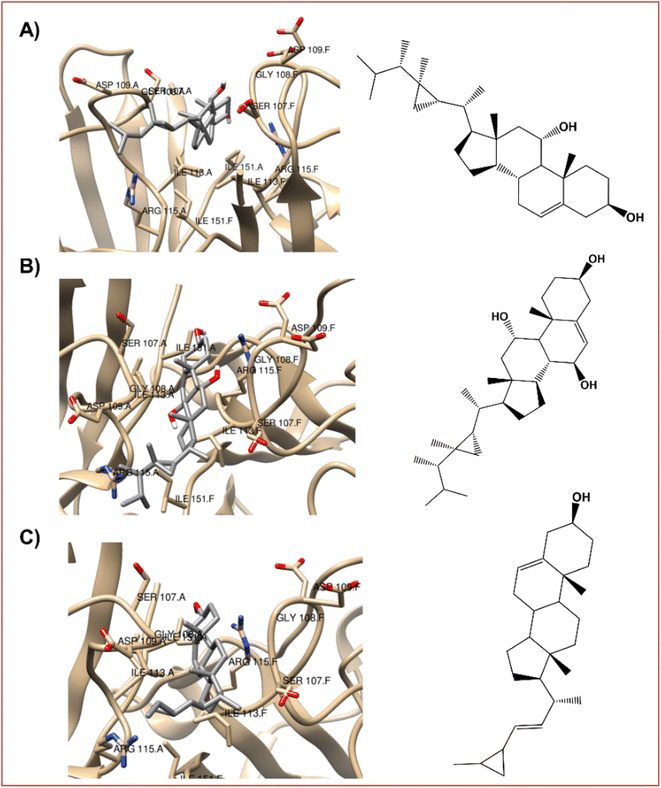 | ||
| Fig. 5 The best-docked poses interaction profiles of (A) 11α-hydroxygorgosterol (12), (B) klyflaccisteroids G (14), and (C) glaucasterol (24). | ||
It is verified that the three selected steroid derivatives (12, 14 and 24) interact with SOD in a different binding site than that obtained for the positive control, silymarin, as show in Fig. 5 and 3B, respectively. However, they all bind at the A and F interface sidechain in SOD. For all the steroid derivatives there is a clear interaction with the residues Gly108-Ile113-Ile151 of the A and F interface sidechain in SOD (Table 7 and Fig. 5) rather than the residues that interact with the positive control, silymarin, Val7-Asn53-Val148 (Fig. 3B).
3.4. In silico prediction of pharmacokinetics, toxicity and druglikeness (ADME/Tox)
In order to assess our compounds' pharmacokinetics, SwissADME (http://www.swissadme.ch//), an online free tool, was used to evaluate the compounds' properties. It was found that the most promise steroids (12, 14, 17, and 24) had only 1 Lipinski rule violation each, while the positive control (silymarin) also had one Veber's rule violation (Table 8).| ADME/Tox | 12 | 14 | 17 | 24 | Silymarin |
|---|---|---|---|---|---|
| a Drug likeness.b Medicinal chemistry.c Lipophilicity.d Water solubility.e Pharmacokinetics. | |||||
| Lipinski #violationsa | 1 | 1 | 1 | 1 | 0 |
| Veber #violationsa | 0 | 0 | 0 | 0 | 1 |
| Bioavailability scorea | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 |
| PAINS #alertsb | 0 | 0 | 0 | 0 | 0 |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w (XLOGP3)c Po/w (XLOGP3)c |
7.59 | 6.48 | 5.42 | 7.26 | 1.90 |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S (ESOL)d S (ESOL)d |
−7.10 | −6.50 | −5.93 | −6.59 | −4.14 |
| GI absorptione | Low | High | High | Low | Low |
| BBB permeante | No | No | No | No | No |
| P-gp substratee | No | No | Yes | No | No |
| CYP inhibitorse | No | No | No | Yes | Yes |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp (skin permeation)e Kp (skin permeation)e |
−3.61 | −4.50 | −5.35 | −3.48 | −7.89 |
None of the steroids or the positive control were predicted to have PAINS alert. The Abbot bioavailability Score predicts the probability of a compound to have at least 10% oral bioavailability in rat or measurable Caco-2 permeability, all four steroides and the positive control were predicted with a score of 55% which is quite acceptable. The parameters lipophilicity, water solubility, gastrointestinal absorption (GI), skin permeability, and P-glycoprotein (P-gp) substrate and cytochrome P450 (CYP) inhibitors were used to predict the absorption level of the four selected steroids and the positive control (silymarin). As it is known that lipid-soluble drugs are less well absorbed than those that are water-soluble, acceptable parameters are achieved for log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S not higher than 6 and lipophilicity between −0.7 and 5. All four steroid derivatives are predicted to have adequate water solubility characteristics but high lipophilicity (>5). It was suggested that P-gp and CYP have a relevant role in the protection of tissues and organisms, thus the interaction with these targets is seen in a positive way. Thus, the steroids (14 and 17) are predicted to be P-gp substrates and the steroid (24) and the positive control (silymarin) are predicted to inhibit at least one CYP type. The more negative the log
S not higher than 6 and lipophilicity between −0.7 and 5. All four steroid derivatives are predicted to have adequate water solubility characteristics but high lipophilicity (>5). It was suggested that P-gp and CYP have a relevant role in the protection of tissues and organisms, thus the interaction with these targets is seen in a positive way. Thus, the steroids (14 and 17) are predicted to be P-gp substrates and the steroid (24) and the positive control (silymarin) are predicted to inhibit at least one CYP type. The more negative the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp, the less skin permeant is the molecule, therefore the least skin permeant predicted is the positive control (silymarin) and the most skin permeant predicted is the steroid (24). Only the steroids (14 and 17) had expected high GI absorption, and no blood–brain barrier penetration (related to distribution properties) was predicted for any of four steroids and the positive control.
Kp, the less skin permeant is the molecule, therefore the least skin permeant predicted is the positive control (silymarin) and the most skin permeant predicted is the steroid (24). Only the steroids (14 and 17) had expected high GI absorption, and no blood–brain barrier penetration (related to distribution properties) was predicted for any of four steroids and the positive control.
4. Conclusions
Drug and xenobiotic metabolism occur mostly in the liver and is profoundly affected by medicine and xenobiotic-mediated toxicity. Underreporting, difficulties in detection or diagnosis, and inadequate surveillance of exposure make it difficult to accurately assess the prevalence of drug-induced hepatotoxicity. Acetaminophen is one of the most commonly drug-induced liver failure in many countries when used at high doses. In summary, soft coral extracts have been shown to modulate the activity of TNF-α and TGF-β by inhibiting their expression and downstream signaling pathways. These anti-inflammatory and anti-fibrotic properties make soft corals potential sources of novel therapeutics for the treatment of inflammatory and fibrotic diseases, including liver fibrosis. These activities seem to be related to the modulation of GST and SOD enzymes by marine steroids isolated from soft coral extracts. A molecular docking study of 26 steroid derivatives revealed two promising steroid modulators (14 and 24) according to their prominent ligand-protein energy scores and relevant binding affinities with the GST and SOD pocket residues. The presence of a double bond in the B ring of the tetracyclic steroid system and the cyclopropane ring in the side chain seem to be key structural elements to potentiate the hepatoprotective activity.Data availability
The data presented in this study are available in the present article and the ESI.†Author contributions
Conceptualization: M. A. T. and A.-E. D.; methodology: M. A. T., F. P., O. A., A. M., and A.-E. D.; software: M. A. T., F. P., and O. A.; formal analysis: M. A. T., F. P., O. A., and A.-E. D.; investigation: M. A. T., F. P., O. A., and A.-E. D.; resources: M. A. T., F. P., O. A., and A.-E. D.; data curation: M. A. T., F. P., O. A., and A.-E. D.; writing original draft preparation: M. A. T., F. P., O. A., M. S, Y. M. D., and A.-E. D.; writing review and editing: M. A. T., F. P., O. A., and A.-E. D.; visualization: M. A. T. and A.-E. D. All authors have read and agreed to the published version of the manuscript.Institutional review board
The animal study protocol was approved by the Ethics Committee of National Research Center, Dokki, Cairo, Egypt, approval number (11236072023).Conflicts of interest
The authors declare that they have no known competing commercial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
Mohamed A. Tammam wishes to thank Prof. E. Ioannou and Prof. V. Roussis for assistance and permission to use facilities of their laboratories at the Department of Pharmacy, National and Kapodistrian University of Athens. Mohamed A. Tammam is humbly dedicating this work to the soul of his sister Dr Mai A. Tammam who passed away on 19 of March 2022, she was always a kind supporter in all aspects of his life. Florbela Pereira gratefully acknowledges FCT – Fundação para a Ciência e a Tecnologia, I. P., for an Assistant Research Position (CEECIND/01649/2021) and the project UIDB/50006/2020 of the Associated Laboratory for Green Chemistry (LAQV) of the Network of Chemistry and Technology (REQUIMTE). Amr El-Demerdash is immensely grateful to the John Innes Centre, Norwich Research Park, United Kingdom for the post-doctoral fellowship. Amr El-Demerdash is thankful to his home university, Mansoura University, Egypt for the unlimited support, inside and outside.References
- O. Aly, D. M. Abouelfadl, O. G. Shaker, G. A. Hegazy, A. M. Fayez and H. H. Zaki, Egypt. J. Med. Hum. Genet., 2020, 21, 1–9 CrossRef.
- J. Boeckmans, R. M. Rodrigues, T. Demuyser, D. Piérard, T. Vanhaecke and V. Rogiers, Arch. Toxicol., 2020, 94, 1367–1369 CrossRef CAS.
- S. Agrawal and B. Khazaeni, Toxicology Cases for the Clinical and Forensic Laboratory, 2023, vol. 1, pp. 75–77 Search PubMed.
- Y. M. Diab, M. A. Tammam, A. M. Emam, M. A. Mohamed, M. E. Mahmoud, W. M. Semida, O. Aly and A. El-Demerdash, Egypt. J. Chem., 2022, 65, 723–733 Search PubMed.
- H. Özbek, S. Uǧraş, H. Dülger, I. Bayram, I. Tuncer, G. Öztürk and A. Öztürk, Fitoterapia, 2003, 74, 317–319 CrossRef PubMed.
- Marine Pharmacology, https://www.marinepharmacology.org/, accessed 23 June 2023 Search PubMed.
- R. Thilagavathi, S. S. Begum, S. D. Varatharaj, A. kumar Balasubramaniam, J. S. George and C. Selvam, Phytother. Res., 2023, 37, 2102–2118 CrossRef PubMed.
- A. J. Oakley, M. Lo Bello, M. Nuccetelli, A. P. Mazzetti and M. W. Parker, J. Mol. Biol., 1999, 291, 913–926 CrossRef CAS PubMed.
- R. W. Strange, S. V. Antonyuk, M. A. Hough, P. A. Doucette, J. S. Valentine and S. S. Hasnain, J. Mol. Biol., 2006, 356, 1152–1162 CrossRef CAS PubMed.
- Y. C. Kim, J. D. Na, D. Y. Kwon and J. H. Park, J. Funct. Foods, 2018, 49, 235–240 CrossRef CAS.
- Z. W. Liu, Y. M. Zhang, L. Y. Zhang, T. Zhou, Y. Y. Li, G. C. Zhou, Z. M. Miao, M. Shang, J. P. He, N. Ding and Y. Q. Liu, Front. Immunol., 2022, 12, 810286 CrossRef PubMed.
- Z. Guizhen, J. Guanchang, L. Liwen, W. Huifen, R. Zhigang, S. Ranran and Y. Zujiang, Front. Endocrinol., 2022, 13, 918869 CrossRef PubMed.
- S. K. Wang, M. K. Hsieh and C. Y. Duh, Mar. Drugs, 2013, 11, 4318–4327 CrossRef.
- J. W. Blunt, B. R. Copp, W. P. Hu, M. H. G. Munro, P. T. Northcote and M. R. Prinsep, Nat. Prod. Rep., 2009, 26, 170–244 RSC.
- A. El-Demerdash, M. A. Tammam, A. G. Atanasov, J. N. A. A. Hooper, A. Al-Mourabit and A. Kijjoa, Mar. Drugs, 2018, 16, 214 CrossRef PubMed.
- A. F. Ahmed, C. R. Tsai, C. Y. Huang, S. Y. Wang and J. H. Sheu, Mar. Drugs, 2017, 15, 23 CrossRef PubMed.
- Approved Marine Drugs, https://www.marinepharmacology.org/approved, accessed 23 June 2023 Search PubMed.
- M. A. Ghareeb, M. A. Tammam, A. El-Demerdash and A. G. Atanasov, Curr. Res. Biotechnol., 2020, 2, 88–102 CrossRef.
- W. Fenical, Calif. Agric., 1997, 51, 45–49 CrossRef.
- Y. T. Song, D. D. Yu, M. Z. Su, H. Luo, J. G. Cao, L. F. Liang, F. Yang and Y. W. Guo, Mar. Drugs, 2023, 21, 69 CrossRef CAS PubMed.
- Y. A. Elkhawas, A. M. Elissawy, M. S. Elnaggar, N. M. Mostafa, E. M. Kamal, M. M. Bishr, A. N. B. Singab and O. M. Salama, Mar. Drugs, 2020, 18, 41 CrossRef CAS PubMed.
- MarinLit – a database of the marine natural products literature, https://marinlit.rsc.org/, accessed 26 April 2023 Search PubMed.
- L. F. Liang and Y. W. Guo, Chem. Biodiversity, 2013, 10, 2161–2196 CrossRef CAS.
- S. K. Wang, M. K. Hsieh and C. Y. Duh, Mar. Drugs, 2012, 10, 1433–1444 CrossRef CAS.
- N. T. Ngoc, T. T. H. Hanh, T. H. Quang, N. X. Cuong, N. H. Nam, D. T. Thao, D. C. Thung, P. Van Kiem and C. Van Minh, Steroids, 2021, 176, 108932 CrossRef CAS PubMed.
- M. S. Zubair, K. O. Al-Footy, S. E. N. Ayyad, S. S. Al-Lihaibi and W. M. Alarif, Nat. Prod. Res., 2016, 30, 869–879 CrossRef CAS PubMed.
- T. Y. Huang, C. Y. Huang, S. R. Chen, J. R. Weng, T. H. Tu, Y. Bin Cheng, S. H. Wu and J. H. Sheu, Mar. Drugs, 2020, 19, 8 CrossRef PubMed.
- M. A. Tammam, M. Sebak, C. Greco, A. Kijjoa and A. El-Demerdash, J. Mol. Struct., 2022, 1268, 133711 CrossRef CAS.
- A. El-Demerdash, A. G. Atanasov, O. K. Horbanczuk, M. A. Tammam, M. Abdel-Mogib, J. N. A. Hooper, N. Sekeroglu, A. Al-Mourabit and A. Kijjoa, Mar. Drugs, 2019, 17, 115 CrossRef CAS PubMed.
- M. A. Tammam, M. I. Gamal El-Din, A. Abood and A. El-Demerdash, RSC Adv., 2023, 13, 8049–8089 RSC.
- F. Pereira, L. Bedda, M. A. Tammam, A. K. Alabdullah, R. Arafa and A. El-Demerdash, J. Biomol. Struct. Dyn., 2023, 1, 1–19 CrossRef.
- M. A. Abdel-Wahhab, A. A. El-Nekeety, N. S. Hassan, M. S. El-Hefnawy, M. M. Kotb, S. A. El-Mekkawy, N. A. Khalil and A. G. Hanna, Global Vet., 2012, 8, 244–253 Search PubMed.
- S. Reitman and S. Frankel, Am. J. Clin. Pathol., 1957, 28, 56–63 CrossRef CAS PubMed.
- A. Belfield and D. M. Goldberg, Enzyme, 1971, 12, 561–573 CrossRef CAS PubMed.
- Y. Wang, Y. Xiong, A. Zhang, N. Zhao, J. Zhang, D. Zhao, Z. Yu, N. Xu, Y. Yin, X. Luan and Y. Xiong, Food Sci. Nutr., 2020, 8, 3872–3881 CrossRef CAS PubMed.
- R. J. Chandler, L. E. Venturoni, J. Liao, B. T. Hubbard, J. L. Schneller, V. Hoffmann, S. Gordo, S. Zang, C. W. Ko, N. Chau, K. Chiang, M. A. Kay, A. Barzel and C. P. Venditti, Hepatology, 2021, 73, 2223 CrossRef CAS PubMed.
- C. J. Abeyounis, J. G. Kim, S. A. Wilhelm, K. R. Diakun and F. Milgrom, Immunol. Invest, 1989, 18, 143–157 CrossRef CAS PubMed.
- W. H. Habig, M. J. Pabst and W. B. Jakoby, J. Biol. Chem., 1974, 249, 7130–7139 CrossRef CAS PubMed.
- M. Nishikimi, N. Appaji Rao and K. Yagi, Biochem. Biophys. Res. Commun., 1972, 46, 849–854 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- A. Corti, G. Fassina, F. Marcucci, E. Barbanti and G. Cassani, Biochem. J., 1992, 284(Pt 3), 905–910 CrossRef CAS PubMed.
- S. J. Kim, D. Romeo, Y. Do Yoo and K. Park, Horm. Res., 1994, 42, 5–8 CrossRef CAS PubMed.
- G. Paget and R. Thomson, Standard operating procedures in pathology, MTP Press, Lancaster, 1979, pp. 134–139 Search PubMed.
- Gaussian 09 Citation | Gaussian.com, https://gaussian.com/g09citation/, accessed 8 January 2023 Search PubMed.
- A. D. Becke, J. Chem. Phys., 1993, 98, 1372–1377 CrossRef CAS.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- N. M. O'Boyle, M. Banck, C. A. James, C. Morley, T. Vandermeersch and G. R. Hutchison, J. Cheminf., 2011, 3, 1–14 Search PubMed.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CAS.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- A. C. Wallace, R. A. Laskowski and J. M. Thornton, Protein Eng., 1995, 8, 127–134 CrossRef CAS PubMed.
- A. Daina, O. Michielin and V. Zoete, Sci. Rep., 2017, 7, 1–13 CrossRef.
- J. Hussein, M. El-Banna, K. F. Mahmoud, S. Morsy, Y. Abdel Latif, D. Medhat, E. Refaat, A. R. Farrag and S. M. El-Daly, Biomed. Pharmacother., 2017, 90, 880–887 CrossRef CAS PubMed.
- F. L. Yen, T. H. Wu, L. T. Lin and C. C. Lin, J. Ethnopharmacol., 2007, 111, 123–128 CrossRef PubMed.
- T. Swaroop and S. Gowda, Int. J. Pharm. Chem. Sci., 2012, 1, 675–682 Search PubMed.
- S. A. Zidan, M. A. Orabi, M. A. Mustafa, M. AAl-Hammady, M. S. Kamel and C. A. Sabry Zidan, J. Pharmacogn. Phytochem., 2016, 5, 247–251 Search PubMed.
- T. Koltai and L. Fliegel, J. Evidence-Based Integr. Med., 2022, 27, 2515690X211068826 CAS.
- S. Landi, Mutation Research/Reviews in Mutation Research, 2000, 463, 247–283 CrossRef CAS PubMed.
- T. Fukai and M. Ushio-Fukai, Antioxid. Redox Signaling, 2011, 15, 1583 CrossRef CAS PubMed.
- A. M. Elsharkawy and D. A. Mann, Hepatology, 2007, 46, 590–597 CrossRef CAS PubMed.
- N. O. Bawakid, H. S. Alorfi, N. M. Alqarni, A. B. Abdel-Naim and W. M. Alarif, Naunyn-Schmiedeberg's Arch. Pharmacol., 2023, 396, 289–300 CrossRef CAS PubMed.
- M. P. Rahelivao, T. Lübken, M. Gruner, O. Kataeva, R. Ralambondrahety, H. Andriamanantoanina, M. P. Checinski, I. Bauer and H. J. Knölker, Org. Biomol. Chem., 2017, 15, 2593–2608 RSC.
- M. E. F. Hegazy, T. A. Mohamed, A. I. Elshamy, A. A. Hassanien, N. S. Abdel-Azim, M. A. Shreadah, I. I. Abdelgawad, E. M. Elkady and P. W. Paré, Nat. Prod. Res., 2016, 30, 340–344 CrossRef CAS PubMed.
- S. C. C. Hsueh, M. Nijland, X. Peng, B. Hilton and S. S. Plotkin, Front. Mol. Biosci., 2022, 9, 845013 CrossRef CAS PubMed.
- A. Arya, N. Azarmehr, M. Mansourian and A. H. Doustimotlagh, Arch. Physiol. Biochem., 2019, 127, 557–564 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra04843h |
| ‡ These authors are equally contributed. |
| This journal is © The Royal Society of Chemistry 2023 |