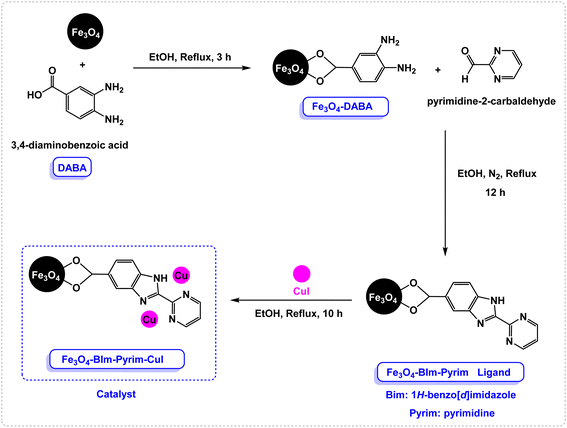
Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Retracted Article: Magnetic nanoparticles modified with a copper(I) complex as a novel and efficient reusable catalyst for A3 coupling leading to C–N bond formation†
Wei Li*a,
Jinlong Yana,
Wenjing Xua and
Li Yan Zhang *b
*b
aCollege of Science and Engineering, Jiaozuo Normal College, Jiaozuo, Henan 454000, China. E-mail: 1295007008@jzsz.edu.cn
bInstitute Chemical Technology, Guangzhou, China. E-mail: liyanzhang9090@gmail.com
First published on 3rd October 2023
Abstract
Propargylamines are an important and valuable family of nitrogen-containing compounds with many applications in the fields of medical, industrial, and chemical processes. One-pot multicomponent A3 coupling reactions of aldehydes, amines, and alkynes in the presence of transition metals as catalysts is an efficient strategy for preparing propargylamines. In this study, we fabricated a novel magnetically reusable copper nanocatalyst [Fe3O4–BIm–Pyrim–CuI] through the immobilization of the copper(I) complex on the surface of the magnetic nanoparticles modified with benzimidazole–pyrimidine ligand and evaluated its catalytic activity in the preparation of propargylamines through one-pot multicomponent A3 coupling reactions of aldehydes, amines, and alkynes. Under this catalytic system, aryl substrates with both electron-donating and electron-withdrawing substituents also gave the desired products in excellent yields under standardized conditions. The Fe3O4–BIm–Pyrim–CuI catalyst was easily separated using an external magnet, and the recovered catalyst was reused in 8 cycles without significant loss of activity.
Introduction
Propargylamines are an important class of alkyne-coupled amine compounds used in heterocyclic chemistry and pharmaceutical chemistry and have a large impact as pharmacophores in medicinal chemistry.1–4 Propargylamines are an important and valuable family of nitrogen-containing compounds with many applications in the fields of medicinal, industrial, and chemical processes.2,5 These compounds have the structure of numerous drugs, antibiotics, and natural products.6–8 One-pot multicomponent A3 coupling reactions of aldehydes, amines, and alkynes in the presence of transition metals as catalysts is an efficient strategy for preparing propargylamines.9,10 Therefore, the development of new, eco-friendly, and efficient catalytic systems for the synthesis of propargylamines via the multicomponent A3 coupling reactions of aldehydes, amines, and alkynes is an important challenge for synthetic chemists. Recently, numerous methods have been reported for the synthesis of propargylamines via the multicomponent A3 coupling reactions of aldehydes, amines, and alkynes using homogeneous transition metals as catalysts.11–13 However, homogeneous transition metal catalysts often suffer from high costs associated with their synthesis and recovery, making them less economically viable for large-scale industrial applications.14 Additionally, their tendency to undergo deactivation or decomposition over time can limit their catalytic activity and longevity, requiring frequent replacement and maintenance.The interrelationship between the magnetic properties and physicochemical characteristics of catalytic materials is a burgeoning field in materials science.15–19 This study explores the intricate connections between the catalytic performance of a material with its magnetic behavior, chemical composition, and physical attributes. This understanding is crucial for optimizing catalysis in various applications, from sustainable energy production to environmental remediation.20–22 By leveraging these connections, researchers aim to design catalytic materials with improved reactivity and efficiency, offering innovative solutions to pressing global challenges.23,24 During the last decade, nanocatalysts have attracted the attention of many researchers and have shown remarkable progress.25–28 The use of nanomaterials as support for catalysts is considered an extensive research field in organic synthesis.29–32 However, it is not easy to separate these nanocatalysts from the reaction mixture, and conventional separation methods, such as filtering are not effective due to the nano size of these catalysts.17,33–36 These limitations hinder the economic use of these nanocatalysts. To overcome this problem, the use of magnetic nanoparticles appears to be a suitable solution.37–41 Magnetic nanoparticles have high catalytic activity and a high degree of chemical stability.42,43 The paramagnetic nature and insolubility of magnetic nanoparticles make it easy to separate this catalyst from the reaction mixture using an external magnet.44,45 In this regard, the use of nanocatalysts could emerge as a promising and relevant approach to overcome obstacles in the synthesis of propargyl amines, offering numerous advantages for this challenging chemical transformation.16
In this study, we fabricated a novel magnetically reusable catalyst [Fe3O4–BIm–Pyrim–CuI] through the immobilization of a copper(I) complex on the surface of magnetic nanoparticles modified with benzimidazole–pyrimidine ligand and evaluated its catalytic activity in the synthesis of propargylamines via multicomponent A3 coupling reactions of aldehydes, amines, and alkynes under eco-friendly conditions.
Results and discussion
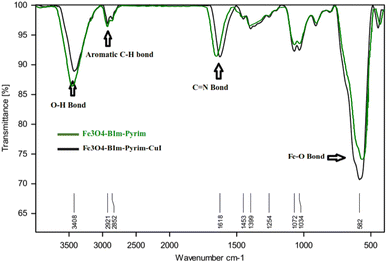
In the first step, Fe3O4 nanoparticles were synthesized using the previously described procedure. Then, 3,4-diaminobenzoic acid (DABA) was grafted on the surface of the magnetic Fe3O4 nanoparticles. The Fe3O4–BIm–Pyrim nanocomposite was fabricated through the reaction of Fe3O4–DABA with pyrimidine-2-carbaldehyde at refluxing ethanol. Finally, the as-fabricated Fe3O4–BIm–Pyrim nanocomposite was used as a ligand in order to afford the Fe3O4–BIm–Pyrim–CuI nanocatalyst via the immobilization of CuI by refluxing with ethanol. Scheme 1 shows the details of the preparation of the Fe3O4–BIm–Pyrim–CuI nanocatalyst.The structure of the Fe3O4–BIm–Pyrim–CuI nanocomposite was studied using Fourier transform infrared spectroscopy (FT-IR), scanning electron microscopy (SEM), transmission electron microscopy (TEM), energy-dispersive X-ray spectroscopy (EDX), thermogravimetric analysis (TGA), X-ray diffraction (XRD), vibrating sample magnetometer (VSM), and inductively coupled plasma optical emission spectroscopy (ICP-OES). FT-IR spectra of Fe3O4–BIm–Pyrim, and Fe3O4–BIm–Pyrim–CuI nanocomposite are shown in Fig. 1. The peak at 582 cm−1 is attributed to the Fe–O bond vibration of Fe3O4.46 The bands at 3400 cm−1 are assigned to the stretching vibration of the O–H groups. The presence of peaks at 2800–3000 are related to C–H aromatic bonds. As shown in Fig. 1, the observed shift in the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N characteristic peak from 1636 cm−1 to 1616 cm−1 confirms the successful complexation of Cu ions with the immobilized BIm–Pyrim ligand on the surface of Fe3O4 nanoparticles (link CuI with a bond).47–49
N characteristic peak from 1636 cm−1 to 1616 cm−1 confirms the successful complexation of Cu ions with the immobilized BIm–Pyrim ligand on the surface of Fe3O4 nanoparticles (link CuI with a bond).47–49
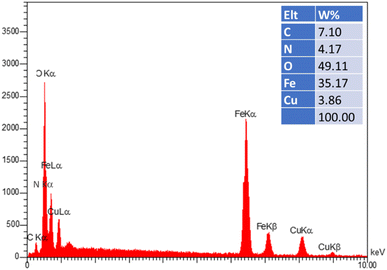
To confirm the support of the Cu complex on the surface of MNPs, EDX analysis of the as-constructed Fe3O4–BIm–Pyrim–CuI nanocomposite was performed (Fig. 2). The results exhibited the existence of Fe, O, N, C, and Cu in the structure of the Fe3O4–BIm–Pyrim–CuI nanocomposite. The ICP-OES technique was used to determine the amount of Cu loaded on the Fe3O4–BIm–Pyrim nanocomposite, which was found to be 15.12 × 10−5 mmol g−1.
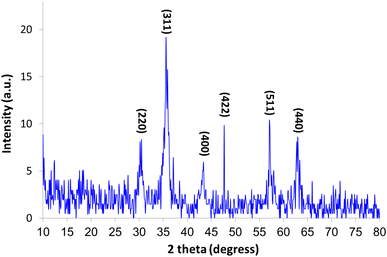
The crystalline structures of the Fe3O4–BIm–Pyrim–CuI nanocomposite were investigated by XRD. As shown in Fig. 3, the characteristic peaks at 2θ = 30.3°, 35.1°, 42.9°, 48.2°, 57.8°, and 63.1°, were indexed to (220), (311), (400), (422), (511), and (440) planes of the Fe3O4 nanoparticles cubic spinel crystal structure (JCPDS No. 75-0449)50,51 with a good match. SEM and TEM images of Fe3O4–BIm–Pyrim–CuI nanocomposite are shown in Fig. 4a and b. SEM and TEM images demonstrate that the catalyst was regular spherical nanoparticles, in which organic groups surround the nanomagnetic nucleus. In addition, based on particle size distribution (Fig. 4c), the catalyst was formed with a size distribution ranging from 17 to 45 nm with an average size of 28.62 nm.
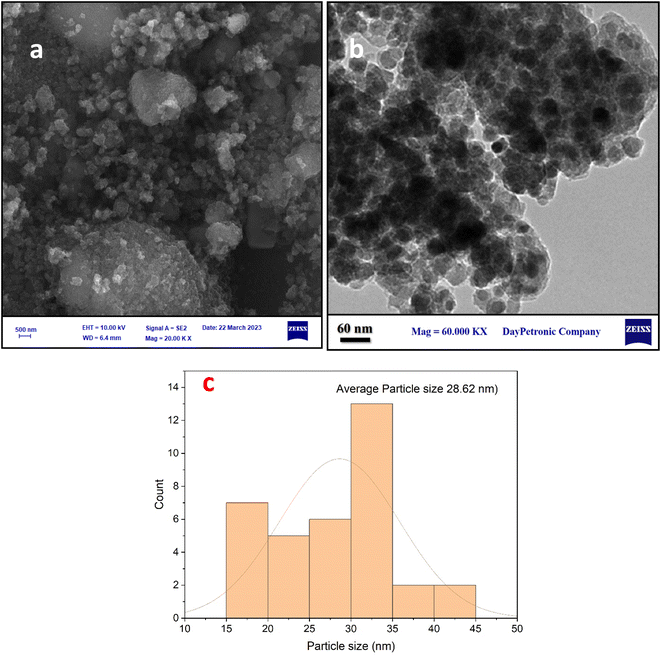 | ||
| Fig. 4 (a) SEM, (b) TEM images, and (c) particle size distribution histogram of the Fe3O4–BIm–Pyrim–CuI nanocatalyst. | ||
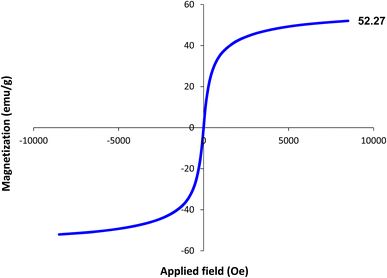
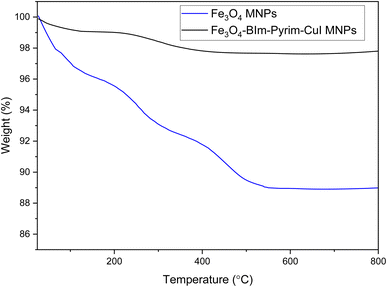
The magnetic properties of the Fe3O4–BIm–Pyrim–CuI nanocomposite were studied by VSM analysis at room temperature. As shown in Fig. 5, the saturation magnetization for the Fe3O4–BIm–Pyrim–CuI nanocomposite was found to be 52.27 emu g−1. The thermal behavior of the Fe3O4–BIm–Pyrim–CuI nanocomposite was investigated by TGA, and the results are shown in Fig. 6. The first weight losses in both of the curves are related to the adsorbed water and solvents that evaporated below 200 °C. The next weight loss for Fe3O4–BIm–Pyrim–CuI nanocomposite (about 9%) is related to the decomposition of functional groups and the complex. The results of the TGA analysis confirmed that organic compounds were present in the structure and showed the successful synthesis of the target catalyst.
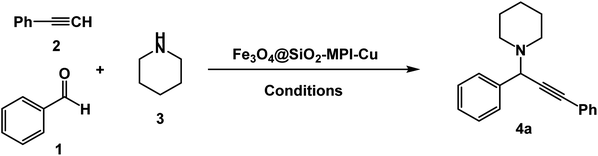
In order to optimize the standardized conditions, the effect of a number of factors such as catalyst loading, solvent, and temperature on the model reaction of benzaldehyde, piperidine, and phenylacetylene was examined. The model reaction was not accomplished when the reaction was carried out in the absence of the catalyst. Next, the model reaction was performed in the presence of various amounts of Fe3O4–BIm–Pyrim–CuI, and the results are summarized in Table 1. As seen in Table 1, no improvement of the reaction was observed by increasing the amount of the catalyst higher than 20 mg, and in terms of the yields and also according to the fact that using a minimum amount of the catalyst, 20 mg was considered as the best and optimized amount of the catalyst. To find the best reaction medium, the model reaction was carried out in different solvents. The best results in terms of the yields and reaction times were observed in the presence of the catalytic amount of Fe3O4–BIm–Pyrim–CuI (20 mg) in water at 100 °C for 3 h (Table 1, entry 13). To study the extent of the catalyst application, the reaction of a diverse range of aromatic aldehydes with phenyl acetylene and piperidine or pyrrolidine was also investigated under optimal reaction conditions, and results are shown in Table 2. In all cases, the three-component coupling reactions were successfully performed under the optimized conditions, and the nitrile products were obtained in high to excellent yields. It is noteworthy that aromatic aldehydes with both electron-donating and electron-withdrawing substituents also gave the desired products in excellent yields under standardized conditions. As shown in Table 2, pyrrolidine was found to be less reactive and afforded good yields of the desired propargylamine products.
| Entry | Catalyst (mg) | Solvent (Tem: °C) | Time (h) | Yield (%) |
|---|---|---|---|---|
| a Isolated yield. | ||||
| 1 | — | DMF (reflux) | 24 | NR |
| 2 | 3 | DMF (reflux) | 3 | 63 |
| 3 | 5 | DMF (reflux) | 3 | 72 |
| 4 | 10 | DMF (reflux) | 3 | 81 |
| 5 | 15 | DMF (reflux) | 3 | 89 |
| 6 | 20 | DMF (reflux) | 3 | 94 |
| 7 | 25 | DMF (reflux) | 3 | 94 |
| 8 | 20 | Toluene (reflux) | 3 | 47 |
| 9 | 20 | EtOH (reflux) | 3 | 91 |
| 10 | 20 | DMSO (reflux) | 3 | 89 |
| 11 | 20 | H2O (reflux) | 3 | 98 |
| 12 | 20 | H2O/EtOH (1/1) (80 °C) | 3 | 93 |
| 13 | 20 | PEG (100 °C) | 3 | 92 |
| 14 | 20 | CH3CN (reflux) | 3 | 81 |
| 15 | 20 | THF (reflux) | 3 | 22 |
| 16 | 20 | Solvent-free (100 °C) | 3 | 13 |
| 17 | 20 | H2O (90 °C) | 3 | 94 |
| a Isolated yield. |
|---|
 |
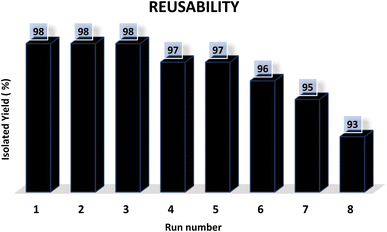
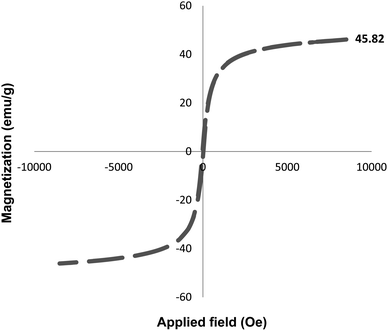
Finally, the reusability of the Fe3O4–BIm–Pyrim–CuI catalyst was also investigated. In this regard, the Fe3O4–BIm–Pyrim–CuI catalyst was separated from the obtained product of the model reaction (4a), washed with ethyl acetate several times and dried at 80 °C for 4 h, and reused in the same reaction. This reaction was repeated 8 times. The results of these experiments are shown in Fig. 7. In order to find the stability of the recovered catalyst, VSM and ICP-OES techniques were used. Also, as shown in Fig. 8, the saturation magnetization for the reused Fe3O4–BIm–Pyrim–CuI catalyst after 9 runs was found to be 45.82 emu/g. ICP-OES analysis of the reused catalyst after 8 runs was used to determine the amount of Cu loading on the Fe3O4–BIm–Pyrim nanocomposite, which was found to be 15.03 × 10−5 mmol g−1.
Comparison
Table 3 presents data on the comprehensive evaluation of the performance of the Fe3O4–BIm–Pyrim–CuI nanocatalyst compared to various other catalysts. Our results demonstrate a yield that surpasses the previously documented outcomes. Moreover, in our investigation, a diverse range of aldehydes, amines, and alkynes were subjected to eco-friendly conditions, yielding remarkable results within a mere 3 hour timeframe. In contrast to previously examined catalysts, our method offers several distinct advantages: it exhibits remarkable activity, is easily manufacturable, remains cost-effective, and can be reused for up to seven cycles without any discernible loss in catalytic efficiency.Summary and outlook
In summary, we show that the Fe3O4–BIm–Pyrim–CuI nanocomposite is a novel, eco-friendly, and efficient catalyst for the preparation of the propargylamines through one-pot multicomponent A3 coupling reactions of aldehydes, amines, and alkynes under eco-friendly conditions. Under this catalytic system, aromatic aldehydes with both electron-donating and electron-withdrawing substituents also gave the desired products in excellent yields under standardized conditions. It is noteworthy that pyrrolidine was found to be less reactive and afforded good yields of the desired propargylamine products. The structure of the Fe3O4–BIm–Pyrim–CuI nanocomposite was well analyzed by a series of spectroscopic techniques. The Fe3O4–BIm–Pyrim–CuI catalyst was easily separated using an external magnet, and the recovered catalyst was reused in 8 cycles without significant loss of activity.Experimental
Materials
All reagents and solvents used in this study were purchased from Sigma-Aldrich, Fluka, or Merck Chemical Companies and were used without further purification.General procedure for the synthesis of propargylamines catalyzed by Fe3O4–BIm–Pyrim–CuI nanocomposite
In a 10 mL-round-bottomed flask, a mixture of aldehyde (1 mmol), amine (1 mmol), and Fe3O4–BIm–Pyrim–CuI catalyst (20 mg) was stirred for 10 min. Phenylacetylene (1 mmol), and 3 mL water were added to the above mixture and stirred under reflux conditions for 3 h. The progress of the reaction was monitored by thin-layer chromatography (TLC). After completion of the reaction, the catalyst was separated using the magnetic stirring bar. After evaporation of the solvent, the desired product was isolated by silica gel flash column chromatography using a mixture of petroleum ether/ethyl acetate as the eluent. All products were well-known and identified by 1HNMR and 13CNMR spectroscopy.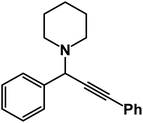 .
.
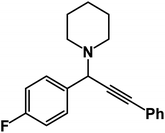
1-(1,3-Diphenylprop-2-yn-1-yl)piperidine: (4a). Yield 98%; 1H NMR (400 MHz, CDCl3): δ = 7.68–7.63 (m, 2H), 7.58–7.52 (m, 2H), 7.46–7.29 (m, 6H), 4.83 (s, 1H), 2.60–2.58 (m, 4H), 1.66–1.60 (m, 4H), 1.56–1.48 (m, 2H); 13C NMR (100 MHz, CDCl3): δ = 141.18, 132.11, 131.52, 130.67, 129.78, 128.64, 127.98, 126.50, 123.08, 122.43, 89.36, 86.41, 62.12, 50.36, 26.57, 24.36.
1-(1-(4-Fluorophenyl)-3-phenylprop-2-yn-1-yl)piperidine: (4b). Yield 93%; 1H-NMR (CDCl3, 400 MHz): δ = 7.63–7.55 (m, 2H), 7.54–7.51 (m, 2H), 7.41–7.32 (m, 5H), 4.83 (s, 1H), 2.59 (d, 4H), 1.67–1.61 (m, 4H), 1.53–1.46 (m, 2H); 13C-NMR (CDCl3, 100 MHz): δ = 137.12, 133.37, 132.41, 130.87, 129.12, 128.14, 123.43, 86.36, 85.21, 62.34, 51.24, 26.64, 25.37.
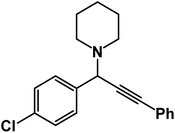
1-[1-(4-Chlorophenyl)-3-phenyl-2-propynyl]piperidine: (4c). Yield 99%; 1H-NMR (CDCl3, 400 MHz): δ = 7.64–7.58 (m, 2H), 7.56–7.51 (m, 2H), 7.40–7.29 (m, 5H), 4.80 (s, 1H), 2.58 (d, 4H), 1.66–1.60 (m, 4H), 1.50–1.47 (m, 2H); 13C-NMR (CDCl3, 100 MHz): δ = 138.02, 134.12, 132.36, 130.41, 129.35, 128.68, 127.11, 123.18, 88.29, 86.32, 62.37, 50.23, 26.30, 25.31.
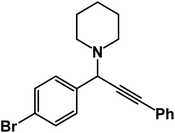
1-(1-(4-Bromophenyl)-3-phenylprop-2-yn-1-yl)piperidine: (4d). Yield 99%; 1H-NMR (CDCl3, 400 MHz): δ = 7.65–7.60 (m, 2H), 7.57–7.55 (m, 2H), 7.41–7.32 (m, 5H), 4.79 (s, 1H), 2.61 (t, 4H), 1.64–1.60 (m, 4H), 1.59–1.46 (m, 2H); 13C-NMR (CDCl3, 100 MHz): δ = 140.23, 132.38, 131.84, 130.36, 129.14, 128.54, 127.02, 126.31, 87.23, 86.30, 61.34, 51.21, 26.27, 25.39.
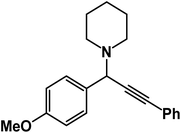
1-(1-(4-Methoxyphenyl)-3-phenylprop-2-yn-1-yl)piperidine: (4e). Yield 96%; 1H-NMR (CDCl3, 400 MHz): δ = 7.61–7.57 (m, 2H), 7.56–7.40 (m, 2H), 7.40–7.37 (m, 2H), 7.36–7.33 (m, 3H), 4.83 (s, 1H), 3.95 (s, 3H), 2.58 (m, 4H), 1.60–1.55 (m, 4H), 1.49–1.45 (m, 2H); 13C-NMR (CDCl3,100 MHz): δ = 159.34, 138.13, 136.25, 134.12, 132.61, 131.96, 126.64, 122.71, 87.32, 86.74, 60.01, 56.36, 51.12, 27.87, 23.71.
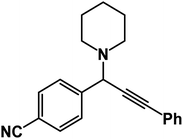
1-[1-(4-Cyanophenyl)-3-phenyl-2-propynyl]piperidine: (4f). Yield 90%; 1H-NMR (CDCl3, 400 MHz): δ = 7.68–7.57 (m, 2H), 7.56–7.52 (m, 2H), 7.39–7.35 (m, 2H), 7.34–7.28 (m, 3H), 4.83 (s, 1H), 2.60 (m, 4H), 1.65–1.48 (m, 4H), 1.47–1.28 (m, 2H); 13C-NMR (CDCl3,100 MHz): δ = 143.85, 132.12, 129.64, 128.37, 128.03, 127.56, 117.97, 112.53, 88.98, 85.32, 63.28, 51.39, 27.36, 24.75.
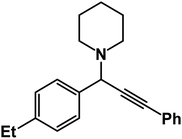
1-(1-(4-Ethylphenyl)-3-phenylprop-2-yn-1-yl)piperidine: (4g). Yield 97%; 1H-NMR (CDCl3, 400 MHz): δ = 7.63–7.61 (m, 2H), 7.58–7.55 (m, 2H), 7.38–7.35 (m, 2H), 7.35–7.29 (m, 3H), 4.84 (s, 1H), 2.59 (m, 6H), 2.44 (d, 3H), 1.62–1.55 (m, 4H), 1.54–1.44 (m, 2H); 13C-NMR (CDCl3,100 MHz): δ = 137.09, 133.89, 129.13, 128.54, 127.97, 125.76, 87.89, 86.47, 59.38, 50.23, 26.89, 24.56, 21.54, 19.36.
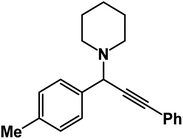
1-(3-Phenyl-1-(p-tolyl)prop-2-yn-1-yl)piperidine: (4h). Yield 96%; 1H-NMR (CDCl3, 400 MHz): δ = 7.63–7.55 (m, 2H), 7.54–7.51 (m, 2H), 7.39–7.36 (m, 2H), 7.35–7.06 (m, 3H), 4.85 (s, 1H), 2.59 (m, 4H), 1.62–1.57 (m, 4H), 1.56–1.46 (m, 2H); 13C-NMR (CDCl3,100 MHz): δ = 138.34, 137.11, 134.02, 132.26, 131.76, 129.24, 128.37, 127.96, 126.31, 123.87, 88.16, 85.97, 59.87, 50.57, 27.55, 24.32, 21.37.
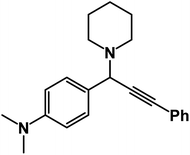
N,N-Dimethyl-4-(3-phenyl-1-(piperidin-1-yl)prop-2-yn-1-yl)aniline: (4i). Yield 92%; 1H-NMR (CDCl3, 400 MHz): δ = 7.60–7.57 (m, 2H), 7.55–7.40 (m, 2H), 7.39–7.36 (m, 2H), 7.36–7.31 (m, 3H), 4.82 (s, 1H), 3.89 (s, 6H), 2.57 (m, 4H), 1.58–1.48 (m, 4H), 1.47–1.41 (m, 2H); 13C-NMR (CDCl3,100 MHz): δ = 138.32, 137.30, 133.98, 131.51, 130.02, 129.64, 128.76, 127.13, 125.76, 123.58, 88.32, 86.94, 59.96, 50.24, 26.64, 24.37, 21.14, 19.07.
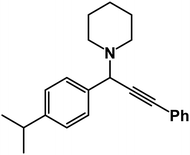
1-(1-(4-Isopropylphenyl)-3-phenylprop-2-yn-1-yl)piperidine: (4j). Yield 88%; 1H-NMR (CDCl3, 400 MHz): δ = 7.62–7.59 (m, 2H), 7.58–7.50 (m, 2H), 7.48–7.38 (m, 2H), 7.36–7.26 (m, 3H), 4.80 (s, 1H), 2.60 (m, 4H), 2.35 (m, 1H), 1.67–1.60 (m, 4H), 1.55–1.50 (m, 2H), 1.19 (s, 6H); 13C-NMR (CDCl3,100 MHz): δ = 147.23, 138.54, 129.52, 128.36, 127.02, 125.91, 88.02, 85.27, 61.22, 50.54, 36.26, 26.19, 24.62, 19.85.
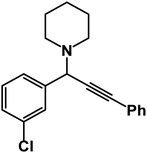
1-(1-(3-Chlorophenyl)-3-phenylprop-2-yn-1-yl)piperidine: (4k). Yield 94%; 1H-NMR (CDCl3, 400 MHz): δ = 7.65–7.62 (m, 1H), 7.61–7.58 (m, 1H), 7.57–7.7.52 (m, 2H), 7.51–7.45 (m, 1H), 7.44–7.35 (m, 3H), 7.33–7.29 (m, 1H), 4.81 (s, 1H), 2.59 (m, 4H), 1.66–1.60 (m, 4H), 1.59–1.52 (m, 2H); 13C-NMR (CDCl3,100 MHz): δ = 142.12, 133.65, 132.71, 131.36, 130.73, 129.23, 128.68, 127.34, 124.06, 123.16, 87.96, 86.36, 61.85, 50.67, 26.72, 24.10.
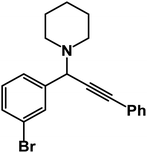
1-(1-(3-Bromophenyl)-3-phenyl-2-propynyl)piperidine: (4l). Yield 95%; 1H-NMR (CDCl3, 400 MHz): δ = 7.83–7.61 (m, 1H), 7.60–7.55 (m, 1H), 7.54–7.51 (m, 2H), 7.50–7.43 (m, 1H), 7.38–7.32 (m, 3H), 7.29–7.23 (m, 1H), 4.79 (s, 1H), 2.60–2.57 (m, 4H), 1.67–1.63 (m, 4H), 1.62–1.50 (m, 2H); 13C-NMR (CDCl3,100 MHz): δ = 141.20, 132.23, 131.46, 130.98, 129.64, 128.38, 128.21, 127.37, 123.78, 122.64, 88.47, 85.12, 62.03, 50.36, 26.97, 24.16.
1-(1-(1-Naphthyl)-3-phenyl-2-propynyl)piperidine: (4m). Yield 97%; 1H-NMR (CDCl3, 400 MHz): δ = 8.10 (s, 1H), 7.90–7.85 (m, 3H), 7.81–7.75 (m, 1H), 7.64–7.55 (m, 2H), 7.53–7.46 (m, 2H), 7.44–7.31 (m, 3H), 4.97 (s, 1H), 2.68–2.61 (m, 4H), 1.721.54 (m, 4H), 1.54–1.40 (m, 2H); 13C-NMR (CDCl3, 100 MHz): δ = 136.22, 133.08, 132.96, 131.87, 128.33, 128.13, 127.76, 127.58, 127.29, 126.71, 125.95, 125.86, 123.35, 88.13, 86.02, 62.53, 50.84, 26.19, 24.46.
1-(3-Phenyl-1-(o-tolyl)prop-2-yn-1-yl)piperidine: (4n). Yield 89%; 1H-NMR (CDCl3, 400 MHz): δ = 7.63–7.60 (m, 1H), 7.59–7.55 (m, 1H), 7.53–7.50 (m, 2H), 7.48–7.43 (m, 1H), 7.38–7.34 (m, 3H), 7.33–7.26 (m, 1H), 4.80 (s, 1H), 2.61 (m, 4H), 2.35 (s, 3H), 1.68–1.61 (m, 4H), 1.60–1.50 (m, 2H); 13C-NMR (CDCl3,100 MHz): δ = 142.18, 132.03, 131.65, 130.85, 129.74, 128.36, 127.45, 125.32, 88.41, 85.16, 62.21, 50.95, 26.17, 24.83, 19.75.
1-[1-(2-Furfuryl)-3-phenyl-prop-2-ynyl]-piperidine: (4o). Yield 90%; 1H-NMR (CDCl3, 400 MHz): δ = 7.55–7.50 (m, 2H), 7.49–7.40 (m, 1H), 7.37–7.32 (m, 3H), 6.51–6.49 (m, 1H), 6.39–6.35 (m, 1H), 4.90 (s, 1H), 2.62–2.58 (m, 4H), 1.76–1.65 (m, 4H), 1.63–1.45 (m, 2H); 13C-NMR (CDCl3, 100 MHz): δ = 152.32, 141.26, 132.02, 129.11, 122.98, 109.98, 109.15, 86.40, 83.20, 56.64, 50.82, 29.87, 25.97.
1-(3-Phenyl-1-(thiophen-3-yl)prop-2-yn-1-yl)piperidine: (4p). Yield 92%; 1H-NMR (CDCl3, 400 MHz): δ = 7.53–7.50 (m, 2H), 7.49–7.40 (m, 1H), 7.37–7.29 (m, 3H), 6.52–6.49 (m, 1H), 6.39–6.33 (m, 1H), 4.85 (s, 1H), 2.60–2.53 (m, 4H), 1.72–1.60 (m, 4H), 1.58–1.40 (m, 2H); 13C-NMR (CDCl3, 100 MHz): δ = 151.64, 140.95, 131.05, 128.64, 121.61, 109.34, 109.04, 85.37, 83.19, 55.16, 50.74, 29.23, 25.68.
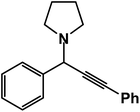
1-(1,3-Diphenylprop-2-yn-1-yl)pyrrolidine: (4q). Yield 95%; 1H-NMR (CDCl3, 400 MHz): δ = 7.67–7.54 (m, 2H), 7.54–7.50 (m, 2H), 7.39–7.30 (m, 6H), 4.82 (s, 1H), 2.80–2.74 (m, 4H), 1.49–1.40 (m, 4H); 13C-NMR (CDCl3, 100 MHz): δ = 139.65, 129.36, 128.67, 127.41, 122.95, 88.50, 84.36, 67.18, 61.85, 23.12.
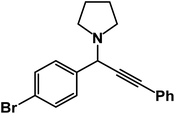
1-(1-(4-Bromophenyl)-3-phenylprop-2-yn-1-yl)pyrrolidine: (4r). Yield 97%; 1H-NMR (CDCl3, 400 MHz): δ = 7.83–7.80 (m, 2H), 7.71–7.56 (m, 2H), 7.54–7.40 (m, 2H), 7.37–7.30 (m, 3H), 4.85 (s, 1H), 2.78–2.74 (m, 4H), 1.70–1.63 (m, 4H); 13C-NMR (CDCl3, 100 MHz): δ = 142.98, 131.65, 129.03, 128.64, 127.11, 122.61, 88.64, 85.37, 66.38, 61.25, 24.19.
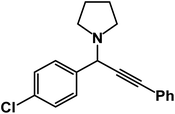
1-(1-(4-Chlorophenyl)-3-phenylprop-2-yn-1-yl)pyrrolidine: (4s). Yield 96%; 1H-NMR (CDCl3, 400 MHz): δ = 7.84–7.80 (m, 2H), 7.67–7.55 (m, 2H), 7.54–7.40 (m, 2H), 7.39–7.31 (m, 3H), 4.85 (s, 1H), 2.81–2.72 (m, 4H), 1.89–1.84 (m, 4H); 13C-NMR (CDCl3, 100 MHz): δ = 1420.1, 131.20, 128.76, 125.36, 122.81, 87.32, 84.95, 63.02, 59.51, 24.94.
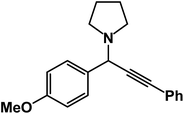
1-(1-(4-Methoxyphenyl)-3-phenylprop-2-yn-1-yl)pyrrolidine: (4t). Yield 90%; 1H-NMR (CDCl3, 400 MHz): δ = 7.55–7.49 (m, 2H), 7.45–7.41 (m, 2H), 7.38–7.31 (m, 2H), 7.27–7.22 (m, 3H), 4.83 (s, 1H), 3.67 (s, 3H), 2.79–2.73 (m, 4H), 1.93–1.89 (m, 4H); 13C-NMR (CDCl3, 100 MHz): δ = 156.38, 131.82, 129.49, 128.33, 122.87, 121.26, 115.73, 88.50, 85.21, 61.12, 57.36, 55.95, 21.54.
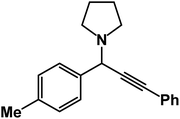
1-(3-Phenyl-1-(p-tolyl)prop-2-yn-1-yl)pyrrolidine: (4u). Yield 90%; 1H-NMR (CDCl3, 400 MHz): δ = 7.57–7.54 (m, 2H), 7.48–7.41 (m, 2H), 7.38–7.31 (m, 2H), 7.27–7.20 (m, 3H), 4.81 (s, 1H), 2.78–2.75 (m, 4H), 2.69 (s, 3H), 1.91–1.88 (m, 4H); 13C-NMR (CDCl3, 100 MHz): δ = 156.38, 131.82, 129.49, 128.33, 122.87, 121.26, 115.73, 88.50, 85.21, 61.12, 57.36, 55.95, 21.54.
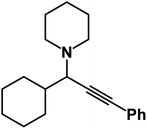
1-(1-Cyclohexyl-3-phenylprop-2-yn-1-yl)piperidine: (4v). Yield 86%; 1H-NMR (CDCl3, 400 MHz): δ = 7.47–7.40 (m, 2H), 7.35–7.27 (m, 3H), 3.82–3.72 (m, 4H), 3.06 (d, 1H), 3.01–370 (m, 2H), 2.69 (m, 2H), 2.57 (m, 1H), 1.99 (t, 3H); 13C-NMR (CDCl3, 100 MHz): δ = 131.70, 128.26, 127.96, 87.12, 86.51, 66.35, 57.15, 35.09, 26.12, 20.01, 15.56.
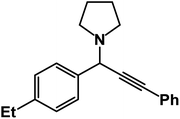
1-(1-(4-Ethylphenyl)-3-phenylprop-2-yn-1-yl)pyrrolidine: (4w). Yield 90%; 1H-NMR (CDCl3, 400 MHz): δ = 7.65–7.60 (m, 2H), 7.56–7.51 (m, 2H), 7.38–7.31 (m, 2H),7.28–7.26 (m, 3H), 4.82 (s, 1H), 2.80–2.71 (m, 6H), 1.59–1.47 (m, 7H); 13C-NMR (CDCl3, 100 MHz): δ = 138.23, 131.81, 128.50, 128.21, 127.89, 123.02, 88.50, 85.23, 61.23, 58.12, 23.56, 20.13, 19.02.
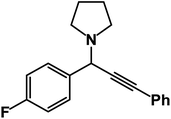
1-(1-(4-Fluorophenyl)-3-phenylprop-2-yn-1-yl)pyrrolidine: (4x). Yield 92%; 1H-NMR (CDCl3, 400 MHz): δ = δ 7.85 (d, J = 8.3 Hz, 2H), 7.64 (d, J = 8 Hz, 2H), 7.57–7.52 (m, 2H),7.38–7.32 (m, 3H), 5.09 (s, 1H), 2.65–2.53 (m, 4H), 1.14 (t, J = 3.3 Hz, 6H); 13C-NMR (CDCl3, 100 MHz, ppm): δ 154.13, 151.25, 135.64, 132.84, 131.50, 129.45, 128.74, 127.31, 124.39, 118.82, 118.23, 79.24, 73.61, 45.85, 44.09, 14.89.
Conflicts of interest
There are no conflicts to declare.References
- B. Udaykumar and M. Periasamy, ACS Omega, 2019, 4, 21587–21595 CrossRef.
- K. Lauder, A. Toscani, N. Scalacci and D. Castagnolo, Chem. Rev., 2017, 117, 14091–14200 CrossRef.
- P.-B. Chen, J.-W. Yang, Z.-X. Rao, Q. Wang, H.-T. Tang, Y.-M. Pan and Y. Liang, J. Colloid Interface Sci., 2023, 652, 866–877 CrossRef.
- Y. Liu, B. Fan, B. Xu and B. Yang, Mater. Lett., 2023, 337, 133979 CrossRef.
- T. Li, H. Pang, Q. Wu, M. Huang, J. Xu, L. Zheng, B. Wang and Y. Qiao, Int. J. Mol. Sci., 2022, 23, 6259 CrossRef PubMed.
- S. Ghosh and K. Biswas, RSC Adv., 2021, 11, 2047–2065 RSC.
- H. Feng, F. Peng, H. Xi, L. Zhong and L. Huang, Asian J. Org. Chem., 2021, 10, 762–765 CrossRef CAS.
- A. Singh and A. Kumar Narula, Results Chem., 2022, 4, 100279 CrossRef CAS.
- M. Wünsch, D. Schröder, T. Fröhr, L. Teichmann, S. Hedwig, N. Janson, C. Belu, J. Simon, S. Heidemeyer, P. Holtkamp, J. Rudlof, L. Klemme, A. Hinzmann, B. Neumann, H.-G. Stammler and N. Sewald, Beilstein J. Org. Chem., 2017, 13, 2428–2441 CrossRef.
- S. Zárate-Roldán, M. C. Gimeno and R. P. Herrera, Catalysts, 2021, 11, 513 CrossRef.
- T. K. Saha and R. Das, ChemistrySelect, 2018, 3, 147–169 CrossRef CAS.
- H. Eshghi, G. H. Zohuri and S. Damavandi, Eur. J. Chem., 2011, 2, 100–103 CrossRef CAS.
- S. Sarkar, R. Chatterjee, A. Mukherjee, S. Santra, G. V. Zyryanov and A. Majee, AIP Conf. Proc., 2020, 050049 CrossRef.
- S. Kumar, B. Mohan, Z. Tao, H. You and P. Ren, Catal. Sci. Technol., 2021, 11, 5734–5771 RSC.
- Y. Peng, H. Tang, B. Yao, X. Gao, X. Yang and Y. Zhou, Chem. Eng. J., 2021, 414, 128800 CrossRef CAS.
- M. Abedi, M. Hosseini, A. Arabmarkadeh and M. Kazemi, Synth. Commun., 2021, 51, 835–855 CAS.
- M. Kazemi and M. Ghobadi, Nanotechnol. Rev., 2017, 6(6), 549–571 CAS.
- F. Casti, F. Basoccu, R. Mocci, L. De Luca, A. Porcheddu and F. Cuccu, Molecules, 2022, 27, 1988 CrossRef CAS PubMed.
- X. Guo, P. Tian, Y. Sun, D. Ding, J. Zhang and Y. Han, Chem. Ind. Eng. Prog., 2021, 40, 605–620 CAS.
- M. Liu, Y. Ye, J. Ye, T. Gao, D. Wang, G. Chen and Z. Song, Magnetochemistry, 2023, 9, 110 CrossRef CAS.
- K. Wang, F. Zhang, K. Xu, Y. Che, M. Qi and C. Song, RSC Adv., 2023, 13, 6713–6736 RSC.
- P. Thakur, S. Taneja, D. Chahar, B. Ravelo and A. Thakur, J. Magn. Magn. Mater., 2021, 530, 167925 CrossRef CAS.
- E. M. Materón, C. M. Miyazaki, O. Carr, N. Joshi, P. H. S. Picciani, C. J. Dalmaschio, F. Davis and F. M. Shimizu, Appl. Surf. Sci. Adv., 2021, 6, 100163 CrossRef.
- H. Ahmad and M. K. Hossain, Mater. Adv., 2022, 3, 859–887 RSC.
- M. Ghobadi, M. Kargar Razi, R. Javahershenas and M. Kazemi, Synth. Commun., 2021, 51(5), 647–669 CrossRef CAS.
- L. Tang, F. Qin, F. Huang, D. Xu, Q. Hu and W. Zhang, Appl. Organomet. Chem., 2022, 36(7), e6723 CrossRef CAS.
- S.-X. Mao, J.-Y. Song, W.-S. Zhu, H.-M. Li, J.-Y. Pang, D.-B. Dang and Y. Bai, Fuel, 2023, 352, 128982 CrossRef CAS.
- Z. Liu, B. Fan, J. Zhao, B. Yang and X. Zheng, Corros. Sci., 2023, 212, 110957 CrossRef CAS.
- M. Kidwai, A. Jain and S. Bhardwaj, Mol. Diversity, 2012, 16, 121–128 CrossRef CAS.
- R. K. Sharma, S. Dutta, S. Sharma, R. Zboril, R. S. Varma and M. B. Gawande, Green Chem., 2016, 18, 3184–3209 RSC.
- H. Li, S. Zhao, W. Zhang, H. Du, X. Yang, Y. Peng, D. Han, B. Wang and Z. Li, Fuel, 2023, 342, 127786 CrossRef CAS.
- Z. Song, D. Han, M. Yang, J. Huang, X. Shao and H. Li, Appl. Surf. Sci., 2023, 607, 155067 CrossRef CAS.
- D. Dharmendra, P. Chundawat, Y. Vyas and C. Ameta, J. Chem. Sci., 2022, 134, 47 CrossRef CAS.
- L. Shiri, A. Ghorbani-Choghamarani and M. Kazemi, Aust. J. Chem., 2017, 70, 9 CrossRef CAS.
- Y. Dou, A. Wang, L. Zhao, X. Yang, Q. Wang, M. Shire Sudi, W. Zhu and D. Shang, J. Colloid Interface Sci., 2023, 650, 943–950 CrossRef CAS.
- J. Xia, Y. Li, C. He, C. Yong, L. Wang, H. Fu, X.-L. He, Z.-Y. Wang, D.-F. Liu and Y.-Y. Zhang, ACS Infect. Dis., 2023, 9, 1711–1729 CrossRef CAS.
- S. Shylesh, V. Schünemann and W. R. Thiel, Angew. Chem., Int. Ed., 2010, 49, 3428–3459 CrossRef CAS.
- M. Lakshman, J. Synth. Chem., 2023, 1, 148–154 Search PubMed.
- X. Feng, B. Wang, G. Gao, S. Gao, C. Xie and J.-W. Shi, Fuel, 2023, 352, 129159 CrossRef CAS.
- Y. Liang, J. Li, Y. Xue, T. Tan, Z. Jiang, Y. He, W. Shangguan, J. Yang and Y. Pan, J. Hazard. Mater., 2021, 420, 126584 CrossRef CAS PubMed.
- W. Chen, W. Liu, H. Liang, M. Jiang and Z. Dai, Ocean Eng., 2023, 270, 113646 CrossRef.
- N. A. Frey, S. Peng, K. Cheng and S. Sun, Chem. Soc. Rev., 2009, 38, 2532 RSC.
- M.-S. Shafik, J. Synth. Chem., 2022, 1, 132–136 Search PubMed.
- R. Hudson, Y. Feng, R. S. Varma and A. Moores, Green Chem., 2014, 16, 4493–4505 RSC.
- L. M. Rossi, N. J. S. Costa, F. P. Silva and R. Wojcieszak, Green Chem., 2014, 16, 2906 RSC.
- H. Veisi, A. Nikseresht, A. Rostami and S. Hemmati, Res. Chem. Intermed., 2019, 45, 507–520 CrossRef CAS.
- S. Molaei and M. Ghadermazi, Sci. Rep., 2023, 13, 15146 CrossRef CAS.
- M. Mohammadi, A. Ghorbani-Choghamarani and N. Hussain-Khil, J. Phys. Chem. Solids, 2023, 177, 111300 CrossRef CAS.
- C. A. Ferretti, L. G. Gutierrez, V. A. Guntero, P. J. Noriega and M. N. Kneeteman, C. R. Chim., 2021, 24, 385–396 CrossRef CAS.
- N. Rismana, A. R. Astuti, Y. H. Suselo, M. Anwar and T. E. Saraswati, in IOP Conference Series: Materials Science and Engineering, IOP Publishing, 2018, vol. 333, p. 12026 Search PubMed.
- S. Lotfi, A. Nikseresht and N. Rahimi, Polyhedron, 2019, 173, 114–148 CrossRef.
- M. Kantam, J. Yadav, S. Laha and S. Jha, Synlett, 2009, 2009, 1791–1794 CrossRef.
- K. Jayaramulu, K. K. R. Datta, M. V. Suresh, G. Kumari, R. Datta, C. Narayana, M. Eswaramoorthy and T. K. Maji, ChemPlusChem, 2012, 77, 743–747 CrossRef CAS.
- F. M. Moghaddam, S. E. Ayati, S. H. Hosseini and A. Pourjavadi, RSC Adv., 2015, 5, 34502–34510 RSC.
- C. Mukhopadhyay and S. Rana, Catal. Commun., 2009, 11, 285–289 CrossRef CAS.
- F. Saadati, M. Gholinejad, H. Janmohammadi and S. Shaybanizadeh, Lett. Org. Chem., 2018, 15(2), 79–86 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra04871c |
| This journal is © The Royal Society of Chemistry 2023 |