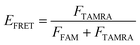Open Access Article
Open Access ArticleInsect multimeric G-quadruplexes fold into antiparallel structures of different compactness and stability in K+ and Na+ solutions†
Chao Gao‡
 a,
Jixin Chen‡a,
Naureen Anwarb,
Jieya Denga,
Zhangqian Wang*a,
Muhammad Umer*c and
Yi He*a
a,
Jixin Chen‡a,
Naureen Anwarb,
Jieya Denga,
Zhangqian Wang*a,
Muhammad Umer*c and
Yi He*a
aNational R&D Center for Se-rich Agricultural Products Processing, Hubei Engineering Research Center for Deep Processing of Green Se-rich Agricultural Products, School of Modern Industry for Selenium Science and Engineering, Wuhan Polytechnic University, Wuhan 430023, China. E-mail: wzqsnu@whpu.edu.cn; yi.he@whpu.edu.cn
bDepartment of Biological Sciences, Faculty of Science and Technology, Virtual University of Pakistan, Punjab 54000, Pakistan
cInstitute for Forest Resources and Environment of Guizhou and Forestry College, Research Center of Forest Ecology, Guizhou University, Guiyang, 550025, China. E-mail: umer@gzu.edu.cn
First published on 11th December 2023
Abstract
Human telomere sequences (TTAGGG)n fold into G-quadruplexes with different conformations in K+ and Na+ solutions, which are highlighted for their potential as antitumor drug targets. Moreover, human multimeric G-quadruplexes have been broadly studied potentially for screening ligands with higher selectivity than monomeric G-quadruplexes. Most insects have telomeres consisting of pentanucleotide (TTAGG) repeats, which fold into an antiparallel structured G-quadruplex with a two-layer G-planar in a K+ solution. However, the structure of insect telomeric G-quadruplexes in Na+ solutions and their higher-order structures have not been explored. The quinoline derivative BMPQ-1 has been reported to bind human multimeric G-quadruplex. This study compared the stability and compactness of insect monomeric and multimeric G-quadruplex structures in K+ and Na+ solutions and further validated the interaction between BMPQ-1 and insect multimeric G-quadruplexes. Circular dichroism (CD) spectral scanning analysis revealed that although the insect telomeric G-quadruplex folds into an antiparallel structure in both K+ and Na+ solutions, all the insect telomeric G-quadruplexes are more stable in Na+ solutions. Fluorescence resonance energy transfer (FRET) analysis indicated insect telomeric G-quadruplexes have a more compact structure in Na+ solutions. BMPQ-1 exhibited higher selectivity for insect multimeric G-quadruplex Bom37 than monomeric G-quadruplex Bom17, and had a different binding pattern to Bom37 G-quadruplex in K+ and Na+ solutions. Finally, BMPQ-1 was found to have a significant inhibitory effect on the proliferation of pest cells. This study contributes to our comprehensive understanding of insect telomeric G-quadruplexes.
Introduction
G-quadruplexes are non-classical high-level structures formed by guanine-rich nucleic acid sequences.1–3 Hoogsteen hydrogen bonds can stabilize the G-quadruplex structures in the presence of metal cations.4–7 G-quadruplexes-forming oligonucleotides always exist in the eukaryotic telomeres and the promoter gene regions.7,8 The formation of the G-quadruplex significantly impacts several biological events.9–11Telomeres are important structures that protect the ends of chromosomes,12 and telomere DNA sequences of humans consist of double-stranded TTAGGG repeats and terminate with around 200 nt guanine-rich single-stranded overhangs beyond the double-stranded region,13–15 which has a high tendency to fold into multimeric G-quadruplexes connected by TTA linkers. Human multimeric G-quadruplexes are promising targets for developing antitumor drugs,16–18 and the monovalent cations in the cell that stabilize the G-quadruplex are mainly K+ and Na+19,20 Human telomeric G-quadruplexes have been discovered for a long time, and those can fold into multiple topologies, including (3 + 1) hybrid-1/-2 structures, parallel structures in K+ solution and antiparallel structure in Na+ solution.21–25 Compared with monomeric G-quadruplexes, the human multimeric G-quadruplexes are probably biologically more related to the structure of long telomeric DNA sequences in vivo.16,17,26,27
However, not only human telomere sequences but also telomere sequences in plants and even in insects can fold into G-quadruplex structures. Furthermore, the structure of these non-human telomeric G-quadruplexes has only been reported to some extent.28–31 The conservation of telomere structure in insects is similarly significant for the integrity of the genome within the insect cell.29,30 Telomeres of most insects are composed of repeated sequences of TTAGG, which are folded into an antiparallel structured G-quadruplex with a two-layer G-planar in K+ solutions.31,32 However, there is limited knowledge of the conformation of insect telomeric G-quadruplexes in the Na+ solutions and insect multimeric G-quadruplexes. Studying the higher-order structure formed by a long chain of insect telomere sequences facilitates understanding insect telomere structure and screening ligands targeting insect multimeric G-quadruplexes.
In this study, we compared the conformation of insect telomeric monomeric G-quadruplexes (Bom17) and multimeric G-quadruplexes (Bom37, Bom57, Bom77, and Bom97) by CD spectral scanning analysis and used CD melting to evaluate their stability in K+ and Na+ solutions. Comparison of the compactness of insect telomeric G-quadruplexes in K+ and Na+ solutions, respectively, based on the FRET efficiency between fluorescent pairs (5′-FAM and 3′-TAMRA) labeled with insect telomeric G-quadruplexes was also investigated. Moreover, we further analyzed the interactive strength between telomeric G-quadruplexes and the ligand BMPQ-1 and the binding modes in K+ and Na+ solutions by fluorescence spectroscopy.
Results
Structure and stability of insect telomeric G-quadruplexes in K+ and Na+ solutions
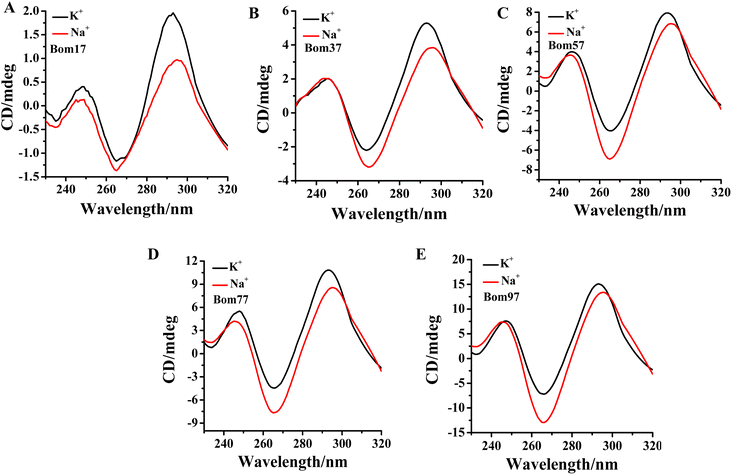
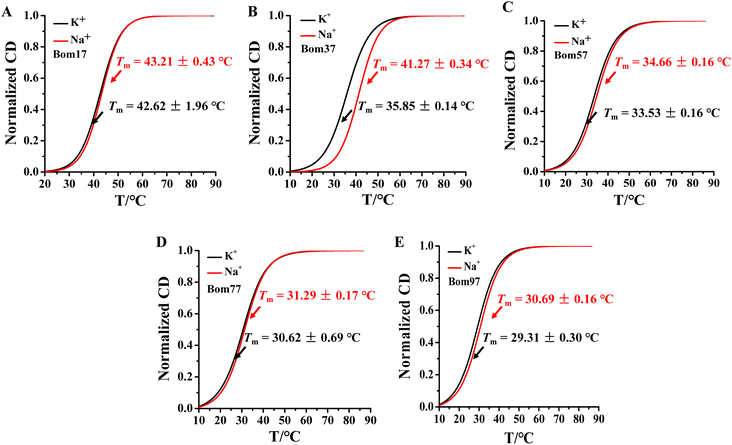
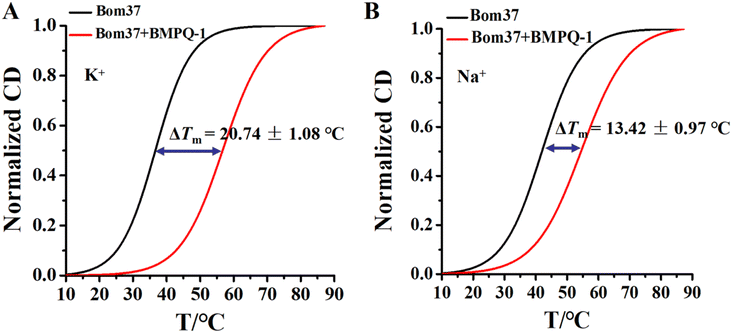
Human telomeric G-quadruplexes fold into different conformations in monovalent cationic K+ and Na+ solutions. To investigate the conformation of insect telomeric G-quadruplexes in the K+ and Na+ solutions, CD spectroscopy was performed to analyze the conformation of insect telomeric monomeric G-quadruplex Bom17 and multimeric G-quadruplexes Bom37, Bom57, Bom77, and Bom97 (Table S1†) in the K+ and Na+ solutions (Fig. 1). In the CD spectrum of insect telomeric G-quadruplexes in both K+ and Na+ solutions, there are two characteristic positive peaks at ∼245 nm and 295 nm, with a strong minimum peak at about 265 nm, which is associated with the characteristic peaks of antiparallel structured G-quadruplexes. The intensity of the signal peak of the insect telomeric G-quadruplex increases as the sequence is stretched. All telomeric G-quadruplexes showed a stronger CD signal peak at about 265 nm and a weaker signal peak at about 295 nm in Na+ solution than in K+ solution, suggesting insect telomeric G-quadruplexes may possess additional stability in Na+ solutions.The thermal stability of insect telomeric G-quadruplexes in K+ and Na+ solutions was further analyzed through CD melting (Fig. 2). Bom17, Bom37, Bom57, Bom77, and Bom97 showed a melting temperature (Tm) values of 42.62 ± 1.96 °C, 35.85 ± 0.14 °C, 33.53 ± 0.16 °C, 30.62 ± 0.69 °C, and 29.31 ± 0.30 °C in K+ solution, respectively, and they presented a Tm value of 43.21 ± 0.43 °C, 41.27 ± 0.34 °C, 34.66 ± 0.16 °C, 31.29 ± 0.17 °C, and 30.69 ± 0.16 °C in Na+ solution, respectively. These results shown that all the insect telomeric G-quadruplex folds into a more stable structure in Na+ solution, and the structural stability of insect telomeric G-quadruplex in both K+ and Na+ solutions drops as the sequence is extended.
Insect telomeric G-quadruplexes have a more compact conformation in Na+ solutions
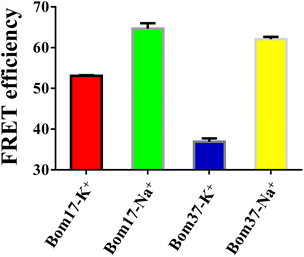
As mentioned above, the results specified that the insect telomere sequence folds into an antiparallel structured G-quadruplex with fluctuating stability in K+ and Na+ solutions. To further investigate the differences in insect telomeric G-quadruplexes in K+ and Na+ solutions, the 5′ and 3′ ends of G-quadruplex-forming oligonucleotides of Bom17 and Bom37 were labeled with FAM and TAMRA tags, respectively. The increase in FRET efficiency between FAM and TAMRA indicates that the distance of FAM and TAMRA at the ends of Bom17 and Bom37 becomes shorter. The FRET efficiencies of Bom17 in K+ and Na+ solutions were 53.08 ± 0.15% and 64.70 ± 1.29%, respectively, which increased by 21.89%. Moreover, the FRET efficiencies of Bom37 in K+ and Na+ solutions were 36.93 ± 0.79% and 62.08 ± 0.57%, respectively, which increased by 61.80% (Fig. 3 and S1†). Since insect telomeric G-quadruplexes have a more stable structure in the Na+ solutions, the significant increase in FRET efficiency is more likely to be triggered by the folding of the insect telomeric G-quadruplex into a more compact structure in the Na+ solution. We also assessed c-MYC G-quadruplex with parallel structures and found that c-MYC both revealed a more stable and compact conformation in the K+ solution (Fig. S2 and S3†). This proposed that the insect telomeric G-quadruplexes in antiparallel conformation and the c-MYC G-quadruplex in parallel conformation have different folding mechanisms in the K+ and Na+ solutions, respectively.The compactness of the structures of Bom17 and Bom37 G-quadruplexes in K+ and Na+ solutions was further analyzed by native polyacrylamide gel electrophoresis. The results revealed that the migration rate of insect telomeric G-quadruplexes in the Na+ solution was significantly greater than that in the K+ solution (Fig. S4†). This result suggested insect telomeric G-quadruplexes fold into a more compact structure in Na+ solution.
BMPQ-1 is more selective for the insect dimeric G-quadruplex Bom37 than the monomeric G-quadruplex Bom17
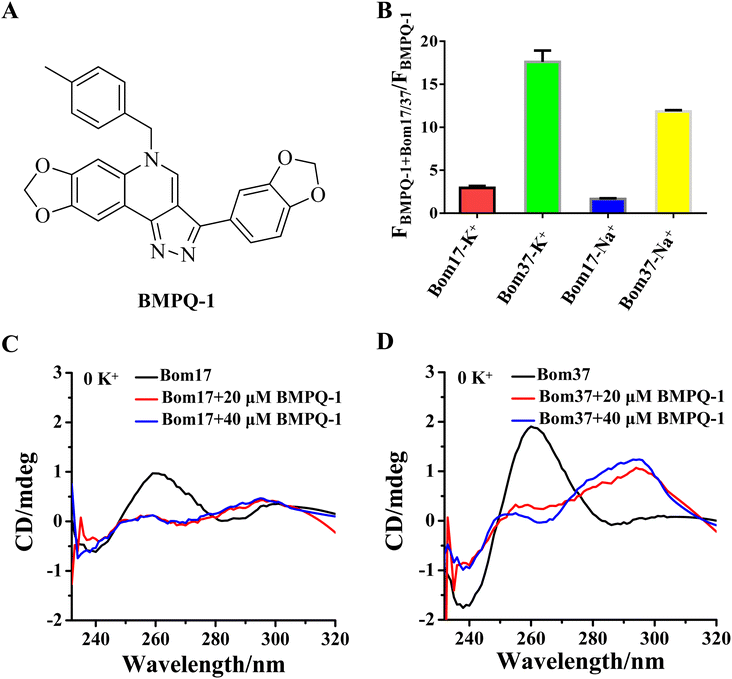
To test whether BMPQ-1 could specifically recognize insect telomeric multimeric G-quadruplexes, we first studied the fluorescence response of BMPQ-1 and insect monomer G-quadruplex (Bom17) and dimeric G-quadruplex (Bom37) (Table S1†). With the addition of BMPQ-1 to the Bom37 in K+ and Na+ solutions, the fluorescence intensity of BMPQ-1 increased significantly by 17.60 folds and 11.84 folds, respectively, and the fluorescence spectrum of BMPQ-1 has a significant blue shift. In contrast, the fluorescence intensity of BMPQ-1 increased by only 2.96 folds and 1.66 folds after adding Bom17 in K+ and Na+ solutions, respectively (Fig. 4B). This result indicated that BMPQ-1 exhibited higher fluorescence selectivity for telomeric multimeric G-quadruplexes in K+ and Na+ conditions and had a higher fluorescence response with Bom37 in K+ solution. In contrast, a relatively weak fluorescence intensity boost was observed when BMPQ-1 was titrated with Bom17 G-quadruplex in K+ and Na+ solutions, even at 10 μM (Fig. S5†). The dissociation constants of BMPQ-1 with Bom17 and Bom37 in K+ or Na+ solutions were analyzed by fluorescence titration (Fig. S6†). Compared with Bom17 in K+ or Na+ solutions (KD = 12.72 ± 2.52 μM, 34.49 ± 6.55 μM), Bom37 in K+ or Na+ solutions exhibited higher binding affinity with BMPQ-1 (KD = 1.46 ± 0.16 μM, 5.15 ± 1.09 μM). These results indicated that BMPQ-1 has a stronger affinity for Bom37 G-quadruplex.The ability of BMPQ-1 to induce the folding of Bom17 and Bom37 into G-quadruplex structures was further assessed by CD spectroscopy (Fig. 4C and D). Oligonucleotides Bom17 and Bom37 were found to fold into a G-quadruplex with a similar parallel structure (It exhibited two negative peaks at about 240 nm and 285 nm and a positive peak at about 260 nm, respectively) in 10 mM Tris–HCl buffer without monovalent cations. However, the CD signal peak intensity of Bom37 was significantly higher than that of Bom17. With the addition of BMPQ-1, the positive signal peak at 260 nm for oligonucleotides Bom17 and Bom37 was changed to a negative signal peak at 265 nm, and the negative signal peak at 285 nm was altered to a positive signal peak at 295 nm, and there was a weak trend towards a positive signal peak at a wavelength of 248 nm. This is a typical antiparallel G-quadruplex CD signal peak. However, the ability of BMPQ-1 to induce the folding of oligonucleotide Bom37 into an antiparallel G-quadruplex was significantly higher than that for Bom17. These results indicated that BMPQ-1 is more selective for the insect dimeric G-quadruplex Bom37 than the monomeric G-quadruplex Bom17.
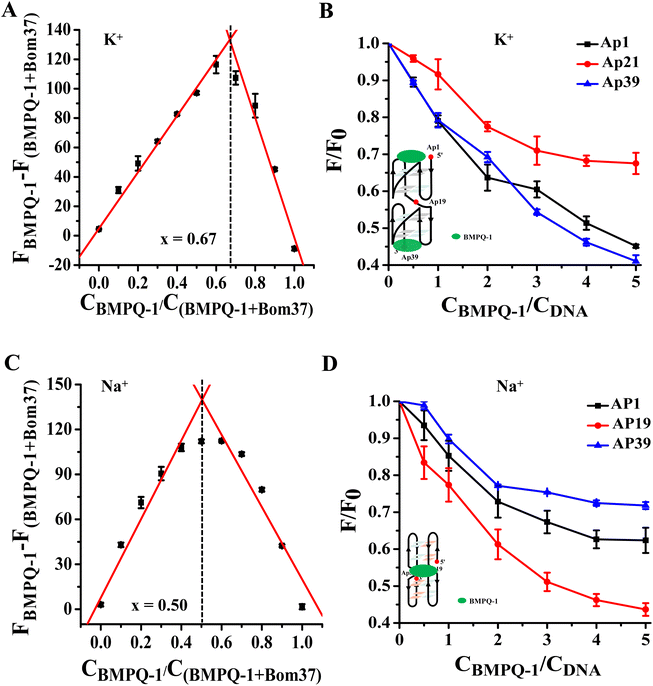
Analysis of the binding mechanism of BMPQ-1 to Bom37 G-quadruplex in K+/Na+ solutions
The results of the experiments described above suggested that BMPQ-1 showed higher capability in stabilizing insect multimeric G-quadruplex. This attracted our interest in studying the recognition mechanism of BMPQ-1 with Bom37 in K+ and Na+ solutions. The job's plot analysis explored the binding stoichiometric ratio of BMPQ-1 to Bom37 G-quadruplex in two cationic solutions. The best-fit lines of the job plot intersect at about x = 0.67, indicating BMPQ-1 bound with the Bom37 G-quadruplex at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry in K+ solutions (Fig. 5A).
1 stoichiometry in K+ solutions (Fig. 5A).
2-Aminopurine (2-Ap)-labeled oligonucleotides Bom37 at the 5′-end (Ap1), 3′-end (Ap39), and the loop between adjacent G-quadruplex units (Ap19) (Table S1†) were used to evaluate the interaction positions of BMPQ-1 with Bom37 G-quadruplex. It was found that the fluorescence intensity of Ap1 and Ap39 was significantly disturbed upon the addition of BMPQ-1. The fluorescence intensity of Ap19 was weakly affected (Fig. 5B). It indicated that BMPQ-1 had close contact with the 5′-end and the 3′-end. Combining the results of stoichiometry results, it can be noted that BMPQ-1 is stacked on the 5′ and 3′-ends of Bom37 G-quadruplex in the K+ solution.
In contrast, the job plot analysis explored that BMPQ-1 bound with the Bom37 G-quadruplex at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 rate (the best-fit lines of the job plot intersect at about x = 0.50) in the Na+ condition (Fig. 5C). Moreover, fluorescence quenching assay results showed that the fluorescence intensity of Ap19 was significantly disturbed upon the addition of BMPQ-1. The fluorescence of Ap1 and Ap39 was weakly affected (Fig. 5D). The above results indicated that BMPQ-1 might intercalate into the pocket between two adjacent G-quadruplex units of Bom37 in Na+ solution.
1 rate (the best-fit lines of the job plot intersect at about x = 0.50) in the Na+ condition (Fig. 5C). Moreover, fluorescence quenching assay results showed that the fluorescence intensity of Ap19 was significantly disturbed upon the addition of BMPQ-1. The fluorescence of Ap1 and Ap39 was weakly affected (Fig. 5D). The above results indicated that BMPQ-1 might intercalate into the pocket between two adjacent G-quadruplex units of Bom37 in Na+ solution.
Effect of BMPQ-1 on the conformation of Bom37 G-quadruplex in K+/Na+ solutions
The effect of BMPQ-1 on the conformation of Bom37 G-quadruplex in K+ and Na+ solutions was further assessed by CD spectroscopy (Fig. 6). The addition of BMPQ-1 significantly decreased the negative CD signal peak at 265 nm and the positive signal peak at about 248 nm of Bom37 in the K+ solution. Additionally, BMPQ-1 causes a significant blue shift in the CD signal peak at 265 nm for Bom37 in K+ solutions. However, BMPQ-1 showed negligible effects on the CD spectra of Bom37 in the Na+ solution. It suggested that BMPQ-1 has a more substantial impact on the conformation of Bom37 in K+ solutions, which is consistent with the results that BMPQ-1 has a higher fluorescence response with the addition of Bom37 in K+ solution.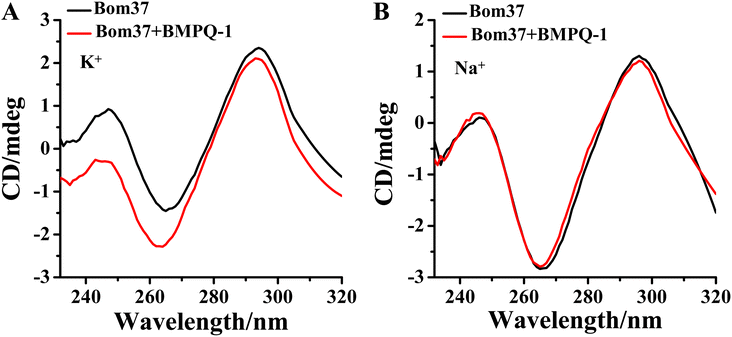 | ||
| Fig. 6 Effect of BMPQ-1 on CD spectra of Bom37 G-quadruplex in 10 mM Tris–HCl with 100 mM (A) K+ and (B) Na+. The concentration of Bom37 was 5.0 μM. | ||
The ability of BMPQ-1 to stabilize the structure of the Bom37 G-quadruplex in K+/Na+ solutions
The ideal G-quadruplex ligand should have the characteristics of high G-quadruplex recognition specificity and high G-quadruplex stability. The above results demonstrated that BMPQ-1 selectively recognized insect multimeric G-quadruplexes against monomeric G-quadruplexes. To test the ability of BMPQ-1 to stabilize insect multimeric G-quadruplexes, the CD melting temperatures (Tm) of Bom37 G-quadruplexes with and without BMPQ-1 in K+/Na+ solutions were measured. As shown in Fig. 7, BMPQ-1 exhibited more robust stabilization of Bom37 in K+ and Na+ solutions, with ΔTm values of about 20.05 ± 1.08 °C and 13.42 ± 0.97 °C, respectively. This result demonstrated that BMPQ-1 has a higher stabilizing capacity for Bom37 G-quadruplex in K+ solution.BMPQ-1 inhibits the proliferation of Spodoptera frugiperda 9 cells (SF9)
The above results suggest that BMPQ-1 targets insect multimeric G-quadruplexes, which stimulated us further to evaluate the inhibitory activity of BMPQ-1 on insect cells. SF9 is the ovipositor cell of Spodoptera frugiperda, a lepidopteran pest (the telomere sequence is a GGTTA repeat) native to tropical America that has become a global super-pest threatening agricultural production. BMPQ-1 was found to have a significant dose-dependent inhibition of SF9 cell proliferation with an IC50 of 1.80 ± 0.36 μM (Fig. S7†). This result further indicated that insect telomeric G-quadruplexes are potential targets for screening insecticidal drugs.Discussions and conclusions
Human telomere sequences (TTAGGG)n have been extensively studied for folding into G-quadruplex structures with multiple topologicals in the K+ and Na+ solutions.33–36 In contrast, the insect telomere sequence (TTAGG)n differs from the telomere sequences of humans by a guanine deletion in each repeat, folding into an antiparallel structured G-quadruplex in K+ and Na+ solutions.Compared to monomeric G-quadruplexes, multimeric G-quadruplexes have a higher-order structure and are effective targets for screening for ligands with high selectivity, making them more worthy of further study. BMPQ-1 has a higher selectivity for insect multimeric G-quadruplexes than monomeric G-quadruplexes, and it was found to have a significant inhibitory effect on the proliferation of SF9 cells, suggesting insect multimeric G-quadruples can be used as potential targets for insecticide screening.
Compared to Bom37 in the K+ solution, insect multimeric telomeric G-quadruplexes in Na+ solutions exhibited additional stability, which may be due to their different compactness. The different compactness of insect telomeric G-quadruplexes may result from different folding patterns, although both exhibit antiparallel conformation. Phan AT discovered that although both insect telomeric G-quadruplexes and aptamer sequences fold into antiparallel structured G-quadruplexes with two planar G-quartets and three loops, they differ by loop arrangements.31 It indicated that the tandem formation of antiparallel G-quadruplex units of varying degrees of compactness may form telomeric G-quadruplexes in insect cells. The stability and compactness of multimeric G-quadruplexes in Na+ solutions are higher than in K+ solutions, and the DNA sequence d(GTTAGG) was found to assemble preferentially into a trimolecular G-quadruplex with two-stacked G-tetrads in Na+ solutions,37 suggesting that telomeric G-quadruplexes may fold more readily into G-quadruplex structures in the Na+ solutions.
Maintaining the stability of chromosome ends is an important role of telomeres, and the human multimeric G-quadruplex is an effective target for screening anti-tumour drugs.38,39 Compounds targeting the human telomeric G-quadruplex can cause damage to telomeric DNA. Insect multimeric G-quadruplex is worth exploring whether insect telomeric G-quadruplexes can be used as an insecticide target.
Materials and methods
Materials
BMPQ-1 was purchased from Shanghai Topscience Co., Ltd (China). The oligonucleotides with PAGE or HPLC purification were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) and TSINGKE Biological Technology Co., Ltd. (Beijing, China). The concentration of oligonucleotides was measured by Nanodrop (DeNovix, USA).Fluorescence spectroscopy
Fluorescence spectra were recorded on a fluorescence spectrophotometer (Hitachi, Japan) with a 1.0 mm path length, 5 nm excitation slit, and emission slit. The DNA solutions were denatured at 95 °C for 5 min and then slowly cooled to room temperature. After each addition of the BMPQ-1, the DNA-complex samples were stirred and allowed to equilibrate at 25 °C for 10 min. The concentration of DNA samples and BMPQ-1 was 4.0 μM. The emission spectra in the wavelength range of 400–750 nm were measured at λex = 395 nm.The fluorescence variation of BMPQ-1 with the titration of Bom17/Bom37 was used to derive the dissociation constant (KD) values according to the method of Benesi-Hildebrand, using the following equation: 1/(ΔF) = 1/(b[DNA][H]0Ka) + 1/(b[H]0). ΔF represents the change in the fluorescence intensity (ΔF = F − F0); F denotes the fluorescence intensity of BMPQ-1 at 525 nm in the presence of a different concentration of Bom17/Bom37; F0 is the fluorescence intensity of the free BMPQ-1 at 525 nm. [DNA] is the total concentration of Bom17/Bom37 G-quadruplexes, and [H]0 is the total concentration of BMPQ-1. Moreover, Ka is the binding constant, while KD is equal to 1/Ka.
2-Ap titration experiments
2-Ap-labeled Bom37 samples were dissolved in Tris–HCl (10 mM, pH 7.5) buffer in 100 mM KCl or NaCl. The samples were denatured at 95 °C for 5 min and then cooled slowly to room temperature. The concentration of BMPQ-1 was in the range of 0–5.0 μM. The complex solution was stirred and equilibrated at 25 °C for 10 min. The fluorescence spectra were recorded at λem = 375 nm and λex = 305 nm with ex/em = 5/5 nm. The concentration of 2-Ap-labeled oligonucleotides was 1.0 μM. F indicates the fluorescence intensity of the mixture of BMPQ-1 and 2-AP-labeled Bom37. F0 shows the fluorescence intensity of 2-AP-labeled Bom37.Job's plot analysis
Job plot assays were conducted by recording the variation of the molar fraction of BMPQ-1 and Bom37. The sum concentration of Bom37 and BMPQ-1 was kept constant at 5.0 μM. Different volumes of Bom37 and BMPQ-1 stock solutions were mixed to give different molar fractions of BMPQ-1. The fluorescence intensity of BMPQ-1 at λex = 511 nm and λem = 550 nm was determined by its molar ratio to Bom37. The binding stoichiometry between Bom37 and BMPQ-1 corresponded to the plot's intersection points.FRET efficiency analysis
The oligonucleotides were labeled with 5′-FAM and 3′-TAMRA and dissolved in Tris–HCl buffer (10 mM, pH 7.5) in the presence of 100 mM KCl or NaCl. All the DNA samples were denatured at 95 °C for 5 min, then slowly cooled to room temperature and incubated at 4 °C overnight. The data was used to record the spectra over a 490–750 nm wavelength range at λex = 488 nm. The fluorescence intensities of FAM and TAMRA were measured at the emission wavelengths of 525 nm and 585 nm, respectively, from which the FRET efficiency was calculated. The concentration of oligonucleotides was 0.5 μM.The efficiency of FRET (EFRET) is given by
Here, FFAM and FTAMRA are the values of fluorescence intensities of FAM and TAMRA, respectively.
Circular dichroism spectroscopy
Circular dichroism (CD) spectra were recorded using a CD spectrophotometer (Jasco, Japan) with 1.0 mm path-length quartz cells. The data was measured from 220–320 nm with a 1.0 nm bandwidth. The data of melting curves data were collected at 265 nm during heating across the 4–95 °C range with intervals of 1.5 °C. All DNA samples were annealed in Tris–HCl buffer (10 mM, pH 7.5) in the presence of 100 mM KCl/NaCl and were denatured at 95 °C for 5 min, then gradually cooled to room temperature and incubated at 4 °C. The concentration of oligonucleotides was 10 μM. The analysis of the CD spectra was conducted using the software Origin Pro9.0.Gel electrophoresis
Native polyacrylamide gel electrophoresis was conducted on non-denaturing gels containing 20% (w/v) polyacrylamide gels in 0.5× TBE buffer with 50 mM KCl. DNA samples (6.0 μM) in 10 mM Tris–HCl (pH 7.5) and 100 mM KCl/NaCl were loaded and analyzed by electrophoresis. The gels were stained with SYBR® Gold (Thermo Fisher Scientific) and imaged using a fluorescent imager (Bio-Rad). Before the measurement, the samples were heated at 95 °C for 5 min, then cooled to room temperature and incubated at 4 °C overnight. The gels were run at 100 V and 25 °C for 180 min.Cell cytotoxicity assay
SF9 cells were cultured in Sf-900™ III SFM medium at 27 °C (Gibco, USA). Cells were seeded on 96-well plates (5 × 103 cells per well) and grown for 24 h for attachment. Then the cells were exposed to various concentrations of BMPQ-1 (0.0625, 0.125, 0.25, 0.5, 1.0, 2.0, 4.0, 8.0, 16.0, and 32.0 μM) and incubated at 27 °C for 72 h. Cell survival was evaluated using a Cell Counting Kit-8 assay (DOJINDO, Japan). After incubation, the old medium was discarded, and added fresh medium was added. Then 100 μL of the Cell Counting Kit-8 assay was added to each well and further incubated for 4 h at 27 °C. The optical density (O. D.) was detected at 450 nm by an automated microplate reader (Bio-Rad). The half maximal inhibitory concentration (IC50) values were derived from the curves of the mean percentage of survival against the drug concentration. All the experiments were performed in five replicates.Author contributions
Chao Gao: conceptualization, investigation, writing – original draft, formal analysis, funding acquisition. Jixin Chen: investigation, writing, formal analysis Naureen Anwar: writing. Jieya Deng: investigation. Zhangqian Wang: resources, writing – review & editing, project administration, funding acquisition. Muhammad Umer: writing – review & editing, project administration. Yi He: conceptualization, resources, project administration, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The National Natural Science Foundation of China (Grant No. 31900011), Ganzhou General Inspection and Testing Institute (Grant No. GZSZJYKF-202209), and Research and Innovation Initiatives of WHPU (2023Y82) supported this research.References
- G. N. Parkinson, M. P. Lee and S. Neidle, Crystal structure of parallel quadruplexes from human telomeric DNA, Nature, 2002, 417(6891), 876–880 CrossRef CAS PubMed.
- E. Cadoni, L. De Paepe, A. Manicardi and A. Madder, Beyond small molecules: targeting G-quadruplex structures with oligonucleotides and their analogues, Nucleic Acids Res., 2021, 49(12), 6638–6659 CrossRef CAS.
- C. J. Lech, B. Heddi and A. T. Phan, Guanine base stacking in G-quadruplex nucleic acids, Nucleic Acids Res., 2013,(3), 2034–2046 CrossRef CAS.
- Q. Xu, M. Yang, Y. Chang, S. Peng, D. Wang, X. Zhou and Y. Shao, Switching G-quadruplex to parallel duplex by molecular rotor clustering, Nucleic Acids Res., 2022, 50(18), 10249–10263 CrossRef CAS PubMed.
- D. Pavc, B. Wang, L. Spindler, I. Drevenšek-Olenik, J. Plavec and P. Šket, GC ends control topology of DNA G-quadruplexes and their cation-dependent assembly, Nucleic Acids Res., 2020, 48(5), 2749–2761 CrossRef CAS PubMed.
- M. Mieczkowski, C. Steinmetzger, I. Bessi, A. K. Lenz, A. Schmiedel, M. Holzapfel, C. Lambert, V. Pena and C. Höbartner, Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine, Nat. Commun., 2021, 12(1), 3549 CrossRef CAS PubMed.
- R. Hänsel-Hertsch, M. Di Antonio and S. Balasubramanian, DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential, Nat. Rev. Mol. Cell Biol., 2017, 18(5), 279–284 CrossRef.
- E. Gazanion, L. Lacroix, P. Alberti, P. Gurung, S. Wein, M. Cheng, J. L. Mergny, A. R. Gomes and J. J. Lopez-Rubio, Genome wide distribution of G-quadruplexes and their impact on gene expression in malaria parasites, PLoS Genet., 2020, 16(7), e1008917 CrossRef CAS.
- S. Chaudhary, M. Kumar and M. Kaushik, Interface of G-quadruplex with both stabilizing and destabilizing ligands for targeting various diseases, Int. J. Biol. Macromol., 2022, 219, 414–427 CrossRef CAS PubMed.
- G. Liu, W. Du, X. Sang, Q. Tong, Y. Wang, G. Chen, Y. Yuan, L. Jiang, W. Cheng, D. Liu, Y. Tian and X. Fu, RNA G-quadruplex in TMPRSS2 reduces SARS-CoV-2 infection, Nat. Commun., 2022, 13(1), 1444 CrossRef CAS PubMed.
- Y. Zhang, S. Liu, H. Jiang, H. Deng, C. Dong, W. Shen, H. Chen, C. Gao, S. Xiao, Z. F. Liu and D. Wei, G2-quadruplex in the 3'UTR of IE180 regulates Pseudorabies virus replication by enhancing gene expression, RNA Biol., 2020, 17(6), 816–827 CrossRef CAS PubMed.
- M. Armanios, The Role of Telomeres in Human Disease, Annu. Rev. Genomics Hum. Genet., 2022, 23, 363–381 CrossRef CAS.
- A. J. Zaug, K. J. Goodrich, J. J. Song, A. E. Sullivan and T. R. Cech, Reconstitution of a telomeric replicon organized by CST, Nature, 2022, 608(7924), 819–825 CrossRef CAS PubMed.
- F. Rossiello, D. Jurk, J. F. Passos and F. d'Adda di Fagagna, Telomere dysfunction in ageing and age-related diseases, Nat. Cell Biol., 2022, 24(2), 135–147 CrossRef CAS.
- R. J. O'Sullivan and J. Karlseder, Telomeres: protecting chromosomes against genome instability, Nat. Rev. Mol. Cell Biol., 2010, 11(3), 171–181 CrossRef.
- C. Gao, Z. Liu, H. Hou, J. Ding, X. Chen, C. Xie, Z. Song, Z. Hu, M. Feng, H. I. Mohamed, S. Xu, G. N. Parkinson, S. Haider and D. Wei, BMPQ-1 binds selectively to (3 + 1) hybrid topologies in human telomeric G-quadruplex multimers, Nucleic Acids Res., 2020, 48(20), 11259–11269 CrossRef CAS.
- M. H. Hu, S. B. Chen, B. Wang, T. M. Ou, L. Q. Gu, J. H. Tan and Z. S. Huang, Specific targeting of telomeric multimeric G-quadruplexes by a new triaryl-substituted imidazole, Nucleic Acids Res., 2017, 45(4), 1606–1618 CrossRef CAS.
- S. Raje and R. Barthwal, Molecular recognition of 3+1 hybrid human telomeric G-quadruplex DNA d-AGGG(TTAGGG)(3) by anticancer drugs epirubicin and adriamycin leads to thermal stabilization, Int. J. Biol. Macromol., 2019, 139, 1272–1287 CrossRef CAS PubMed.
- F. R. Winnerdy, B. Bakalar, P. Das, B. Heddi, A. Marchand, F. Rosu, V. Gabelica and A. T. Phan, Unprecedented hour-long residence time of a cation in a left-handed G-quadruplex, Chem. Sci., 2021, 12(20), 7151–7157 RSC.
- L. Olejko, A. Dutta, K. Shahsavar and I. Bald, Influence of Different Salts on the G-Quadruplex Structure Formed from the Reversed Human Telomeric DNA Sequence, Int. J. Mol. Sci., 2022, 23(20), 12206 CrossRef CAS PubMed.
- C. Zhao, L. Wu, J. Ren, Y. Xu and X. Qu, Targeting Human Telomeric Higher-Order DNA: Dimeric G-Quadruplex Units Serve as Preferred Binding Site, J. Am. Chem. Soc., 2013, 135(50), 18786–18789 CrossRef CAS.
- M. Bončina, G. Vesnaver, J. B. Chaires and J. Lah, Unraveling the Thermodynamics of the Folding and Interconversion of Human Telomere G-Quadruplexes, Angew. Chem., Int. Ed., 2016, 55(35), 10340–10344 CrossRef.
- C. Lin, G. Wu, K. Wang, B. Onel, S. Sakai, Y. Shao and D. Yang, Molecular Recognition of the Hybrid-2 Human Telomeric G-Quadruplex by Epiberberine: Insights into Conversion of Telomeric G-Quadruplex Structures, Angew. Chem., Int. Ed., 2018, 57(34), 10888–10893 CrossRef CAS.
- H. L. Bao, H. S. Liu and Y. Xu, Hybrid-type and two-tetrad antiparallel telomere DNA G-quadruplex structures in living human cells, Nucleic Acids Res., 2019, 47(10), 4940–4947 CrossRef CAS PubMed.
- R. C. Monsen, S. Chakravarthy, W. L. Dean, J. B. Chaires and J. O. Trent, The solution structures of higher-order human telomere G-quadruplex multimers, Nucleic Acids Res., 2021, 49(3), 1749–1768 CrossRef CAS PubMed.
- J. Abraham Punnoose, Y. Ma, Y. Li, M. Sakuma, S. Mandal, K. Nagasawa and H. Mao, Adaptive and Specific Recognition of Telomeric G-Quadruplexes via Polyvalency Induced Unstacking of Binding Units, J. Am. Chem. Soc., 2017, 139(22), 7476–7484 CrossRef CAS PubMed.
- I. Frasson, V. Pirota, S. N. Richter and F. M. Doria, G-quadruplexes: a review on their biological roles and targeting, Int. J. Biol. Macromol., 2022, 204, 89–102 CrossRef CAS.
- D. Bai, S. W. Shan, X. Zhang, Y. Li, J. Xie and W. Q. Wu, Comprehensive insights into the structures and dynamics of plant telomeric G-quadruplexes, Int. J. Biol. Macromol., 2023, 231, 123281 CrossRef CAS PubMed.
- L. F. Rosin, J. Gil Jr, I. A. Drinnenberg and E. P. Lei, Oligopaint DNA FISH reveals telomere-based meiotic pairing dynamics in the silkworm, Bombyx mori, PLoS Genet., 2021, 17(7), e1009700 CrossRef CAS.
- K. On, G. Crevel, S. Cotterill, M. Itoh and Y. Kato, Drosophila telomere capping protein HOAP interacts with DSB sensor proteins Mre11 and Nbs, Genes Cells, 2021, 26(4), 219–229 CrossRef CAS.
- S. Amrane, R. W. Ang, Z. M. Tan, C. Li, J. K. Lim, J. M. Lim, K. W. Lim and A. T. Phan, A novel chair-type G-quadruplex formed by a Bombyx mori telomeric sequence, Nucleic Acids Res., 2009, 37(3), 931–938 CrossRef CAS.
- A. Kettani, S. Bouaziz, W. Wang, R. A. Jones and D. J. Patel, Bombyx mori single repeat telomeric DNA sequence forms a G-quadruplex capped by base triads, Nat. Struct. Biol., 1997, 4(5), 382–389 CrossRef CAS PubMed.
- C. Saintomé, P. Alberti, N. Guinot, P. Lejault, J. Chatain, P. Mailliet, J. F. Riou and A. Bugaut, Binding properties of mono- and dimeric pyridine dicarboxamide ligands to human telomeric higher-order G-quadruplex structures, Chem. Commun., 2018, 54(15), 1897–1900 RSC.
- A. Ambrus, D. Chen, J. Dai, T. Bialis, R. A. Jones and D. Yang, Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution, Nucleic Acids Res., 2006, 34(9), 2723–2735 CrossRef CAS PubMed.
- J. Dai, C. Punchihewa, A. Ambrus, D. Chen, R. A. Jones and D. Yang, Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence, Nucleic Acids Res., 2007, 35(15), 4927–4940 CrossRef CAS PubMed.
- J. Dai, C. Punchihewa, A. Ambrus, D. Chen, R. A. Jones and D. Yang, Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation, Nucleic Acids Res., 2007, 35, 2440–2450 CrossRef CAS.
- H. Jing, W. Fu, W. Hu, S. Xu, X. Xu, M. He, Y. Liu and N. Zhang, NMR structural study on the self-trimerization of d(GTTAGG) into a dynamic trimolecular G-quadruplex assembly preferentially in Na+ solution with a moderate K+ tolerance, Nucleic Acids Res., 2021, 49(4), 2306–2316 CrossRef CAS PubMed.
- M. H. Hu, X. T. Lin, B. Liu and J. H. Tan, Dimeric aryl-substituted imidazoles may inhibit ALT cancer by targeting the multimeric G-quadruplex in telomere, Eur. J. Med. Chem., 2020, 186, 111891 CrossRef CAS.
- T. C. Liao, T. Z. Ma, S. B. Chen, A. Cilibrizzi, M. J. Zhang, J. H. Li and C. Q. Zhou, Human telomere double G-quadruplex recognition by berberine-bisquinolinium imaging conjugates in vitro and cells, Int. J. Biol. Macromol., 2020, S0141-8130, 33034–33038 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra06805f |
| ‡ The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors. |
| This journal is © The Royal Society of Chemistry 2023 |