Open Access Article
Open Access ArticleTargeting multidrug resistant Staphylococcus infections with bacterial histidine kinase inhibitors†
Adeline
Espinasse‡
 a,
Manibarsha
Goswami‡
a,
Junshu
Yang
b,
Onanong
Vorasin
ac,
Yinduo
Ji
a,
Manibarsha
Goswami‡
a,
Junshu
Yang
b,
Onanong
Vorasin
ac,
Yinduo
Ji
 *b and
Erin E.
Carlson
*b and
Erin E.
Carlson
 *adef
*adef
aDepartment of Chemistry, University of Minnesota, 225 Pleasant St. SE, Minneapolis, 55454, MN, USA. E-mail: carlsone@umn.edu
bDepartment of Veterinary and Biomedical Sciences, University of Minnesota, 1971 Commonwealth Ave, Falcon Heights 55108, MN, USA. E-mail: jixxx002@umn.edu
cDepartment of Chemistry, Faculty of Science, Mahidol University, Rama 6 Road, Bangkok 10400, Thailand
dDepartment of Medicinal Chemistry, University of Minnesota, 208 Harvard Street SE, Minneapolis 55454, Minnesota, USA
eDepartment of Biochemistry, Molecular Biology, and Biophysics, University of Minnesota, 321 Church St SE, Minneapolis 55454, Minnesota, USA
fDepartment of Pharmacology, University of Minnesota, 321 Church St SE, Minneapolis 55454, Minnesota, USA
First published on 11th April 2023
Abstract
The emergence of drug-resistant bacteria, such as methicillin-resistant Staphylococcus aureus (MRSA), which are not susceptible to current antibiotics has necessitated the development of novel approaches and targets to tackle this growing challenge. Bacterial two-component systems (TCSs) play a central role in the adaptative response of bacteria to their ever-changing environment. They are linked to antibiotic resistance and bacterial virulence making the proteins of the TCSs, histidine kinases and response regulators, attractive for the development of novel antibacterial drugs. Here, we developed a suite of maleimide-based compounds that we evaluated against a model histidine kinase, HK853, in vitro and in silico. The most potent leads were then assessed for their ability to decrease the pathogenicity and virulence of MRSA, resulting in the identification of a molecule that decreased the lesion size caused by a methicillin-resistant S. aureus skin infection by 65% in a murine model.
Introduction
Staphylococcus aureus is one of the deadliest pathogens in the world, and the rise in resistant strains of this organism has led to many life-threatening medical conditions. This Gram-positive coccus causes a range of maladies, from minor skin infections to severe infections such as toxic shock syndrome or endocarditis and is responsible for more deaths in the United States than any other drug-resistant pathogen.1,2 Approximately 11.6 million outpatient and emergency room visits and 464![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 hospital admissions occur annually in the US due to staphylococcus infections.3 As the use of antibiotics is rising and multidrug-resistant strains in hospitals are emerging, the most notable being methicillin-resistant S. aureus (MRSA), treatment of infections with traditional antibiotics is proving to be futile. At present, MRSA accounts for more than 60% of S. aureus infections in hospitals (HA-MRSA) in the US and has increasingly spread beyond healthcare facilities and emerged as a community-associated pathogen (CA-MRSA).4–6 To fight against MRSA and other deadly pathogens that are impervious to existing antibiotics, discovery of alternative antibacterial agents has become extremely crucial.7,8
000 hospital admissions occur annually in the US due to staphylococcus infections.3 As the use of antibiotics is rising and multidrug-resistant strains in hospitals are emerging, the most notable being methicillin-resistant S. aureus (MRSA), treatment of infections with traditional antibiotics is proving to be futile. At present, MRSA accounts for more than 60% of S. aureus infections in hospitals (HA-MRSA) in the US and has increasingly spread beyond healthcare facilities and emerged as a community-associated pathogen (CA-MRSA).4–6 To fight against MRSA and other deadly pathogens that are impervious to existing antibiotics, discovery of alternative antibacterial agents has become extremely crucial.7,8
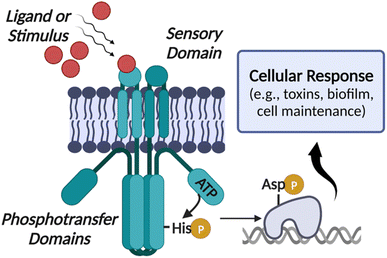
Traditional antibiotics inherently lead to resistant mutant development as they are intended to kill the bacteria, placing a high level of evolutionary pressure on them to adapt and survive. To combat resistant pathogens, an alternative approach that has gained significant attention in recent years is controlling their “virulence” or ability to spread infections instead of directly killing them.9–11 Most bacteria have evolved complex networks for virulence regulation relying on the two-component systems (TCSs), enzymes central to the pathogenicity of many organisms, including S. aureus.12–14 A prototypical TCS consists of a sensor kinase, known as a histidine kinase (HK), and a cognate partner called the response regulator (RR). The main function of the TCSs is to sense various environmental stimuli, including temperature, nutrients, pH changes, and even antibiotic exposure and to modulate cellular response and virulence-associated gene expression. Upon receiving a given signal, the membrane-bound HK undergoes autophosphorylation on a catalytic histidine residue using ATP, and subsequent phosphoryl group transfer to the RR, which is often a transcription factor, leads to cellular response (Fig. 1).15,16
To identify inhibitors that will be broadly applicable to a range of HKs in different bacteria, we targeted the highly-conserved catalytic ATP-binding (CA) domain of these proteins. Within this domain, HKs utilize a Bergerat fold to bind their ATP substrate. While the Bergerat fold is not found in eukaryotic kinases, we reasoned that some compounds that have been developed to mimic ATP, such as mammalian-kinase inhibitors, could provide useful starting points for the identification of HK-specific molecules. Almost all bacteria have TCSs and most possess 20–100 distinct protein pairs per organism with an average of 30.17,18 While some TCSs or HKs are essential for growth and survival such as WalK in Gram-positive bacteria, they are quintessential in virulence processes for both Gram-positive and Gram-negative bacteria.13 The ubiquity of the HKs and their importance in key pathogenesis and virulence mechanisms suggests that a therapy targeting these enzymes may have a widespread and profound effect.
The ability of S. aureus to resist antibiotics and to cause infection is partially due to the coordinated regulation of gene expression mediated by the TCSs. Staphylococcus, including MRSA strains, have 17 TCSs including the quorum-sensing Agr, the regulator of staphylococcal accessory exoproteins SaeRS, autolysin-related locus ArlRS, oxygen-sensing and redox-signaling AirSR (previously known as YhcS), and WalRK, which is essential for growth (also named YycFG or VicRK).19,20 Previously, we have developed small molecule inhibitors that can inactivate multiple HKs simultaneously and more recently, we have shown that benzothiazole-based compounds are effective at reducing the virulence of Pseudomonas aeruginosa, another dangerous nosocomial pathogen.21–23 These molecules dramatically reduced several virulence behaviors of P. aeruginosa, such as swarming and the production of toxic metabolites. Taking this together, we hypothesized that inhibitors that target the virulence-associated HKs in S. aureus will be effective alternative antibacterial agents against MRSA infections. However, this previously identified scaffold had no activity against MRSA necessitating the discovery of new chemical matter (data not shown).
Results and discussion
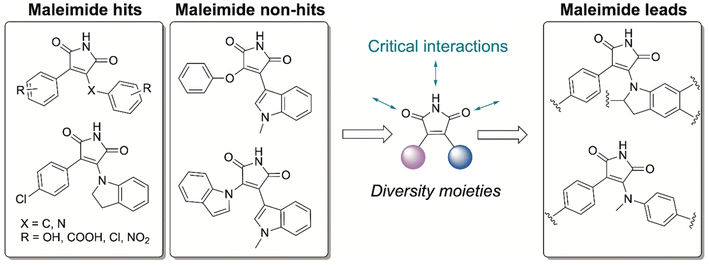
The pharmaceutical industry has put enormous effort into targeting of the eukaryotic kinases and a plethora of good candidates and their pharmacological data remains underutilized. Hence, companies like GlaxoSmithKIine (GSK) have made their unsuccessful molecules freely available.24–26 We screened the Published Kinase Inhibitor Set (PKIS), a collection of 367 small molecule ATP-competitive kinase inhibitors,27 to determine if these Ser/Thr/Tyr mammalian kinase binders can also interact with the CA domain of bacterial HKs, despite substantial differences in the amino acid residues and geometry. We hypothesized that these molecules could act as starting points from which we would improve HK potency and identify key binding events for selective HK inhibition.Previously, we reported a high-throughput fluorescence polarization assay to examine compound libraries, including a mammalian kinase inhibitor set from Roche, to identify pan-HK inhibitors.21,22 Using this assay, we tested the GSK compound library for competitive binding of HKs, specifically HK853, a model protein from the marine bacterium Thermotoga maritima. This protein was used as it is drastically easier to produce in large quantities and is substantially more stable than other HK proteins.21,22 Of the 367 compounds, only a small subset of molecules (12) displayed strong affinity for HK853 further demonstrating that bacterial and eukaryotic kinases bind ATP through dramatically different interactions. Next, we performed dose-response studies with these 12 hits to determine their binding affinities and to rule out false positives. We found that 7 out of the 12 molecules possessed micromolar affinity for HK853 (Fig. S1†) and surprisingly, all 7 of these molecules contained the same core structure, a maleimide functionalized with ring substituents on both ends of the alkene (Fig. 2). Despite the presence of a maleimide, we do not anticipate that these molecules are covalent inhibitors as there are no cysteine residues in or near the active site. The maleimide “hits” all possess a phenyl substituent and either a N-methylanilino or indolinyl group on the other side. We also identified related compounds that failed to bind to HK853 (non-hits; Fig. 2 and S1†). These maleimide-based molecules are not PAN-kinase inhibitors but instead only interact with one kinase, Glycogen Synthase Kinase-3 (GSK-3),24–26 which is implicated in neurogenerative diseases and type-2 diabetes.28 This observation was very promising as it implies that there is a high probability of optimizing the selectivity of our leads for bacterial versus mammalian kinases. Compounds containing anilino-, indolo-, phenol-type substituents are potent inhibitors of GSK-3, while N-methylanilino and indolinyl-containing molecules are weak binders or gave inconclusive results for GSK-3 inhibition.24,26 Taking this structure activity relationship (SAR) information into account, we pursued maleimide-containing structures with indolinyl- or N-methylanilino- and phenyl-substituents (Scheme 1, PMI-series), as these are strong HK binders and showed minimal to no activity towards mammalian kinases.
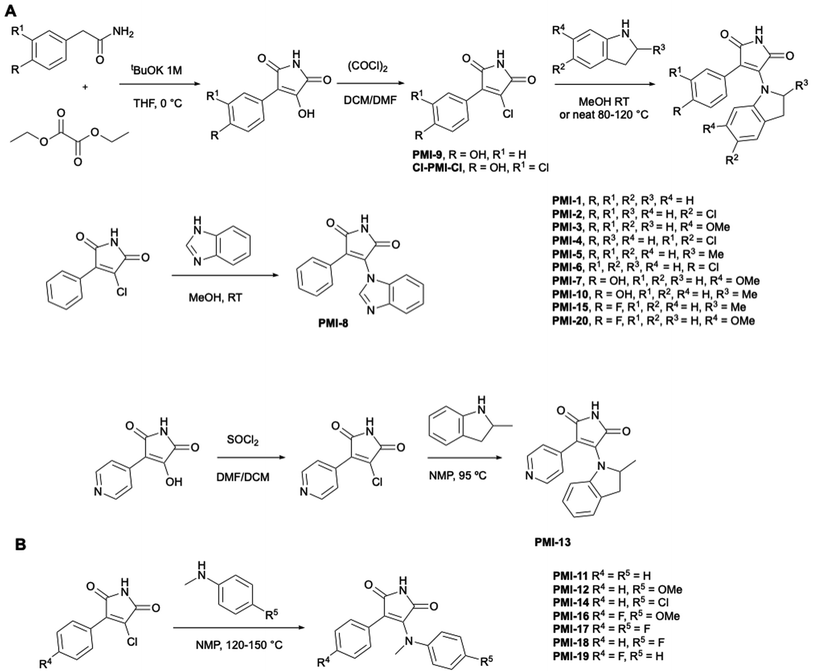
To further study this compound class, we devised a 3-step modular synthetic route by which maleimide derivatives (Scheme 1, PMI-1–20) could be easily generated from cheap starting materials. By condensation of various phenylacetamides with diethyl oxalate, the core maleimide-alcohol was synthesized, which was further converted to the chloro-derivatives using oxalyl chloride or thionyl chloride (pyridinyl ring). PMI-Cl derivatives were then reacted with various indolines or N-methyl anilines to yield the desired maleimide-containing products. Preliminary degradation studies of PMI-5 in human and mouse liver microsomes demonstrated that this molecule was quickly metabolized in both systems (half-life of 18.2 and <2.3 min, respectively; full data in ESI†).
To potentially prevent fast metabolism by cytochrome P450 enzymes at the para-position of the phenyl, we designed analogues bearing substituents (hydroxy, chlorine or fluorine atom) at this position (indoline: PMI-6, 7, 10, 15, 20; N-methyl aniline: PMI-16, 17, 19). Additionally, we replaced the phenyl group with a 4-pyridinyl group to block this metabolism site (PMI-13). These substituents were also designed to enhance membrane permeation and to potentially mediate additional non-covalent interactions with the protein.29,30
We tested the ability of these lead molecules to inhibit HK autophosphorylation, and thereby their ability to arrest bacterial signaling events, using an activity-based assay that was previously developed by our research group (Table S1†).31 This assay uses a fluorophore labeled ATP-based probe to enable rapid read-out of the ability of a molecule to prevent autophosphorylation and thus, fluorescent labeling of the HK. We examined two HKs, our model protein HK853 and AirS (formerly YhcS) from S. aureus, which is important for its survival32,33 and involved in oxygen and redox sensing,34 nitrate metabolism,35 and virulence.34,36 Results were consistent with the fluorescence polarization binding assay (Table S1†) in that all of the compounds that were decorated with phenyl and indoline substituents strongly inhibited HK853 activity in the low-mid micromolar range (Fig. S2†). The N-methyl anilino analogues had poor potency against HK853 (Fig. S2†). Overall, the maleimide derivatives were not as potent against AirS with either IC50 values in the low-mid micromolar range or showing no inhibition (Table S1 and Fig. S3†). The analogues containing a combination of phenyl/indolinyl groups (PMI-1, PMI-5), fluorophenyl/indolinyl groups (PMI-15, PMI-20) or phenyl/N-methylanilino groups with an electron-donating moiety (PMI-12) were the most promising for AirS inhibition. Nevertheless, we observed a correlation between the most potent lead compounds for both HK853 and AirS.
Given the activity of these molecules in our CA-domain targeted assays, it is likely that the active molecules interact with the ATP-binding domains of HK853 and AirS. To evaluate the potential role of the ring structures in crucial binding interactions, we tested synthetic intermediate PMI-9 for HK inhibition, which showed weak activity (Table S1†) indicating the importance of the indoline ring or another large aromatic substituent. However, we noted that substituents on the phenyl or indolinyl rings did not enhance the activity of the molecules (e.g., PMI-1vs.PMI-2–8). Similarly, the presence of a pyrimidyl moiety (PMI-13), a hydrogen bonding acceptor, resulted in worse inhibition. For the N-methylaniline series, we observed that the presence of an electron-donating group on the para-position of the aniline promoted potency (PMI-11vs.PMI-12), whereas a chlorine atom decreased activity (PMI-11vs.PMI-14) and a fluorine atom yielded an inactive molecule (PMI-11vs.PMI-18). Next, we investigated the impact of a fluorine atom on the para-position of the phenyl moiety. Decreased potency was observed with the N-methyl aniline series (PMI-11vs.PMI-19; PMI-12vs.PMI-16) whereas with substituents on the indolinyl group, a similar or enhanced activity was seen (PMI-3vs.PMI-20; PMI-5vs.PMI-15). The presence of a chlorine atom on the phenyl or indolinyl rings drastically decreased AirS inhibition (PMI-1vs.PMI-2 and PMI-6), while potency for HK853 was similar. Interestingly, PMI-9 was the only analogue to show superior potency against AirS compared to HK853. Finally, we investigated how fluorine atoms modulated compound activity when they were present on one or both aromatic rings. Addition of fluorine to the phenyl ring recovered activity for HK853 (PMI-17vs.PMI-18) and its removal from the anilino ring further improved this result (PMI-19vs.PMI-17). Overall, the presence of any fluorine atom resulted in complete lack of inhibition for AirS (PMI-16, 17, 18, 19) in the N-methylaniline series. Taken together, these results demonstrated that HK853 and AirS have substantially different ligand binding preferences but also that the indoline analogues represent one of the most potent classes of bacterial HK inhibitors known. While a structure of AirS is not available, differences can be noted in a predicted structure (AlphaFold) such as a tyrosine that is likely important for compound binding in HK853 (Tyr 384; see docking results below) instead being a histidine residue in AirS (His296).
Using a native gel-based assay, we confirmed that PMI-3, 5, 11, 12, 15, 18 and 20 do not cause aggregation of HK853, which would indicate that their activity resulted from protein aggregation instead of specific molecule–protein interactions (Fig. S4†). These molecules were selected for evaluation based on preliminary inhibition and biological results indicating their promise in cell-based assays (see below).
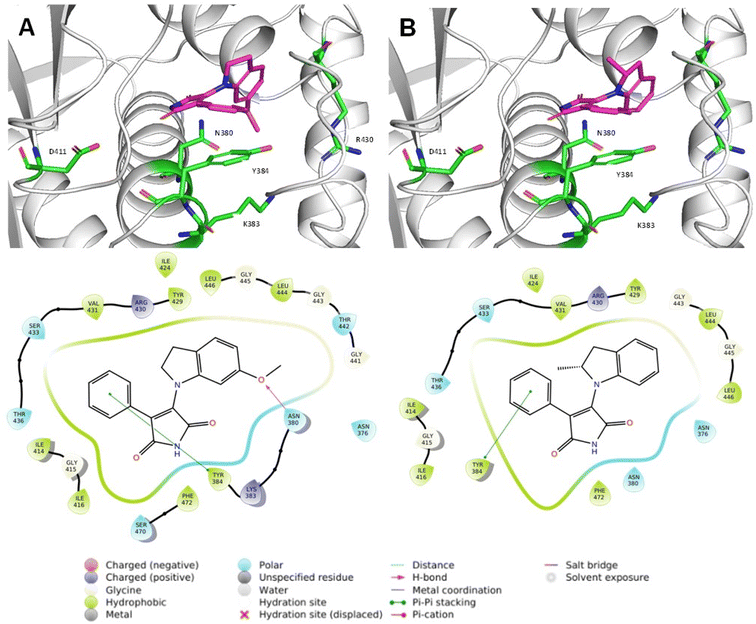
Next, we utilized molecular docking to further our understanding of the ligand–receptor interactions within HK853 (PDB-3DGE; PMI-3, 5, 11, 12, 15, 20). We found that the lead compounds interacted with the ATP-binding pocket through π–π interactions with Tyr384 of the N-box, a conserved motif in the ATP-binding pocket (Fig. 3 and S5†). PMI-3 and PMI-20 formed hydrogen bonds with Asn380 of the N-box through the oxygen atom of the methoxy group. We also noted that the molecules bound within the glycine-rich flexible loops, especially the G1- and G2-boxes.22 All of the studied molecules were docked with the maleimide moiety oriented towards the highly conserved residue, Asp411, which is critical for ATP binding through interactions with the exocyclic amine of adenine. Even so, no interactions with this residue were observed (Fig. 3 and S5†). Asp411 is also required for the interactions of several HK inhibitors such as guanine-based compounds and benzothiazole derivatives with the HK active site.21,22PMI-18, which did not inhibit HK853 activity, displayed similar interactions to the lead molecules but had the lowest docking score, which is consistent with its lack of potency (Table S2†). Interestingly, PMI-11 was oriented in the opposite direction compared to compounds with similar structures such as PMI-12 (Fig. S5†).
To further evaluate the differences in potency and identify the molecular features important for activity, we performed an AutoQSAR using the entire set of compounds (Table S3†).37 AutoQSAR is an automated machine-learning application used to develop quantitative SAR models based on experimental results.37 The model was built using the molecular structures and their corresponding −log(IC50)HK853 values to establish correlation between these two parameters followed by analysis and visualization in Canvas,38 which enabled us to assess the importance of each atom in protein binding. We found that the phenyl, maleimido, and indolinyl moieties were important for activity whereas N-methylation of the aniline decreased potency. The presence of electron-withdrawing atoms (fluorine, chlorine) on the aromatic groups, methyl groups on the indoline, and O-methylation of a phenol also negatively affected activity.
Finally, we used an ADP-glo™ assay to evaluate our lead compounds for activity against GSK-3β, one of the isoforms of GSK-3 which is the known target of the original hits from the GSK library. We found that lead compounds PMI-3, 5, 11, 12, 15 and 20 were at least 100 times less potent than the positive control staurosporine. However, they inhibit GSK-3β with similar efficacy to the activity observed in HK853 suggesting that future work will be required to achieve selectivity (Fig. S6†). The N-aniline analogues were less active than the indoline compounds, in line with previously published results.26 These results, combined with initial metabolism data, indicate that these compounds would likely be most promising in a skin infection model, which largely avoids issues of selectivity and metabolism (see below).
To determine whether these HK inhibitors are able to impede virulence in S. aureus, we examined their effect on the hemolytic activity of MRSA culture supernatant on sheep red blood cells.39 Our studies showed that some inhibitors, PMI-1, 2, 3, 5, and 6 (50 μM), significantly reduced staphylococcal hemolytic activity in both HA-MRSA WCUH29 (ref. 40) and USA300 CA-MRSA 923 (Table 1 and Fig. 4A).41PMI-4 was bactericidal at 50 μM making it a poor candidate for the development of an anti-virulence agent. We also found that PMI-1, 2, 3, or 5 alone had only limited hemolytic activity at 500 μM (Table 1). Some analogs, including PMI-7, 8, 9, 10, and 14 (50 μM), had no influence on the hemolytic activity of either staphylococcal strain (Table 1). Interestingly, PMI-11, 12, and 13 selectively inhibited the hemolytic activity of WCUH29 (Table 1). This is likely due to the differential expression of hemolysins between the HA-MRSA WCUH29 and USA300 CA-MRSA 923 strains.42,43 Together, these results indicate that PMI-1, 2, 3, and 5 are the most promising lead compounds against staphylococcal virulence. To further validate these potential lead compounds, we conducted cytotoxicity assays using the human lung epithelial A549 cells.44PMI-1, 2, 4, and 6 led to significant cell death at 250 μM, whereas PMI-3 and 5 (500 μM) had no cytotoxic effect on A549 cells (Fig. 4B), indicating the selectivity of these analogs against bacterial hemolytic factors.
| Molecule | % Hemolytic activity (molecule alone) | % A549 cell viability (molecule alone) | % Decrease in hemolysis by S. aureus WCUH29 | % Decrease in hemolysis by S. aureus CA-MRSA 923 |
|---|---|---|---|---|
| a Hemolytic activity of the PMI compounds evaluated at 500 μM in sheep red blood cells. Cell viability evaluated with A549 cells exposed to compounds (250 μM). Hemolysis activity of S. aureus evaluated in WCUH29 and CA-MRSA 923 pre-exposed to inhibitors (50 μM) and the supernatants used for cytotoxicity assays in sheep red blood cells. DMSO-treated WCUH29 was positive control, PBS-only was negative control. % Hemolysis = [(A450 test sample − A450 negative control)/(A450 positive control − A450 negative control)] × 100. Values of lysed cells obtained from three independent experiments. ND: not determined. | ||||
| PMI-1 | 3.37 | >99 | 66.1 | 72.3 |
| PMI-2 | 1.53 | 2.6 | 86.9 | 91.6 |
| PMI-3 | 0.79 | >99 | 74.2 | 12.7 |
| PMI-4 | 8.59 | 2.6 | Bactericidal | Bactericidal |
| PMI-5 | 2.42 | >99 | 69.5 | 85.6 |
| PMI-6 | 9.20 | >99 | 86.5 | 85.0 |
| PMI-7 | 12.5 | >99 | 0 | 0 |
| PMI-8 | 9.84 | >99 | 0 | 0 |
| PMI-9 | 8.16 | 30.2 | 0 | 4.34 |
| PMI-10 | 9.76 | >99 | 0 | 0 |
| PMI-11 | 0 | >99 | 72.0 | 0 |
| PMI-12 | 0 | >99 | 51.2 | 0 |
| PMI-13 | 0 | >99 | 41.9 | 0.85 |
| PMI-14 | 0 | >99 | 0 | 0 |
| PMI-15 | 14.1 | ND | 95.4 | 0 |
| PMI-16 | 34.9 | ND | 94.8 | 0 |
| PMI-17 | 11.5 | ND | 43.3 | 4.16 |
| PMI-18 | 11.6 | ND | 7.36 | 4.27 |
| PMI-19 | 21.6 | ND | 60.2 | 4.06 |
| PMI-20 | 11.6 | ND | 32.5 | 1.54 |
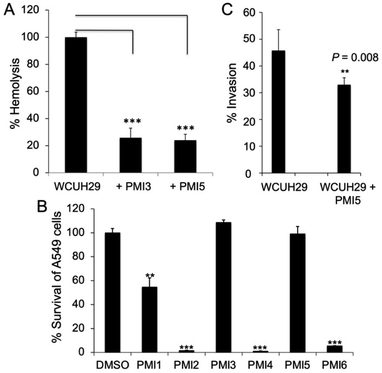 | ||
| Fig. 4 HK inhibitor effects in cell models. (A) S. aureus hemolytic activity. S. aureus WCUH29 was pre-exposed to inhibitors (50 μM) and the supernatants from stationary cultures were used for cytotoxicity assays in sheep red blood cells. Untreated WCUH29 is positive control, PBS-only is negative control. % Hemolysis = [(A450 test sample − A450 negative control)/(A450 positive control − A450 negative control)] × 100. Values of lysed cells were obtained from three independent experiments. (B) Effect of HK inhibitors on cell viability. A549 cells were exposed to a PMI inhibitor (500 μM; note that values in Tables 1 and S4† are for 250 μM treatment) for 24 h at 37 °C in 5% CO2 and the viability of cells was measured using the Cell-Glo kit.44 Values of percent survival are average from three independent experiments. (C) Effect of PMI-5 on WCUH29 invasion of the human lung epithelial A549 cells. Staphylococcal WCUH29 cells grown to exponential phase were added into monolayers of A549 cells in the presence or absence of PMI-5 (50 μM). After 2 h of incubation, the extracellular bacteria were killed with lysostaphin and gentamycin. The internalized bacteria were released by lysis of A549 cells and cultured for CFU counting by plating of the cell lysates onto TSA plates. Results statistically analysed using the T-test (n = 6). Experiment performed at least three times. **P < 0.01, ***P < 0.001. Error bars represent the STDEV. | ||
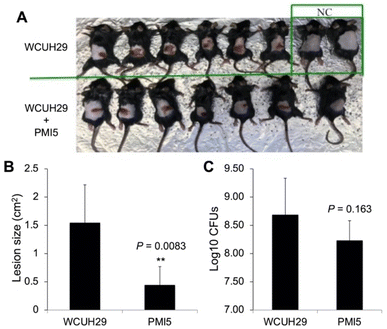
The capacity of S. aureus to cause various infections is attributable to its ability to adhere to and invade host cells. To elucidate whether the lead compounds affect bacterial invasion of host cells, we determined the impact of PMI-5 on invasion using the human lung epithelial A549 cells.45 We prioritized PMI-5 over PMI-3 given its superior activity in strain USA300 CA-MRSA 923. Remarkably, addition of PMI-5 (50 μM) decreased the staphylococcal invasion of A549 cells by 25% (Fig. 4C). Finally, to determine whether a lead compound could be effective in vivo during bacterial infection, we tested the effect of PMI-5 on the pathogenicity of WCUH29 in a mouse model of S. aureus skin infection. The bacterial cells (∼108 CFU) of exponential phase were used to subcutaneously infect mice in the presence of PMI-5 or vehicle control. Lesions appeared in the control group (n = 6) after three days of infection (Fig. 5A and B). However, the lesion size was significantly reduced, by 65%, in the PMI-5 treatment group (n = 7). Additionally, PMI-5 treatment slightly decreased (10%) the bacterial burden in the infected skin compared to the vehicle control (Fig. 5C). These data indicate that PMI-5 likely diminishes bacterial virulence in vivo during infection given that the damage caused to the host following treatment was dramatically decreased. A data summary table for PMI-1, 2, 3 and 5 is provided as ESI (Table S4†).
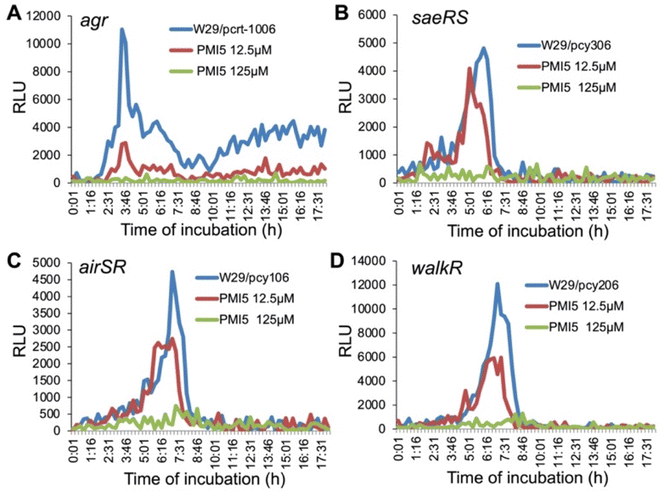
To elucidate the mode of action of the lead compound PMI-5 against staphylococcal pathogenicity, we explored the impact of this molecule on the expression of several S. aureus TCSs. Phosphorylation of RRs can cause positive regulation of their own transcription, as well as of their corresponding HK generating a feedback loop. Therefore, inhibition of HKs and, consequently, of the phosphorylation cascade can decrease the transcription of TCSs. The discovered molecules are expected to inhibit several or perhaps many HKs given their ability to interact with a shared domain. In order to determine which TCSs are affected by these novel HK inhibitors in S. aureus, we conducted preliminary studies to examine their impact on the transcription of several key TCSs in S. aureus using a promoter-lux report system.32,36 These investigations demonstrated that PMI-5 dramatically inhibited the transcription of agr (Fig. 6A), saeRS (Fig. 6B), airSR (yhcSR) (Fig. 6C), and walRK (Fig. 6D) in a dose-dependent manner, and had no obvious effect on bacterial growth (data not shown). PMI-3 exhibited a similar effect on the transcription of these genes (Fig. S7†). Combined with the fact that PMI-5 eliminated the hemolytic activity of MRSA, profoundly affected the staphylococcal capacity for invasion of A549 cells and attenuated the pathogenicity of MRSA in a murine skin infection, these data indicate that PMI-5 interferes with bacterial virulence. A key feature of the efficacy of this molecule may be its ability to inhibit various HKs that play important roles in MRSA pathogenicity, compared with known HK inhibitors.46 Interestingly, although PMI-5 causes downregulation of walRK transcription, an essential TCS for growth, no impact on growth was observed. This apparent contradiction will be explored in future studies.
Conclusions
Taken together, the results presented here demonstrate that inhibition of TCS signaling by targeting the HK CA domain is a viable strategy to decrease bacterial virulence. Moreover, we have identified a novel class of compounds that decrease MRSA pathogenicity. Further studies will be pursued to evaluate these molecules in other Gram-positive bacteria. Current antibiotics promote cell death and, consequently, pressure pathogens to develop resistance mechanisms. Given that our compounds do not kill bacteria but are instead anti-virulence agents, we anticipate that this evolutionary pressure may be considerably decreased. We envision using these anti-virulence compounds as adjuvants with currently utilized antibiotics or as a stand-alone therapy, augmenting the immune system response to fight bacterial infectivity.Data availability
The datasets supporting this article have been uploaded as part of the ESI.†Author contributions
A. E., M. G., and J. Y. conceived, planned, and carried out the experiments. O. V. carried out the experiments. Y. J. and E. E. C. supervised the project. All authors discussed the results and contributed to the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank GlaxoSmithKline for providing the Published Kinase Inhibitor Set (PKIS) for the initial screen. We thank Prof. Taeok Bae for providing AirS plasmid. We thank Prof. Bhagi-Damodaran, Dr Sanyal and L. Rander for use of TECAN instrumentation. We thank J. Smith for help on synthesis of the indoline derivatives. This work was supported by the University of Minnesota, the College of Veterinary Medicine Research Office UMN Ag Experiment Station General Ag Research Funds (MIN-63-075 to Y. J.), the UMN NIH Biotechnology Training Grant (T32GM008347 to A. E.), NIH DP2OD008592, and R01GM134538 (E. E. C.). The authors acknowledge the Minnesota Supercomputing Institute (MSI) at the University of Minnesota.References
- S. Y. C. Tong, J. S. Davis, E. Eichenberger, T. L. Holland and V. G. Fowler, Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management, Clin. Microbiol. Rev., 2015, 28(3), 603–661 CrossRef CAS PubMed.
- T. A. Taylor and C. G. Unakal, National Library of Medicine: Staphylococcus aureus entry, https://www.ncbi.nlm.nih.gov/books/NBK441868/.
- L. F. McCaig, L. C. McDonald, S. Mandal and D. B. Jernigan, Staphylococcus aureus-associated skin and soft tissue infections in ambulatory care, Emerging Infect. Dis., 2006, 12(11), 1715–1723 CrossRef PubMed.
- A. Hassoun, P. K. Linden and B. Friedman, Incidence, prevalence, and management of MRSA bacteremia across patient populations-a review of recent developments in MRSA management and treatment, Crit. care, 2017, 21(1), 211 CrossRef PubMed.
- E. J. Choo, Community-Associated Methicillin-Resistant Staphylococcus aureus in Nosocomial Infections, Infect. Chemother., 2017, 49(2), 158–159 CrossRef PubMed.
- E. F. Kong, J. K. Johnson and M. A. Jabra-Rizk, Community-Associated Methicillin-Resistant Staphylococcus aureus: An Enemy amidst Us, PLoS Pathog., 2016, 12(10), e1005837 CrossRef PubMed.
- H. K. Allen, Alternative to Antibiotics: Why and How, NAM Perspectives, Washington, DC, 2017 Search PubMed.
- C. Ghosh, P. Sarkar, R. Issa and J. Haldar, Alternatives to Conventional Antibiotics in the Era of Antimicrobial Resistance, Trends Microbiol., 2019, 27(4), 323–338 CrossRef CAS PubMed.
- O. Fleitas Martínez, M. H. Cardoso, S. M. Ribeiro and O. L. Franco, Recent Advances in Anti-virulence Therapeutic Strategies With a Focus on Dismantling Bacterial Membrane Microdomains, Toxin Neutralization, Quorum-Sensing Interference and Biofilm Inhibition, Front. Cell. Infect. Microbiol., 2019, 9, 74 CrossRef PubMed.
- P. F. Vale, L. McNally, A. Doeschl-Wilson, K. C. King, R. Popat, M. R. Domingo-Sananes, J. E. Allen, M. P. Soares and R. Kümmerli, Beyond killing: Can we find new ways to manage infection?, Evol. Med. Public Health, 2016, 2016(1), 148–157 CrossRef PubMed.
- S. V. Lynch and J. P. Wiener-Kronish, Novel strategies to combat bacterial virulence, Curr. Opin. Crit. Care, 2008, 14(5), 593–599 CrossRef PubMed.
- J. F. Barrett and J. A. Hoch, Two-component signal transduction as a target for microbial anti-infective therapy, Antimicrob. Agents Chemother., 1998, 42(7), 1529–1536 CrossRef CAS PubMed.
- S. Tiwari, S. B. Jamal, S. S. Hassan, P. V. S. D. Carvalho, S. Almeida, D. Barh, P. Ghosh, A. Silva, T. L. P. Castro and V. Azevedo, Two-Component Signal Transduction Systems of Pathogenic Bacteria As Targets for Antimicrobial Therapy: An Overview, Front. Microbiol., 2017, 8, 1878 CrossRef PubMed.
- A. Delauné, S. Dubrac, C. Blanchet, O. Poupel, U. Mäder, A. Hiron, A. Leduc, C. Fitting, P. Nicolas, J.-M. Cavaillon, M. Adib-Conquy and T. Msadek, The WalKR System Controls Major Staphylococcal Virulence Genes and Is Involved in Triggering the Host Inflammatory Response, Infect. Immun., 2012, 80(10), 3438–3453 CrossRef PubMed.
- S. Wang, Bacterial Two-Component Systems: Structures and Signaling Mechanisms, in Protein Phosphorylation in Human Health, ed. C. Huang, IntechOPen, 2012 Search PubMed.
- F. Jacob-Dubuisson, A. Mechaly, J.-M. Betton and R. Antoine, Structural insights into the signalling mechanisms of two-component systems, Nat. Rev. Microbiol., 2018, 16(10), 585–593 CrossRef CAS PubMed.
- J. M. Skerker, M. S. Prasol, B. S. Perchuk, E. G. Biondi and M. T. Laub, Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis, PLoS Biol., 2005, 3(10), e334 CrossRef PubMed.
- G. E. Schaller, S.-H. Shiu and J. P. Armitage, Two-Component Systems and Their Co-Option for Eukaryotic Signal Transduction, Curr. Biol., 2011, 21(9), R320–R330 CrossRef CAS PubMed.
- T. L. Kane, K. E. Carothers and S. W. Lee, Virulence Factor Targeting of the Bacterial Pathogen Staphylococcus aureus for Vaccine and Therapeutics, Curr. Drug Targets, 2018, 19(2), 111–127 CrossRef CAS PubMed.
- S. Wu, K. Lin, Y. Liu, H. Zhang and L. Lei, Two-component signaling pathways modulate drug resistance of Staphylococcus aureus (Review), Biomed. Rep., 2020, 13(2), 5 CAS.
- K. E. Wilke, S. Francis and E. E. Carlson, Inactivation of Multiple Bacterial Histidine Kinases by Targeting the ATP-Binding Domain, ACS Chem. Biol., 2015, 10(1), 328–335 CrossRef CAS PubMed.
- M. Goswami, K. E. Wilke and E. E. Carlson, Rational Design of Selective Adenine-Based Scaffolds for Inactivation of Bacterial Histidine Kinases, J. Med. Chem., 2017, 60(19), 8170–8182 CrossRef CAS PubMed.
- M. Goswami, A. Espinasse and E. E. Carlson, Disarming the virulence arsenal of Pseudomonas aeruginosa by blocking two-component system signaling, Chem. Sci., 2018, 9(37), 7332–7337 RSC.
- M. P. Coghlan, A. A. Culbert, D. A. E. Cross, S. L. Corcoran, J. W. Yates, N. J. Pearce, O. L. Rausch, G. J. Murphy, P. S. Carter, L. Roxbee Cox, D. Mills, M. J. Brown, D. Haigh, R. W. Ward, D. G. Smith, K. J. Murray, A. D. Reith and J. C. Holder, Selective small molecule inhibitors of glycogen synthase kinase-3 modulate glycogen metabolism and gene transcription, Chem. Biol., 2000, 7(10), 793–803 CrossRef CAS PubMed.
- M. P. Coghlan, A. E. Fenwick, D. Haigh, J. C. Holder, R. J. Ife, A. D. Reith, D. G. Smith and R. W. Ward, Pyrrole-2,5-diones as GSK-3 inhibitors, WO-0021927-A3, 2000.
- D. G. Smith, M. Buffet, A. E. Fenwick, D. Haigh, R. J. Ife, M. Saunders, B. P. Slingsby, R. Stacey and R. W. Ward, 3-Anilino-4-arylmaleimides: potent and selective inhibitors of glycogen synthase kinase-3 (GSK-3), Bioorg. Med. Chem. Lett., 2001, 11(5), 635–639 CrossRef CAS PubMed.
- D. H. Drewry, T. M. Willson and W. J. Zuercher, Seeding collaborations to advance kinase science with the GSK Published Kinase Inhibitor Set (PKIS), Curr. Top. Med. Chem., 2014, 14(3), 340–342 CrossRef CAS PubMed.
- Z. Yang, H. Liu, B. Pan, F. He and Z. Pan, Design and synthesis of (aza)indolyl maleimide-based covalent inhibitors of glycogen synthase kinase 3β, Org. Biomol. Chem., 2018, 16(22), 4127–4140 RSC.
- P. Zhou, J. Zou, F. Tian and Z. Shang, Fluorine Bonding — How Does It Work In Protein–Ligand Interactions?, J. Chem. Inf. Model., 2009, 49(10), 2344–2355 CrossRef CAS PubMed.
- S. Purser, P. R. Moore, S. Swallow and V. Gouverneur, Fluorine in medicinal chemistry, Chem. Soc. Rev., 2008, 37(2), 320–330 RSC.
- K. E. Wilke, S. Francis and E. E. Carlson, Activity-Based Probe for Histidine Kinase Signaling, J. Am. Chem. Soc., 2012, 134(22), 9150–9153 CrossRef CAS PubMed.
- M. Yan, J. W. Hall, J. Yang and Y. Ji, The essential yhcSR two-component signal transduction system directly regulates the lac and opuCABCD operons of Staphylococcus aureus, PLoS One, 2012, 7(11), e50608 CrossRef CAS PubMed.
- J. Sun, L. Zheng, C. Landwehr, J. Yang and Y. Ji, Identification of a Novel Essential Two-Component Signal Transduction System, YhcSR, in Staphylococcus aureus, J. Bacteriol., 2005, 187(22), 7876–7880 CrossRef CAS PubMed.
- F. Sun, Q. Ji, M. B. Jones, X. Deng, H. Liang, B. Frank, J. Telser, S. N. Peterson, T. Bae and C. He, AirSR, a [2Fe-2S] Cluster-Containing Two-Component System, Mediates Global Oxygen Sensing and Redox Signaling in Staphylococcus aureus, J. Am. Chem. Soc., 2012, 134(1), 305–314 CrossRef CAS PubMed.
- M. Yan, C. Yu, J. Yang and Y. Ji, The Essential Two-Component System YhcSR Is Involved in Regulation of the Nitrate Respiratory Pathway of Staphylococcus aureus, J. Bacteriol., 2011, 193(8), 1799–1805 CrossRef CAS PubMed.
- J. W. Hall, J. Yang, H. Guo and Y. Ji, The AirSR two-component system contributes to Staphylococcus aureus survival in human blood and transcriptionally regulates sspABC operon, Front. Microbiol., 2015, 6, 682 Search PubMed.
- S. L. Dixon, J. Duan, E. Smith, C. D. Von Bargen, W. Sherman and M. P. Repasky, AutoQSAR: an automated machine learning tool for best-practice quantitative structure–activity relationship modeling, Future Med. Chem., 2016, 8(15), 1825–1839 CrossRef CAS PubMed.
- Academy S, Machine Learning for Materials Science, 2020, https://www.schrodinger.com/system/files/ml_materialsscience_2020-3.pdf, accessed August, 10th 2020.
- C. Tang, Q. Li and T. Lin, Lycopene attenuates Staphylococcus aureus-induced inflammation via inhibiting α-hemolysin expression, Microbes Infect., 2021, 23(9–10), 104853 CrossRef CAS PubMed.
- T. Lei, Y. Zhang, J. Yang, K. Silverstein and Y. Ji, Complete Genome Sequence of Hospital-Acquired Methicillin-Resistant Staphylococcus aureus Strain WCUH29, Microbiol. Resour. Announce., 2019, 8(23), 005511 Search PubMed.
- C. P. Montgomery, S. Boyle-Vavra and R. S. Daum, Importance of the Global Regulators Agr and SaeRS in the Pathogenesis of CA-MRSA USA300 Infection, PLoS One, 2010, 5(12), e15177 CrossRef CAS PubMed.
- H. Guo, J. W. Hall, J. Yang and Y. Ji, The SaeRS Two-Component System Controls Survival of Staphylococcus aureus in Human Blood through Regulation of Coagulase, Front. Cell. Infect. Microbiol., 2017, 7, 204 CrossRef PubMed.
- Y. Liu, W. Gao, J. Yang, H. Guo, J. Zhang and Y. Ji, Contribution of Coagulase and Its Regulator SaeRS to Lethality of CA-MRSA 923 Bacteremia, Pathogens, 2021, 10(11), 1396 CrossRef CAS PubMed.
- X. Liang and Y. Ji, Involvement of α5β1-integrin and TNF-α in Staphylococcus aureus α-toxin-induced death of epithelial cells, Cell. Microbiol., 2007, 9(7), 1809–1821 CrossRef CAS PubMed.
- N. Ji, J. Yang and Y. Ji, Determining Impact of Growth Phases on Capacity of Staphylococcus aureus to Adhere to and Invade Host Cells, Methods Mol. Biol., 2020, 2069, 187–195 CrossRef CAS PubMed.
- N. Velikova, S. Fulle, A. S. Manso, M. Mechkarska, P. Finn, J. M. Conlon, M. R. Oggioni, J. M. Wells and A. Marina, Putative histidine kinase inhibitors with antibacterial effect against multi-drug resistant clinical isolates identified by in vitro and in silico screens, Sci. Rep., 2016, 6, 26085 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Full experimental details including molecule synthesis and characterization, biochemical and computational methods, protein production and purification, aggregation analysis, metabolism data. See DOI: https://doi.org/10.1039/d2sc05369a |
| ‡ Adeline Espinasse and Manibarsha Goswami are the co-first authors. |
| This journal is © The Royal Society of Chemistry 2023 |