Open Access Article
Open Access ArticleDevelopment of synthetic, self-adjuvanting, and self-assembling anticancer vaccines based on a minimal saponin adjuvant and the tumor-associated MUC1 antigen†
Carlo
Pifferi‡
 a,
Leire
Aguinagalde
a,
Leire
Aguinagalde
 a,
Ane
Ruiz-de-Angulo
a,
Nagore
Sacristán
a,
Priscila Tonon
Baschirotto
a,
Ana
Poveda
a,
Ane
Ruiz-de-Angulo
a,
Nagore
Sacristán
a,
Priscila Tonon
Baschirotto
a,
Ana
Poveda
 b,
Jesús
Jiménez-Barbero
b,
Jesús
Jiménez-Barbero
 bcde,
Juan
Anguita
bcde,
Juan
Anguita
 *cf and
Alberto
Fernández-Tejada
*cf and
Alberto
Fernández-Tejada
 *ac
*ac
aChemical Immunology Laboratory, Center for Cooperative Research in Biosciences (CIC BioGUNE), Basque Research and Technology Alliance (BRTA), Biscay Technology Park, Building 801A, 48160 Derio, Spain. E-mail: afernandeztejada@cicbiogune.es
bChemical Glycobiology Laboratory, CIC BioGUNE, BRTA, Spain
cIkerbasque, Basque Foundation for Science, Maria Diaz de Haro 13, 48009 Bilbao, Spain
dDepartment of Organic Chemistry II, Faculty of Science & Technology, University of the Basque Country, 48940 Leioa, Spain
eCentro de Investigación Biomédica En Red de Enfermedades Respiratorias, Av. Monforte de Lemos, 3-5, 28029 Madrid, Spain
fInflammation and Macrophage Plasticity Laboratory, CIC BioGUNE, BRTA, Spain. E-mail: janguita@cicbiogune.es
First published on 1st March 2023
Abstract
The overexpression of aberrantly glycosylated tumor-associated mucin-1 (TA-MUC1) in human cancers makes it a major target for the development of anticancer vaccines derived from synthetic MUC1-(glyco)peptide antigens. However, glycopeptide-based subunit vaccines are weakly immunogenic, requiring adjuvants and/or additional immunopotentiating approaches to generate optimal immune responses. Among these strategies, unimolecular self-adjuvanting vaccine constructs that do not need coadministration of adjuvants or conjugation to carrier proteins emerge as a promising but still underexploited approach. Herein, we report the design, synthesis, immune-evaluation in mice, and NMR studies of new, self-adjuvanting and self-assembling vaccines based on our QS-21-derived minimal adjuvant platform covalently linked to TA-MUC1-(glyco)peptide antigens and a peptide helper T-cell epitope. We have developed a modular, chemoselective strategy that harnesses two distal attachment points on the saponin adjuvant to conjugate the respective components in unprotected form and high yields via orthogonal ligations. In mice, only tri-component candidates but not unconjugated or di-component combinations induced significant TA-MUC1-specific IgG antibodies able to recognize the TA-MUC1 on cancer cells. NMR studies revealed the formation of self-assembled aggregates, in which the more hydrophilic TA-MUC1 moiety gets exposed to the solvent, favoring B-cell recognition. While dilution of the di-component saponin–(Tn)MUC1 constructs resulted in partial aggregate disruption, this was not observed for the more stably-organized tri-component candidates. This higher structural stability in solution correlates with their increased immunogenicity and suggests a longer half-life of the construct in physiological media, which together with the enhanced antigen multivalent presentation enabled by the particulate self-assembly, points to this self-adjuvanting tri-component vaccine as a promising synthetic candidate for further development.
Introduction
Vaccination represents one of the key achievements in the history of medicine, making it possible to reduce the burden of life-threatening viral and bacterial diseases, and having saved millions of lives to date.1–3 More recently, in the context of cancer immunotherapy, impressive advances have been made over the last decades, enabling the development of a variety of strategies devoted to the selective targeting and clearance of malignant cells.4,5 Among these approaches, anticancer vaccines are a type of antigen-specific active immunotherapy6 aimed at triggering the patient's immune system to mount a tumor-selective, adaptive response that can lead to eradication of the tumor with minimal impact on neighboring healthy cells.7 The vast majority of cancer vaccines reported to date have incorporated tumor-associated antigens (TAAs) in their formulations.8 TAAs include differentiation antigens expressed only on tumor cells and the tissue of origin (e.g. prostate-specific antigen “PSA”),9 overexpressed antigens found on normal cells only at low levels (e.g. breast and ovarian “HER2/neu”),10 and cancer testis antigens, which are aberrantly expressed in a wide variety of cancer types restricted to reproductive tissues (e.g. melanoma antigen gene “MAGE”).11 Because of their shared expression profiles across many tumors and their ability to elicit cancer-specific cellular and humoral immunity, TAAs represent attractive targets in cancer immunotherapy. In particular, the well-known tumor-associated mucin-1 (TA-MUC1) oncoprotein (and its glycoforms),12–16 regarded as a high-ranked antigen by the National Cancer Institute pilot project,17 is being included in a growing number of experimental vaccines.18–20 Thus, established antigens such as the TA-MUC1 glycoprotein represent a gold standard to design improved strategies for the development of potent and safe anticancer vaccines that can lead to long-lasting, adaptive immune responses. A widely used approach to generate subunit vaccines based on homogeneous antigenic fragments exploits the use of immunogenic carrier proteins to enhance the immunogenicity of the covalently attached hapten as well as to activate helper T lymphocytes.21–23 On the other hand, fully-synthetic vaccines rely on modular chemical approaches whereby subunit components can be obtained separately and then subsequently assembled.24–26 These subunit vaccines are molecularly-defined minimal constructs, devoid of unnecessary elements that could negatively influence the immunological outcome.27,28 They are tractable and monodisperse entities compared to protein conjugates, enabling a more reliable characterization via benchmark laboratory techniques (e.g. mass spectrometry, liquid chromatography, nuclear magnetic resonance), while drastically minimizing batch-to-batch variations. Leveraging on synthetic building blocks and orthogonal ligation chemistries, unimolecular multicomponent anticancer vaccines can benefit from efficient “plug-and-play” design approaches that facilitate in vivo evaluation of various B-cell antigens,29,30 T-cell epitopes,31–33 adjuvants,34–36 linkers/spacers,37,38 and scaffolds.39,40Over the last few years, growing attention has been paid to the role of vaccine adjuvants, not only as enhancers but also as “directors” of the immune response.41 In addition to increasing the immunogenicity of the antigen, adjuvants can indeed shape the fate of the immune response either by unspecific means (e.g. emulsions, controlled release, nanomedicine),42–44 or via receptor-mediated mechanisms involving the innate immune system.45–47 While the large majority of vaccines are simply coformulated with adjuvants, self-adjuvanting vaccines are state-of-the-art constructs in which the antigen and the adjuvant modules are covalently attached within the same molecule, thus enabling their simultaneous uptake by the same antigen-presenting cell (e.g. B lymphocytes, dendritic cells),48–51 ultimately leading to enhanced antigen-directed immune responses.52 Pioneering examples of such fully-synthetic self-adjuvanting vaccines featuring the MUC1 tumor-associated antigen include those reported by Boons and co-workers based on the TLR1/TLR2 agonist Pam3CysSK4,53–55 a design that inspired other research groups to develop further MUC1-based synthetic vaccine candidates with mixed outcomes.56–59
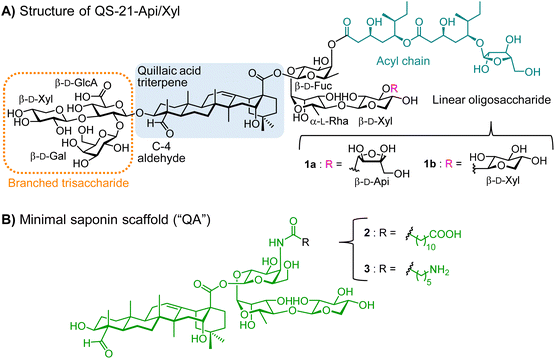
In the framework of our research program on adjuvant development, we have applied a versatile semisynthetic strategy to develop a streamlined saponin platform inspired by the potent QS-21 natural product adjuvant (Fig. 1A). This includes one of our saponin lead compounds (2)60 and its free amine containing analogue 3 (“QA”),61,62 which is amenable to late-stage chemoselective functionalization (Fig. 1B).60–66 QS-21 is a purified saponin fraction extracted from the bark of the Quillaja saponaria (QS) tree67 that consists of a ≈ 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
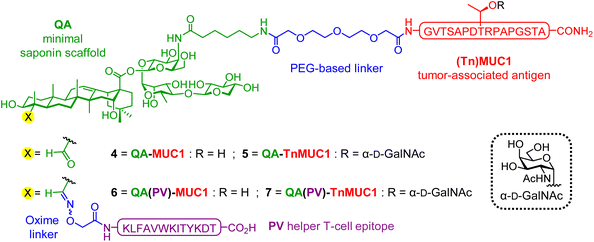
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of isomers sharing the four main structural domains but differing at the terminal apiose (QS-21Api = 1a), or xylose (QS-21Xyl = 1b) residue (Fig. 1A). Despite its potent adjuvant activity and clinical promise,68,69 the scarcity, heterogeneity and dose-limiting toxicity of natural QS-21 prompted us to develop optimized synthetic saponin adjuvants overcoming such constraints.47,60,70,71 In a recent report, we exploited our minimal saponin platform 3, featuring a 6-aminohexanoic acyl chain, to covalently link the Tn carbohydrate antigen as a preliminary glycoconjugate design.72 Herein, we have gone one step further with the development of di- and tri-component, self-adjuvanting vaccine candidates that incorporate both peptide (MUC1) and glycopeptide (TnMUC1) TAAs (Fig. 2), investigating the immunological and structural properties of the synthetic conjugates. The target constructs (4–7) were assembled using an expedient and efficient synthetic strategy involving one (for 4 and 5) or two (for 6 and 7) late-stage conjugation steps for final coupling of advanced, unprotected saponin and peptide building blocks. The immunological evaluation in mice showed that administration of tri-component vaccines 6 and 7 alone induced significantly higher antibody levels than their respective di-component constructs and/or combinations of their non-conjugated admixed components, eliciting antibodies that recognized the native TA-MUC1 antigen on the cancer cell surface. NMR studies provided early insights into the structural features of the conjugates in solution, showing the presence of self-assembled aggregates with a solvent-exposed multivalent (Tn)MUC1 display, and different concentration-dependent aggregation behaviors that correlated with their in vivo immunogenicity.
1 mixture of isomers sharing the four main structural domains but differing at the terminal apiose (QS-21Api = 1a), or xylose (QS-21Xyl = 1b) residue (Fig. 1A). Despite its potent adjuvant activity and clinical promise,68,69 the scarcity, heterogeneity and dose-limiting toxicity of natural QS-21 prompted us to develop optimized synthetic saponin adjuvants overcoming such constraints.47,60,70,71 In a recent report, we exploited our minimal saponin platform 3, featuring a 6-aminohexanoic acyl chain, to covalently link the Tn carbohydrate antigen as a preliminary glycoconjugate design.72 Herein, we have gone one step further with the development of di- and tri-component, self-adjuvanting vaccine candidates that incorporate both peptide (MUC1) and glycopeptide (TnMUC1) TAAs (Fig. 2), investigating the immunological and structural properties of the synthetic conjugates. The target constructs (4–7) were assembled using an expedient and efficient synthetic strategy involving one (for 4 and 5) or two (for 6 and 7) late-stage conjugation steps for final coupling of advanced, unprotected saponin and peptide building blocks. The immunological evaluation in mice showed that administration of tri-component vaccines 6 and 7 alone induced significantly higher antibody levels than their respective di-component constructs and/or combinations of their non-conjugated admixed components, eliciting antibodies that recognized the native TA-MUC1 antigen on the cancer cell surface. NMR studies provided early insights into the structural features of the conjugates in solution, showing the presence of self-assembled aggregates with a solvent-exposed multivalent (Tn)MUC1 display, and different concentration-dependent aggregation behaviors that correlated with their in vivo immunogenicity.
Results and discussion
Design and synthesis
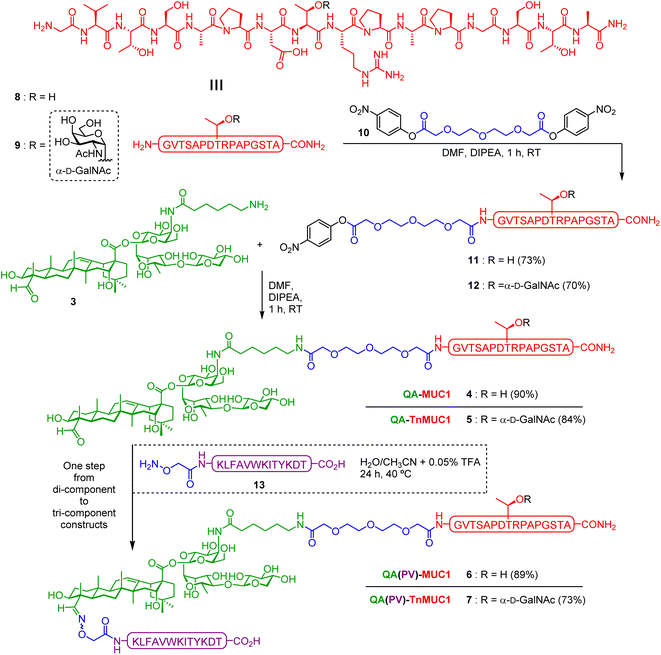
In our previous di-component design, covalent linkage of the simplest tumor-associated Tn carbohydrate antigen (GalNAc-α-O-Thr) to our minimal saponin scaffolds provided saponin–Tn conjugates that induced modest Tn-specific antibody responses in mice.72 With the aim of enhancing immunogenicity, we reasoned that presentation of the Tn antigen in a context more closely resembling the TA-MUC1 glycoprotein (i.e. as a TnMUC1 glycopeptide antigen) would result in a better mucin mimic and therefore in a more immunogenic construct. Further, as a well-established cancer biomarker,73,74 the underlying MUC1 peptide would also provide additional antigenic epitopes. In principle, this could lead to higher levels of antigen-specific antibodies that could recognize better the native MUC1 antigen expressed on cancer cells. Thus, we designed two di-component saponin (“QA”)–MUC1 conjugates incorporating part of the MUC1 tandem repeat sequence, both non-glycosylated (QA–MUC1, 4) and displaying the Tn antigen on its most immunogenic and protective SAPDTRPAP epitope (QA–TnMUC1, 5) (Fig. 2).30,75Moreover, with a view to achieving efficient isotype switching to IgG antibodies via helper T-cell (Th) activation, we also designed the corresponding tri-component conjugates including the well-established mouse MHC class II-restricted Th epitope (PV103–115, KLFAVWKITYKDT) derived from poliovirus (PV).76 Processing and presentation of this CD4+ peptide fragment in complex with MHC-II molecules on the antigen presenting cell surface should lead to Th cell priming, which in turn activates B-cells, resulting in class switch from IgM to high-affinity IgG antibody production. Thus, inclusion of this additional immunostimulatory element should increase the immunogenicity of our constructs, leading to stronger T-cell dependent immune responses. With the above design criteria in mind, we developed a streamlined synthetic strategy to access the multicomponent, self-adjuvanting constructs building upon our optimized, quillaic acid-based saponin adjuvant platform (“QA”).61,72 Di-component conjugates 4 [QA–MUC1] and 5 [QA–TnMUC1], as well as tri-component candidates 6 [QA(PV)–MUC1] and 7 [QA(PV)–TnMUC1] were obtained via a convergent synthetic approach that exploits the unique versatility of our streamlined saponin scaffold featuring two distal conjugation points (Scheme 1). For the attachment of the corresponding B-cell antigens [i.e. the MUC1 peptide 8 (Scheme S1, ESI†) and TnMUC1 glycopeptide 9 (Scheme S2, ESI†)], we chose a diethylene glycol-based spacer as a flexible linker to minimize the potential steric congestion around the saponin domain and to favor antigen accessibility and presentation. Thus, both antigenic fragments were first reacted with an excess of homobifunctional, p-nitrophenyl-activated spacer 10 (Scheme S3, ESI†) to afford the respective intermediates 11 and 12 (Scheme 1) in ca. 70% yield and with excellent hydrolytic stability after RP-HPLC purification and lyophilization.
At this stage, key saponin scaffold 3 (“QA”)61 was reacted through its acyl chain primary amine with 11 and 12 to generate QA–MUC1 (4) and QA–TnMUC1 (5) constructs, respectively (Scheme 1). In addition to these conjugates, we synthesized the corresponding tri-component candidates by incorporating the PV103–115 Th epitope into the saponin scaffold via chemoselective oxime ligation at the triterpene C4-aldehyde. Thus, starting from di-component constructs 4 and 5, facile oxime condensation with an aminooxy-bearing PV peptide 13 generated tri-component candidates 6 and 7 in very good yields after HPLC purification and lyophilization (89% and 73%, respectively).
For the functionalization of the PV epitope, resin-bound side-chain protected peptide S7 (synthesized by SPPS) was reacted at its N-terminus with a slight excess of (Boc-aminooxy)acetic acid NHS ester (Scheme S4, ESI†). Acidic cleavage from the resin in the presence of scavengers afforded ready-to-use, free-aminooxy peptide 13 upon HPLC purification and lyophilization.
Thus, by applying our versatile and modular synthetic approach involving facile sequential conjugation of the spacer-containing B-cell epitopes 11/12 and the aminooxy Th peptide 13 to the saponin scaffold 3, we were able to efficiently prepare the complex tri-component constructs 6 [QA(PV)–MUC1] and 7 [QA(PV)–TnMUC1] over two steps in 80% and 61% overall yield, respectively (Scheme 1).
Immunological evaluation
With the successful synthesis of this saponin-based adjuvant–antigen platform incorporating MUC1 and TnMUC1 (glyco)peptides, we next assessed the self-adjuvanting properties of the di- and tri-component conjugates to induce robust immune responses in vivo. In our initial immunological evaluation in mice, we first investigated the effect of conjugation versus co-administration of the discrete vaccine components using the synthetic constructs bearing the MUC1 peptide antigen. Groups of five mice (C57BL/6) were administered with equimolar amounts of the relevant MUC1 compounds (based on a 50 μg dose of MUC1 peptide 8 as standard for each administration) in three bi-weekly subcutaneous injections (Fig. 3A). As controls, mice from Group A received MUC1 peptide 8 alone in PBS, whereas Group B mice were injected an admixture of the same MUC1 peptide with the PV103–115 peptide (compound S8, Scheme S5, ESI†). As test groups, Group C was immunized with di-component conjugate 4 [QA–MUC1] co-administered with PV, while Group D was vaccinated with the tricomponent MUC1 construct 6 [QA(PV)–MUC1], probing the importance of covalent attachment of the Th peptide in the resulting immune response, as assessed in terms of isotype-switched, anti-MUC1 IgG antibody production. It is worth emphasizing the absence of any particular formulation of the constructs (e.g. emulsification or liposomal preparation) before their administration into mice. Immunization doses were readily prepared from stock solutions of the compounds at ≈2 mg mL−1 in sterile PBS (pH 7.4, 10 mM) and diluted to the required volume with the same buffer.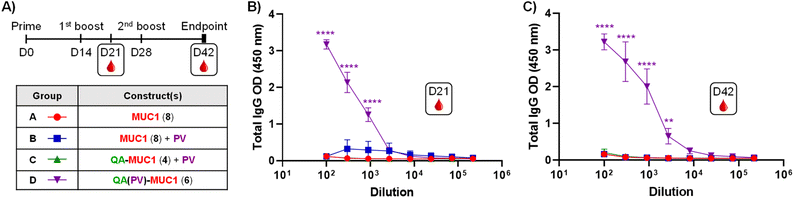 | ||
| Fig. 3 (A) Immunization schedule: three bi-weekly injections at day (D)0, D14, and D28. Sera bleeding performed on D21, end-point sera collection on D42. Immunization groups of five C57BL/6 mice (A–D) were administered different constructs as shown in the inset table. (B) Anti-MUC1 antibody levels from middle-point (D21), and (C) end-point (D42) sera. ELISA dilution curves were plotted including post-immunization sera from each individual mouse and represented as mean values of five mice; the respective error bars indicate variability between those mice for each group. ELISA plates were coated with a BSA–MUC1 conjugate (Scheme S6, ESI†). Statistical significance across the different antibody response curves (OD) at the various dilutions was assessed by comparing to the group administered the MUC1 construct (8) (Group A) using two-way ANOVA analysis with Dunnett's multiple comparisons test. *p ≤ 0.05; ***p ≤ 0.001; ****p ≤ 0.0001. | ||
Mice were bled one week before (day 21) and two weeks after (day 42) the third immunization (Fig. 3A), and the sera was analyzed for anti-MUC1 antibodies by ELISA using a BSA–MUC1 conjugate (Scheme S6, ESI†) (0.05 μg per well) for coating. Serial serum dilutions plotted against IgG optical density (OD) values at 450 nm clearly showed dilution-dependent curves for all mice immunized with tricomponent candidate 6 [QA(PV)–MUC1, Group D] (Fig. 3B and C), indicating considerable induction of anti-MUC1 IgG antibodies by these mice. Notably, good levels of IgG antibodies were already reached just one week after the second immunization (day 21) (Fig. 3B), which were similar at the 1/100 dilution to those observed with the endpoint sera (day 42) (Fig. 3C), thus suggesting the prospect of shortening the immunization schedule and reducing the amount of construct administered with consequent dose sparing (see immunization series below with TnMUC1-based constructs). In contrast, all other groups of mice, including that injected with a combination of di-component conjugate 4 [QA–MUC1] plus PV peptide (Group C), did not show any significant OD signal above the baseline, indicating that the three components (i.e. saponin adjuvant, MUC1 B-cell antigen, and PV Th epitope) must be covalently linked to elicit high IgG antibody levels. In addition, subtyping of the IgG antibodies induced from Group D mice resulted in OD values consistent with those observed for total IgGs with the following trend IgG1 > IgG2b > IgG2c > IgG3, where IgG1 and IgG2b subtypes were clearly predominant almost to the same extent (see Fig. S49, ESI†). ELISA analysis for anti-PV IgG antibodies showed negligible OD signal in all cases, indicating that the IgG antibody response was directed towards the MUC1 antigen as intended, without interference or immune suppression caused by the Th epitope (see Fig. S51, ESI†).
Encouraged by these initial results with the tri-component MUC1 peptide conjugate 6, we next performed in vivo immunological studies with an extended set of compounds based on the relevant TnMUC1 glycopeptide antigen 9 (Scheme 1). This epitope resembles more closely the aberrantly glycosylated, cancer-related MUC1 protein that is characterized by the presence of truncated tumor associated glycans such as the Tn antigen, which has been proposed to be important for antibody induction and to enhance the immunogenicity of the MUC1 antigen,30,54 presumably due to potential conformational effects of the carbohydrate on the underlying peptide.77–79
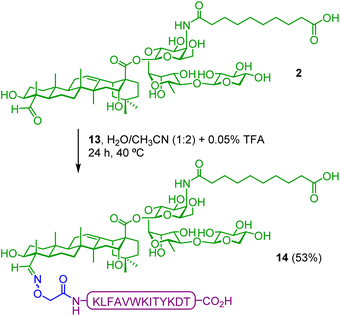
In addition to the QA–TnMUC1 conjugate (5) and the PV-containing construct QA(PV)–TnMUC1 (7), we included another control group to investigate the effect of covalent linkage of the different modules of the vaccine candidate on the antibody response. As key previous studies have consistently demonstrated that a two-component conjugate involving attachment of the PV Th peptide to a TnMUC1 epitope elicits very low IgG antibodies in mice [53, 54], we prepared the saponin–PV conjugate 14 as a control (Scheme 2). Such di-component molecule lacking the B-cell epitope, referred to as QA(PV), was synthesized by linking the PV Th peptide 13 to the C4-aldehyde of adjuvant-active saponin 2 (Scheme 2),60 and was administered in combination with the TnMUC1 antigen 9.
Based on our first study above, in vivo immunological evaluation of the saponin–TnMUC1 constructs 5 and 7 followed a shorter immunization schedule involving two subcutaneous vaccinations (a prime injection on day 0 and a single boost injection on day 14), with endpoint sera collection on day 24, ten days after the boost (Fig. 4A). Groups of five mice were administered with equimolar amounts of the compounds (based on a 50 μg dose of TnMUC1 glycopeptide 9 as standard for each administration) as follows. As controls, Group A received PBS alone whereas two groups were injected mixtures of saponin adjuvant 2 and TnMUC1 B-cell antigen (Groups B and C), the latter coadministered with the PV Th epitope as well. Two groups received the saponin–TnMUC1 conjugate, either alone (Group D) or in combination with PV (Group E), while another control group was injected with the saponin–PV conjugate, QA(PV) 14, coadministered with TnMUC1 antigen 9 (Group F). Finally, Group G was immunized with the tri-component construct QA(PV)–TnMUC1 (7) alone.
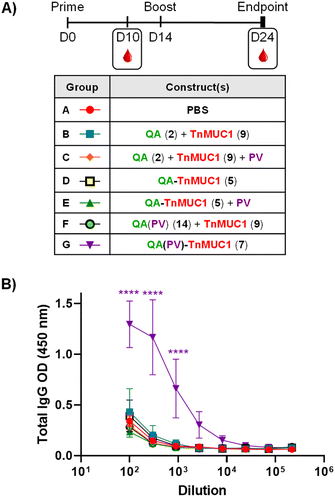 | ||
| Fig. 4 (A) Immunization schedule: two bi-weekly injections at day (D)0 and D14. Sera bleeding performed on D10, endpoint sera collection on D24. Immunization groups (A–G) were administered with different constructs as reported in the table. (B) Anti-MUC1 antibody titers from end-point (D24) sera. ELISA dilution curves were plotted including post-immunization sera from each individual mouse and represented as mean values of five mice; the respective error bars indicate variability between those mice for each group. ELISA plates were coated with a BSA–TnMUC1 conjugate (Scheme S6, ESI†). Statistical significance across the different antibody response curves (OD) at the various dilutions was assessed by comparing to the group administered PBS (Group A) using two-way ANOVA analysis with Dunnett's multiple comparisons test. *p ≤ 0.05; ***p ≤ 0.001; ****p ≤ 0.0001. | ||
As in our previously described assay, we checked the presence of TnMUC1-specific IgG antibodies in mouse sera by ELISA (Fig. 4B) using the corresponding BSA–TnMUC1 conjugate (Scheme S6, ESI†) for coating. Generally, IgG antibody responses were in line with those observed for the MUC1-based synthetic constructs above (Fig. 3B and C). Likewise, a further experiment including an additional boost injection (three in total) following the same MUC1 immunization scheme did not result in higher anti-TnMUC1 antibody levels (data not shown).
Compared to the other groups, mice from Group G immunized with tri-component conjugate 7 generated significantly higher levels of anti-TnMUC1 IgG antibodies (Fig. 4B), highlighting the requirement for covalent attachment of all three vaccine components. Finally, IgG subtyping showed the prevalence of IgG1 and IgG2b subclasses (see Fig. S52, ESI†), as in the case of the MUC1 peptide-based constructs.
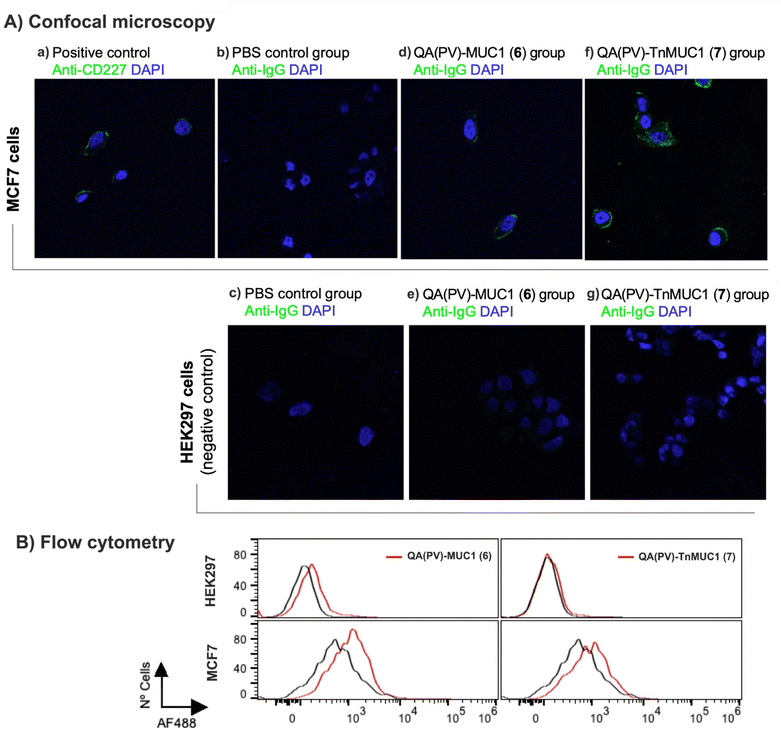
Next, we tested whether the anti-sera from mice immunized with tri-component vaccine constructs QA(PV)–MUC1 (6) and QA(PV)–TnMUC1 (7) were able to recognize the TA-MUC1 antigen in its native context, i.e. displayed on the surface of cancer cells. For this purpose, we performed a fluorescence immunoassay by confocal microscopy (Fig. 5A) using the MCF7 human cancer cell line, which is well-known to express TA-MUC1 on their surface.80 The antisera from mice from both groups (immunized with 6 and 7 respectively) stained MCF7 cells (surrounding green color; Fig. 5A, panels (d) and (f)) but not negative control HEK293 cells lacking TA-MUC1 (Fig. 5A, panels (e) and (g)). Likewise, sera from PBS-administered mice (Fig. 5A, panel (b)) did not provide any signal under the same conditions. As expected, a surface pattern of labeling was observed when the cells were incubated with the CD227 mAb as positive control (green color; Fig. 5A, panel (a)). These confocal microscopy results are in agreement with the results obtained by flow cytometry (see Fig. 5B), showing that immunizations with both tri-component constructs induce antibodies able to selectively recognize the native TA-MUC1 on the cancer cell surface.
NMR studies
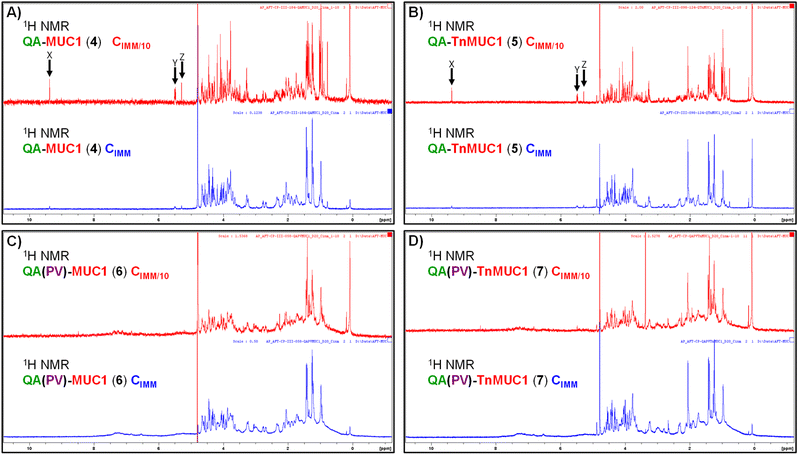
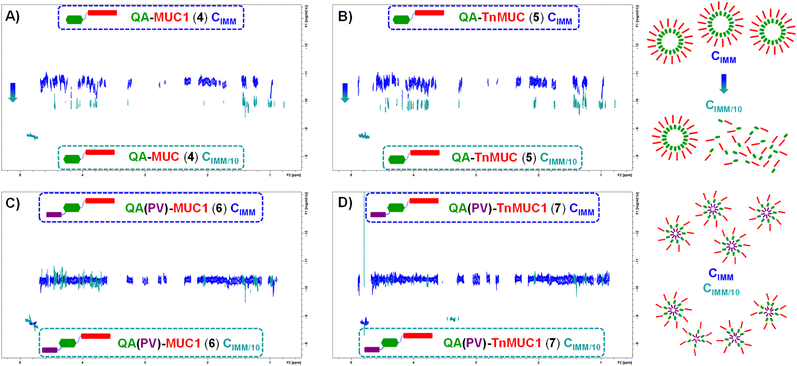
To gain insights into the structural features and molecular behavior of the di- and tri-component constructs 4–7 in aqueous media, especially their ability to form particulate/three-dimensional structures (e.g. aggregates) that could be important for multivalent antigen presentation, we next performed the analysis of the compounds in solution by conducting NMR experiments in aqueous conditions (i.e. PBS/D2O). We first recorded the 1H-NMR spectra of QA–MUC1 (4), QA–TnMUC1 (5), QA(PV)–MUC1 (6), and QA(PV)–TnMUC1 (7) in PBS (pD 7.4)/D2O at the concentration of immunization (CIMM = 0.3 mM) (Fig. 6, blue spectra). The low intensity (Fig. 6A and B, blue spectra) or absence (Fig. 6C and D, blue spectra) of characteristic signals from the saponin fragment at this CIMM suggested aggregation in all cases. Indeed, the 1H-NMR spectrum acquired with ten-fold diluted samples (CIMM/10, red spectra) provided sharper signals for the di-component constructs 4 and 5, as well as the increase/emergence of diverse saponin signals within the conjugate, from which some isolated resonance signals were identified: X (triterpene C4–CHO), Y (rhamnose and 4-amido-4-deoxygalactose anomeric protons) and Z (triterpene H-12) (Fig. 6A and B, red spectra). This strongly suggests the partial disruption of the aggregates with the concomitant appearance of monomeric species. On the other hand, no substantial changes were evident in the 1H-NMR spectra of the tricomponent vaccines 6 and 7 upon 10-fold dilution (CIMM/10) (Fig. 6C and D, red spectra). In addition, the aromatic amino acid signals corresponding to the PV peptide (Trp, Tyr) were always nearly invisible at both concentrations (see 1H NMR spectra of the free PV peptide S8 in Fig. S38, ESI†). Thus, these data point out that the aggregation observed with these conjugates is more likely to be disrupted in di-component constructs QA–MUC1 (4) and QA–TnMUC1 (5) compared to tri-component vaccines QA(PV)–MUC1 (6) and QA(PV)–TnMUC1 (7), which are still aggregated even at lower concentrations. Moreover, whereas the saponin and the PV peptide are the key domains within the conjugates in forming the aggregate core, at the concentration of immunization (CIMM) the (Tn)MUC1 B-cell antigen appears to be more exposed to the solvent, which would facilitate multivalent antigen presentation and interactions with B-cell receptors.Next, diffusion-ordered spectroscopy (DOSY NMR) experiments on the above samples (compounds 4–7, both at CIMM and CIMM/10) were carried out (Fig. 7) to obtain molecular diffusion parameters that provide key information on the hydrodynamic properties of the different species coexisting in solution (see Table S3, ESI†). At CIMM, the di-component constructs 4 and 5 generate large molecular weight aggregates that are disrupted upon ten-fold dilution (CIMM/10), as deduced from the observed marked changes in the estimated diffusion coefficients. This strongly suggests the presence of monodisperse, free monomers of the di-component constructs at this diluted CIMM/10 (Fig. 7A and B). In contrast, ten-fold dilution of the tri-component vaccine samples (6 and 7) did not translate into any significant variation of the measured diffusion coefficients when compared to those estimated for their respective CIMM samples (Fig. 7C and D), indicating that the aggregation degree stays similar. As deduced from the 1H-NMR experimental data, the aggregation state of the tri-component vaccines 6 and 7 is less sensitive to dilution, whereas for the di-component constructs 4 and 5 the ten-fold dilution led to a partially aggregated system with an increased and significant fraction of monomeric conjugate species in solution.
Overall, these NMR studies demonstrate the existence of distinct aggregation properties and states for the di-component constructs relative to the tri-component conjugates (see proposed model in Fig. 7, right side). While all the molecules showed tendency to aggregate, the PV-containing constructs QA(PV)–MUC1 (6) and QA(PV)–TnMUC1 (7) exhibited superior aggregation and a more compact and stable particulate structure, which is not disrupted upon 10-fold dilution.
On the other hand, although the di-component conjugates lacking the Th peptide, QA–MUC1 (4) and QA–TnMUC1 (5), generated larger aggregates than 6 and 7, they were also less stable and could be disrupted upon dilution. Considering the above NMR-based observations, we propose the schematic representation in Fig. 7 (right side) as a putative model for the formation of aggregated/particulate structures for the corresponding constructs (4/5 and 6/7, respectively), whereby the most hydrophilic portion of the conjugates [i.e. the (Tn)MUC1 (glyco)peptide, red rectangle in Fig. 7] encloses the hydrophobic core that contains the more aggregation-prone saponin and PV peptide units. In this way, the solvent accessible area is reduced, with the concomitant stabilization of this three-dimensional structure. In addition to potentially increasing the conjugates half-life and stability against degradation, the more compact molecular organization of the tri-component vaccine candidates could also be connected to their superior immunogenicity compared to the di-component molecules as shown in our mouse in vivo studies.
Conclusions
In conclusion, leveraging our saponin adjuvant platform 3 as a core scaffold, we have successfully developed tri-component synthetic vaccine candidates (6 and 7), via a streamlined two-step chemoselective strategy, providing immunogenic constructs with self-adjuvanting and self-assembling properties. In this work, we focused on conjugates containing the tumor-associated (Tn)MUC1 (glyco)peptides as important cancer antigens and assessed in vivo their ability to elicit antibody responses in mouse immunization studies. The incorporation of a promiscuous Th peptide epitope (PV103–115) in the tri-component constructs 6 and 7 was key for inducing good levels of anti-(Tn)MUC1 antibodies, which were able to recognize the native tumor antigen on the cancer cell surface as assessed in immunofluorescence and flow cytometry assays. In contrast, mice injected with the three components (i.e. saponin adjuvant, (Tn)MUC1 B-cell antigen, and PV Th epitope) either unconjugated (as an admixture) or in partially conjugated combinations failed to elicit anti-(Tn)MUC1 antibody levels as high as those induced by tri-component vaccines QA(PV)–MUC1 (6) and QA(PV)–TnMUC1 (7).It is worth highlighting the versatility and modularity of our synthetic strategy, in which unprotected saponin 3 can be subjected to a first conjugation via its acyl chain terminal amine to provide the respective di-component constructs 4 and 5. In a second, straightforward step, the corresponding tri-component constructs 6 and 7 can be readily accessed in very good yields via facile oxime condensation at the triterpene C4 aldehyde of the parent compounds (4 and 5, respectively). Despite their amphiphilic character, these compounds showed optimal solubility in aqueous media and were easily isolated via standard semi-preparative HPLC procedures. Notably, this modular approach is generalizable and enables the rapid, divergent synthesis of a variety of di-component and, additionally, complex tri-component conjugates incorporating different combinations of antigens, epitopes and further immunoactive moieties. This aspect is particularly appealing in light of recent advances in cancer immunotherapy focusing on tumor-specific antigens (TSAs), which unlike tumor-associated antigens (viz TA-MUC1), are not found in normal cells, but can originate from oncogenic viral proteins, somatic mutations, translocation or aberrant splicing of normal proteins (neoantigens). While individual oncoviral antigens are expressed in specific tumor types (e.g. human papillomavirus E6 and E7 antigens in cervical cancer), most neoantigens are unique to specific tumors of individual patients (referred to as “private neoantigens”) and therefore require customized approaches. Recently, important steps have been made towards the development of personalized anticancer vaccines,81–84 emphasizing the scope of a personalized immunogenomic strategy, combined with state-of-the-art vaccine design.85,86 In such scenario, modular approaches such the one presented herein are attractive for their potential to be implemented in translational pipelines that originate from neoantigen identification.
With a view to rationalize the above immunological results, we investigated the physicochemical properties of constructs 4–7 by performing NMR experiments at the concentrations used for mouse immunization and in ten-fold diluted samples. These structural studies revealed the self-assembly of the molecules into aggregates, forming particulate structures with implications in the immunogenicity and immunostimulatory properties of the constructs. For instance, in terms of enhanced, multivalent antigen presentation as well as improved delivery of the vaccine to the lymph nodes, leading to increased uptake by antigen-presenting cells. In the case of the di-component constructs, the aggregates were partially disrupted upon dilution with the emergence of some free/monomeric species. On the other hand, the aggregated structures were more stable for the tri-component conjugates, with a molecular organization presenting the more hydrophilic B-cell antigen (glyco)peptides on the surface as multivalent clusters, and the less polar PV peptide and the triterpene saponin forming the core. The increased stability of these particles with the surface-exposed clustered display of the (Tn)MUC1 antigen could translate into higher resistance to physiological degradation as well as enhanced trafficking/internalization of the particulate system and improved multivalent antigen presentation to B-cell receptors. Overall, these structural insights provide a correlation and a potential molecular rationale for the superior immunogenicity observed for the tri-component candidates.
Finally, it is remarkable how these synthetic tri-component vaccines, QA(PV)–MUC1 (6) and QA(PV)–TnMUC1 (7), were able to elicit good levels of anti-(Tn)MUC1 antibodies recognizing the naturally occurring antigen on tumor cells without the need for any additional formulation step and only upon 3 and 2 administrations, respectively. In contrast, other self-adjuvanting synthetic vaccine strategies based on TA-MUC1 (glyco)peptide antigens52–59 but incorporating the highly lipophilic Pam3CysSK4 suffer from challenging ligations as well as purification and isolation issues associated to the low solubility of the resulting conjugates. Moreover, immunological evaluations of most of these candidates have been carried out through extended protocols involving 3 to 5 bi-weekly injections in preformed liposomal formulations. Overall, the broadly applicable approach and significant results presented in this study highlight the promise of our synthetic, saponin-based modular platform to gain access to tri-component vaccine constructs with great translational potential.
Data availability
Experimental details for chemical syntheses, HPLC and MS analytical data, 1H-NMR spectra for synthetic intermediates and final constructs; immunological evaluation procedures including mouse immunization protocol, antibody quantification and subtyping, antibody reactivity against human cancer cell line analyzed by confocal microscopy and flow cytometry;† experimental information on NMR structural studies, including 1H-NMR and DOSY experiments and their spectra at immunization concentration and 10-fold dilution. See the ESI for complete details.†Author contributions
Conceptualization, C. P., J. A., A. F. T.; chemical synthesis, C. P.; immunological evaluation, C. P., L. A., A. R. A., N. S., P. T. B., J. A.; NMR studies, A. P, J. J. B.; data analysis, C. P., L. A., A. R. A., N. S., P. T. B., A. P, J. J. B., J. A., A. F. T.; schemes & figures, C. P.; project management, C. P., J. A., A. F. T.; funding acquisition, J. J. B., J. A., A. F. T.; writing – original draft, C. P., A. F. T.; writing – review & editing, C. P., A. P, J. J. B., J. A., A. F. T.Conflicts of interest
C. P., J. A., and A. F. T. are inventors on patents and/or patent applications that include saponin constructs presented in this work.Acknowledgements
Funding from the European Research Council (ERC-2016-STG-716878 to A. F.-T.; ERC-2017-AdG-788143 to J. J. B.) and the Spanish Ministry of Science and Innovation MCIN/AEI (PID2020-117911RB-I00, CTQ2017-87530-R, RYC-2015-17888 to A. F.-T.; RTI2018-096494-B-100 to J. A.; RTI2018-094751-B-C21 to J. J. B) is gratefully acknowledged. We thank Felix Elortza and Ibon Iloro from the CIC bioGUNE Proteomics Platform and Javier Calvo from the CIC biomaGUNE Mass Spectrometry Platform for their support with MALDI and HRMS analyses. A. F. T. thanks Raquel Fernandez for inspiration.Notes and references
- M. R. Hilleman, Vaccine, 2000, 18, 1436–1447 CrossRef CAS.
- A. J. Pollard and E. M. Bijker, Nat. Rev. Immunol., 2021, 21, 83–100 CrossRef CAS PubMed.
- J. Varadé, S. Magadán and Á. González-Fernández, Cell. Mol. Immunol., 2021, 18, 805–828 CrossRef.
- I. Mellman, G. Coukos and G. Dranoff, Nature, 2011, 480, 480–489 CrossRef CAS PubMed.
- L. Galluzzi, E. Vacchelli, J.-M. B.-S. Pedro, A. Buqué, L. Senovilla, E. E. Baracco, N. Bloy, F. Castoldi, J.-P. Abastado, P. Agostinis, R. N. Apte, F. Aranda, M. Ayyoub, P. Beckhove, J.-Y. Blay, L. Bracci, A. Caignard, C. Castelli, F. Cavallo, E. Celis, V. Cerundolo, A. Clayton, M. P. Colombo, L. Coussens, M. V. Dhodapkar, A. M. Eggermont, D. T. Fearon, W. H. Fridman, J. Fučíková, D. I. Gabrilovich, J. Galon, A. Garg, F. Ghiringhelli, G. Giaccone, E. Gilboa, S. Gnjatic, A. Hoos, A. Hosmalin, D. Jäger, P. Kalinski, K. Kärre, O. Kepp, R. Kiessling, J. M. Kirkwood, E. Klein, A. Knuth, C. E. Lewis, R. Liblau, M. T. Lotze, E. Lugli, J.-P. Mach, F. Mattei, D. Mavilio, I. Melero, C. J. Melief, E. A. Mittendorf, L. Moretta, A. Odunsi, H. Okada, A. K. Palucka, M. E. Peter, K. J. Pienta, A. Porgador, G. C. Prendergast, G. A. Rabinovich, N. P. Restifo, N. Rizvi, C. Sautès-Fridman, H. Schreiber, B. Seliger, H. Shiku, B. Silva-Santos, M. J. Smyth, D. E. Speiser, R. Spisek, P. K. Srivastava, J. E. Talmadge, E. Tartour, S. H. Van Der Burg, B. J. Van Den Eynde, R. Vile, H. Wagner, J. S. Weber, T. L. Whiteside, J. D. Wolchok, L. Zitvogel, W. Zou and G. Kroemer, Oncotarget, 2014, 5, 12472–12508 CrossRef PubMed.
- I. Melero, G. Gaudernack, W. Gerritsen, C. Huber, G. Parmiani, S. Scholl, N. Thatcher, J. Wagstaff, C. Zielinski, I. Faulkner and H. Mellstedt, Nat. Rev. Clin. Oncol., 2014, 11, 509–524 CrossRef CAS PubMed.
- M. A. Morse, W. R. Gwin and D. A. Mitchell, Target. Oncol., 2021, 16, 121–152 CrossRef.
- C. J. M. Melief, T. Van Hall, R. Arens, F. Ossendorp and S. H. Van Der Burg, J. Clin. Invest., 2015, 125, 3401–3412 CrossRef.
- H. Lilja, D. Ulmert and A. J. Vickers, Nat. Rev. Cancer, 2008, 8, 268–278 CrossRef CAS PubMed.
- J. Wang and B. Xu, Signal Transduct. Target. Ther., 2019, 4, 34 CrossRef PubMed.
- P. G. Coulie, B. J. Van den Eynde, P. van der Bruggen and T. Boon, Nat. Rev. Cancer, 2014, 14, 135–146 CrossRef CAS PubMed.
- S. Nath and P. Mukherjee, Trends Mol. Med., 2014, 20, 332–342 CrossRef CAS PubMed.
- F. G. Hanisch, T. R. E. Stadie, F. Deutzmann and J. Peter-Katalinic, Eur. J. Biochem., 1996, 236, 318–327 CrossRef CAS PubMed.
- M. A. Tarp, A. L. Sørensen, U. Mandel, H. Paulsen, J. Burchell, J. Taylor-Papadimitriou and H. Clausen, Glycobiology, 2007, 17, 197–209 CrossRef CAS PubMed.
- S. Rangappa, G. Artigas, R. Miyoshi, Y. Yokoi, S. Hayakawa, F. Garcia-Martin, H. Hinou and S.-I. Nishimura, Med. Chem. Commun., 2016, 7, 1102–1122 RSC.
- Y. Singh, M. C. Rodriguez Benavente, M. H. Al-Huniti, D. Beckwith, R. Ayyalasomayajula, E. Patino, W. S. Miranda, A. Wade and M. Cudic, J. Org. Chem., 2020, 85, 1434–1445 CrossRef CAS PubMed.
- M. A. Cheever, J. P. Allison, A. S. Ferris, O. J. Finn, B. M. Hastings, T. T. Hecht, I. Mellman, S. A. Prindiville, J. L. Viner, L. M. Weiner and L. M. Matrisian, Clin. Cancer Res., 2009, 15, 5323–5337 CrossRef.
- D. Feng, A. S. Shaikh and F. Wang, ACS Chem. Biol., 2016, 11, 850–863 CrossRef CAS PubMed.
- D. M. Beckwith and M. Cudic, Semin. Immunol., 2020, 47, 101389 CrossRef CAS PubMed.
- M. Anderluh, F. Berti, A. Bzducha-Wróbel, F. Chiodo, C. Colombo, F. Compostella, K. Durlik, X. Ferhati, R. Holmdahl, D. Jovanovic, W. Kaca, L. Lay, M. Marinovic-Cincovic, M. Marradi, M. Ozil, L. Polito, J. J. Reina, C. A. Reis, R. Sackstein, A. Silipo, U. Švajger, O. Vaněk, F. Yamamoto, B. Richichi and S. J. van Vliet, FEBS J., 2021, 289, 4251–4303 CrossRef.
- F. Micoli, R. Adamo and P. Costantino, Molecules, 2018, 23, 1451 CrossRef PubMed.
- R. Rappuoli, Sci. Transl. Med., 2018, 10, eaat4615 CrossRef PubMed.
- R. Rappuoli, E. De Gregorio and P. Costantino, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 14–16 CrossRef CAS PubMed.
- T. Buskas, P. Thompson and G. J. Boons, Chem. Commun., 2009, 5335–5349 RSC.
- R. M. Wilson and S. J. Danishefsky, J. Am. Chem. Soc., 2013, 135, 14462–14472 CrossRef CAS PubMed.
- R. Mettu, C. Y. Chen and C. Y. Wu, J. Biomed. Sci., 2020, 27, 9 CrossRef CAS PubMed.
- M. P. Schutze, C. Leclerc, M. Jolivet, F. Audibert and L. Chedid, J. Immunol., 1985, 135, 2319–2322 CrossRef CAS.
- A. Jegerlehner, M. Wiesel, K. Dietmeier, F. Zabel, D. Gatto, P. Saudan and M. F. Bachmann, Vaccine, 2010, 28, 5503–5512 CrossRef CAS PubMed.
- N. Martínez-Sáez, J. M. Peregrina and F. Corzana, Chem. Soc. Rev., 2017, 46, 7154–7175 RSC.
- X. Wu, Z. Yin, C. McKay, C. Pett, J. Yu, M. Schorlemer, T. Gohl, S. Sungsuwan, S. Ramadan, C. Baniel, A. Allmon, R. Das, U. Westerlind, M. G. Finn and X. Huang, J. Am. Chem. Soc., 2018, 140, 16596–16609 CrossRef CAS PubMed.
- H. Cai, M. S. Chen, Z. Y. Sun, Y. F. Zhao, H. Kunz and Y. M. Li, Angew. Chem., Int. Ed., 2013, 52, 6106–6110 CrossRef CAS PubMed.
- B. Palitzsch, S. Hartmann, N. Stergiou, M. Glaffig, E. Schmitt and H. Kunz, Angew. Chem., Int. Ed., 2014, 53, 14245–14249 CrossRef CAS PubMed.
- V. Lakshminarayanan, N. T. Supekar, J. Wei, D. B. McCurry, A. C. Dueck, H. E. Kosiorek, P. P. Trivedi, J. M. Bradley, C. S. Madsen, L. B. Pathangey, D. B. Hoelzinger, M. A. Wolfert, G. J. Boons, P. A. Cohen and S. J. Gendler, PLoS One, 2016, 11, 1–21 CrossRef PubMed.
- J. Gao and Z. Guo, Med. Res. Rev., 2018, 38, 556–601 CrossRef CAS PubMed.
- N. R. M. Reintjens, E. Tondini, A. R. De Jong, N. J. Meeuwenoord, F. Chiodo, E. Peterse, H. S. Overkleeft, D. V. Filippov, G. A. Van Der Marel, F. Ossendorp and J. D. C. Codée, J. Med. Chem., 2020, 63, 11691–11706 CrossRef CAS PubMed.
- S. Jin, H. T. Vu, K. Hioki, N. Noda, H. Yoshida, T. Shimane, S. Ishizuka, I. Takashima, Y. Mizuhata, K. Beverly Pe, T. Ogawa, N. Nishimura, D. Packwood, N. Tokitoh, H. Kurata, S. Yamasaki, K. J. Ishii and M. Uesugi, Angew. Chem., Int. Ed., 2021, 60, 961–969 CrossRef CAS PubMed.
- T. Buskas, Y. Li and G.-J. Boons, Chem. - Eur. J., 2004, 10, 3517–3524 CrossRef CAS PubMed.
- Z. Yin, S. Chowdhury, C. McKay, C. Baniel, W. S. Wright, P. Bentley, K. Kaczanowska, J. C. Gildersleeve, M. G. Finn, L. BenMohamed and X. Huang, ACS Chem. Biol., 2015, 10, 2364–2372 CrossRef CAS PubMed.
- F. Peri, Chem. Soc. Rev., 2013, 42, 4543–4556 RSC.
- C. Pifferi, N. Berthet and O. Renaudet, Biomater. Sci., 2017, 5, 953–965 RSC.
- E. Bergmann-Leitner and W. Leitner, Vaccines, 2014, 2, 252–296 CrossRef PubMed.
- C. H. Huang, C. Y. Huang, C. P. Cheng, S. H. Dai, H. W. Chen, C. H. Leng, P. Chong, S. J. Liu and M. H. Huang, Sci. Rep., 2016, 6, 36732 CrossRef CAS PubMed.
- Y. Ishii-Mizuno, Y. Umeki, Y. Onuki, H. Watanabe, Y. Takahashi, Y. Takakura and M. Nishikawa, Int. J. Pharm., 2017, 516, 392–400 CrossRef CAS PubMed.
- D. J. Irvine and E. L. Dane, Nat. Rev. Immunol., 2020, 20, 321–334 CrossRef CAS PubMed.
- R. L. Coffman, A. Sher and R. A. Seder, Immunity, 2010, 33, 492–503 CrossRef CAS PubMed.
- S. G. Reed, M. T. Orr and C. B. Fox, Nat. Med., 2013, 19, 1597–1608 CrossRef CAS PubMed.
- C. Pifferi, R. Fuentes and A. Fernández-Tejada, Nat. Rev. Chem., 2021, 5, 197–216 CrossRef CAS PubMed.
- F. D. Batista and N. E. Harwood, Nat. Rev. Immunol., 2009, 9, 15–27 CrossRef CAS PubMed.
- T. Kambayashi and T. M. Laufer, Nat. Rev. Immunol., 2014, 14, 719–730 CrossRef CAS PubMed.
- C. E. Hughes, R. A. Benson, M. Bedaj and P. Maffia, Front. Immunol., 2016, 7, 481 Search PubMed.
- S. K. Wculek, F. J. Cueto, A. M. Mujal, I. Melero, M. F. Krummel and D. Sancho, Nat. Rev. Immunol., 2020, 20, 7–24 CrossRef CAS PubMed.
- Y. Manabe, T. C. Chang and K. Fukase, Drug Discov. Today Technol., 2020, 37, 61–71 CrossRef PubMed.
- S. Ingale, M. A. Wolfert, J. Gaekwad, T. Buskas and G. J. Boons, Nat. Chem. Biol., 2007, 3, 663–667 CrossRef CAS PubMed.
- V. Lakshminarayanan, P. Thompson, M. A. Wolfert, T. Buskas, J. M. Bradley, L. B. Pathangey, C. S. Madsen, P. A. Cohen, S. J. Gendler and G. J. Boons, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 261–266 CrossRef CAS PubMed.
- P. Thompson, V. Lakshminarayanan, N. T. Supekar, J. M. Bradley, P. A. Cohen, M. A. Wolfert, S. J. Gendler and G. J. Boons, Chem. Commun., 2015, 51, 10214–10217 RSC.
- H. Cai, Z.-Y. Sun, M.-S. Chen, Y.-F. Zhao, H. Kunz and Y.-M. Li, Angew. Chem., Int. Ed., 2014, 53, 1699–1703 CrossRef CAS PubMed.
- B. L. Wilkinson, S. Day, L. R. Malins, V. Apostolopoulos and R. J. Payne, Angew. Chem., Int. Ed., 2011, 50, 1635–1639 CrossRef CAS PubMed.
- W. H. Li and Y. M. Li, Chem. Rev., 2020, 120, 11420–11478 CrossRef CAS PubMed.
- N. Stergiou, M. Urschbach, A. Gabba, E. Schmitt, H. Kunz and P. Besenius, Chem. Rec., 2021, 21, 3313–3331 CrossRef CAS PubMed.
- M. Ghirardello, A. Ruiz-De-Angulo, N. Sacristan, D. Barriales, J. Jiménez-Barbero, A. Poveda, F. Corzana, J. Anguita and A. Fernández-Tejada, Chem. Commun., 2020, 56, 719–722 RSC.
- A. Fernández-Tejada, E. K. Chea, C. George, N. Pillarsetty, J. R. Gardner, P. O. Livingston, G. Ragupathi, J. S. Lewis, D. S. Tan and D. Y. Gin, Nat. Chem., 2014, 6, 635–643 CrossRef PubMed.
- A. Fernández-Tejada, E. K. Chea, C. George, J. R. Gardner, P. O. Livingston, G. Ragupathi, D. S. Tan and D. Y. Gin, Bioorganic Med. Chem., 2014, 22, 5917–5923 CrossRef PubMed.
- A. Fernández-Tejada, Pure Appl. Chem., 2017, 89, 1359–1378 CrossRef.
- A. Fernández-Tejada, D. S. Tan and D. Y. Gin, Acc. Chem. Res., 2016, 49, 1741–1756 CrossRef PubMed.
- A. Fernández-Tejada, D. S. Tan and D. Y. Gin, Chem. Commun., 2015, 51, 13949–13952 RSC.
- A. Fernández-Tejada, W. E. Walkowicz, D. S. Tan and D. Y. Gin, in Methods in Molecular Biology, ed. C. B. Fox, Springer New York, New York, NY, 2017, vol. 1494, pp. 45–71 Search PubMed.
- C. R. Kensil, U. Patel, M. Lennick and D. Marciani, J. Immunol., 1991, 146, 431–437 CrossRef CAS.
- RTS,S Clinical Trials Partnership, Lancet, 2015, 386, 31–45 CrossRef PubMed.
- H. Lal, A. L. Cunningham, O. Godeaux, R. Chlibek, J. Diez-Domingo, S.-J. Hwang, M. J. Levin, J. E. McElhaney, A. Poder, J. Puig-Barberà, T. Vesikari, D. Watanabe, L. Weckx, T. Zahaf and T. C. Heineman, N. Engl. J. Med., 2015, 372, 2087–2096 CrossRef PubMed.
- R. Fuentes, A. Ruiz-de-Angulo, N. Sacristán, C. D. Navo, G. Jiménez-Osés, J. Anguita and A. Fernández-Tejada, Chem. – Eur. J., 2021, 27, 4731–4737 CrossRef CAS PubMed.
- R. Fuentes, L. Aguinagalde, C. Pifferi, A. Plata, N. Sacristán, D. Castellana, J. Anguita and A. Fernández-Tejada, Front. Immunol., 2022, 13, 865507 CrossRef CAS PubMed.
- R. Fuentes, L. Aguinagalde, N. Sacristán and A. Fernández-Tejada, Chem. Commun., 2021, 57, 11382–11385 RSC.
- S. Zhang, L. A. Graeber, F. Helling, G. Ragupathi, S. Adluri, K. O. Lloyd and P. O. Livingston, Cancer Res., 1996, 56, 3315–3319 CAS.
- T. Gilewski, S. Adluri, G. Ragupathi, S. Zhang, T.-J. Yao, K. Panageas, M. Moynahan, A. Houghton, L. Norton and P. O. Livingston, Clin. Cancer Res., 2000, 6, 1693–1701 CAS.
- U. Karsten, N. Serttas, H. Paulsen, A. Danielczyk and S. Goletz, Glycobiology, 2004, 14, 681–692 CrossRef CAS PubMed.
- C. Leclerc, E. Deriaud, V. Mimic and S. van der Werf, J. Virol., 1991, 65, 711–718 CrossRef CAS PubMed.
- D. M. Coltart, A. K. Royyuru, L. J. Williams, P. W. Glunz, D. Sames, S. D. Kuduk, J. B. Schwarz, X.-T. Chen, S. J. Danishefsky and D. H. Live, J. Am. Chem. Soc., 2002, 124, 9833–9844 CrossRef CAS PubMed.
- N. Martínez-Sáez, J. Castro-Lõpez, J. Valero-González, D. Madariaga, I. Compañõn, V. J. Somovilla, M. Salvadõ, J. L. Asensio, J. Jiménez-Barbero, A. Avenoza, J. H. Busto, G. J. L. Bernardes, J. M. Peregrina, R. Hurtado-Guerrero and F. Corzana, Angew. Chem., Int. Ed., 2015, 54, 9830–9834 CrossRef PubMed.
- I. A. Bermejo, I. Usabiaga, I. Compañón, J. Castro-López, A. Insausti, J. A. Fernández, A. Avenoza, J. H. Busto, J. Jiménez-Barbero, J. L. Asensio, J. M. Peregrina, G. Jiménez-Osés, R. Hurtado-Guerrero, E. J. Cocinero and F. Corzana, J. Am. Chem. Soc., 2018, 140, 9952–9960 CrossRef CAS PubMed.
- K. Lavrsen, C. B. Madsen, M. G. Rasch, A. Woetmann, N. Ødum, U. Mandel, H. Clausen, A. E. Pedersen and H. H. Wandall, Glycoconj. J., 2013, 30, 227–236 CrossRef CAS PubMed.
- T. Schumacher, L. Bunse, S. Pusch, F. Sahm, B. Wiestler, J. Quandt, O. Menn, M. Osswald, I. Oezen, M. Ott, M. Keil, J. Balß, K. Rauschenbach, A. K. Grabowska, I. Vogler, J. Diekmann, N. Trautwein, S. B. Eichmüller, J. Okun, S. StevanoviÄ, A. B. Riemer, U. Sahin, M. A. Friese, P. Beckhove, A. Von Deimling, W. Wick and M. Platten, Nature, 2014, 512, 324–327 CrossRef CAS PubMed.
- S. Kreiter, M. Vormehr, N. van de Roemer, M. Diken, M. Löwer, J. Diekmann, S. Boegel, B. Schrörs, F. Vascotto, J. C. Castle, A. D. Tadmor, S. P. Schoenberger, C. Huber, Ö. Türeci and U. Sahin, Nature, 2015, 520, 692–696 CrossRef CAS PubMed.
- S. Stevanović, A. Pasetto, S. R. Helman, J. J. Gartner, T. D. Prickett, B. Howie, H. S. Robins, P. F. Robbins, C. A. Klebanoff, S. A. Rosenberg and C. S. Hinrichs, Science, 2017, 356, 200–205 CrossRef PubMed.
- P. A. Ott, Z. Hu, D. B. Keskin, S. A. Shukla, J. Sun, D. J. Bozym, W. Zhang, A. Luoma, A. Giobbie-Hurder, L. Peter, C. Chen, O. Olive, T. A. Carter, S. Li, D. J. Lieb, T. Eisenhaure, E. Gjini, J. Stevens, W. J. Lane, I. Javeri, K. Nellaiappan, A. M. Salazar, H. Daley, M. Seaman, E. I. Buchbinder, C. H. Yoon, M. Harden, N. Lennon, S. Gabriel, S. J. Rodig, D. H. Barouch, J. C. Aster, G. Getz, K. Wucherpfennig, D. Neuberg, J. Ritz, E. S. Lander, E. F. Fritsch, N. Hacohen and C. J. Wu, Nature, 2017, 547, 217–221 CrossRef CAS PubMed.
- J. Banchereau and K. Palucka, Nat. Rev. Clin. Oncol., 2018, 15, 9–10 CrossRef PubMed.
- R. E. Hollingsworth and K. Jansen, npj Vaccines, 2019, 4, 7 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sc05639a |
| ‡ C. P. present address: Centre de Biophysique Moléculaire, CNRS UPR 4301, Rue Charles Sadron, 45071 Orléans Cedex 2, France. |
| This journal is © The Royal Society of Chemistry 2023 |