Open Access Article
Open Access ArticleThe expedient, CAET-assisted synthesis of dual-monoubiquitinated histone H3 enables evaluation of its interaction with DNMT1†
Zichen
Li‡
ab,
Zebin
Tong‡
a,
Qingyue
Gong‡
bc,
Huasong
Ai
 *a,
Shuai
Peng
b,
Cong
Chen
b,
Guo-Chao
Chu
a and
Jia-Bin
Li
*a,
Shuai
Peng
b,
Cong
Chen
b,
Guo-Chao
Chu
a and
Jia-Bin
Li
 *b
*b
aDepartment of Chemistry, Tsinghua-Peking Center for Life Sciences, Tsinghua University, Beijing 100084, China. E-mail: ahs17@tsinghua.org.cn
bJiangsu Key Laboratory of Neuropsychiatric Diseases and College of Pharmaceutical Sciences, Jiangsu Key Laboratory of Preventive and Translational Medicine for Geriatric Diseases, Suzhou Key Laboratory of Drug Research for Prevention and Treatment of Hyperlipidemic Diseases, Soochow University, Suzhou, 215123, China. E-mail: lijiabin@suda.edu.cn
cSchool of Life Sciences, University of Science and Technology of China, Hefei 230026, China
First published on 1st May 2023
Abstract
Site-selective conjugation chemistry has proven effective to synthesize homogenously ubiquitinated histones. Recently, a powerful strategy using 2-((2-chloroethyl) amino) ethane-1-thiol (CAET) as a bifunctional handle was developed to generate chemically stable ubiquitin chains without racemization and homodimerization. Herein, we extend this strategy to the expedient synthesis of ubiquitinated histones, exemplifying its utility to not only synthesize single-monoubiquitinated histones, but dual-monoubiquitinated histones as well. The synthetic histones enabled us to evaluate the binding of DNMT1 to ubiquitinated nucleosomes and map the hotspots of this interaction. Our work highlights the potential of modern chemical protein synthesis to synthesize ubiquitinated histones for epigenetic studies.
Introduction
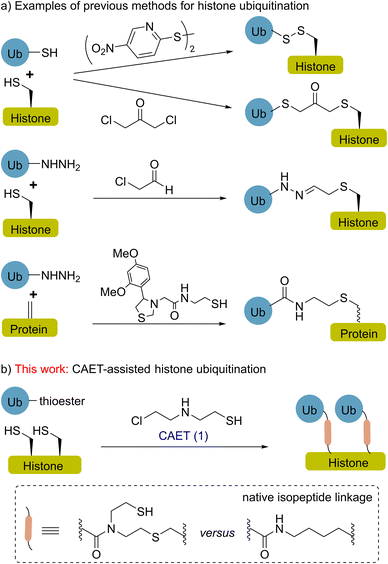
Histone ubiquitination is a critical post-translational modification (PTM) affecting a diverse array of cellular signals that are implicated in almost all chromatin-templated processes including DNA replication, transcription, and damage repair.1,2 Our understanding of these processes necessitates the use of homogenously modified histones to assign functional consequences to site-specific ubiquitin marks.3–11 In the past decade, site-selective conjugation chemistry using recombinant proteins as starting materials has been an attractive technology for making ubiquitinated histones, thereby facilitating the functional and mechanistic studies of histone ubiquitination in a chemically defined manner.3,8,12–18 For instance (Fig. 1a), disulfide-exchange and α-halogen ketone-directed methods have been developed for histone site-specific monoubiquitination,12–14 and the resulting histones were widely used in biochemical and structural studies.3,19–22 In a recent study, hydrazide mimics of acylated histones were synthesized to evaluate their effects on nucleosome dynamics and assess deubiquitinase action.17 The functionalization of dehydroalanine, a method that allows the installation of various natural PTMs and unnatural labels,23–27 also shows potential for the chemical synthesis of homogenously ubiquitinated histones.28,29More recently, Pan et al. reported a strategy to prepare ubiquitin chains using 2-((2-chloroethyl) amino) ethane-1-thiol (1, CAET in Fig. 1b) as a bifunctional handle, avoiding the formation of racemic isomers or homodimers in the synthesis and generating a chemically stable isopeptide bond mimic.30,31 This strategy also allows the generation of customized ubiquitin tools (e.g., E2-conjugated ubiquitin chains) for exploring the catalytic processes of ubiquitin machinery.31,32 Nevertheless, the CAET-assisted site-specific installation of ubiquitin onto protein substrates (e.g., histones) remains unexplored. Herein, we report the expedient chemical synthesis of K18 and/or K23 monoubiquitinated histone H3 (namely H3K18Cub, H3K23Cub and H3K18CubK23Cub) through the CAET-assisted strategy (Fig. 1b), exemplifying the utility of this strategy to not only prepare single-monoubiquitinated histones, but dual-monoubiquitinated histones as well. The resulting ubiquitinated histones enabled us to evaluate their interaction with DNA methyltransferase 1 (DNMT1) at the nucleosome level, yielding new insights into this interaction.
Results and discussion
CAET-assisted synthesis of single-monoubiquitinated H3
Our study was instigated by the recent cell biology finding that the adjacent K18 and K23 of histone H3 are monoubiquitinated by the E3 ligase Uhrf1.33,34 The resulting ubiquitinated H3 is essential to recruit and activate DNMT1, a major maintenance DNA methyltransferase responsible for the inheritance of DNA methylation patterns during replication.35,36 Recent studies, especially the structural studies of DNMT1 bound to ubiquitinated H3 peptides, revealed the direct interaction of ubiquitin with DNMT1 and suggested that H3 ubiquitination may allosterically activate the enzymatic activity of DNMT1.35,36 However, the regulatory mechanism of DNMT1's recruitment and activation by H3 ubiquitination at the nucleosome level are not fully understood, and its study requires expedient access to structurally defined H3 ubiquitinated at the K18 and/or K23 positions.First, we explored the chemical synthesis of K18 or K23 single-monoubiquitinated H3 by the CAET-assisted conjugation between a ubiquitin thioester (2) and a histone H3 mutant bearing a Lys-to-Cys mutation at K18 or K23 (Fig. 2a and S1†). Ubiquitin thioester 2 was generated by E1-catalyzed thiolysis of the recombinant ubiquitin (2a, Fig. 2b).30,37 We cloned the gene of full-length ubiquitin into the vector and expressed it in E. coli. After ultrasonic lysis and centrifugation, the lysate was treated with 1% HClO4 to precipitate most of the impurities. The crude 2a in the supernatant was dialyzed into an activation buffer (25 mM HEPES, pH 7.5, 100 mM NaCl), concentrated to 5 mg mL−1, and activated by the E1 enzyme UBA1 (0.5 μM) in the presence of TCEP (5 mg mL−1), MgCl2 (10 mM) and ATP (10 mM). The in situ thiolysis was accomplished by the addition of sodium 2-mercaptoethanesulfonate (MesNa) (Fig. 2b), and after stirring at 37 °C for 12–18 hours, >15 mg of ubiquitin thioester 2 was generated from 1L LB expression after HPLC purification (Fig. S2†). H3 mutants were generated by mutating the native Cys residues at positions 96 and 110 and Lys residues at positions 18 or 23 to Ser and Cys, respectively, followed by cloning of the corresponding genes into the pET22b vector and expressing them in E. coli. After lysis and centrifugation, the desired H3K18C (3a) and H3K23C (3b) in the inclusion body were purified by RP-HPLC (Fig. S3†).
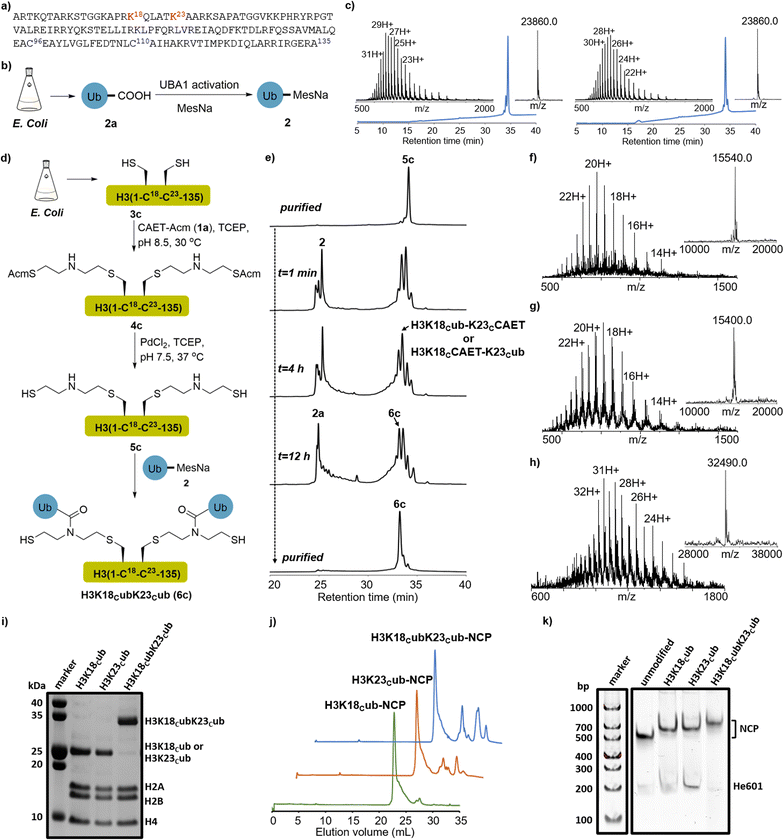 | ||
Fig. 2 CAET-assisted synthesis of single- and dual-monoubiquitinated H3 proteins. (a) Histone H3 amino acid sequence. The ubiquitination sites (K18 and K23) are marked in red, and the native Cys residues (C96 and C110) that were mutated to Ser are coloured in blue. (b) E1-catalyzed synthesis of recombinant ubiquitin thioester 2. (c) RP-HPLC (214 nm) and ESI-MS characterization of H3K18Cub (6a, left. obs. 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 860.0 Da, calc. 23 860.0 Da, calc. 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 864.5 Da) and H3K23Cub (6b, right. obs. 23 864.5 Da) and H3K23Cub (6b, right. obs. 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 860.0 Da, calc. 23 860.0 Da, calc. 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 864.5 Da). The UniDec software39 was used to generate the deconvoluted mass spectra and provided the observed (Obs.) molecular weight. (d) Schematic depiction of CAET-assisted synthesis of H3K18CubK23Cub (6c). (e) Analytic HPLC traces (214 nm) of CAET-assisted ligation of 5c with 2. (f–h) RP-HPLC (214 nm) and ESI-MS characterization of purified 4c (Obs. 15 864.5 Da). The UniDec software39 was used to generate the deconvoluted mass spectra and provided the observed (Obs.) molecular weight. (d) Schematic depiction of CAET-assisted synthesis of H3K18CubK23Cub (6c). (e) Analytic HPLC traces (214 nm) of CAET-assisted ligation of 5c with 2. (f–h) RP-HPLC (214 nm) and ESI-MS characterization of purified 4c (Obs. 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 540.0 Da, Calc. 15 540.0 Da, Calc. 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 539.2 Da), 5c (Obs. 15 539.2 Da), 5c (Obs. 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400.0 Da, Calc. 15 400.0 Da, Calc. 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399.1 Da), and H3K18CubK23Cub (6c, Obs. 32 399.1 Da), and H3K18CubK23Cub (6c, Obs. 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 490.0 Da, Calc. 32 490.0 Da, Calc. 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 491.1 Da), respectively. (i) SDS-PAGE analysis of the purified ubiquitinated histone octamers, stained with Coomassie brilliant blue. (j, k) The anion-exchange chromatography and native gels of reconstituted nucleosome core particle (NCPs), stained with SYBR gold. 491.1 Da), respectively. (i) SDS-PAGE analysis of the purified ubiquitinated histone octamers, stained with Coomassie brilliant blue. (j, k) The anion-exchange chromatography and native gels of reconstituted nucleosome core particle (NCPs), stained with SYBR gold. | ||
Next, segments 2 and 3a (or 3b) were conjugated using the CAET-assisted method.30,31 First, acetamidomethyl-protected CAET precursor (1a, CAET-Acm) was installed onto the sulfhydryl group of histone H3 by Cys-directed alkylation (Fig. S1†). Taking the synthesis of H3K18Cub as an example, segment 3a was treated with a solution of 100 equiv. of 1a in the reaction buffer (6.0 M Gn·HCl, 100 mM Na2HPO4, 5 mg mL−1 TCEP, pH 8.5) at 37 °C for 2 hours. The intermediate 4a was then purified by RP-HPLC (Fig. S4†). Subsequently, palladium-catalyzed Acm deprotection38 was conducted to activate the sulfhydryl group of 4a, generating intermediate 5a for the subsequent native chemical ligation (NCL) with segment 2 (Fig. S5†). Peptides 2 and 5a were then dissolved in the ligation buffer (6.0 M Gn·HCl, 100 mM Na2HPO4, pH 6.4) at final concentrations of 1.2 mM and 1.0 mM, respectively, and 200 mM 4-mercaptophenylacetic acid (MPAA) was added to accelerate the reaction. After stirring at room temperature for 12 hours, the ligation was terminated and yielded the desired H3K18Cub in an isolated yield of 42%. The purity and identity of H3K18Cub were confirmed by RP-HPLC and ESI-MS analyses (Fig. 2c). Using the same protocols, we further synthesized H3K23Cub from recombinant mutant H3K23C (Fig. 2c).
CAET-assisted synthesis of dual-monoubiquitinated H3
Having demonstrated the applicability of the CAET-assisted strategy to make single-monoubiquitinated histones, we then examined its potential to generate a dual-monoubiquitinated histone, a synthesis of which, to the best of our knowledge, has yet to be reported. The dual-monoubiquitinated histone we targeted was H3K18CubK23Cub. A gene corresponding to histone H3 bearing Lys-to-Cys mutations at positions 18 and 23 and Cys-to-Ser mutations at positions 96 and 110 was expressed in E. coli, generating mutant H3K18CK23C (3c, Fig. S6†). The purified 3c was treated with 200 equiv. of CAET-Acm and the reaction was kept for at least 12 hours. To avoid the potential oxidation of amino acids (e.g., Met) caused by the prolonged exposure of H3 at 37 °C, the reaction was conducted at 30 °C or room temperature (Fig. 2d). 3c was completely consumed and transformed to 4c with a conversion yield of approximately 90% (estimated by ESI-MS), accompanied by byproducts bearing one molecular of CAET-Acm. The desired 4c was obtained with an isolated yield of 40% (Fig. 2f). Note: the byproducts exhibited identical retention times as 4c in RP-HPLC, and could not be eliminated by either increasing reaction time or CAET-Acm concentration, but could be fortunately removed in the next purification steps. Then, palladium-catalyzed removal of the two Acm groups of 4c yielded 5c with an isolated yield of 45% (Fig. 2e and g).The ligation of two molecules of 2 with 5c to generate H3K18CubK23Cub was next attempted (Fig. 2d). Using similar protocols to those described above, ubiquitin thioester 2 and 5c were mixed in a molecular ratio of 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in the presence of 200 mM MPAA. After stirring at 30 °C or room temperature for 16 h, the reaction produced almost equal amounts of the desired H3K18CubK23Cub (6c) and single-monoubiquitinated H3 species bearing a CAET handle (H3K18Cub-K23CCAET or H3K18CCAET-K23Cub), while any unreacted 2 was completely hydrolyzed (Fig. 2e). The failure of almost half of the single-monoubiquitinated H3 to convert to H3K18CubK23Cub suggested the installation of the second ubiquitin to be an inefficient process—perhaps a consequence of the steric hindrance associated with the newly-installed first ubiquitin. The result reminded us that when multiple unfolded peptides are ligated to the adjacent lysines of a substrate, their steric hindrance may also largely affect the efficiency of NCL. Nonetheless, a yield of H3K18CubK23Cub of 12% was obtained—sufficient for subsequent biochemical experiments. The purity and identity of H3K18CubK23Cub were confirmed by RP-HPLC and ESI-MS analyses (Fig. 2e and h).
1 in the presence of 200 mM MPAA. After stirring at 30 °C or room temperature for 16 h, the reaction produced almost equal amounts of the desired H3K18CubK23Cub (6c) and single-monoubiquitinated H3 species bearing a CAET handle (H3K18Cub-K23CCAET or H3K18CCAET-K23Cub), while any unreacted 2 was completely hydrolyzed (Fig. 2e). The failure of almost half of the single-monoubiquitinated H3 to convert to H3K18CubK23Cub suggested the installation of the second ubiquitin to be an inefficient process—perhaps a consequence of the steric hindrance associated with the newly-installed first ubiquitin. The result reminded us that when multiple unfolded peptides are ligated to the adjacent lysines of a substrate, their steric hindrance may also largely affect the efficiency of NCL. Nonetheless, a yield of H3K18CubK23Cub of 12% was obtained—sufficient for subsequent biochemical experiments. The purity and identity of H3K18CubK23Cub were confirmed by RP-HPLC and ESI-MS analyses (Fig. 2e and h).
Synthetic H3 proteins enable the evaluation of DNMT1 binding to nucleosomes bearing different ubiquitination marks
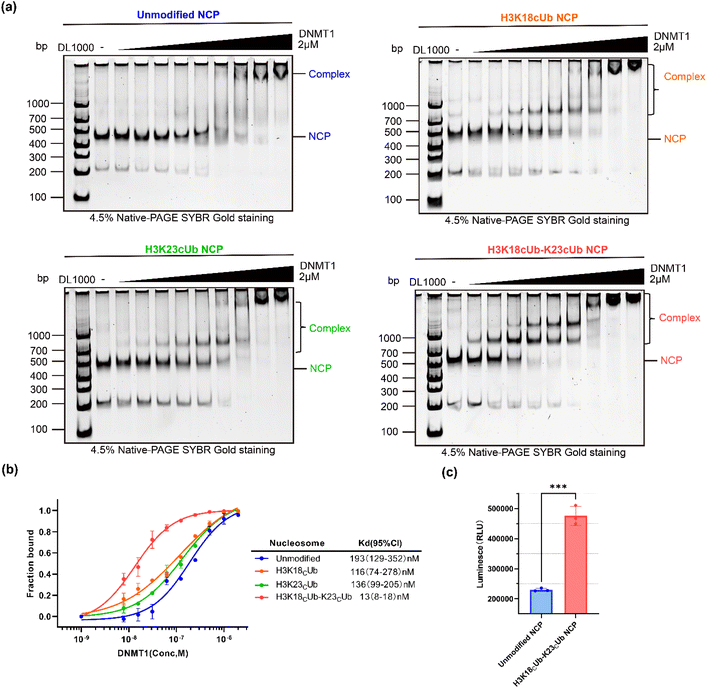
The CAET-directed chemistry results in the linkage between the ubiquitin and substrate having the same length as the native linkage, and N-alkylation of the isopeptide bond. Such mimics have been demonstrated to be deubiquitinase (DUB)-resistant and suitable for capturing their interactors including DUBs and “reader” proteins.30,32 In previous studies, ubiquitinated H3 peptide mimics that are structurally similar to the CAET-directed linkage, were prepared to explore the effects of H3 ubiquitination on DNMT1 activity, and only showed slight differences from the natural structure.15,35 Given that the CAET-directed mimic should retain the properties of ubiquitinated histones recognized by chromatin-related factors, we then used our single- and dual-monoubiquitinated H3 proteins to evaluate their effects on the binding of DNMT1 with the nucleosome.First, the synthetic H3KC18ub, H3K23Cub, H3K18CubK23Cub and recombinant H3 were individually reconstituted into histone octamers together with recombinant H2A, H2B and H4. After purification by size-exclusion chromatography (SEC), SDS-PAGE analysis confirmed that the histones were stoichiometrically incorporated into octamers (Fig. 2i and S7a†). Meanwhile, a 207-base pair (bp) of Widom 601 DNA40 flanked on each side by 30 bp of hemi-methylated CpG-containing sequence (He601) was prepared for nucleosome core particle (NCP) reconstitution (Fig. S8†), since the hemi-methylated DNA is the preferred substrate of DNMT1. The purified octamers bearing the single- or dual-monoubiquitinated H3 were reconstituted into NCPs with He601 by gradient dialysis (Fig. 2j). Native gel analyses indicated these NCPs were correctly assembled and homogenous (Fig. 2k and S7b†).
Next, the N-terminal truncated mouse DNMT1 (residues 291–1620, hereafter mDNMT1), comprising the C-terminal catalytic domain, the pair of BAH domains, CXXC zinc finger and RFTS domain, was expressed and used in the following experiments (Fig. S9†). The binding abilities of mDNMT1 to NCPs with or without ubiquitination were measured using the electrophoretic mobility shift assay (EMSA). Using the double dilution method, a series of different final concentrations (from 2000 to 7.8 nM) of mDNMT1 were incubated with 20 nM of the desired NCPs. After incubation at 4 °C for 15 min, the samples were analysed by native gel, in which the formed mDNMT1-NCP complexes migrated slower than the free NCPs. With increasing mDNMT1, the shifted bands of H3K18Cub23Cub-NCP began to appear at a much lower mDNMT1 concentration than the H3K18Cub-, H3K23Cub- and unmodified NCP (Fig. 3a). Notably, when the concentration of mDNMT1 was 250 nM, almost all the H3K18Cub23Cub-NCPs (>95%) and about 70% of the monoubiquitinated NCPs migrated upward, while only half of the unmodified NCPs were shifted (Fig. 3a and b).
Based on binding curves drawn by measuring changes of NCP bands corresponding to mDNMT1 concentrations, the binding affinity (Kd) of mDNMT1 to the H3K18Cub23Cub-NCP was measured to be 13 nM, about 8.9, 10.5 and 14.8 times higher than that of H3K18Cub- (116 nM), H3K23Cub- (136 nM) and unmodified NCPs (193 nM), respectively (Fig. 3b). These results indicated that H3K18Cub and H3K23Cub could promote mDNMT1 binding to the nucleosomes to similar extents, but their promoting effects were much weaker than that of H3K18CubK23Cub. It confirmed that H3 dual-monoubiquitination is essential for the stable binding of DNMT1 with the nucleosome.
We further examined whether H3K18CubK23Cub can activate DNMT1 activity. mDNMT1 was incubated with the unmodified- or H3K18CubK23Cub-NCPs in the presence of SAM. We found that the enzymatic activity of mDNMT1 on H3K18CubK23Cub-NCPs was much higher than that on unmodified-NCPs (Fig. 3c). It was consistent with the previous results that were measured on ubiquitinated H3 peptides.35 Collectively, these experiments confirmed that H3 dual-monoubiquitination directly recruits and activates DNMT1 in the context of nucleosome, thereby being crucial for DNA methylation maintenance.33–36
Mapping mDNMT1-H3K18CubK23Cub-NCP interaction with CXMS
To further identify the interacting interfaces between mDNMT1 and the H3K18CubK23Cub-NCP, we conducted a chemical cross-linking of proteins coupled with mass spectrometry analysis (CXMS) on this complex. The mDNMT1-H3K18CubK23Cub-NCP complex was cross-linked by Bis (sulfosuccinimidyl) suberate sodium salt (BS3, Fig. S9c†), a bifunctional cross-linker that is primary amine reactive with a maximum cross-linking distance of 34 Å.41 A band corresponding to the desired complex was digested by trypsin and subsequently analysed by LC-MS/MS (Fig. S9d†). The cross-linked peptides that were detected twice in three replicates using pLink-2 software with a false-discovery rate (FDR) of less than 0.05, were used in the following analysis.As shown in Fig. 4a, a total of 101 lysine–lysine cross-links were identified. Notably, we found some expected cross-links of lysine pairs that are located at histone–histone interfaces. Their distances in the nucleosome structure4 (for example, H3K56–H2AK74, 15 Å) were consistent with the spacer length of BS3 (Fig. S10†). For comparison, we also conducted the CXMS analysis on the complex of mDNMT1 binding to the unmodified NCP (mDNMT1-NCP, Fig. S11†). The observed cross-links in mDNMT1-NCP complex were much less than that in mDNMT1-H3K18CubK23Cub-NCP complex (Fig. 4a and S11b†). For example, the ubiquitinated complex showed 43 cross-links of mDNMT1 with histones, while only 12 cross-links were found in the unmodified complex. Particularly, no cross-link between H3 and mDNMT1 was observed in mDNMT1-NCP complex.
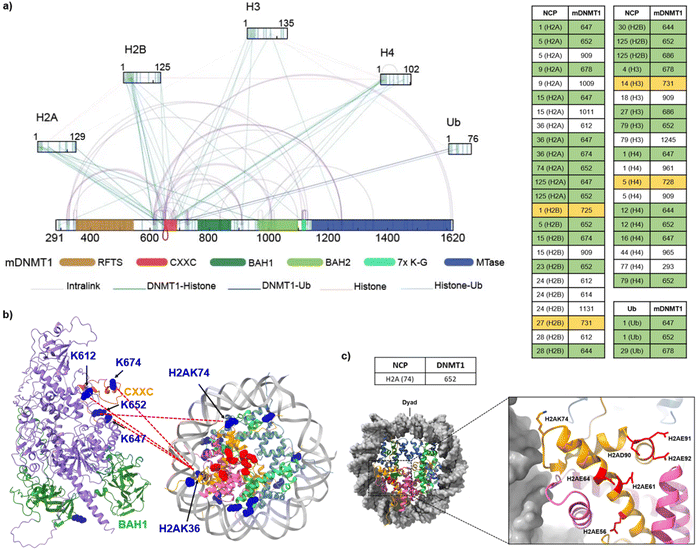 | ||
| Fig. 4 Cross-links of mDNMT1 in complex with the H3K18CubK23Cub-NCP. (a) Interprotein cross-links and intraprotein cross-links were indicated by lines (Left) and cross-linked Lys pairs between mDNMT1 and histones or ubiquitin were listed on the right. The crosslinked Lys residues located in and near the CXXC domain and CXXC-BAH1 linker were highlighted by green and yellow, respectively. (b) The representative cross-linked Lys residues on the CXXC domain and the BAH-CXXC linker were mapped into the predicted mDNMT1 structure from the AlphaFold database (AF-P13864-F1-model_v3) and the corresponding histone Lys residues were mapped into the structure of NCP (PDB: 7XD1).4 The crosslinked Lys residues were marked in blue and the amino acids constituting the acidic patch were marked in red. (c) The representative histone Lys residues (e.g., H2AK74) in (b) are adjacent to the acid patch in the nucleosome structure (PDB: 7XD1). | ||
Focusing on the cross-links of mDNMT1 with histones and ubiquitin, most of the cross-linked lysines of mDNMT1 in the ubiquitinated complex (e.g., K647, K652, K674, K725, K728 and K731) are located in or near the CXXC domain and the CXXC-BAH1 linker (Fig. 4a and b). By contrast, the unmodified NCP showed few cross-links with the two regions (Fig. S11b†). Giving that these regions contribute to the autoinhibition of mDNMT1 activity,42,43 we suggested that ubiquitination may induce their conformational changes, thereby facilitating the activation of DNMT1. Interestingly, some corresponding cross-linked histone lysines (e.g., H2AK36, H2AK74) are located near the acidic patch of the nucleosome (Fig. 4b and c), suggesting the possible interaction of mDNMT1 with the acidic patch. This is consistent with recent findings that DNMT1 is an acidic patch-dependent nucleosome binding protein.44 Moreover, several Lys residues close to the RFTS domain (K343, K348 and K353) formed cross-links with the catalytic domain in the mDNMT1-NCP complex, but not in the ubiquitinated complex (Fig. 4a and S11b†). It suggested that ubiquitination may induce the RFTS domain being out of contact with the catalytic domain. The observations were consistent with the previous results that H3 ubiquitination promotes the dissociation of the RFTS domain from the catalytic domain, thereby inducing an open and active conformation of DNMT1.35,36 Collectively, the CXMS analysis revealed different cross-linking patterns of mDNMT1 between the ubiquitinated and unmodified nucleosomes, suggesting that the recruitment and activation of DNMT1 may require its H3 ubiquitination-induced interaction and conformational changes.
Conclusions
In summary, single- and dual-monoubiquitinated histones have been expediently synthesized via the CAET-assisted conjugation of recombinant proteins, and used to evaluate and map the interaction between mDNMT1 and ubiquitinated NCPs in a chemically defined manner. The results provided important protein samples and valuable clues for further mechanistic studies on H3 ubiquitination-dependent modulation of DNMT1 activity. Our study proved the applicability of the CAET-assisted strategy to synthesize diverse ubiquitinated substrates, including histones monoubiquitinated at two adjacent sites, and enlarged the toolbox of methods to access ubiquitinated histones. The application of this strategy to the synthesis of other dual-monoubiquitinated histones (e.g., the combinations of monoubiquitination that are recently identified at the sites 125, 127 and 129 of H2A)45,46 is anticipated. Its combination with other site-specific conjugation chemistries may further facilitate the installation of more complex ubiquitin units onto other protein substrates.47,48Data availability
All detailed experimental methods and supplementary figures can be found in the ESI.†Author contributions
Z. L., Q. G., H. A. and J. B. L. conceived the work and designed the experiments. Z. L., G. C. C., S. P. and C. C. conducted the chemical synthesis. Z. L. Z. T. and H. A. performed the biochemical assays. Z. L., Z. T. and Q. G. conducted the CXMS experiments. Z. L., Z. T., Q. G. and J. B. L. wrote the manuscript. The manuscript was proofread by all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key R&D Program of China (2022YFA0913200), National Natural Science Foundation of China (22177085, 21977090), Jiangsu Key Laboratory of Neuropsychiatric Diseases (BM2013003), and a project funded by the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.Notes and references
- R. M. Vaughan, A. Kupai and S. B. Rothbart, Trends Biochem. Sci., 2020, 46, 258–269 CrossRef.
- F. Mattiroli and L. Penengo, Trends Genet., 2021, 37, 566–581 CrossRef CAS PubMed.
- H. Ai, G.-C. Chu, Q. Gong, Z.-B. Tong, Z. Deng, X. Liu, F. Yang, Z. Xu, J.-B. Li, C. Tian and L. Liu, J. Am. Chem. Soc., 2022, 144, 18329–18337 CrossRef CAS.
- H. Ai, M. Sun, A. Liu, Z. Sun, T. Liu, L. Cao, L. Liang, Q. Qu, Z. Li, Z. Deng, Z. Tong, G. Chu, X. Tian, H. Deng, S. Zhao, J.-B. Li, Z. Lou and L. Liu, Nat. Chem. Biol., 2022, 18, 972–980 CrossRef CAS.
- C. Zuo, R. Ding, X. Wu, Y. Wang, G. Chu, L. Liang, H. Ai, Z. Tong, J. Mao, Q. Zheng, T. Wang, Z. Li, L. Liu and D. Sun, Angew. Chem., Int. Ed., 2022, 61, e202201887 CAS.
- A. C. Conibear, Nat. Rev. Chem., 2020, 4, 674–695 CrossRef CAS PubMed.
- K. Nakatsu, G. Hayashi and A. Okamoto, Curr. Opin. Chem. Biol., 2020, 58, 10–19 CrossRef CAS.
- M. Jbara, H. Sun, G. Kamnesky and A. Brik, Curr. Opin. Chem. Biol., 2018, 45, 18–26 CrossRef CAS.
- A. Dhall, C. E. Weller, A. Chu, P. M. Shelton and C. Chatterjee, ACS Chem. Biol., 2017, 12, 2275–2280 CrossRef CAS PubMed.
- J.-B. Li, Y.-K. Qi, Q.-Q. He, H.-S. Ai, S. Liu, J.-X. Wang, J.-S. Zheng, L. Liu and C. Tian, Cell Res., 2017, 28, 257–260 CrossRef.
- R. K. McGinty, J. Kim, C. Chatterjee, R. G. Roeder and T. W. Muir, Nature, 2008, 453, 812–816 CrossRef CAS.
- C. Chatterjee, R. K. McGinty, B. Fierz and T. W. Muir, Nat. Chem. Biol., 2010, 6, 267–269 CrossRef CAS PubMed.
- L. Long, M. Furgason and T. Yao, Methods, 2014, 70, 134–138 CrossRef CAS.
- M. T. Holt, Y. David, S. Pollock, Z. Tang, J. Jeon, J. Kim, R. G. Roeder and T. W. Muir, Proc. Natl. Acad. Sci. U.S.A., 2015, 112, 10365–10370 CrossRef CAS PubMed.
- T. Kawakami, Y. Mishima, H. Hojo and I. Suetake, J. Pept. Sci., 2017, 23, 532–538 CrossRef CAS PubMed.
- J. Liang, Q. Gong, Y. Li, Y. Zheng, J.-S. Zheng, C. Tian and J.-B. Li, Chem. Commun., 2019, 55, 12639–12642 RSC.
- S. Bhat, Y. Hwang, M. D. Gibson, M. T. Morgan, S. D. Taverna, Y. Zhao, C. Wolberger, M. G. Poirier and P. A. Cole, J. Am. Chem. Soc., 2018, 140, 9478–9485 CrossRef CAS PubMed.
- G.-C. Chu, M. Pan, J. Li, S. Liu, C. Zuo, Z.-B. Tong, J.-S. Bai, Q. Gong, H. Ai, J. Fan, X. Meng, Y.-C. Huang, J. Shi, H. Deng, C. Tian, Y.-M. Li and L. Liu, J. Am. Chem. Soc., 2019, 141, 3654–3663 CrossRef CAS PubMed.
- B. Fierz, C. Chatterjee, R. K. McGinty, M. Bar-Dagan, D. P. Raleigh and T. W. Muir, Nat. Chem. Biol., 2011, 7, 113–119 CrossRef CAS PubMed.
- L. Zhou, M. T. Holt, N. Ohashi, A. Zhao, M. M. Müller, B. Wang and T. W. Muir, Nat. Commun., 2016, 7, 10589 CrossRef CAS.
- G. T. Debelouchina, K. Gerecht and T. W. Muir, Nat. Chem. Biol., 2016, 13, 105–110 CrossRef.
- M. T. Morgan, M. Haj-Yahya, A. E. Ringel, P. Bandi, A. Brik and C. Wolberger, Science, 2016, 351, 725–728 CrossRef CAS.
- J. M. Chalker, L. Lercher, N. R. Rose, C. J. Schofield and B. G. Davis, Angew. Chem., Int. Ed., 2012, 51, 1835–1839 CrossRef CAS PubMed.
- T. H. Wright, B. J. Bower, J. M. Chalker, G. J. L. Bernardes, R. Wiewiora, W.-L. Ng, R. Raj, S. Faulkner, M. R. J. Vallée, A. Phanumartwiwath, O. D. Coleman, M.-L. Thézénas, M. Khan, S. R. G. Galan, L. Lercher, M. W. Schombs, S. Gerstberger, M. E. Palm-Espling, A. J. Baldwin, B. M. Kessler, T. D. W. Claridge, S. Mohammed and B. G. Davis, Science, 2016, 354, aag1465 CrossRef.
- T. H. Wright and B. G. Davis, Nat. Protoc., 2017, 12, 2243–2250 CrossRef PubMed.
- J. Dadová, S. R. Galan and B. G. Davis, Curr. Opin. Chem. Biol., 2018, 46, 71–81 CrossRef PubMed.
- B. Josephson, C. Fehl, P. G. Isenegger, S. Nadal, T. H. Wright, A. W. J. Poh, B. J. Bower, A. M. Giltrap, L. Chen, C. Batchelor-McAuley, G. Roper, O. Arisa, J. B. I. Sap, A. Kawamura, A. J. Baldwin, S. Mohammed, R. G. Compton, V. Gouverneur and B. G. Davis, Nature, 2020, 585, 530–537 CrossRef CAS.
- M. Pan, Q. Zheng, S. Ding, L. Zhang, Q. Qu, T. Wang, D. Hong, Y. Ren, L. Liang, C. Chen, Z. Mei and L. Liu, Angew. Chem., Int. Ed., 2019, 58, 2627–2631 CrossRef CAS.
- M. Pan, Q. Zheng, S. Gao, Q. Qu, Y. Yu, M. Wu, H. Lan, Y. Li, S. Liu, J. Li, D. Sun, L. Lu, T. Wang, W. Zhang, J. Wang, Y. Li, H.-G. Hu, C. Tian and L. Liu, CCS Chem., 2019, 1, 476–489 CrossRef CAS.
- Q. Zheng, T. Wang, G. Chu, C. Zuo, R. Zhao, X. Sui, L. Ye, Y. Yu, J. Chen, X. Wu, W. Zhang, H. Deng, J. Shi, M. Pan, Y. Li and L. Liu, Angew. Chem., Int. Ed., 2020, 59, 13496–13501 CrossRef CAS PubMed.
- Q. Zheng, T. Wang, J. Mao, G. Chu, L. Liang, Y. Jing, C. Zuo, Y. Yu, H. Hu and M. Pan, Nat. Protoc., 2023, 18, 530–554 CrossRef CAS PubMed.
- M. Pan, Q. Zheng, T. Wang, L. Liang, J. Mao, C. Zuo, R. Ding, H. Ai, Y. Xie, D. Si, Y. Yu, L. Liu and M. Zhao, Nature, 2021, 600, 334–338 CrossRef CAS PubMed.
- A. Nishiyama, L. Yamaguchi, J. Sharif, Y. Johmura, T. Kawamura, K. Nakanishi, S. Shimamura, K. Arita, T. Kodama, F. Ishikawa, H. Koseki and M. Nakanishi, Nature, 2013, 502, 249–253 CrossRef CAS.
- W. Qin, P. Wolf, N. Liu, S. Link, M. Smets, F. Mastra, I. Forné, G. Pichler, D. Hörl, K. Fellinger, F. Spada, I. Bonapace, A. Imhof, H. Harz and H. Leonhardt, Cell Res., 2015, 25, 911–929 CrossRef CAS PubMed.
- S. Ishiyama, A. Nishiyama, Y. Saeki, K. Moritsugu, D. Morimoto, L. Yamaguchi, N. Arai, R. Matsumura, T. Kawakami, Y. Mishima, H. Hojo, S. Shimamura, F. Ishikawa, S. Tajima, K. Tanaka, M. Ariyoshi, M. Shirakawa, M. Ikeguchi, A. Kidera, I. Suetake, K. Arita and M. Nakanishi, Mol. Cell, 2017, 68, 350–360 CrossRef CAS PubMed.
- A. Kikuchi, H. Onoda, K. Yamaguchi, S. Kori, S. Matsuzawa, Y. Chiba, S. Tanimoto, S. Yoshimi, H. Sato, A. Yamagata, M. Shirouzu, N. Adachi, J. Sharif, H. Koseki, A. Nishiyama, M. Nakanishi, P.-A. Defossez and K. Arita, Nat. Commun., 2022, 13, 7130 CrossRef PubMed.
- X. A. Wang, Y. Kurra, Y. Huang, Y. Lee and W. R. Liu, ChemBioChem, 2014, 15, 37–41 CrossRef CAS.
- S. K. Maity, M. Jbara, S. Laps and A. Brik, Angew. Chem., Int. Ed., 2016, 55, 8108–8112 CrossRef CAS PubMed.
- M. T. Marty, A. J. Baldwin, E. G. Marklund, G. K. A. Hochberg, J. L. P. Benesch and C. V. Robinson, Anal. Chem., 2015, 87, 4370–4376 CrossRef CAS PubMed.
- P. T. Lowary and J. Widom, J. Mol. Biol., 1998, 276, 19–42 CrossRef CAS PubMed.
- S. Rahman, N. A. Hoffmann, E. J. Worden, M. L. Smith, K. E. W. Namitz, B. A. Knutson, M. S. Cosgrove and C. Wolberger, Proc. Natl. Acad. Sci. U.S.A., 2022, 119, e2205691119 CrossRef CAS PubMed.
- J. Song, O. Rechkoblit, T. H. Bestor and D. J. Patel, Science, 2011, 331, 1036–1040 CrossRef CAS PubMed.
- K. Takeshita, I. Suetake, E. Yamashita, M. Suga, H. Narita, A. Nakagawa and S. Tajima, Proc. Natl. Acad. Sci. U.S.A., 2011, 108, 9055–9059 CrossRef CAS.
- A. Skrajna, D. Goldfarb, K. M. Kedziora, E. M. Cousins, G. D. Grant, C. J. Spangler, E. H. Barbour, X. Yan, N. A. Hathaway, N. G. Brown, J. G. Cook, M. B. Major and R. K. McGinty, Nucleic Acids Res., 2020, 48, gkaa544 CrossRef.
- R. Kalb, D. L. Mallery, C. Larkin, J. T. J. Huang and K. Hiom, Cell Rep., 2014, 8, 999–1005 CrossRef CAS.
- M. Uckelmann, R. M. Densham, R. Baas, H. H. K. Winterwerp, A. Fish, T. K. Sixma and J. R. Morris, Nat. Commun., 2018, 9, 229 CrossRef PubMed.
- S. K. Singh, I. Sahu, S. M. Mali, H. P. Hemantha, O. Kleifeld, M. H. Glickman and A. Brik, J. Am. Chem. Soc., 2016, 138, 16004–16015 CrossRef CAS PubMed.
- H. Ai, Z. Tong, Z. Deng, J. Tian, L. Zhang, M. Sun, Y. Du, Z. Xu, Q. Shi, L. Liang, Q. Zheng, J.-B. Li, M. Pan and L. Liu, Chem, 2023, 9 DOI:10.1016/j.chempr.2023.01.012.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc00332a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |