Open Access Article
Open Access ArticleCascade nanozymatic network mimicking cells with selective and linear perception of H2O2†
Caixia
Zhu
a,
Zhixin
Zhou
a,
Xuejiao J.
Gao
 b,
Yanhong
Tao
b,
Xuwen
Cao
a,
Yuan
Xu
a,
Yanfei
Shen
b,
Yanhong
Tao
b,
Xuwen
Cao
a,
Yuan
Xu
a,
Yanfei
Shen
 a,
Songqin
Liu
a,
Songqin
Liu
 a and
Yuanjian
Zhang
a and
Yuanjian
Zhang
 *a
*a
aJiangsu Engineering Laboratory of Smart Carbon-Rich Materials and Device, Jiangsu Province Hi-Tech Key Laboratory for Bio-Medical Research, School of Chemistry and Chemical Engineering, Medical School, Southeast University, Nanjing 211189, China. E-mail: Yuanjian.Zhang@seu.edu.cn
bCollege of Chemistry and Chemical Engineering, Jiangxi Normal University, Nanchang 330022, China
First published on 25th May 2023
Abstract
A single stimulus leading to multiple responses is an essential function of many biological networks, which enable complex life activities. However, it is challenging to duplicate a similar chemical reaction network (CRN) using non-living chemicals, aiming at the disclosure of the origin of life. Herein, we report a nanozyme-based CRN with feedback and feedforward functions for the first time. It demonstrates multiple responses at different modes and intensities upon a single H2O2 stimulus. In the two-electron cascade oxidation of 3,3′,5,5′-tetramethylbenzidine (TMB), the endogenous product H2O2 competitively inhibited substrates in the first one-electron oxidation reaction on a single-atom nanozyme (Co-N-CNTs) and strikingly accelerated the second one-electron oxidation reaction under a micellar nanozyme. As a proof-of-concept, we further confined the nanozymatic network to a microfluidic chip as a simplified artificial cell. It exhibited remarkable selectivity and linearity in the perception of H2O2 stimulus against more than 20 interferences in a wide range of concentrations (0.01–100 mM) and offered an instructive platform for studying primordial life-like processes.
Introduction
The creation and evolution of life are one of the most important scientific questions. Simulating a true evolutionary process as well as a primordial life-like process, and then answering one of the most intriguing and fascinating questions of humankind, has been the research topic of scientists' passion. As Lehn envisioned more than a decade ago,1 possible answers to these challenging questions come from chemistry, in particular “systems chemistry”, a field that aims to mimic the functional behavior of living systems using reaction networks. Considerable efforts have already been made in this direction. Purified biochemical components are used as building blocks to form chemical reaction networks (CRN) triggered by auxiliary chemical or physical stimuli, leading to complex and programmed reaction pathways. These chemical reaction networks reveal dissipative network,2,3 adaptation,4,5 signaling network,6 switching,7,8 feedback,9–11 communication,12,13 and oscillator processes.14 However, further development of chemical reaction networks suffers from fundamental limitations of natural matter such as nucleic acids and enzymes in the scalability for implementing batch experiments or large-scale production, which impedes their broad applications. Moreover, according to the theory of chemical evolution, under the conditions of the primitive earth, inorganics are the origin of evolution.15,16 Therefore, there is an urgent need to fill the gap in the field of reaction networks based on nonbiological matters.As the basis of enzymatic reaction networks, cascade reaction systems play a crucial role in mediating biological processes such as signal transduction and amplification, and metabolic and catabolic reactions. Owing to their low cost, ease of production, and high stability against biodegradation and denaturation,17–20 nanomaterials with enzyme-like activity (termed nanozymes) have drawn increasing attention in various fields, such as biosensors, immunoassays, and cancer therapeutics.21–29 Along this line, nanozyme-based cascade systems have already been reported for signal amplification and selective biosensing.30,31 However, to engineer more advanced behaviors, such as metabolic regulation and signal transduction, it is necessary to use a single stimulus to trigger diverse responses for a cascade system. For example, in the muscle, glycogen phosphorylase is inhibited by the metabolites glucose and glucose-6-phosphate (G6P); however, glycogen synthase is activated by glucose and G6P. Similarly, insulin acts on glucose metabolism through the activation of glycogen synthase and inhibition of phosphorylase kinase and phosphorylase a.32 The reaction characteristics are finally manifested as the flexibility and tunability of the reaction network. Nonetheless, it is challenging to achieve such a metabolic response using nanozymes because of the lack of allosteric regulatory functions as enzymes. Interestingly, considering the intrinsic characteristics of nanozymes, their regulatory ability may be alternatively obtained in kinetics by the competition of different substrates at the same active site. Despite numerous unprecedented successes of nanozymes in biological catalysis and medical applications,33–39 nanozyme-based reaction networks have not yet been explored.
Here, we report a cascade nanozymatic network based on Co-N-doped carbon nanotubes (Co-N-CNTs) with active sites competing with different substrates. H2O2, an important short-lived product in biochemical processes, was devised as the endogenous product of a 3,3′,5,5′-tetramethylbenzidine (TMB) oxidation cascade via two-electron transfer. It was found that H2O2 competitively inhibited the first one-electron oxidation of TMB at the active site Co-N4 of Co-N-CNTs nanozymes but accelerated the second one-electron oxidation driven by a micellar nanozyme. The cascade reaction network with diverse responses to a single stimulus was further incorporated into a microfluidic flow reactor as a simplified artificial cell. Interestingly, highly selective recognition and linear perception of H2O2 over a wide range of concentrations were successfully achieved using the proposed cell mimic.
Results and discussion
Synthesis and characterization of Co-N-CNTs
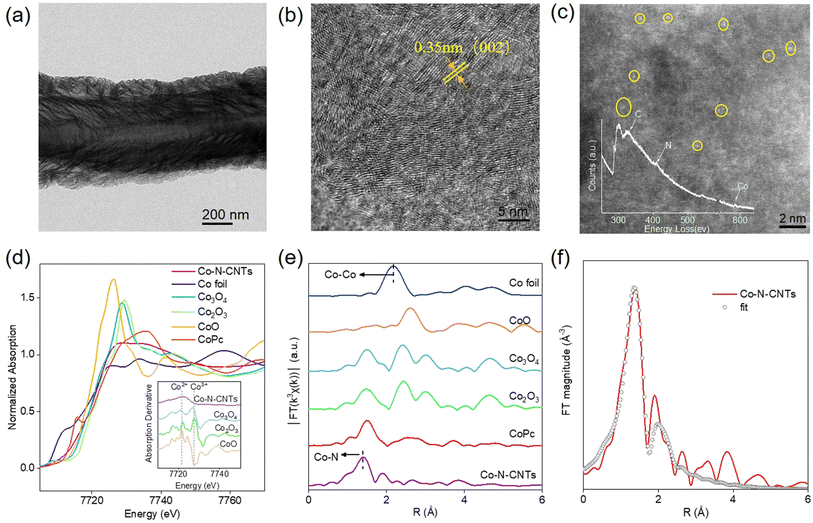
The atomically dispersed metal centers of single-atom nanozymes made it easy to distinguish the actual active sites and search for the source of enzyme-like activity.37,40–45 Given the interaction of a single atom with a ligand, single-atom Co-N-CNTs were prepared by two-step pyrolysis, in which the product of the first reaction was used as the catalyst for the second chemical vapor deposition (Fig. S1†).46–49 As shown in Fig. 1a and S1† (inset), scanning electron microscopy (SEM) and transmission electron microscopy (TEM) images indicated that Co-N-CNTs consisted of nanotubes with multiple interlaced walls. High-resolution TEM images were obtained to ascertain the detailed texture of Co-N-CNTs (Fig. 1b), depicting the (002) graphitic striations. Consistently, the (002) peak at 2θ = 25.9° was observed in the XRD pattern (Fig. S2†). The energy-dispersive X-ray spectroscopy (EDS) mapping demonstrated a uniform distribution of Co, N, and C without any evident phase separation (Fig. S3†). High-angle annular dark-field scanning transmission electron microscopy (HAADF-STEM) revealed the state of the metal center in Co-N-CNTs. As shown in Fig. 1c, single Co atoms, highlighted by yellow circles were dispersed across the entire nanotube. Meanwhile, a single Co atom surrounded by the N atoms was observed using the electron energy loss spectrum (Fig. 1c, inset).50 X-ray photoelectron spectroscopy (XPS) analysis of Co-N-CNTs suggested that the valence state of Co was between +2 and +3 (see Fig. S4 and ESI† for more details). The content of Co ions in Co-N-CNTs measured by inductively coupled plasma-mass spectrometry (ICP-MS) was approximately 2.5 wt%. These results suggest that Co-N-CNTs with a single-atom cobalt dispersion were successfully synthesized by secondary pyrolytic chemical vapor deposition.To further confirm the electronic structure and coordination environment of Co-N-CNTs at the atomic level, X-ray absorption near-edge structure (XANES) and extended X-ray absorption fine structure (EXAFS) analysis were performed. The absorption edge position of Co-N-CNTs was located between that of CoO and Co3O4 (Fig. 1d), suggesting that a single Co atom carries a positive charge, and the valence state of Co was between +2 and +3. The Fourier transform (FT) k3-weighted EXAFS spectra in Fig. 1e illustrated that, compared with the reference CoPc, Co-N-CNTs exhibited only a prominent peak located at ≈1.4 Å, which was mainly attributed to the Co-N first coordination shell rather than the Co–Co bond (2.2 Å). This result was complementary to the HAADF-STEM images, confirming that the Co species in Co-N-CNTs were atomically dispersed. The quantitative coordination configuration of the Co atom was obtained by EXAFS fitting (Fig. 1f, Table S1†). The bond length was 1.86 Å and the coordination number of Co was approximately 4. The above structural analysis determined that the isolated Co atoms were atomically dispersed in a carbon matrix and coordinated by four N atoms.
Competitive inhibition at the active site Co-N4 of Co-N-CNTs
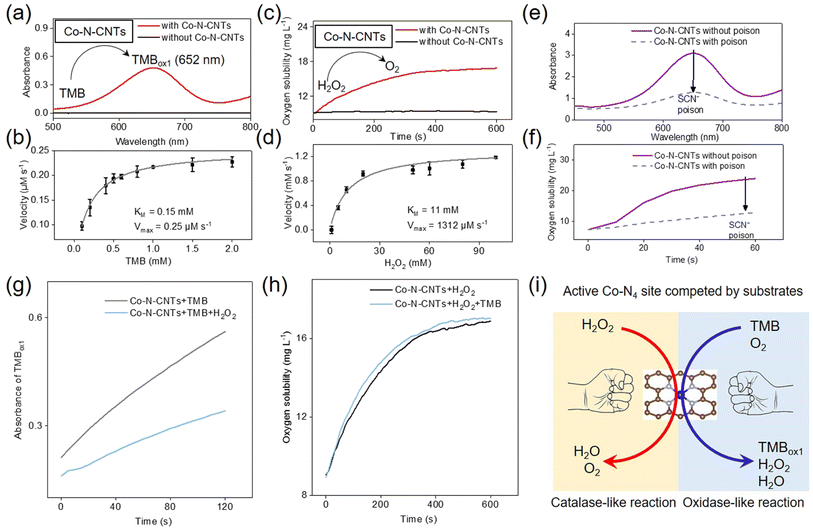
In kinetics, the activation energy of the intermediates formed on the active site of the catalyst determines the preferred reaction pathway on the catalyst. In the field of nanozyme research, oxidoreductase reactions are widely studied as the most common type of enzymatic reactions. H2O2 plays an important role as a substrate for oxidation reactions and sometimes as an intermediate product of redox reactions. H2O2 is an important short-lived product of biochemical processes that is essential for the normal activities of living organisms. Therefore, to regulate the reaction kinetics, in addition to O2, the basic substrate of oxidoreductase, H2O2 was selected as an important competitive substrate to investigate the enzyme-like activity of Co-N-CNTs.The oxidase-like catalytic activity of Co-N-CNTs was first investigated using molecular O2 and standard 3,3′,5,5′-tetramethylbenzidine (TMB) as substrates. As shown in Fig. 2a, a typical one-electron oxidized product of TMB (i.e., TMBox1) was observed (see the detailed reaction equation in Fig. S5a†). At the same time, the control experiments verified that O2 participated in the oxidation of TMB (Fig. S6†). In addition to the oxidase-like catalytic activity, the catalase-like catalytic activity of Co-N-CNTs was experimentally confirmed by comparative experiments. As shown in Fig. 2c, the addition of Co-N-CNTs significantly increased the O2 production rate (red line) compared to that of the blank sample without catalysis (black line). Detailed kinetic studies demonstrated that the oxidase (Table S2†) and catalase-like (Table S3†) activities of Co-N-CNTs were comparable to those of previous nanozymes, indicating the outstanding enzyme-mimicking activity of Co-N-CNTs (Fig. 2b and d). Based on the molar concentration of Co-N-CNTs,51 the Co-N-CNT nanozyme catalyzed the oxidation of TMB with a turnover number (kcat) of ≈0.012 s−1 and kcat/KM ≈ 0.08 mM s−1. For the catalase-like activity of Co-N-CNTs, the values were found to be kcat ≈ 61.89 s−1 and kcat/KM ≈ 5.63 mM s−1. Notably, for catalase-like activity, Co-N-CNTs showed a lower KM and a higher Vmax than other reported catalase-like nanozymes, indicative of an excellent affinity and catalytic activity for H2O2 (Table S3†). In addition, Co-N-CNTs also demonstrated high stability in cycling tests and long-term storage (see further discussion in Fig. S7†).
More interestingly, it was found that Co-N-CNTs could not oxidize TMB using H2O2; instead, the oxidation of TMB was inhibited by H2O2 (Fig. S5b†). This is distinctive from the well-known Fe–N–C single-atom nanozyme that had both peroxidase and oxidase-like activities.22,52–54 In essence, this competition phenomenon was supposed to be closely related to the catalytic active site. To further understand the origin of the catalytic activity of Co-N-CNTs, the thiocyanate ion (SCN−) was used as a poison probe to investigate the active sites of single-atom Co-N-CNTs.55–57 It was found that both oxidase and catalase-like activities of Co-N-CNTs were irreversibly inhibited by 59% and 47%, respectively, after SCN− poisoning (Fig. 2e and f), indicating that the single-atom Co sites (Co-N4) of Co-N-CNTs served as both oxidase- and catalase-like active sites. Therefore, a competitive reaction between H2O2 and O2 (TMB) occurred at the Co-N4 active site. In addition, the O2 formation rate remained almost the same with and without TMB, whereas the catalytic oxidation rate of TMB by Co-N-CNTs decreased upon the addition of H2O2 (Fig. 2g and h). In biology, this phenomenon is known as competitive inhibition. H2O2 and O2 (TMB) competed for the same binding site—the active site (Co-N4)—of the nanozyme (Fig. 2i), which hindered the combination of O2 (TMB) and the nanozyme, resulting in the reduction of the oxidase-like catalytic reaction rate of the nanozyme. In addition, H2O2 was detected during the oxidation of TMB by Co-N-CNTs (Fig. S5c†), which indicated that the reduction product of O2 also included H2O2. As an intrinsic factor, the intermediate product H2O2 would, in turn, compete with TMB and reduce the velocity of the oxidase-like reaction, thus becoming a foundation for constructing the nanozyme network with an intrinsic feedback feature (vide infra).
To explore the kinetics of the TMB oxidation reaction catalyzed by Co-N-CNTs, Lineweaver–Burk plots were constructed (Fig. S8a and b†). The two groups of lines displayed characteristic intersecting lines, suggesting that the catalytic reaction underwent a symbolic sequence Bi–Bi mechanism.58–60 It was supposed that both O2 and TMB were bound to Co-N-CNTs to form a ternary intermediate complex. To identify the above kinetic mechanism, electron spin resonance (ESR) spectra were obtained to monitor the possible intermediate reactive oxygen species (ROS) in TMB-involved redox reactions (Fig. S8c†). No signal was observed for any ROS-trapping agent adduct during the expression of oxidase-like activity by the Co-N-CNTs nanozyme. Furthermore, SOD and isopropyl alcohol (IPA) were selected as scavengers for monitoring superoxide and hydroxyl radicals. As shown in Fig. S8d,† the addition of a trapping agent also had no significant effect on the oxidation of TMB by Co-N-CNTs. This indicated that the oxidase-like activity of Co-N-CNTs was not derived from the generation of free radicals, which conformed to the sequence Bi–Bi mechanism. To gain insights into the catalase-mimicking activities of Co-N-CNTs, electron spin resonance (ESR), and optical trapping experiments were performed to detect possible intermediate ROS (see more discussion in Fig. S8e and f†). It was found that there was no detectable free radical generation during the catalase-like reaction by the Co-N-CNTs nanozyme.
Density functional theory studies of competitive inhibition at the active site Co-N4
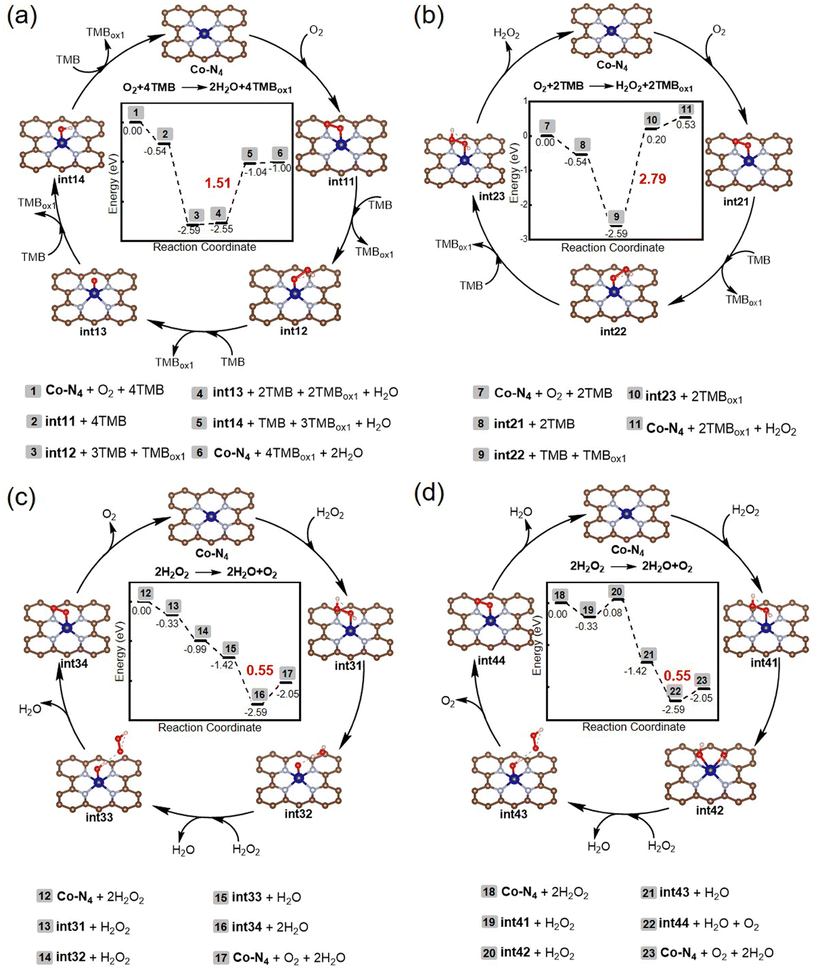
DFT calculations were conducted to gain further insight into the underlying mechanisms of the oxidase- and catalase-mimicking activities of Co-N-CNTs, in which the Co-N4 patches were considered as active centers. We aimed to solve two problems using DFT calculations: one was the chemical change process behind enzyme-mimicking activity, and the other was the competitive mechanism of oxidase- and catalase-mimicking catalytic processes. The proposed mechanisms and calculated energy profiles are shown in Fig. 3.For the oxidase-mimicking activity, two possible mechanisms for the four- and two-electron O2 reductions were investigated, and the results are shown in Fig. 3a and b, respectively. From the calculated potential energy surface, it can be seen that whether in the 4-electron or the 2-electron process, the oxidation of the first TMB by OO* was easy, accompanied by a significant energy release of −2.05 eV (2 to 3 in Fig. 3a and 8 to 9 in Fig. 3b). Then, the O on the outside of the OOH*, rather than the O on the adsorption end, is preferred to capture the reductive H from TMB, because there was only an energy increase of 0.04 eV from int12 to int13, while it was necessary to overcome 2.79 eV from int22 to int23. The rise in energy values for the oxidation of TMB by O* and OH* were 1.51 and 0.04 eV, respectively. The above results indicated that the four-electron catalytic cycle was more thermodynamically favorable than the two-electron process at room temperature because its maximum energy rise (1.51 eV) was less than that of the two-electron process (2.79 eV). Nonetheless, considering the observation of H2O2 in experiments with a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to the TMBox1 product (see Fig. S5 and more discussion in ESI†), the oxidation of TMB by O2via the 2-electron pathway also occurred significantly, presumably owing to favorable kinetics.
1 to the TMBox1 product (see Fig. S5 and more discussion in ESI†), the oxidation of TMB by O2via the 2-electron pathway also occurred significantly, presumably owing to favorable kinetics.
For catalase-mimicking activity, two possible mechanisms regarding the heterolytic and homolytic cleavage of H2O2 were investigated, and the results are shown in Fig. 3c and d, respectively. The difference between these two mechanisms lies in the decomposition mode of H2O2. The step from int31 to int32 was the heterolytic cleavage of the O–O bond, while that from int41 to int42 was the homogeneous cleavage of the O–O bond. From the energy point of view, heterolytic cleavage was more advantageous with an energy decrease of −0.66 eV, while the homogeneous cleavage was accompanied by an energy increase of 0.41 eV. Moreover, for these two mechanisms, the highest energy-rising step was the desorption of O2, and the energy increase was only 0.55 eV. This indicated that Co-N-CNTs had superior catalase-mimicking activity compared to the oxidase-mimicking activity, as the highest energy increase for the latter was 1.51 or 2.79 eV. In this regard, DFT calculations indicated that Co-N-CNTs had both oxidase- and catalase-mimicking activities. For oxidase-mimicking activity, the 4-electron reduction of O2 was thermodynamically more favorable than the 2-electron reduction pathway. However, considering the results of the experiment, both pathways were equally likely to occur (see Fig. S5 and further discussion in the ESI†). Reminiscent of the oxygen reduction reaction at electrocatalysts, the kinetics of the combinations of 4- and 2-electron pathways is common.57 For the catalase-mimicking activity, both heterolytic and homolytic cleavage of the O–O bond of H2O2 might occur, and the former was more advantageous.
More importantly, the selectivity of catalase-mimicking activity was higher than that of oxidase-mimicking activity. In other words, when H2O2 and O2 were present simultaneously, Co-N-CNTs were preferentially catalyzed by H2O2 instead of O2. These results agreed well with the experimental observations. Therefore, a unique competition for the same active site was created between different substrates, catalyzed by different enzyme-like activities of Co-N-CNTs. At Co-N4, the reaction of O2 (TMB) was completed not only by exogenous H2O2 but also by endogenous H2O2, which provided an opportunity to construct a nanozyme reaction network with intrinsic feedback properties.
Nanozymatic reaction networks using Co-N-CNTs
The metabolic regulation evolved by organisms to adapt to environmental changes mainly depends on the regulation of enzyme activity. As discussed above, the well-defined structure and excellent enzyme-like activity of the single-atom Co-N-CNTs nanozyme provided a good basis for the realization of a biomimetic metabolism network. Although the activity cannot be modulated allosterically like that in an enzyme, Co-N-CNTs with competitive inhibition at the active site Co-N4 would alternatively provide flexible solutions to build complicated chemical reaction networks, meanwhile overcoming the fundamental limitations of natural matter in terms of scalability for implementing batch experiments or large-scale production. Thus, a metabolic reaction network based on Co-N-CNTs is proposed.As the basis of chemical reaction networks, enzyme cascade reactions play an important role in signal transduction and amplification, as well as in metabolic and catalytic pathways. In contrast to other enzymatic substrates, such as o-phenylenediamine (OPD) and 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), the oxidation of TMB is not a one-step reaction but a two-step cascade.61 Therefore, the stepwise oxidation of TMB was chosen as the model for the cascade reaction network. As mentioned above, the oxidation of TMB by the Co-N-CNT nanozyme resulted in a one-electron oxidation product, TMBox1. It is well known that this cascade requires the synergistic action of multiple catalysts. Therefore, to achieve a cascade of reactions, it is necessary to search for catalysts capable of oxidizing TMBox1 to TMBox2 (Fig. 4a).
Indeed, because of the reducing nature of TMB, it is challenging to achieve TMBox1 reoxidation in the presence of TMB (the substrate of the first-step reaction). However, micellar nanozymes have catalytic mechanisms that are different from those of previous oxidoreductases, providing an interesting candidate to construct a cascade reaction of TMB oxidation.62 In brief, similar to enzymes and cell membranes, micelles also consist of hydrophilic and hydrophobic portions and thus could self-assemble into nanoreactors in water to accelerate reactions.63,64 Among them, anionic micellar nanozymes prefer to generate positively charged products due to electrostatic interactions. Therefore, based on the positive electrical properties of TMBox2,65 an SDS micelle nanozyme consisting of a common anionic surfactant was chosen to construct the cascade reaction (Fig. 4a and S9a†). To monitor the transition of the reaction product, the time-dependent absorbances at 652 nm (TMBox1) and 450 nm (TMBox2) were collected. As shown in Fig. 4b, TMB (0.5 mM) was oxidized by Co-N-CNTs (0.05 mg mL−1) to lose an electron and obtain TMBox1, which was accompanied by an increase in absorbance at 652 nm. Upon reaching a reaction time of 300 s, the second step of the cascade transformation was induced by the addition of SDS micellar nanozymes (10 mM). The absorbance of TMBox2 (450 nm) increased sharply, whereas the absorbance of TMBox1 (652 nm) decreased (Fig. 4b). Therefore, the SDS micellar nanozyme catalyzed the oxidation of TMBox1 into TMBox2 (see further discussion in Fig. S9b and c†). Notably, the hydrophobic TMB was supposed to be encapsulated in the hydrophobic chamber of the micelle nanozyme, which eliminated the potential interference to TMBox2 generation.66 In this sense, the cascade reaction could be constructed by using Co-N-CNTs nanozyme (E1) and SDS micelle nanozyme (E2), as summarized in Fig. 4a.
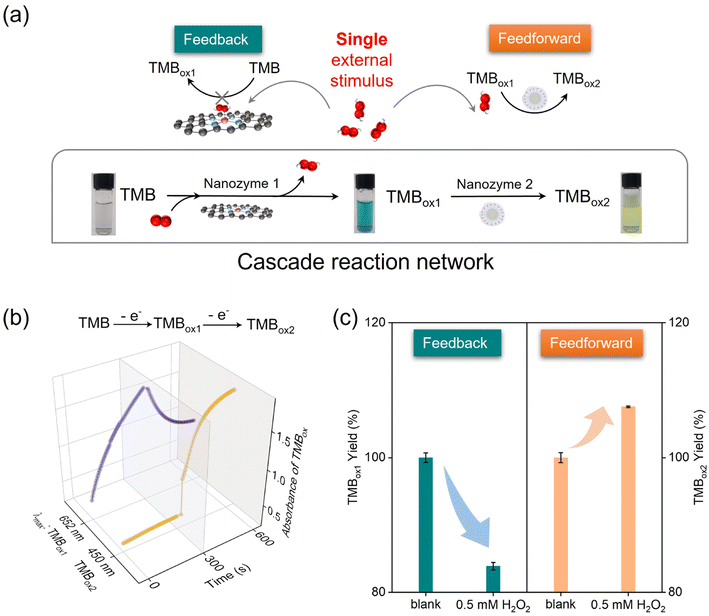
Cascade reactions form the basis of sophisticated metabolic reactions in organisms. Among them, the diverse responses of individual enzymes in the cascade to stimuli ensure the flexibility of cellular metabolic activities. In the cascade reaction involving a series of signal transmissions in the cell, there must be two positive and negative complementary feedback mechanisms for precise control. For example, gluco-6-phosphate has a positive feedforward effect on glycogen synthesis and a negative feedback effect on hexokinase.32 As mentioned before, both theoretical calculations and experiments show that a competitive reaction occurs between the substrate H2O2 and O2 (TMB), which was the basis for achieving a more advanced nanozyme reaction network. Notably, H2O2 (an important short-lived product in biochemical processes) was detected as an intermediate metabolite in the catalytic oxidation of TMB by Co-N-CNTs (Fig. S5c†), which made H2O2 an intrinsic regulator of the nanozyme reaction network (Fig. 4a). In addition, Co-N-CNTs exhibited a more catalase-like activity in the presence of both H2O2 and O2 (TMB), whereas the performance of the oxide-like reaction was inhibited (Fig. 2g and h). This suggested that H2O2 had a competitive inhibitory effect on the O2 (TMB) reaction at the Co-N4 active site, which was the key to realizing feedback in the nanozyme reaction network. Therefore, as shown in Fig. 4c (left) and Fig. S10a,† the addition of H2O2 significantly reduced the catalytic oxidation of TMB to TMBox1 by Co-N-CNTs.
In fact, the intermediate metabolite H2O2, a common oxidant, also has an accelerating effect on the re-oxidation reaction of TMBox1, which provides the possibility of feedforward regulation. As shown in Fig. 4c (right) and Fig. S10b,† the yield and the generation rate of TMBox2 increased with the increase in H2O2 concentration (see the mechanism in another work62). The results showed that H2O2 promoted the rate and efficiency of the second re-oxidation step. In contrast to the inhibition of TMBox1 production, H2O2, as an oxidation species, showed a positive feedforward effect on TMBox2 production, promoting cascaded reaction catalysis. All these results collaboratively showed that the intermediate metabolite H2O2 had a feedback inhibition effect in the first step of the reaction and a distinct feedforward activation effect in the second step of the reaction of the proposed nanozymatic reaction network (Fig. 4a). Moreover, the degrees of feedback inhibition and feedforward activation at the same concentration of H2O2 were also different. This interesting single stimulus enabled different metabolic pathways to communicate and was a prerequisite for a metabolic response network to have a self-regulation function. This demonstrated the feasibility of using nanozymes as basic units to replace natural matter in the design of chemical reaction networks. This finding greatly expands the potential applications of lifelike materials and provide new options and possibilities for designing complicated living systems. In addition, the interesting multi-enzymatic activity and competitive inhibition of the Co-N-CNTs further open up the future possibility of studying substance metabolism and metabolic regulation of the Co-N-CNTs nanozyme in the cellular milieu. Considering the nature of the nanozyme, we assume that it would be located mostly in the cytoplasmic and mitochondrial granules because the constituting metal Co might have an affinity toward cytoplasmic as well as mitochondrial granules.67
Nanozymatic network-based cell mimics enable highly selective and linear responses to H2O2
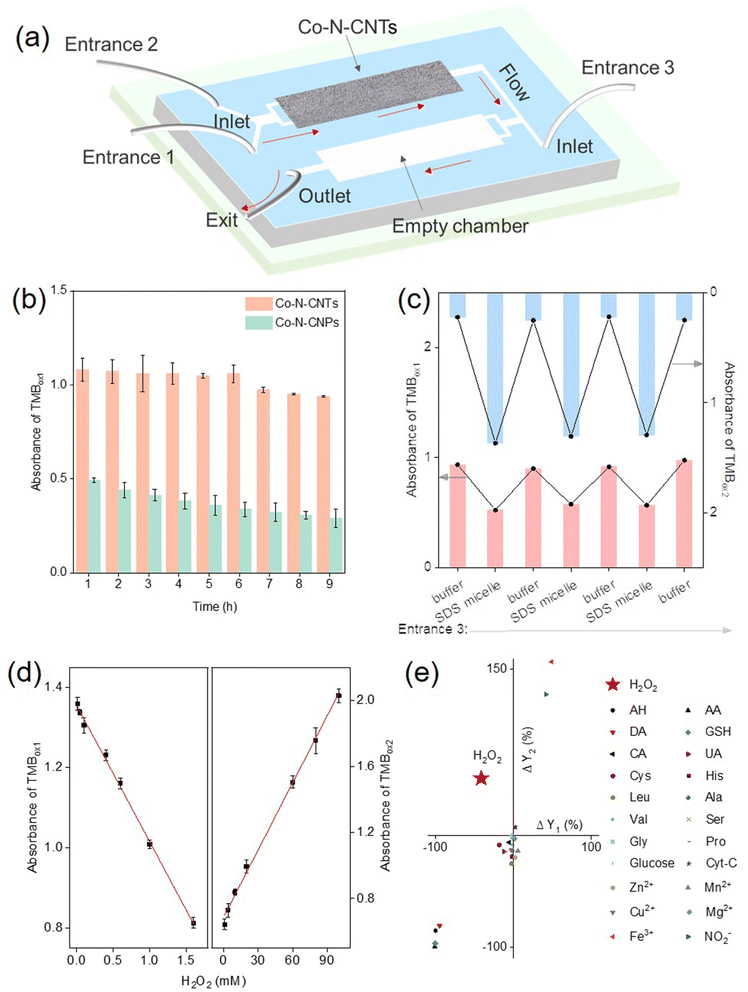
Based on the above analysis, a cascade nanozymatic network with diverse responses to a single stimulus was successfully constructed. A further important step toward engineering biomimetic systems is to confine the reactions in an “artificial cell”.68 In general, enzymes are separated in the cell by endomembrane systems to prevent unwanted interference between the enzymatic reactions. There is also a flow of substances between different organelles for metabolism and signaling. Therefore, the separation of enzyme-like reactions in a single device is of great significance for constructing practical chemical reaction networks. Microfluidic chip technology provides an opportunity to achieve in vitro separation and assembly of enzyme-like reactions (Fig. 5a).30 The control sample, Co- and N-doped carbon nanoparticles (Co-N-CNPs) were synthesized according to a previous ref. 69. An interesting geometric effect on the stability and activity of Co- and N-doped carbon was observed when deposited in microfluidic chips, that is, nanotubular ones were superior to nanoparticular ones. It was supposed that for effective immobilization, each nanoparticle should contact the channels of microfluidic chips; in contrast, the crosslinked nanotube networks would inherently work better, even with fewer contact points. Accordingly, the active sites on the nanotubes would be more exposed, leading to higher activity. As shown in Fig. 5b, the tubular characteristics of Co-N-CNTs favored the assembly and fixation in the chamber of the microfluidic chips compared to their particulate counterparts. Such advantages of Co-N-CNTs are crucial for the successful development of artificial organelles using microfluidic chips as a simplified model for long-term operation. The composition of the product was determined by sampling the outflow at regular intervals using an online UV-vis spectrometer. Precise control of the cascade reaction was achieved by altering the injected factors at entrances 2 and 3 in the microfluidic chip (Fig. 5c and S11†). As mentioned earlier, evolution supposes that life may somehow come from some type of non-living matter millions of years ago. The successful implementation of the proposed cell mimics demonstrated the possibility of a flexible response process for the initial life evolution.Notably, although more and more advanced, yet functional chemical reaction networks are being constructed, the question of practicality, and the suggestion of feasible uses of such systems beyond basic science, are still open. The concentration of H2O2 in different environments varies greatly, typically ranging from micromolar (in vivo conditions, foodstuff, and drinking water) to tens of millimolar (hazardous wastes and bleaching environment).70,71 Therefore, the selective and rapid response of H2O2 over a wide range is an essential feature for cell mimics. By the feedback and feedforward response of the Co-N-CNTs-derived network, a highly selective colorimetric response to H2O2 with a wide range, particularly at high concentrations, was developed. Owing to the stimulus-response of different intensities, the linear response to H2O2 at various concentrations was realized under the feedback (0.01–1.6 mM) and feedforward (1–100 mM) responses (Fig. 5d, see Fig. S12 and more details in ESI†). Compared with previous studies quantitatively responsive to H2O2 (Table S4†), the proposed cell mimics demonstrated a wider linear response range, which was particularly important for a primordial life-like process in mutable and harsh conditions. Owing to the multiple responses of variable intensities for the network to stimulus, the colorimetric response to H2O2 in concentrations from micromoles to tens of millimoles per liter was realized.
Notably, living systems and conventional sensing methods often require receptors that are highly specific to the target (such as a lock and key) for recognition. Interestingly, the unique response characteristics of each step in the reaction network to H2O2 enable a highly selective response from other interfering molecules without any natural biological recognition units (e.g., nucleic acids, enzymes, and antibody/antigen). As shown in Fig. 5e, the decrease in TMBox1 production and the increase in TMBox2 production were due to the addition of H2O2, making H2O2 (the second quadrant) clearly different from other interferents, including other reductive small biomolecules, amino acids, and metal ions. Therefore, without biological components, a cascade nanozymatic reaction network with diverse responses provides an effective way to specifically recognize small molecules. Such a mode that merely uses nonbiological chemicals to achieve substance-specific responses would be a possibility for initial life evolution. Moreover, beyond basic science, nanozymatic reaction networks would also be attractive for building highly selective biosensors with high stability and low cost for practical applications.
Conclusion
In summary, we report a cascade nanozymatic network. Under a single stimulus, that is, using H2O2 (an important short-lived product in biochemical processes), distinctive feedback and feedforward responses of different intensities were observed for the proposed cascade reactions of the two-electron TMB oxidation. The competition between different substrates, that is, H2O2 and O2, at the same Co-N4 active site in Co-N-CNTs nanozyme played a crucial role in successful cascade network construction. The proposed nanozymatic network was further incorporated into a microfluidic flow reactor as a simplified artificial cell. Owing to the unique responses of variable intensities and modes, the proposed cell mimics exhibited highly selective recognition and linear perception of H2O2 over a wide range of concentrations. The proposed nanozymatic network fills a gap in the reaction networks using non-living chemicals. This demonstrates the possibility of initial life evolution and opens a scalable pathway for chemical reaction networks to solve practical application problems beyond basic science. It is worth noting that as a proof of concept, the widely discussed oxidoreductase-like activity was used in the present research, although there is still a long way to synthesize versatile nanozymes in the future for more prominent chemical reaction networks. For example, the present systems operate in an in vitro bulk solution or cell-like containments. Their implementation in cells or animal tissues seems to be the next step to mimic natural systems. It is what the field of chemical reaction networks has been working on thus far.Data availability
The data that supports the findings of this study is available from the corresponding author upon reasonable request.Author contributions
Y. Z. and C. Z. conceived and designed the experiments. C. Z. performed the synthesis, characterization, and activity evaluation of the Co-N-CNTs. Z. Z. assisted in the design of a cascade nanozymatic network. X. G. and Y. T. performed the DFT calculation. X. C. and Y. X. assisted in the synthesis of the Co-N-CNTs. All authors contributed to the analysis and discussion of the results. C. Z. and Z. Z. co-wrote the manuscript, Y. Z., S. L., and Y. S. revised the manuscript. All authors reviewed the manuscript. Y. Z. supervised the project.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (22174014 and 22074015).References
- J. M. Lehn, From supramolecular chemistry towards constitutional dynamic chemistry and adaptive chemistry, Chem. Soc. Rev., 2007, 36, 151–160 RSC.
- F. J. Rizzuto, C. M. Platnich, X. Luo, Y. Shen, M. D. Dore, C. Lachance-Brais, A. Guarné, G. Cosa and H. F. Sleiman, A dissipative pathway for the structural evolution of DNA fibres, Nat. Chem., 2021, 13, 843–849 CrossRef CAS PubMed.
- Z. Zhou, Y. Ouyang, J. Wang and I. Willner, Dissipative Gated and Cascaded DNA Networks, J. Am. Chem. Soc., 2021, 143, 5071–5079 CrossRef CAS PubMed.
- B. Helwig, B. van Sluijs, A. A. Pogodaev, S. G. J. Postma and W. T. S. Huck, Bottom-Up Construction of an Adaptive Enzymatic Reaction Network, Angew. Chem., Int. Ed., 2018, 57, 14065–14069 CrossRef CAS PubMed.
- L. Yue, S. Wang, Z. Zhou and I. Willner, Nucleic Acid Based Constitutional Dynamic Networks: From Basic Principles to Applications, J. Am. Chem. Soc., 2020, 142, 21577–21594 CrossRef CAS PubMed.
- R. Peng, L. Xu, H. Wang, Y. Lyu, D. Wang, C. Bi, C. Cui, C. Fan, Q. Liu, X. Zhang and W. Tan, DNA-based artificial molecular signaling system that mimics basic elements of reception and response, Nat. Commun., 2020, 11, 978 CrossRef CAS PubMed.
- S. Wang, L. Yue, Z. Y. Li, J. Zhang, H. Tian and I. Willner, Light-Induced Reversible Reconfiguration of DNA-Based Constitutional Dynamic Networks: Application to Switchable Catalysis, Angew. Chem., Int. Ed., 2018, 57, 8105–8109 CrossRef CAS PubMed.
- M. Teders, A. A. Pogodaev, G. Bojanov and W. T. S. Huck, Reversible Photoswitchable Inhibitors Generate Ultrasensitivity in Out-of-Equilibrium Enzymatic Reactions, J. Am. Chem. Soc., 2021, 143, 5709–5716 CrossRef CAS PubMed.
- S. N. Semenov, A. S. Wong, R. M. van der Made, S. G. Postma, J. Groen, H. W. van Roekel, T. F. de Greef and W. T. Huck, Rational design of functional and tunable oscillating enzymatic networks, Nat. Chem., 2015, 7, 160–165 CrossRef CAS PubMed.
- A. A. Pogodaev, A. S. Y. Wong and W. T. S. Huck, Photochemical Control over Oscillations in Chemical Reaction Networks, J. Am. Chem. Soc., 2017, 139, 15296–15299 CrossRef CAS PubMed.
- L. Yue, S. Wang, V. Wulf, S. Lilienthal, F. Remacle, R. D. Levine and I. Willner, Consecutive feedback-driven constitutional dynamic networks, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 2843–2848 CrossRef CAS PubMed.
- D. Han, C. Wu, M. You, T. Zhang, S. Wan, T. Chen, L. Qiu, Z. Zheng, H. Liang and W. Tan, A cascade reaction network mimicking the basic functional steps of adaptive immune response, Nat. Chem., 2015, 7, 835–841 CrossRef CAS PubMed.
- C. Wang, L. Yue and I. Willner, Controlling biocatalytic cascades with enzyme-DNA dynamic networks, Nat. Catal., 2020, 3, 941–950 CrossRef CAS.
- N. Srinivas, J. Parkin, G. Seelig, E. Winfree and D. Soloveichik, Enzyme-free nucleic acid dynamical systems, Science, 2017, 358, eaal2052 CrossRef PubMed.
- A. Oparin, Proikhozndenie Zhizni, Izd, Moskovskij Rabochij, Moscow, 1924 Search PubMed.
- J. B. S. Haldane, The origin of life, Rationalist Annual, 1929, 148, pp. 3–10 Search PubMed.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang, S. Perrett and X. Yan, Intrinsic peroxidase-like activity of ferromagnetic nanoparticles, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- J. Wu, X. Wang, Q. Wang, Z. Lou, S. Li, Y. Zhu, L. Qin and H. Wei, Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes (II), Chem. Soc. Rev., 2019, 48, 1004–1076 RSC.
- Y. Huang, J. Ren and X. Qu, Nanozymes: Classification, Catalytic Mechanisms, Activity Regulation, and Applications, Chem. Rev., 2019, 119, 4357–4412 CrossRef CAS PubMed.
- Y. Song, K. Qu, C. Zhao, J. Ren and X. Qu, Graphene oxide: intrinsic peroxidase catalytic activity and its application to glucose detection, Adv. Mater., 2010, 22, 2206–2210 CrossRef CAS PubMed.
- L. Jiao, W. Xu, H. Yan, Y. Wu, C. Liu, D. Du, Y. Lin and C. Zhu, Fe–N–C Single-Atom Nanozymes for the Intracellular Hydrogen Peroxide Detection, Anal. Chem., 2019, 91, 11994–11999 CrossRef CAS PubMed.
- W. Zhen, Y. Liu, W. Wang, M. Zhang, W. Hu, X. Jia, C. Wang and X. Jiang, Specific “Unlocking” of a Nanozyme-Based Butterfly Effect To Break the Evolutionary Fitness of Chaotic Tumors, Angew. Chem., Int. Ed., 2020, 59, 9491–9497 CrossRef CAS PubMed.
- Y. Zhou, C. Liu, Y. Yu, M. Yin, J. Sun, J. Huang, N. Chen, H. Wang, C. Fan and H. Song, An Organelle-Specific Nanozyme for Diabetes Care in Genetically or Diet-Induced Models, Adv. Mater., 2020, 32, 2003708 CrossRef CAS PubMed.
- W. Zhen, S. An, S. Wang, W. Hu, Y. Li, X. Jiang and J. Li, Precise Subcellular Organelle Targeting for Boosting Endogenous-Stimuli-Mediated Tumor Therapy, Adv. Mater., 2021, 33, 2101572 CrossRef CAS PubMed.
- X. Zhang, S. Lin, R. Huang, A. Gupta, S. Fedeli, R. Cao-Milán, D. C. Luther, Y. Liu, M. Jiang and G. Li, Degradable ZnS-Supported Bioorthogonal Nanozymes with Enhanced Catalytic Activity for Intracellular Activation of Therapeutics, J. Am. Chem. Soc., 2022, 144, 12893–12900 CrossRef CAS PubMed.
- A. Adhikari, S. Mondal, M. Das, P. Biswas, U. Pal, S. Darbar, S. S. Bhattacharya, D. Pal, T. Saha-Dasgupta and A. K. Das, Incorporation of a biocompatible nanozyme in cellular antioxidant enzyme cascade reverses huntington's like disorder in preclinical model, Adv. Healthcare Mater., 2021, 10, 2001736 CrossRef CAS PubMed.
- S. Mondal, A. Adhikari, R. Ghosh, M. Singh, M. Das, S. Darbar, S. S. Bhattacharya, D. Pal and S. K. Pal, Synthesis and spectroscopic characterization of a target-specific nanohybrid for redox buffering in cellular milieu, MRS Adv., 2021, 6, 427–433 CrossRef CAS.
- Z. Wang, Y. Zhang, E. Ju, Z. Liu, F. Cao, Z. Chen, J. Ren and X. Qu, Biomimetic nanoflowers by self-assembly of nanozymes to induce intracellular oxidative damage against hypoxic tumors, Nat. Commun., 2018, 9, 3334 CrossRef PubMed.
- Y. Sang, F. Cao, W. Li, L. Zhang, Y. You, Q. Deng, K. Dong, J. Ren and X. Qu, Bioinspired construction of a nanozyme-based H2O2 homeostasis disruptor for intensive chemodynamic therapy, J. Am. Chem. Soc., 2020, 142, 5177–5183 CrossRef CAS PubMed.
- Q. Zhou, H. Yang, X. Chen, Y. Xu, D. Han, S. Zhou, S. Liu, Y. Shen and Y. Zhang, Cascaded Nanozyme System with High Reaction Selectivity by Substrate Screening and Channeling in a Microfluidic Device, Angew. Chem., Int. Ed., 2022, 61, e202112453 CAS.
- W. Xu, L. Jiao, Y. Wu, L. Hu, W. Gu and C. Zhu, Metal–Organic Frameworks Enhance Biomimetic Cascade Catalysis for Biosensing, Adv. Mater., 2021, 33, 2005172 CrossRef CAS PubMed.
- D. Voet, C. W. Pratt, and J. G. Voet, Principles of Biochemistry, International Student Version, 4th edn, 2015 Search PubMed.
- X. Shen, W. Liu, X. Gao, Z. Lu, X. Wu and X. Gao, Mechanisms of Oxidase and Superoxide Dismutation-like Activities of Gold, Silver, Platinum, and Palladium, and Their Alloys: A General Way to the Activation of Molecular Oxygen, J. Am. Chem. Soc., 2015, 137, 15882–15891 CrossRef CAS PubMed.
- Z. Zhang, X. Zhang, B. Liu and J. Liu, Molecular Imprinting on Inorganic Nanozymes for Hundred-fold Enzyme Specificity, J. Am. Chem. Soc., 2017, 139, 5412–5419 CrossRef CAS PubMed.
- P. Zhang, D. Sun, A. Cho, S. Weon, S. Lee, J. Lee, J. W. Han, D. P. Kim and W. Choi, Modified carbon nitride nanozyme as bifunctional glucose oxidase-peroxidase for metal-free bioinspired cascade photocatalysis, Nat. Commun., 2019, 10, 940 CrossRef PubMed.
- M. Li, J. Chen, W. Wu, Y. Fang and S. Dong, Oxidase-like MOF-818 Nanozyme with High Specificity for Catalysis of Catechol Oxidation, J. Am. Chem. Soc., 2020, 142, 15569–15574 CrossRef CAS PubMed.
- Y. Wang, G. Jia, X. Cui, X. Zhao, Q. Zhang, L. Gu, L. Zheng, L. H. Li, Q. Wu, D. J. Singh, D. Matsumura, T. Tsuji, Y.-T. Cui, J. Zhao and W. Zheng, Coordination Number Regulation of Molybdenum Single-Atom Nanozyme Peroxidase-like Specificity, Chem, 2021, 7, 436–449 CAS.
- X. Chen, Y. Wang, X. Dai, L. Ding, J. Chen, G. Yao, X. Liu, S. Luo, J. Shi, L. Wang, R. Nechushtai, E. Pikarsky, I. Willner, C. Fan and J. Li, Single-Stranded DNA-Encoded Gold Nanoparticle Clusters as Programmable Enzyme Equivalents, J. Am. Chem. Soc., 2022, 144, 6311–6320 CrossRef CAS PubMed.
- M. Das, S. Mondal, R. Ghosh, P. Biswas, Z. Moussa, S. Darbar, S. A. Ahmed, A. K. Das, S. S. Bhattacharya and D. Pal, A nano erythropoiesis stimulating agent for the treatment of anemia and associated disorders, iScience, 2022, 25, 105021 CrossRef CAS PubMed.
- F. He, L. Mi, Y. Shen, T. Mori, S. Liu and Y. Zhang, Fe–N–C Artificial Enzyme: Activation of Oxygen for Dehydrogenation and Monooxygenation of Organic Substrates under Mild Condition and Cancer Therapeutic Application, ACS Appl. Mater. Interfaces, 2018, 10, 35327–35333 CrossRef CAS PubMed.
- B. Xu, H. Wang, W. Wang, L. Gao, S. Li, X. Pan, H. Wang, H. Yang, X. Meng, Q. Wu, L. Zheng, S. Chen, X. Shi, K. Fan, X. Yan and H. Liu, A Single-Atom Nanozyme for Wound Disinfection Applications, Angew. Chem., Int. Ed., 2019, 58, 4911–4916 CrossRef CAS PubMed.
- L. Huang, J. Chen, L. Gan, J. Wang and S. Dong, Single-atom nanozymes, Sci. Adv., 2019, 5, eaav5490 CrossRef CAS PubMed.
- L. Jiao, H. Yan, Y. Wu, W. Gu, C. Zhu, D. Du and Y. Lin, When Nanozymes Meet Single-Atom Catalysis, Angew. Chem., Int. Ed., 2020, 59, 2565–2576 CrossRef CAS PubMed.
- F. Cao, L. Zhang, Y. You, L. Zheng, J. Ren and X. Qu, An Enzyme-Mimicking Single-Atom Catalyst as an Efficient Multiple Reactive Oxygen and Nitrogen Species Scavenger for Sepsis Management, Angew. Chem., Int. Ed., 2020, 59, 5108–5115 CrossRef CAS PubMed.
- S. Ji, B. Jiang, H. Hao, Y. Chen, J. Dong, Y. Mao, Z. Zhang, R. Gao, W. Chen, R. Zhang, Q. Liang, H. Li, S. Liu, Y. Wang, Q. Zhang, L. Gu, D. Duan, M. Liang, D. Wang, X. Yan and Y. Li, Matching the kinetics of natural enzymes with a single-atom iron nanozyme, Nat. Catal., 2021, 4, 407–417 CrossRef CAS.
- X. Wang, J. Song, F. Zhang, C. He, Z. Hu and Z. Wang, Electricity Generation based on One-Dimensional Group-III Nitride Nanomaterials, Adv. Mater., 2010, 22, 2155–2158 CrossRef CAS PubMed.
- Y. Yan, J. Miao, Z. Yang, F. Xiao, H. Yang, B. Liu and Y. Yang, Carbon nanotube catalysts: recent advances in synthesis, characterization and applications, Chem. Soc. Rev., 2015, 44, 3295–3346 RSC.
- Y. Cheng, S. Zhao, B. Johannessen, J. P. Veder, M. Saunders, M. R. Rowles, M. Cheng, C. Liu, M. F. Chisholm, R. De Marco, H. Cheng, S. Yang and S. Jiang, Atomically Dispersed Transition Metals on Carbon Nanotubes with Ultrahigh Loading for Selective Electrochemical Carbon Dioxide Reduction, Adv. Mater., 2018, 30, e1706287 CrossRef PubMed.
- D. Xia, X. Yang, L. Xie, Y. Wei, W. Jiang, M. Dou, X. Li, J. Li, L. Gan and F. Kang, Direct Growth of Carbon Nanotubes Doped with Single Atomic Fe–N4 Active Sites and Neighboring Graphitic Nitrogen for Efficient and Stable Oxygen Reduction Electrocatalysis, Adv. Funct. Mater., 2019, 29, 1906174 CrossRef CAS.
- Y. Pan, R. Lin, Y. Chen, S. Liu, W. Zhu, X. Cao, W. Chen, K. Wu, W. C. Cheong, Y. Wang, L. Zheng, J. Luo, Y. Lin, Y. Liu, C. Liu, J. Li, Q. Lu, X. Chen, D. Wang, Q. Peng, C. Chen and Y. Li, Design of Single-Atom Co–N5 Catalytic Site: A Robust Electrocatalyst for CO2 Reduction with Nearly 100% CO Selectivity and Remarkable Stability, J. Am. Chem. Soc., 2018, 140, 4218–4221 CrossRef CAS PubMed.
- S. Ji, B. Jiang, H. Hao, Y. Chen, J. Dong, Y. Mao, Z. Zhang, R. Gao, W. Chen, R. Zhang, Q. Liang, H. Li, S. Liu, Y. Wang, Q. Zhang, L. Gu, D. Duan, M. Liang, D. Wang, X. Yan and Y. Li, Matching the kinetics of natural enzymes with a single-atom iron nanozyme, Nat. Catal., 2021, 4, 407–417 CrossRef CAS.
- K. Fan, J. Xi, L. Fan, P. Wang, C. Zhu, Y. Tang, X. Xu, M. Liang, B. Jiang and X. Yan, In vivo guiding nitrogen-doped carbon nanozyme for tumor catalytic therapy, Nat. Commun., 2018, 9, 1440 CrossRef PubMed.
- J. Xi, R. Zhang, L. Wang, W. Xu, Q. Liang, J. Li, J. Jiang, Y. Yang, X. Yan, K. Fan and L. Gao, A Nanozyme-Based Artificial Peroxisome Ameliorates Hyperuricemia and Ischemic Stroke, Adv. Funct. Mater., 2020, 31, 2007130 CrossRef.
- R. Niu, Y. Liu, Y. Wang and H. Zhang, An Fe-based single-atom nanozyme with multi-enzyme activity for parallel catalytic therapy via a cascade reaction, Chem. Commun., 2022, 58, 7924–7927 RSC.
- Q. Wang, Z. Zhou, Y. Lai, Y. You, J. G. Liu, X. Wu, E. Terefe, C. Chen, L. Song, M. Rauf, N. Tian and S. Sun, Phenylenediamine-based FeN(x)/C catalyst with high activity for oxygen reduction in acid medium and its active-site probing, J. Am. Chem. Soc., 2014, 136, 10882–10885 CrossRef CAS PubMed.
- H. Liang, S. Bruller, R. Dong, J. Zhang, X. Feng and K. Mullen, Molecular metal–Nx centres in porous carbon for electrocatalytic hydrogen evolution, Nat. Commun., 2015, 6, 7992 CrossRef CAS PubMed.
- Y. Xu, J. Xue, Q. Zhou, Y. Zheng, X. Chen, S. Liu, Y. Shen and Y. Zhang, The Fe–N–C Nanozyme with Both Accelerated and Inhibited Biocatalytic Activities Capable of Accessing Drug-Drug Interactions, Angew. Chem., Int. Ed., 2020, 59, 14498–14503 CrossRef CAS PubMed.
- P. Pérez Galende, N. Hidalgo Cuadrado, E. Y. Kostetsky, M. G. Roig, E. Villar, V. L. Shnyrov and J. F. Kennedy, Kinetics of Spanish broom peroxidase obeys a Ping–Pong Bi–Bi mechanism with competitive inhibition by substrates, Int. J. Biol. Macromol., 2015, 81, 1005–1011 CrossRef PubMed.
- X. Liang and L. Han, White Peroxidase-Mimicking Nanozymes: Colorimetric Pesticide Assay without Interferences of O2 and Color, Adv. Funct. Mater., 2020, 30, 2001933 CrossRef CAS.
- C. Zhang, L. Chen, Q. Bai, L. Wang, S. Li, N. Sui, D. Yang and Z. Zhu, Nonmetal Graphdiyne Nanozyme-Based Ferroptosis-Apoptosis Strategy for Colon Cancer Therapy, ACS Appl. Mater. Interfaces, 2022, 14, 27720–27732 CrossRef CAS PubMed.
- P. D. Josephy, T. Eling and R. P. Mason, The horseradish peroxidase-catalyzed oxidation of 3,5,3′,5′-tetramethylbenzidine. Free radical and charge-transfer complex intermediates, J. Biol. Chem., 1982, 257, 3669–3675 CrossRef CAS PubMed.
- C. Zhu, Y. Xu, Q. Hong, X. Cao, K. Wang, Y. Shen, S. Liu and Y. Zhang, Enhancing Reliability of Chromogenic Bioassay with Two-Electron TMB Oxidation Enabled by Self-assembled Surfactants Nanozyme, ChemRxiv, preprint, 2022, DOI:10.26434/chemrxiv-2022-cw9v0.
- D. Piszkiewicz, Micelle Catalyzed Reactions Are Models of Enzyme Catalyzed Reactions Which Show Positive Homotropic Interactions, J. Am. Chem. Soc., 1976, 98, 3053–3055 CrossRef CAS PubMed.
- S. Serrano-Luginbühl, K. Ruiz-Mirazo, R. Ostaszewski, F. Gallou and P. Walde, Soft and dispersed interface-rich aqueous systems that promote and guide chemical reactions, Nat. Rev. Chem., 2018, 2, 306–327 CrossRef.
- Y. Misono, Y. Ohkata, T. Morikawa and K. Itoh, Resonance Raman and absorption spectroscopic studies on the electrochemical oxidation processes of 3,3′,5,5′-tetramethylbenzidine, J. Electroanal. Chem., 1997, 436, 203–212 CrossRef CAS.
- A. Kumar, M. K. Gupta and M. Kumar, Non-ionic surfactant catalyzed synthesis of Betti base in water, Tetrahedron Lett., 2010, 51, 1582–1584 CrossRef CAS.
- T. Porwol, W. Ehleben, K. Zierold, J. Fandrey and H. Acker, The influence of nickel and cobalt on putative members of the oxygen-sensing pathway of erythropoietin-producing HepG2 cells, Eur. J. Biochem., 1998, 256, 16–23 CrossRef CAS PubMed.
- J. Gaitzsch, X. Huang and B. Voit, Engineering Functional Polymer Capsules toward Smart Nanoreactors, Chem. Rev., 2016, 116, 1053–1093 CrossRef CAS PubMed.
- X. Chen, L. Zhao, K. Wu, H. Yang, Q. Zhou, Y. Xu, Y. Zheng, Y. Shen, S. Liu and Y. Zhang, Bound oxygen–atom transfer endows peroxidase–mimic M–N–C with high substrate selectivity, Chem. Sci., 2021, 12, 8865–8871 RSC.
- C. B. Chidambara Raj and H. Li Quen, Advanced oxidation processes for wastewater treatment: optimization of UV/H2O2 process through a statistical technique, Chem. Eng. Sci., 2005, 60, 5305–5311 CrossRef.
- D. G. Soares, F. G. Basso, E. C. V. Pontes, L. D. F. R. Garcia, J. Hebling and C. A. de Souza Costa, Effective tooth-bleaching protocols capable of reducing H2O2 diffusion through enamel and dentine, J. Dent., 2014, 42, 351–358 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc01714a |
| This journal is © The Royal Society of Chemistry 2023 |