Open Access Article
Open Access ArticleA universal orthogonal imaging platform for living-cell RNA detection using fluorogenic RNA aptamers†
Peng
Yin
,
Mingmin
Ge
,
Shiyi
Xie
,
Li
Zhang
,
Shi
Kuang
* and
Zhou
Nie
 *
*
State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Hunan Provincial Key Laboratory of Biomacromolecular Chemical Biology, Hunan University, Changsha 410082, China. E-mail: kuangshi@hnu.edu.cn; niezhou.hnu@gmail.com
First published on 2nd December 2023
Abstract
MicroRNAs (miRNAs) are crucial regulators of gene expression at the post-transcriptional level, offering valuable insights into disease mechanisms and prospects for targeted therapeutic interventions. Herein, we present a class of miRNA-induced light-up RNA sensors (miLS) that are founded on the toehold mediated principle and employ the fluorogenic RNA aptamers Pepper and Squash as imaging modules. By incorporating a sensor switch to disrupt the stabilizing stem of these aptamers, our design offers enhanced flexibility and convertibility for different target miRNAs and aptamers. These sensors detect multiple miRNA targets (miR-21 and miR-122) with detection limits of 0.48 and 0.2 nM, respectively, while achieving a robust signal-to-noise ratio of up to 44 times. Capitalizing on the distinct fluorescence imaging channels afforded by Pepper-HBC620 (red) and Squash-DFHBI-1T (green), we establish an orthogonal miRNA activation imaging platform, enabling the simultaneous visualization of different intracellular miRNAs in living cells. Our dual-color orthogonal miLS imaging platform provides a powerful tool for sequence-specific miRNA imaging in different cells, opening up new avenues for studying the intricate functions of RNA in living cells.
Introduction
MicroRNAs (miRNAs) are small non-coding RNA molecules that engage in complementary base pairing with non-coding regions in mRNA of various eukaryotes. They are pivotal in post-transcriptional regulation of gene expression, affecting over 60% of protein genes and impacting various physiological processes.1–8 Dysregulation of miRNAs has been implicated in multiple diseases, including but not limited to lung, cervical, and breast cancers.6,9,10 Owing to their compact size (19–24 nucleotides) and comparatively more straightforward biological processes than proteins and DNA,11,12 miRNAs are considered valuable biomarkers for disease diagnosis and therapy targets.13–17 However, traditional miRNA analysis methods, such as those involving fluorescent hybridization probes (including electrochemical18–20 and quantitative real-time polymerase chain reaction (qPCR)21 approaches) and MS2 RNA binding protein-fluorescent protein systems,22–27 have limitations in terms of probe degradation, delivery challenges, complex kinetics,21–24 and inability to visualize multiple miRNA targets in a single cell, hindering their effectiveness.28 Besides, the presence of homologous sequences and the low expression levels of miRNAs in living cells pose challenges to their detection.29 Additionally, the visualization of multiple miRNA targets in different living cells remained challenging, necessitating the development of more efficient and orthogonal RNA imaging approaches.Recently, fluorogenic RNA aptamers, also known as fluorescent light-up aptamers, have emerged as a promising alternative for RNA imaging.30,31 Fluorogenic RNA aptamers are discovered using the systematic evolution of ligands by exponential enrichment (SELEX)32,33 that fold into specific shapes and specifically activate the fluorescence of non-fluorescent small molecule dyes.30–33 This property renders fluorogenic RNA aptamers a powerful tool for RNA visualization. Over the past decade, various fluorogenic RNAs, including Spinach,34 Broccoli,35 Corn,36 Mango,37,38 SRB-2,39–42 Pepper,43 Beetroot,44 RhoBAST45–47 and Squash,48 have been developed as genetically encoded RNA sensors, enabling the imaging of metabolites,49–53 metals,54 proteins,55,56 and RNAs in living cells.57–65 Importantly, a genetically encoded fluorogenic RNA sensor for intracellular RNA imaging comes with minimal interference with the normal cellular processes and exhibits a lower background without a target, contributing to its broad and promising applications.
Successful light-up RNA sensors should exhibit a substantial disparity in fluorescence emission before and after binding to the target RNA. To this end, the current light-up RNA sensors often employ split60,66,67 or allosteric59,61,62 design principles to break the RNA aptamer's correct folding conformation, yielding low background fluorescence. Huang et al. and Ying et al. demonstrated RNA aptamer allosteric strategies for RNA imaging and quantitative studies of target RNA in living cells.62,68 Nevertheless, applying these design methods to other RNA aptamers and various targets entails intricate design challenges. In addition, Wang et al.57 developed the iPepper system, a tandem array of inert Pepper with split principles, for imaging diverse endogenous mRNA. Chen et al.69 used a dynamic analysis strategy that introduced a structure-switching aptamer to monitor the spatiotemporal localization of biomolecules in live systems. However, for the complex intracellular environment, these methods have limitations for simultaneous imaging of multiple target RNAs with distinct fluorescence colors in cells. Therefore, there is a need for a simple and versatile RNA sensor platform capable of orthogonal imaging of multiple miRNAs across diverse living cells.
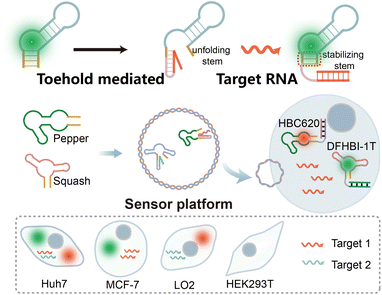
In this study, we present an orthogonal light-up RNA sensor platform designed for the multiplex imaging of miRNAs. The platform utilizes red-emissive Pepper-HBC620 and green-emissive Squash-DFHBI-1T as imaging modules (Scheme 1). We initially validated the sensor design principle using the Pepper RNA aptamer, wherein introducing a complementary miRNA sequence destroys the HBC-binding loop, thereby preventing its interaction with HBC and resulting in diminished fluorescence. However in the presence of the target miRNA, the Pepper-HBC binding core structure recovers, leading to a significant enhancement in fluorescence. We also incorporated multiple sensor units to amplify the detection signal.
Next, building on the versatility of the sensor design principle, we extended this approach to the Squash aptamer48 to achieve orthogonal dual-color imaging. The orthogonal light-up RNA sensor platform exhibits sub-nanomolar detection sensitivity for miRNAs within cells and demonstrates the ability to differentiate between target miRNAs. Due to the powerful operability of the sensor design principle, we further enhance its usability by designing it to bind different targets. Overall, our results showcase the feasibility of this orthogonal light-up RNA sensor platform for detecting and imaging intracellular miRNAs within living cells. The simplicity, high signal-to-noise ratio, and versatility of the platform make it a valuable tool for investigating RNA functions within living systems.
Results and discussion
Design and optimization of the new Pepper aptamer
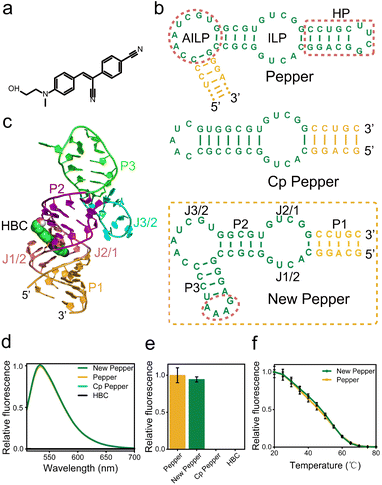
The design of the fluorogenic RNA allosteric sensor is typically fraught with intricate optimization processes aimed at acquiring the optimal transducer sequence. Unfortunately, these efforts are often accompanied by challenges, including a relatively low signal-to-noise ratio and insufficient universality. Therefore, to achieve a highly versatile fluorogenic RNA sensor that could readily accommodate various aptamers and adopt a more straightforward design strategy, we initially attempted to design the fluorogenic RNA sensor by screening from a library source. Previous studies have shown that most aptamer SELEX libraries retain an anchor sequence to enhance screening affinity70 (Fig. S1†). The anchor sequence in the initial random library comprises a 4-base pair stem and a stable UUCG tetraloop. Moreover, this sequence exhibits minimal ligand binding involvement while vital for aptamer folding. Therefore, we propose utilizing this anchor sequence as the switch of the fluorogenic RNA sensor design.To design the miRNA activation sensor platform, we selected the Pepper RNA aptamer, an uncomplicated and compact RNA molecule. The Pepper aptamer activated the bright fluorescence of HBC43 (Fig. 1a). The Pepper aptamer was predicted to form a helix, one asymmetric internal loop structure (AILP), an internal loop structure (ILP), and one hairpin loop region (HP)43 using RNA Structure software (Fig. 1c). Additionally, crystallographic structure determination of the Pepper-HBC revealed that the HBC fluorophore is sandwiched between one non-G-quadruplex quadruple and a noncanonical G·U wobble helical base pair in the ILP structural pocket. Therefore, the ILP structure plays a crucial role in the fluorescent lighting of Pepper-HBC.71 By comparing with the Pepper SELEX library sequence, it was found that HP as a structural role existed in every library member43 (Fig. S1†).
Meanwhile, the HP structure of Pepper has much stronger base interactions relative to other structures from the thermodynamic prediction of the NUPACK software (Fig. S2†).
Thus, we hypothesized that the presence of this HP is essential for facilitating the proper folding of Pepper and holds promise as a trigger in miRNA sensor design72 (Fig. S3 and Table S1†). To explore this potential, we demonstrated two circular permutations to alter the orientation of the 5′ and 3′ ends of the Pepper aptamer. One modification resulted in the direct closure of AILP, thereby opening up the HP structure to form Cp Pepper (Fig. 1b and S1†). However, as determined by fluorescence and the PAGE gel analysis, the Cp Pepper aptamer disrupted effective AILP formation and lacked HBC-binding RNA functionality compared to the original aptamer. Meanwhile, we designed a modified Pepper sequence with a paired stem and stable GAAA tetraloop to padlock the open AILP and create a new Pepper aptamer (Fig. 1b and S1†). The P1 of new Pepper has a higher contribution to the correct formation of the secondary structure of new Pepper (Fig. S2†). The fluorescence and the PAGE gel analyses demonstrated that new Pepper retained in vitro fluorescence properties and thermal stability compared to the original Pepper (Fig. 1d–f, S4 and S5†). In addition, tertiary structures of the HBC binding to RNA aptamer new Pepper in the calculated stable binding state are exhibited in Fig. 1c.
Design of the miRNA sensor using new Pepper
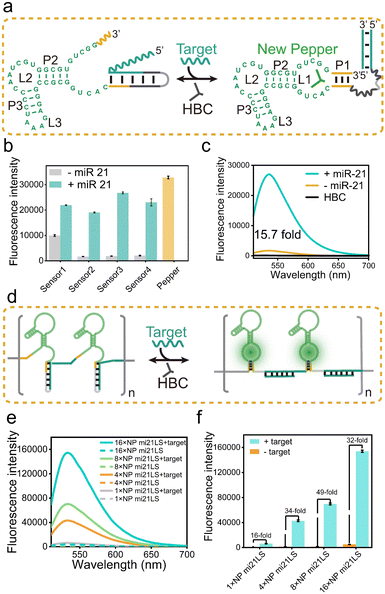
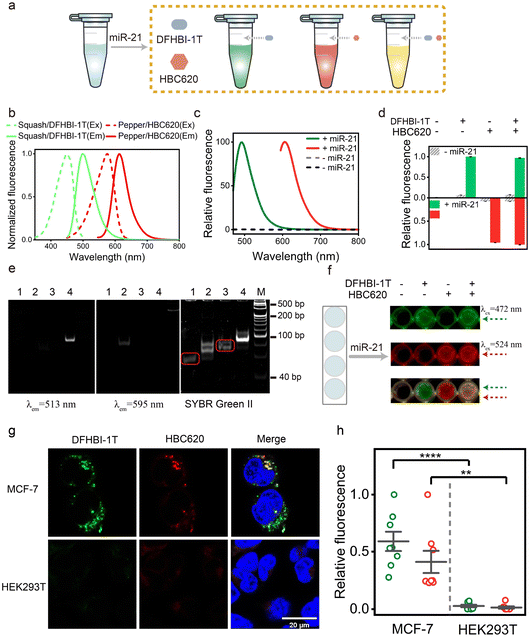
Following the principles elucidated above, we strategically harnessed the P1 stem-loop of the new Pepper aptamer as a folding-dependent domain, integrating it with the complementary sequence of the target miRNA. Employing a toehold-mediated RNA strand displacement,73 we devised a novel light-up RNA sensor, as illustrated in Fig. 2a and S6a.† This design hindered the proper folding of the Pepper aptamer, leading to disruption of the core structure (L1) of the new Pepper-HBC binding site. Upon hybridizing with its complementary toehold sequence, the target miRNA would release the masked stem and restore the P1 stem of the aptamer, promoting the reformation of the fluorophore binding pocket. The sensor design was generated by NUPACK web and RNA Structure software to predict its structure and achieve minimum free energy. As a result, the properly folded Pepper enabled fluorescence activation in HBC, achieving sensitive detection of miRNA. We have named this miRNA-induced light-up RNA sensor miLS.Next, to validate the feasibility of miLS as an in vitro sensing module, we selected miR-21 as the target miRNA due to its overexpression in various tumor cells.74 The sensor was constructed by incorporating a miR-21 responsive sequence into the P1 of new Pepper. After simple optimization of the sensor module, sensor 3 was identified as the optimal RNA sensor, named NP mi21LS (Fig. 2b and S7–S9†). In the absence of miR-21, negligible fluorescence was observed at ∼530 nm when HBC530 was incubated with this sensor. This observation suggests that new Pepper did not format the correct secondary structure and disrupted the HBC-binding site. However, in the presence of miR-21 (1 μM), a significant increase in fluorescence intensity at 530 nm, approximately 16-fold, was observed, suggesting the successful restoration of the Pepper aptamer structure (Fig. 2c). Compared to other detection methods, this light-up RNA sensor exhibited a higher signal-to-noise ratio (Table S3†). One commonly employed strategy for enhancing the imaging sensitivity of the fluorescence RNA aptamer is to fabricate arrays akin to MS2 cassettes. To improve the sensitivity of the light-up RNA sensors, we attempted to connect NP mi21LS in a series of tandem arrays containing 4×, 8× and 16× miR-21 sensors using the concepts mentioned above. To minimize steric hindrance among the sensors, we used an adenine-rich sequence as the linker to fold Pepper in the tandem arrays properly (Fig. 2d and S10†). As anticipated, the fluorescence intensity showed a proportional increase with the number of arrays utilized. Notably, the signal-to-noise ratio exhibited a marked improvement ranging from 16 to 49 as the number of arrays increased from 1× to 8×. However, an extension of the assay to 16× resulted in a reduction of the signal-to-noise ratio, which could be attributed to a higher likelihood of misfolding of the longer sequences (Fig. 2e and f).
Ulteriorly, the fluorescence intensity of the miR-21 sensor was found to be a dose-dependent behavior within a concentration range of 1 nM to 100 nM. The estimated detection limit was 0.48 nM, reflecting the remarkable sensitivity of this sensor (Fig. S11a†). Selective experiments of NP mi21LS demonstrated its excellent selectivity in complex biological environments, including RNAs (miR-122, miR-224, and iRFP mRNA), proteins (mCherry), and LO2 cell lysate (Fig. S11b†). Furthermore, the PAGE gel analysis and tube imaging more intuitively demonstrated the sensing effect of miR-21 (Fig. S11c and d†). To assess the responsiveness of NP mi21LS to tumor cells, we introduced MCF-7 cells (human breast cancer cells) with high endogenous miR-21 expression levels and HEK293T cells (human embryonic kidney cells) exhibiting low miR-21 expression levels to 500 nM NP mi21LS transfection. Confocal microscopy imaging revealed distinct red fluorescent spots in the cellular cytoplasm of MCF-7 during incubation with HBC620 (Fig. S12†). Besides, when miR-21 was hybridized with anti-miRNA, MCF-7 cells exhibited lower HBC fluorescence (Fig. S12†), NP mi21LS transfected into the majority of the cells and was located in the cytoplasm mostly (Fig. S13†), and HBC showed negligible cytotoxicity to the cells under experimental conditions (Fig. S14†). Therefore, these results demonstrate the exceptional detection performance of this NP mi21LS for miR-21 both in vitro and in living cells.
In addition, agarose gel electrophoresis analysis showed that the array sensor and miR-21 mixture migrated slower than the separate array sensor, indicating that the array miR-21 sensor is bound with the target RNA (Fig. S15†). Most importantly, the 16× NP mi21LS showed high fluorescence intensity and sensitivity, which is highly encouraging for subsequent detection of miR-21 in living cells (Fig. S16†). Increasing the number of array sensor repeats resulted in a similar melting temperature (Tm), indicating that thermostability and folding stabilities were maintained even in more extended arrays (Fig. S15†). Collectively, these results proved that the construction of a miLS tandem array can serve as a valuable tool for miRNA imaging.
Engineering of the light-up sensor platform for multiplex miRNA imaging
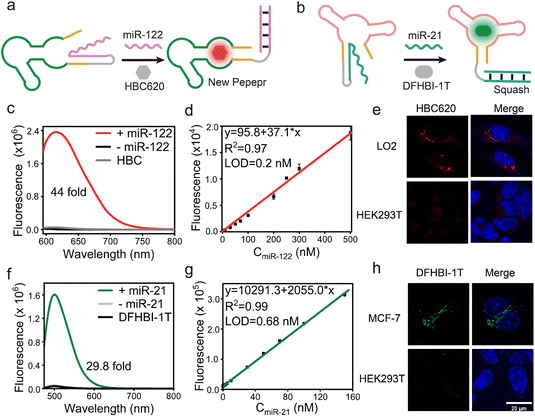
The correct folding of the fluorogenic RNA aptamer is a crucial step, which depends on the formation of a stabilizing structure framework encompassing elements such as a hairpin structure or stem. Drawing inspiration from these concepts, we applied our miLS design principle to develop a multiplex miRNA sensor and different aptamer imaging platforms (Fig. S17†). By optimizing the sensing module, in this case, by substituting the miR-21 responsive sequence with a miR-122 responsive sequence within the P1 region of new Pepper (Fig. 3a and S6b†), sensor 2 was identified as the optimal RNA sensor, named NP mi122LS, achieving a relatively sensitive light-up sensor that responds to miR-122 (Fig. S18†). In contrast, this sensor exhibited a significantly higher signal-to-background ratio of approximately 44-fold when incubated with 0.5 μM miR-122 and showed remarkable fluorescence recovery efficiency (Fig. 3c). The detection limit was estimated at 0.2 nM (Fig. 3d and S19a†). This sensor displayed negligible fluorescence intensity when exposed to other intracellular substances containing RNAs or proteins (Fig. S19†). Furthermore, the PAGE gel analysis and tube imaging analysis further directly proved the binding of the light-up sensor with miR-122 (Fig. S11c and d†). Notably, miR-122 is the most abundant miRNA in hepatocytes (∼40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 copies per hepatocyte).75,76 Consequently, LO2 cells were selected as positive cells, and we observed some red fluorescent foci in the cytoplasm, distinguishing them from the negative HEK293T cells (Fig. 3e and S20†). Moreover, upon treatment with elevated concentrations of anti-miR-122, these cells exhibited diminished fluorescence (Fig. S20†). These results collectively emphasize the ability of NP mi122LS to facilitate the imaging of various RNAs both in vitro and within living cells.
000 copies per hepatocyte).75,76 Consequently, LO2 cells were selected as positive cells, and we observed some red fluorescent foci in the cytoplasm, distinguishing them from the negative HEK293T cells (Fig. 3e and S20†). Moreover, upon treatment with elevated concentrations of anti-miR-122, these cells exhibited diminished fluorescence (Fig. S20†). These results collectively emphasize the ability of NP mi122LS to facilitate the imaging of various RNAs both in vitro and within living cells.
Recently, a fluorescence turn-on aptamer Squash, derived from the adenine riboswitch, showed high-folding efficiency in vitro and living cells. Squash activated the GFP-like fluorophores dye DFHBI-1T, resulting in an emission maximum at 500 nm. The crystal structure of fluorophore-bound Squash revealed that DFHBI-1T was sandwiched between a base triple and a noncanonical quadruple, effectively limiting non-radiative transitions and restoring the fluorescence. Besides, the P1 stem did not participate in molecular recognition but instead assisted in aptamer folding. This hypothesis was confirmed through nucleotide mutations and structure predictions of this stem of Squash (Fig. S21†).
Subsequently, we continue to employ the same toehold sequence of NP miLS, as detailed previously. MiR-21 was selected as the target RNA and a miR-21 responsive sensing module was obtained by breaking the P1 stem of Squash (Fig. 3b and S22†). When incubating with 1 μM target RNA, the sensor exhibited ∼30-fold increased fluorescence intensity at 500 nm (Fig. 3f). This result suggested that the blocking stem sequence hybridized with the target RNA and restored the folding of Squash. The selectivity and concentration-dependent experiment further elucidates the successful construction of Sq mi21LS (Fig. 3g and S23†). Meanwhile, upon incubation of Squash miLS (500 nM) in MCF-7 cells, the imaging results by confocal laser microscopy showed that a majority of green fluorescence spots were observed in living cells (Fig. 3h), and DFHBI-1T showed negligible cytotoxicity (Fig. S14†). Overall, these findings underscore the versatility of the miLS design strategy in enabling the creation of a universal platform for imaging multiple target RNAs and fluorescence aptamers possessing distinct emission wavelengths.
Characterization of orthogonal dual-color fluorogenic sensors
Over the past decade, light-up fluorescence RNA aptamer imaging has provided essential techniques for gaining insight into many RNA functions in living cells. However, the development of light-up RNA aptamer sensors that are characterized by high orthogonality and free from crosstalk is imitated. Furthermore, the characterization of a light-up RNA sensor based on orthogonal aptamers for dual-color imaging of target RNA in mammalian cells is still uncommon, indicating the need for further research in this area (Fig. 4a). Herein, we introduce an orthogonal miLS system using a pair of fluorescence RNA aptamers, Pepper-HBC620, and Squash-DFHBI-1T (Fig. 4b and S24†), thereby enabling dual-color imaging of red Pepper and green Squash.Next, we explored the feasibility of our miLS platform for orthogonal dual-color imaging of miRNA. By utilizing a system comprising NP mi21LS and Sq mi21LS, along with their respective fluorophores, we observed negligible fluorescence in the range of 450–700 nm in the absence of miR-21. By contrast, this platform incubated with miR-21 at 37 °C exhibited intense fluorescence turn-on at 500 nm or 620 nm, respectively (Fig. 4c). Besides, as shown in Fig. 4d, our system exhibited selectivity in activating the corresponding fluorophore, resulting in distinct fluorescence emission. Additionally, the PAGE gel analysis of the orthogonal miLS system revealed distinct dual-color band shift when subjected to staining with different dyes, stained initially with DFHBI-1T (left), followed by HBC620 (middle), and finally with SYBR green II (right) (Fig. 4e). In addition, the orthogonal miLS facilitated the visualization of green and red fluorescence upon incubation with miR-21 in 96-well plates (Fig. 4f). Upon transfection with this system (500 nM) and incubation with the fluorophores, MCF-7 cells exhibited bright red and green fluorescence (Fig. 4g and h), while HEK293T cells displayed significantly lower fluorescence levels. These results indicate that target RNA can be simultaneously detected using orthogonal miLS in this system. The rational design of this approach represents a significant advancement in RNA bioimaging.
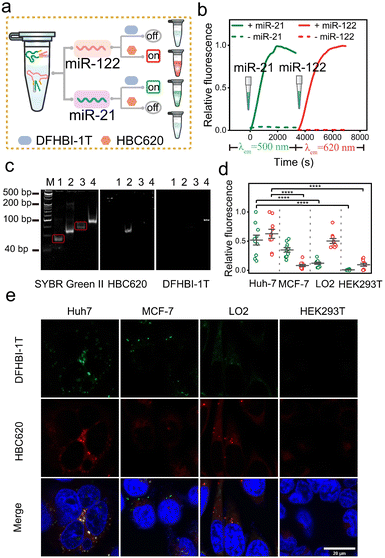
Encouraged by the data mentioned above, we subsequently evaluated the ability of the sensor platform system to discriminate between different miRNAs using a dual-color approach. Specifically, we initially assessed cuvettes containing solutions of 1 μM NP mi122LS-HBC620 pair and Sq mi21LS-DFHBI-1T, each exposed solely to either miR-122 or miR-21 (Fig. 5a). In the presence of miR-21, a slight increase in fluorescence was observed in the green light range. In addition, upon incubation with only miR-122, enhanced fluorescence at 620 nm from HBC620 was detected (Fig. S25†). Real-time kinetics analysis of the fluorescence miLS platform demonstrated gradual increases in green fluorescence from DHFBI-1T upon adding 1 μM miR-21. Meanwhile, incubation with miR-122 prompted a prominent onset of red fluorescence (Fig. 5b).
Next, we analysed the miLS platform using PAGE gel and 96-well imaging to demonstrate its capacity for differential target detection through dual-color orthogonal imaging (Fig. 5c and S25†). Finally, four distinct cell lines (Huh7, MCF-7, LO2, and HEK293T) were selected for testing the miLS platform. Huh7 cells showed overexpression levels of miR-21 and miR-122, while MCF-7 cells displayed high expression of miR-21 and no expression of miR-122. Conversely, LO2 cells exhibited low expression levels of miR-21 and high miR-122, and HEK293T cells served as control cells with no expression of either miR-21 or miR-122. The endogenous expression levels of these two miRNAs in the four cell lines were confirmed via qRT-PCR (Fig. S26†). To validate the functionality of the miLS platform system, the above cell lines were transfected with 500 nM of the miLS platform. Confocal laser microscopy images showed a stronger green and red fluorescence signal in Huh7 cells, while MCF-7 cells and LO2 cells detected only green or red fluorescence, each corresponding to their respective miRNA targets, and almost no signal was detected in HEK293T cells (Fig. 5d and e). These results affirm the successful development of the miLS sensor system capable of activating fluorescence in response to different endogenous target RNA molecules in different cells, which can be visualized using orthogonal dual-color imaging RNA.
Conclusions
In summary, we have developed a novel miRNA-induced light-up RNA sensor (miLS) that enables the visualization of miRNA of interest both in vitro and in living cells. The design concept for miLS was inspired by aptamer screening principles, utilizing the screening anchor hairpin structure as the foundational sensor switch. By introducing a miRNA hybrid sequence into the hairpin loop of the aptamer as the toehold domain, we have effectively modulated the sensor response. MiLS exhibits a significant signal-to-noise ratio (>40 fold) in vitro, thereby enabling remarkable sensitive and accurate detection of multiple target miRNAs using diverse aptamers with distinct fluorescent colours. The versatility of miLS was further enhanced by its capacity for dual-color orthogonal imaging of miR-21, both in vitro and in living cells, by leveraging the integration of Pepper and Squash aptamers with responsive units. Notably, our system exhibits relatively high resolution between the red and green fluorescence channels, thus enabling the dependable differentiation of endogenous miRNA targets in various cellular contexts. By harnessing the potential of this dual-color orthogonal imaging platform, the capability to image different miRNAs as needed, by concurrently employing the corresponding miLS is realized. This flexibility expands the toolbox of light-up imaging techniques and provides a valuable approach for deciphering the intricate physiological processes involving miRNAs in living cells.Data availability
The data that support the findings of this study are available in the main text and the ESI.†Author contributions
P. Yin, S. Kuang, and N. Zhou proposed the idea, designed the experiments, and revised the manuscript. P. Yin performed experimental work and wrote the manuscript. M. Ge, S. Xie, and L. Zhang performed most of the assays. All authors contributed to the manuscript.Conflicts of interest
The author(s) declare that they have no competing interests.Acknowledgements
This work was supported by the National Key R&D Program of China (2021YFA0910100), the National Natural Science Foundation of China (22034002, 21725503, and 22204047), the Natural Science Foundation of Hunan Province (2023ZK1120 and 2023JJ40122) and the Fundamental Research Funds for the Central Universities. The cell lines used in the study were obtained from the American type culture collection (ATCC).Notes and references
- D. P. Bartel, Cell, 2009, 136, 215–233 CrossRef CAS.
- J. Lu, G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz and T. R. Golub, Nature, 2005, 435, 834–883 CrossRef CAS.
- M. N. Poy, L. Eliasson, J. Krutzfeldt, S. Kuwajima, X. Ma, P. E. MacDonald, S. Pfeffer, T. Tuschl, N. Rajewsky, P. Rorsman and M. Stoffel, Nature, 2004, 43, 226–230 CrossRef.
- V. Ambros, Nature, 2004, 43, 350–355 CrossRef.
- L. He and G. J. Hannon, Nat. Rev. Genet., 2004, 5, 522–531 CrossRef CAS.
- D. P. Bartel, Cell, 2004, 11, 281–297 CrossRef.
- T. Jet, G. Gines, Y. Rondelez and V. Taly, Chem. Soc. Rev., 2021, 50, 4141–4161 RSC.
- Z. He, S. Peng, Q. Wei, S. Jia, S. Guo, K. Chen and X. Zhou, CCS Chem., 2020, 2, 89–97 CrossRef CAS.
- B. Mead and S. Tomarev, Neural Regen. Res., 2022, 17, 516–522 CrossRef CAS PubMed.
- L. Yu, P. Verwilst, I. Shim, Y.-Q. Zhao, Y. Zhou and J. S. Kim, CCS Chem., 2020, 3, 2725–2739 CrossRef.
- H. Zhou, Y. Jiang, W. Zhao and S. Zhang, ACS Appl. Mater. Interfaces, 2022, 14, 13070–13078 CrossRef CAS.
- H. Xu, Y. Zheng, D. Chen, Y. Cheng, X. Fang, C. Zhong, X. Huang, Q. Huang, J. Xu, J. Xu and C. Xue, Anal. Chem., 2022, 95, 1210–1218 Search PubMed.
- J. Li, S. Tan, R. Kooger, C. Zhang and Y. Zhang, Chem. Soc. Rev., 2014, 4, 506–517 RSC.
- W. Zhou, D. Li, R. Yuan and Y. Xiang, Anal. Chem., 2019, 91, 3628–3635 CrossRef CAS.
- K. Quan, J. Wang, J. Li, K. Wang and J. Huang, CCS Chem., 2021, 4, 3505–3515 CrossRef.
- J. Wei, J. Shang, S. He, Y. Ouyang, I. Willner and F. Wang, CCS Chem., 2022, 4, 3549–3562 CrossRef CAS.
- Q. Chao, Y. Zhang, Q. Li, L. Jiao, X. Sun, X. Chen, L. Zhu, Q. Yang, C. Shang, R.-M. Kong, G.-C. Fan, Z.-L. Song and X. Luo, Anal. Chem., 2023, 95, 7723–7734 CrossRef CAS.
- A. Roychoudhury, J. W. Dear and T. T. Bachmann, Biosens. Bioelectron., 2022, 212, 114404 CrossRef CAS.
- X.-L. Zhang, S.-S. Li, Y.-J. Liu, W.-W. Liu, L.-Q. Kong, Y.-Q. Chai, X.-L. Luo and R. Yuan, Anal. Chem., 2023, 95, 4077–4085 CrossRef CAS.
- P. Miao and Y. Tang, CCS Chem., 2020, 3, 2331–2339 CrossRef.
- C. Chen, D. A. Ridzon, A. J. Broomer, Z. Zhou, D. H. Lee, J. T. Nguyen, M. Barbisin, N. L. Xu, V. R. Mahuvakar, M. R. Andersen, K. Q. Lao, K. J. Livak and K. J. Guegler, Nucleic Acids Res., 2005, 33, e179 CrossRef.
- E. Tutucci, M. Vera, J. Biswas, J. Garcia, R. Parker and R. H. Singer, Nat. Methods, 2018, 15, 81–89 CrossRef CAS PubMed.
- M. E. Tanenbaum, L. A. Gilbert, L. S. Qi, J. S. Weissman and R. D. Vale, Cell, 2014, 159, 635–646 CrossRef CAS.
- E. Bertrand, P. Chartrand, M. Schaefer, S. M. Shenoy, R. H. Singer and R. M. Long, Mol. Cell, 1998, 2, 437–445 CrossRef CAS.
- H. Y. Park, H. Lim, Y. J. Yoon, A. Follenzi, C. Nwokafor, M. Lopez-Jones, X. Meng and R. H. Singer, Science, 2014, 343, 422–424 CrossRef CAS.
- T. J. Carrocci and A. A. Hoskins, Analyst, 2014, 139, 44–47 RSC.
- S. Han, B. S. Zhao, S. A. Myers, S. A. Carr, C. He and A. Y. Ting, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 22068–22079 CrossRef CAS.
- S. Tyagi, Nat. Methods, 2009, 6, 331–338 CrossRef CAS PubMed.
- E. Varkonyi-Gasic, R. Wu, M. Wood, E. F. Walton and R. P. Hellens, Plant Methods, 2007, 3, 12 CrossRef.
- J. R. Babendure, S. R. Adams and R. Y. Tsien, J. Am. Chem. Soc., 2003, 125, 14716–14717 CrossRef CAS PubMed.
- X. Lu, K. Y. S. Kong and P. Unrau, J. Chem. Soc. Rev., 2023, 52, 4071–4098 RSC.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- B. Hall, J. M. Micheletti, P. Satya, K. Ogle, J. Pollard and A. D. Ellington, Curr. Protoc. Mol. Biol., 2009, 88, 24.2.1–24.2.27 Search PubMed.
- J. S. Paige, K. Y. Wu and S. R. Jaffrey, Science, 2011, 333, 642–646 CrossRef CAS.
- G. S. Filonov, J. D. Moon, N. Svensen and S. R. Jaffrey, J. Am. Chem. Soc., 2014, 136, 16299–16308 CrossRef CAS.
- K. D. Warner, L. Sjekloća, W. Song, G. S. Filonov, S. R. Jaffrey and A. R. Ferré-D’Amaré, Nat. Chem. Biol., 2017, 13, 1195–1201 CrossRef CAS.
- E. V. Dolgosheina, S. C. Y. Jeng, S. S. S. Panchapakesan, R. Cojocaru, P. S. K. Chen, P. D. Wilson, N. Hawkins, P. A. Wiggins and P. J. Unrau, ACS Chem. Biol., 2014, 9, 2412–2420 CrossRef CAS PubMed.
- A. Autour, S. C. Y. Jeng, A. D. Cawte, A. Abdolahzadeh, A. Galli, S. S. S. Panchapakesan, D. Rueda, M. Ryckelynck and P. J. Unrau, Nat. Commun., 2018, 9, 1–12 CrossRef CAS PubMed.
- L. A. Holeman, S. L. Robinson, J. W. Szostak and C. Wilson, Fold. Des., 1998, 3, 423–431 CrossRef CAS PubMed.
- M. Sunbul and A. Jäschke, Nucleic Acids Res., 2018, 46, e110 CrossRef.
- F. Bouhedda, K. T. Fam, M. Collot, A. Autour, S. Marzi, A. Klymchenko and M. Ryckelynck, Nat. Chem. Biol., 2020, 16, 69–76 CrossRef CAS PubMed.
- K. T. Fam, R. Pelletier, F. Bouhedda, M. Ryckelynck, M. Collot and A. S. Klymchenko, Anal. Chem., 2022, 94, 6657–6664 CrossRef CAS.
- X. Chen, D. Zhang, N. Su, B. Bao, X. Xie, F. Zuo, L. Yang, H. Wang, L. Jiang, Q. Lin, M. Fang, N. Li, X. Hua, Z. Chen, C. Bao, J. Xu, W. Du, L. Zhang, Y. Zhao, L. Zhu, J. Loscalzo and Y. Yang, Nat. Biotechnol., 2019, 37, 1287–1293 CrossRef CAS.
- J. Wu, N. Svensen, W. Song, H. Kim, S. Zhang, X. Li and S. R. Jaffrey, J. Am. Chem. Soc., 2022, 144, 5471–5477 CrossRef CAS PubMed.
- B. Bühler, J. Schokolowski, A. Benderoth, D. Englert, F. Grün, A. Jäschke and M. Sunbul, Nat. Chem. Biol., 2023, 19, 478–487 CrossRef.
- M. Sunbul, J. Lackner, A. Martin, D. Englert, B. Hacene, F. Grün, K. Nienhaus, G. U. Nienhaus and A. Jäschke, Nat. Biotechnol., 2021, 39, 686–690 CrossRef CAS.
- D. Englert, E.-M. Burger, F. Grün, M. S. Verma, J. Lackner, M. Lampe, B. Bühler, J. Schokolowski, G. U. Nienhaus, A. Jäschke and M. Sunbul, Nat. Commun., 2023, 14, 3879 CrossRef CAS.
- S. K. Dey, G. S. Filonov, A. O. Olarerin-George, B. T. Jackson, L. W. S. Finley and S. R. Jaffrey, Nat. Chem. Biol., 2022, 18, 180–190 CrossRef CAS.
- H. Kim and S. R. Jaffrey, Cell Chem. Biol., 2019, 26, 1725–1731 CrossRef CAS.
- L. Mi, Q. Yu, A. P. K. K. Karunanayake Mudiyanselage, R. Wu, Z. Sun, R. Zheng, K. Ren and M. You, ACS Sens., 2023, 8, 308–316 CrossRef CAS.
- G. Zheng, L. Zhao, D. Yuan, J. Li, G. Yang, D. Song, H. Miao, L. Shu, X. Mo, X. Xu, L. Li, X. Song and Y. Zhao, Biosens. Bioelectron., 2022, 198, 113827 CrossRef CAS.
- M. You, J. L. Litke and S. R. Jaffrey, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E2756–E2765 CAS.
- M. Fang, H. Li, X. Xie, H. Wang, Y. Jiang, T. Li, B. Zhang, X. Jiang, Y. Cao, R. Zhang, D. Zhang, Y. Zhao, L. Zhu, X. Chen and Y. Yang, Biosens. Bioelectron., 2023, 235, 115411 CrossRef CAS PubMed.
- Q. Yu, J. Shi, A. P. K. K. K. Mudiyanselage, R. Wu, B. Zhao, M. Zhou and M. You, Chem. Commun., 2019, 55, 707–710 RSC.
- I. Masuda, T. Igarashi, R. Sakaguchi, R. G. Nitharwal, R. Takase, K. Y. Han, B. J. Leslie, C. Liu, H. Gamper, T. Ha, S. Sanyal and Y.-M. Hou, Nucleic Acids Res., 2017, 45, 4081–4093 CrossRef CAS.
- Z.-M. Ying, H.-Y. Xiao, H. Tang, R.-Q. Yu and J.-H. Jiang, Chem. Commun., 2018, 54, 8877–8880 RSC.
- Q. Wang, F. Xiao, H. Su, H. Liu, J. Xu, H. Tang, S. Qin, Z. Fang, Z. Lu, J. Wu, X. Weng and X. Zhou, Nucleic Acids Res., 2022, 50, e84 CrossRef CAS.
- D. Li, F. Yang, R. Yuan and Y. Xiang, Anal. Chim. Acta, 2020, 1102, 84–90 CrossRef CAS PubMed.
- S. S. Aw, M. X. Tang, Y. N. Teo and S. M. Cohen, Nucleic Acids Res., 2016, 44, e92 CrossRef.
- A. P. K. K. Karunanayake Mudiyanselage, Q. Yu, M. A. Leon-Duque, B. Zhao, R. Wu and M. You, J. Am. Chem. Soc., 2018, 140, 8739–8745 CrossRef CAS PubMed.
- C.-X. Dou, C. Liu, Z.-M. Ying, W. Dong, F. Wang and J.-H. Jiang, Anal. Chem., 2021, 93, 2534–2540 CrossRef CAS PubMed.
- Z.-M. Ying, Z. Wu, B. Tu, W. Tan and J.-H. Jiang, J. Am. Chem. Soc., 2017, 139, 9779–9782 CrossRef CAS PubMed.
- B. Bühler, J. Schokolowski, A. Jäschke and M. Sunbul, ACS Chem. Biol., 2023, 18, 1838–1845 CrossRef.
- Y. Peng, L. Shu, X. Deng, X. Huang, X. Mo, F. Du and Z. Tang, Anal. Chem., 2023, 95, 13762–13768 CrossRef CAS PubMed.
- Y.-P. Zhang, Z.-G. Wang, Y.-F. Tian, L.-H. Jiang, L. Zhao, D.-M. Kong, X. Li, D.-W. Pang and S.-L. Liu, Angew. Chem., Int. Ed., 2023, 62, e202217230 CrossRef CAS.
- Z. Wang, Y. Luo, X. Xie, X. Hu, H. Song, Y. Zhao, J. Shi, L. Wang, G. Glinsky, N. Chen, R. Lal and C. Fan, Angew. Chem., Int. Ed., 2018, 57, 972–976 CrossRef CAS PubMed.
- K. Ren, R. Wu, A. P. K. K. Karunanayake Mudiyanselage, Q. Yu, B. Zhao, Y. Xie, Y. Bagheri, Q. Tian and M. You, J. Am. Chem. Soc., 2020, 142, 2968–2974 CrossRef CAS.
- K. Huang, F. Doyle, Z. E. Wurz, S. A. Tenenbaum, R. K. Hammond, J. L. Caplan and B. C. Meyers, Nucleic Acids Res., 2017, 45, e130 CrossRef CAS PubMed.
- H.-R. Chen, M.-L. Su, Y.-M. Lei, Z.-X. Ye, Z.-P. Chen, P.-Y. Ma, R. Yuan, Y. Zhuo, C.-Y. Yang and W.-B. Liang, J. Am. Chem. Soc., 2023, 145, 12812–12822 CrossRef CAS PubMed.
- J. H. Davis and J. W. Szostak, Proc. Natl. Acad. Sci. U. S. A., 2002, 9, 11616–11621 CrossRef PubMed.
- K. Huang, X. Chen, C. Li, Q. Song, H. Li, L. Zhu, Y. Yang and A. Ren, Nat. Chem. Biol., 2021, 1, 1289–1295 CrossRef.
- J. S. Paige, T. Nguyen-Duc, W. Song and S. R. Jaffrey, Science, 2012, 335, 1194 CrossRef CAS PubMed.
- Y. Li, X. Teng, K. Zhang, R. Deng and J. Li, Anal. Chem., 2019, 91, 3989–3996 CrossRef CAS.
- D. Bautista-Sánchez, C. Arriaga-Canon, A. Pedroza-Torres, I. A. De La Rosa-Velázquez, R. González-Barrios, L. Contreras-Espinosa, R. Montiel-Manríquez, C. Castro-Hernández, V. Fragoso-Ontiveros, R. M. Álvarez-Gómez and L. A. Herrera, Mol. Ther. Nucleic Acids, 2020, 20, 409–420 CrossRef.
- B. López-Longarela, E. E. Morrison, J. D. Tranter, L. Chahman-Vos, J.-F. Léonard, J.-C. Gautier, S. Laurent, A. Lartigau, E. Boitier, L. Sautier, P. Carmona-Saez, J. Martorell-Marugan, R. J. Mellanby, S. Pernagallo, H. Ilyine, D. M. Rissin, D. C. Duffy, J. W. Dear and J. Díaz-Mochón, J. Anal. Chem., 2020, 92, 3388–3395 Search PubMed.
- D. De Rie, I. Abugessaisa, T. Alam, E. Arner, P. Arner, H. Ashoor, G. Åström, M. Babina, N. Bertin, A. M. Burroughs, A. J. Carlisle, C. O. Daub, M. Detmar, R. Deviatiiarov, A. Fort, C. Gebhard, D. Goldowitz, S. Guhl, T. J. Ha, J. Harshbarger, A. Hasegawa, K. Hashimoto, M. Herlyn, P. Heutink, K. J. Hitchens, C. C. Hon, E. Huang, Y. Ishizu, C. Kai, T. Kasukawa, P. Klinken, T. Lassmann, C.-H. Lecellier, W. Lee, M. Lizio, V. Makeev, A. Mathelier, Y. A. Medvedeva, N. Mejhert, C. J. Mungall, S. Noma, M. Ohshima, M. Okada-Hatakeyama, H. Persson, P. Rizzu, F. Roudnicky, P. Sætrom, H. Sato, J. Severin, J. W. Shin, R. K. Swoboda, H. Tarui, H. Toyoda, K. Vitting-Seerup, L. Winteringham, Y. Yamaguchi, K. Yasuzawa, M. Yoneda, N. Yumoto, S. Zabierowski, P. G. Zhang, C. A. Wells, K. M. Summers, H. Kawaji, A. Sandelin, M. Rehli, Y. Hayashizaki, P. Carninci, A. R. R. Forrest and M. J. L. de Hoon, Nat. Biotechnol., 2017, 35, 872–878 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc04957d |
| This journal is © The Royal Society of Chemistry 2023 |